The Sirtuin 2 (SIRT2) gene was initially identified
as ribonuclease mar1, which acted by preventing the expression of
mating genes in yeast (1). The yeast
silent information regulator 2 (SIR2) was shown to be involved in
transcriptional silencing, ribosomal DNA recombination, life span
and other physiological functions (2–4). It was
initially hypothesized that SIR2 possessed an adenosine diphosphate
(ADP)-ribosyltransferase activity that could transfer ADP-ribose
from nicotinamide adenine dinucleotide (NAD) to histones (5). This activity was subsequently shown to
be a low efficiency side reaction (6). It is well accepted that SIR2 is a
NAD-dependent lysine deacetylase. The SIRT enzymes are highly
conserved from bacteria to humans. Bacteria and archaea only
express one or two SIRT, while mammals have seven SIRT homologs
(SIRT1-7) (7,8). The seven mammalian SIRT have different
subcellular localization patterns, including cytoplasmic (SIRT1 and
2), nuclear (SIRT1, 2, 3, 6 and 7) and mitochondrial (SIRT3, 4 and
5). Among them, SIRT1-3 belong to class I SIRTs and have higher
homology to the yeast silent information regulator 2 (Sir2),
histone deacetylase (Hst)1 and Hst2. All these enzymes exhibit
potent deacetylase activity (7).
SIRT2 is a member of the SIRT family and is also
known as SIR2, SIRT type 2 or Sir2-related protein type 2; it
belongs to the classic type III deacetylases and acts in a
NAD+ dependent manner (9). SIRT2 is unique amongst SIRTs as it is
the only primary cytoplasmic enzyme with robust deacetylase
activity. Recently, SIRT2 has been shown to catalyze the removal of
lysine fatty acylation, including hexanoylation, decanoylation and
myristoylation (10). The newly
characterized de-acylation activities of SIRT2 have improved the
current knowledge on the function of this enzyme and have provided
novel opportunities to study the physiological and pathological
role of SIRT2.
The human SIRT2 gene is located on chromosome 19
with 16 exons spanning 21 kilobases of the genomic DNA. The SIRT2
transcript undergoes alternative splicing and produces three
isoforms with different cellular and tissue distributions, and
different functions (11,12). The full-length isoform 1 is abundant
in the skeletal muscle, while isoform 2, which lacks the N-terminal
37 residue, accumulates in the brain. Both isoforms 1 and 2 are
enzymatically active and are able to shuttle between the nucleus
and the cytoplasm. In contrast to these findings, isoform 5 lacks a
nuclear export signal that contains the entire nuclear export
signal (NES) and a short fragment of the catalytic domain,
resulting in the nuclear enrichment and loss of catalytic activity
(12).
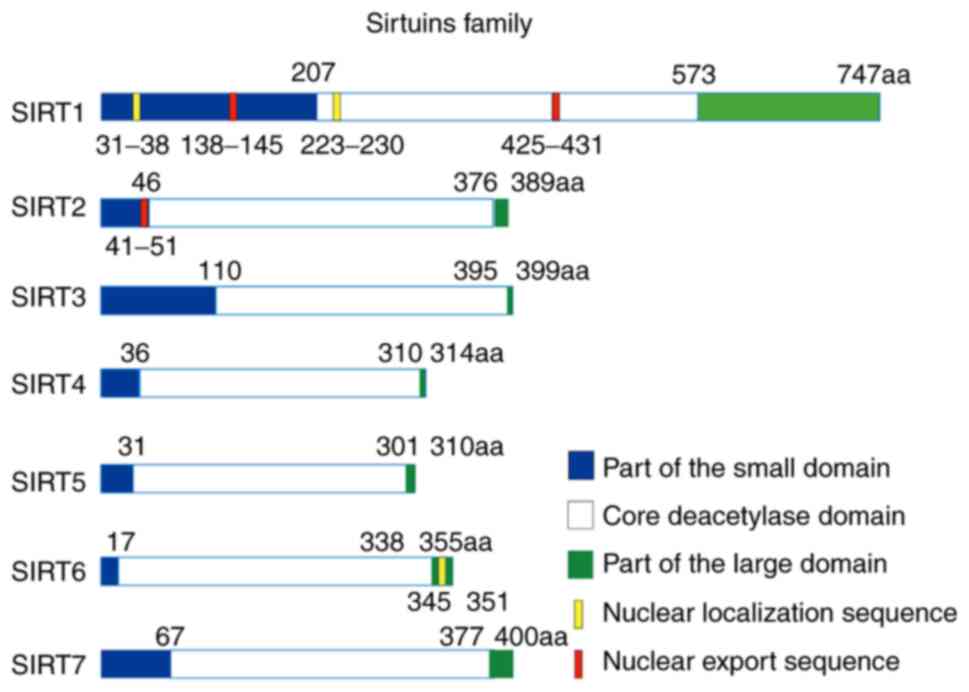
X-ray crystallographic studies have revealed that
the human SIRT family enzymes contain two major functional domains:
A small domain that binds to zinc ions and a large domain that is
responsible for NAD+ binding. A conserved large
substrate binding groove is also presented at the interface of the
two domains (13). The longest SIRT2
isoform contains 389 amino acids, of which the 65–340 amino acid
sequence is the NAD-dependent catalytic domain (14). The 41–51 amino acid sequence at the
N-terminal of SIRT2 is a NES, which is responsible for guiding the
cytoplasmic localization of the protein (15). The detailed schematic diagram of each
domain of the SIRTs family is shown in Fig. 1. Despite their conserved structural
features, human SIRTs diverge significantly in their subcellular
distribution, substrate targets and enzymatic activity
characteristics. For example, the lipoamidase activity of SIRT4 is
superior to its deacetylase activity (16). SIRT6 exhibits preferential activity
for the removal of long chain fatty acyls (10). SIRT7 employs nuclear acid oligos
(double-stranded DNA, ribosomal RNA and transfer RNA) as co-factors
for catalysis (17). SIRT1, SIRT2
and SIRT3 possess a robust deacetylase activity.
Although SIRT2 is predominantly cytoplasmic, it
shuttles into the nucleus under certain circumstances, such as
mitosis or bacterial infection (18). SIRT2 predominantly regulates cellular
processes through its enzymatic activity (18–23).
Previous studies indicated a conserved role of SIR2 in extending
the lifespan of yeast, flies and worms in a deacetylase-dependent
manner (24–26). In mammals, the significant longevity
gene BUB1 mitotic checkpoint serine/threonine kinase B (BubR1) was
shown to be deacetylated by SIRT2. The latter deacetylates BubR1 at
the K668 site, thereby stabilizing the BubR1 protein. As a
consequence, the lifespan of progeroid hypomorphic BubR1 mice was
largely increased by SIRT2 overexpression (27). In general, the anti-aging effect of
SIRT2 seems to be conservative from yeast to mammals.
Mammalian SIRT2 has been shown to restrain cell
cycle progression in a deacetylase activity-dependent manner
(28). SIRT2 is associated with
chromatin during mitosis and facilitates chromatin condensation by
deacetylating Histone H4 lysine (H4K) 16 (29). The deacetylation of H4K16 is in turn
essential for the mitotic deposition of H4K20 methylation (30). The upregulation of the level of
mitotic regulators in SIRT2-knockout mice confirmed the cell cycle
regulatory function of SIRT2 (31).
It was proposed that SIRT2 regulates mitosis by deacetylating
cadherin-1 and cell division cycle protein 20 homolog, which are
two adenomatous polyposis coli/C coactivators (31). In addition to its action as a mitotic
regulator, SIRT2 also deacetylates ribonucleotide reductase
regulatory subunit M2 at the K95 site during the S phase of the
cell cycle. This deacetylation increases the dNTP pool size and
accelerates DNA replication fork progression by enhancing
ribonucleotide reductase activity, which ultimately results in an
increased cancer cell proliferative rate (32).
A number of previous studies have shown that SIRT2
also plays a role in regulating cell metabolism (33,34).
SIRT2 can promote the metastasis of gastric cancer through the
RAS/ERK/JNK/matrix metalloproteinase-9 pathway by increasing
phosphoenolpyruvate carboxykinase 1-associated metabolism (35). SIRT2 promotes glycolysis and tumor
growth by deacetylating the K305 site of pyruvate kinase M2, a
hallmark enzyme that bridges metabolism and immunity (36). A comprehensive understanding of the
function and mechanism of SIRT2 in the progression of various
cancer types is imminent. Clinical data and mechanisms of action
for SIRT2 in cancer are shown in Table
I. Glucose-6-phosphate dehydrogenase (G6PD) is a key enzyme
that is involved in the pentose phosphate pathway and is
responsible for producing NADPH. Deacetylation of the residue K403
of G6PD by SIRT2 increases its activity and enhances NADPH
production (37). It is important to
note that SIRT2 can inhibit glycolysis and metabolic reprogramming
of induced pluripotent stem cells (iPSCs) by deacetylating the
following four key glycolytic enzymes: Aldolase, phosphoglycerate
kinase 1, enolase and GAPDH (19). A
recent study demonstrated that SIRT2 is a master organizer of
T-cell metabolism, since it inhibits T-cell glycolysis and impairs
T-cell effector functions by deacetylating a number of metabolic
enzymes, such as phosphofructokinase, α-ketoglutarate
dehydrogenase, succinate dehydrogenase complex, subunit A,
flavoprotein variant and succinyl-CoA ligase (GDP-forming) subunit
α mitochondrial (38).
In addition to its classical deacetylase activity,
SIRT2 has also been shown to catalyze the removal of long chain
fatty acyls from Kras4a and Ras like proto-oncogene B (RalB)
(39,40). Furthermore, the de-myristoylation
activity of SIRT2 towards ADP-ribosylation factor 6 lysine 3 was
recently identified (41). The
diverse functions of SIRT2 in the nervous system, mitosis, genome
integrity, cell differentiation, cell homeostasis, aging,
infection, inflammation, oxidative stress and autophagy have been
previously reviewed (42–44).
Dysregulation of SIRT2 in cases of gene
amplification and mutation, protein overexpression and
mislocalization has been associated with the progression of various
cancer types. Cross-cancer analysis of The Cancer Genome Atlas
database indicates that the SIRT2 gene is amplified in ~9% (52 out
of 584 cases) of ovarian epithelial tumors and 4% (41 out of 1,053
cases) of NSCLCs (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
The gene locus of SIRT2 is known to be frequently deleted in human
oligodendrogliomas (45). In
addition, a number of somatic mutations within SIRT2 are found in
endometrial carcinoma, melanoma, leukemia and NSCLC (cbioportal
website) (http://www.cbioportal.org/).
Particularly, multiple cancer-associated SIRT2 mutations at
evolutionarily conserved sites have been reported as functionally
significant (46). For example, the
R42L mutation found in lung cancer decreased the protein levels of
SIRT2, while the P128L mutation found in both colon and uterine
cancer types abrogated the enzymatic activity of SIRT2 (46).
Due to tumor heterogeneity, the SIRT2 expression
pattern is not always consistent. According to two recent review
articles, SIRT2 expression was elevated in neuroblastoma, uveal
melanoma, renal cell carcinoma and acute myeloid leukemia, while it
was decreased in glioma, neck squamous cell carcinoma, breast
cancer, prostate cancer and liver cancer (47,48). In
addition, dual SIRT2 expression patterns have been noted in NSCLC.
Li et al (49) demonstrated
that SIRT2 mRNA and protein expression levels were downregulated in
NSCLC. Grbesa et al (50)
demonstrated that the expression levels of the SIRT2 proteins were
significantly higher in lung primary tumors than those noted in
normal tissues. It is notable that Gao et al (51) demonstrated patients with NSCLC with
low SIRT2 expression had longer overall survival (OS) compared with
those with high SIRT2 expression, according to a survival analysis
of 1,926 patients and SIRT2 expression levels were significantly
related to the survival time of patients with lung adenocarcinoma
(ADC) but not squamous cell carcinoma (SCC). Despite nuclear and
cytoplasmic shuttling, SIRT2 is in fact a primary cytoplasmic
protein. High levels of mislocalized nuclear SIRT2 protein were
associated with shorter disease-free survival time in ER-negative
breast cancer (52). Gong et
al (53) revealed that the
combination of SIRT1 and SIRT2 was an improved recurrence-free
survival prediction model for NSCLC. It is also notable that SIRT2
has been identified as a candidate plasma biomarker in invasive
cervical cancer (54).
The role of SIRT2 in cancer is controversial. SIRT2
has been reported to exert either tumor suppressor or oncogenic
functions. It was initially proposed as a tumor suppressor due to
its regulatory role on the mitotic checkpoint and due to its
deacetylase activity on histone H3K56, which is a frequent
modification noted in cancer cells (55,56). The
tumor suppressive role of SIRT2 was supported by genetic
experiments demonstrating that Sirt2-deficient mice exhibited
increased tumor incidence (30). In
contrast to these observations, the tumor promoting activity of
SIRT2 was supported by its ability to deacetylate p53 and
downregulate its transcriptional activity (57). Moreover, SIRT2 deacetylates K5 of
lactate dehydrogenase A and increases its activity and protein
levels, thereby accelerating glycolysis and lactate production,
which in turn leads to increased cancer cell proliferation and
migration (58). Moreover, SIRT2
inhibitors have been shown to have broad anticancer activity
(38,59), suggesting their therapeutic potential
in cancer cells.
These seemingly opposite observations may reflect a
context-specific role of SIRT2 in cancer progression. Researchers
are therefore encouraged to appropriately assess the clinical and
molecular features in order to determine SIRT2 protein
abnormalities in NSCLC.
Kras and epidermal growth factor receptor (EGFR)
mutations, and Myc amplification are among the most common
molecular abnormalities in NSCLC. SIRT2 positively regulates Kras
activity by catalyzing deacetylation of Kras-K104 or fatty
deacylation of K-Ras4a (39,60). The SIRT2 inhibitor JH-T4 has been
described as a potent anticancer agent. The mode of action of JH-T4
possibly involves increased fatty acylation of K-Ras4a (61). In contrast to this evidence, in
previous studies, SIRT2 stabilized Myc oncoprotein by repressing
neuronally expressed developmentally downregulated 4 (NEDD4) E3
ubiquitin-protein ligase gene expression (62). In addition, Kras-mutant NSCLC cells
were sensitive to loss of SIRT2 expression (60). This is consistent with the fact that
degradation of SIRT2 is positively correlated with NSCLC cell
proliferation (63).
In addition, SIRT2 can participate in cancer
progression by regulating physiological processes, including
metabolism and autophagy. Enhanced glycolysis is a distinctive and
prominent feature of cancer cells. Phosphoglycerate mutase (PGAM)
is a glycolytic enzyme that catalyzes the reversible conversion of
3-phosphoglycerate to 2-phosphoglycerate. PGAM is considered
oncogenic, while its inhibition attenuates tumor growth in NSCLC
cells (64). SIRT2 deacetylates the
K100 residue of PGAM and facilitates its activation, resulting in
enhanced NADPH production and accelerated tumor growth (64). The autophagic pathway is associated
with tumor suppression. Forkhead box protein O1 (FoxO1) is involved
in the induction of autophagy and serves as a tumor suppressor,
ectopically expressed FoxO1 interacts with SIRT2 and is
deacetylated by SIRT2 in H1299 NSCLC cells (65). In either lung or colon cancer cells,
inhibition of SIRT2 increases FoxO1 acetylation and promotes the
interaction between FoxO1 and autophagy related 7, which is
required for the induction of autophagy, a process that is
negatively correlated to tumor development (65). Tang et al (66) revealed that the high expression of
SIRT2 contributes to induce the protective autophagy mechanism of
HL-60/A cells, which is closely related with the drug resistance of
patients.
In addition, a clinical survival analysis of 1,926
patients with NSCLC demonstrated that the median survival time of
patients with low SIRT2 expression levels was significantly higher
than that of patients with high SIRT2 expression levels (15.0
versus 14.0 months, P=0.029) (51).
This evidence supports an oncogenic role of SIRT2 in NSCLC.
Conflicting studies have nevertheless shown that
SIRT2 expression is downregulated in NSCLC and that SIRT2 can
inhibit tumor growth (49,67). Zhu et al (68) reported that deacetylation of
aldo-keto reductase family 1 member C1 (AKR1C1) by SIRT2 inhibited
the binding of AKR1C1 to STAT3, therefore decreasing the
transcriptional activity of STAT3 and inhibiting migration of NSCLC
cells. SIRT2 was shown to inhibit migration of A549 lung cancer
cell by the removal of fatty acyls from the RalB protein (40). Similarly, overexpression of SIRT2 in
A549 and H1299 cells caused inhibition of cell proliferation,
induction of cell apoptosis and cell cycle arrest by deacetylating
S-phase kinase-associated protein 2 (49).
Increased lipogenesis plays a critical role in tumor
growth. ATP-citrate lyase (ACLY) exhibits an oncogenic function in
NSCLC. Acetylation of K540, K546 and K554 (3K) residues on ACLY
inhibits its ubiquitylation and degradation, ACLY is a key enzyme
that catalyzes the ATP-dependent conversion of citrate and coenzyme
A (CoA) to oxaloacetate and acetyl-CoA, SIRT2 deacetylates ACLY and
promotes its degradation, leading to decreased fatty acid
synthesis, as well as delayed tumor growth in NSCLC cells (69). SIRT2 attenuates the oncogenic
activity of ACLY by acting as its primary deacetylase in NSCLC
cells (69). In addition, Mu et
al (70) revealed that SIRT1/2
inhibition triggers pro-survival autophagy by increasing
acetylation of HSPA5 and upregulating expression levels of ATF4 and
DDIT4 to obstruct the mTOR signaling pathway in human NSCLC cells.
SIRT2 directly binds to transcription factor EB, to regulate acute
shear stress-induced cell apoptosis by regulating the release of
autophagy components and exosomes, which contributes to the
suppression of tumorigenesis and the metastasis of NSCLC (71). A recent study demonstrated that SIRT2
was readily degraded via homologous recombination repair-mediated
ubiquitination in NSCLC (67),
supporting the anticancer function of SIRT2 in this disease.
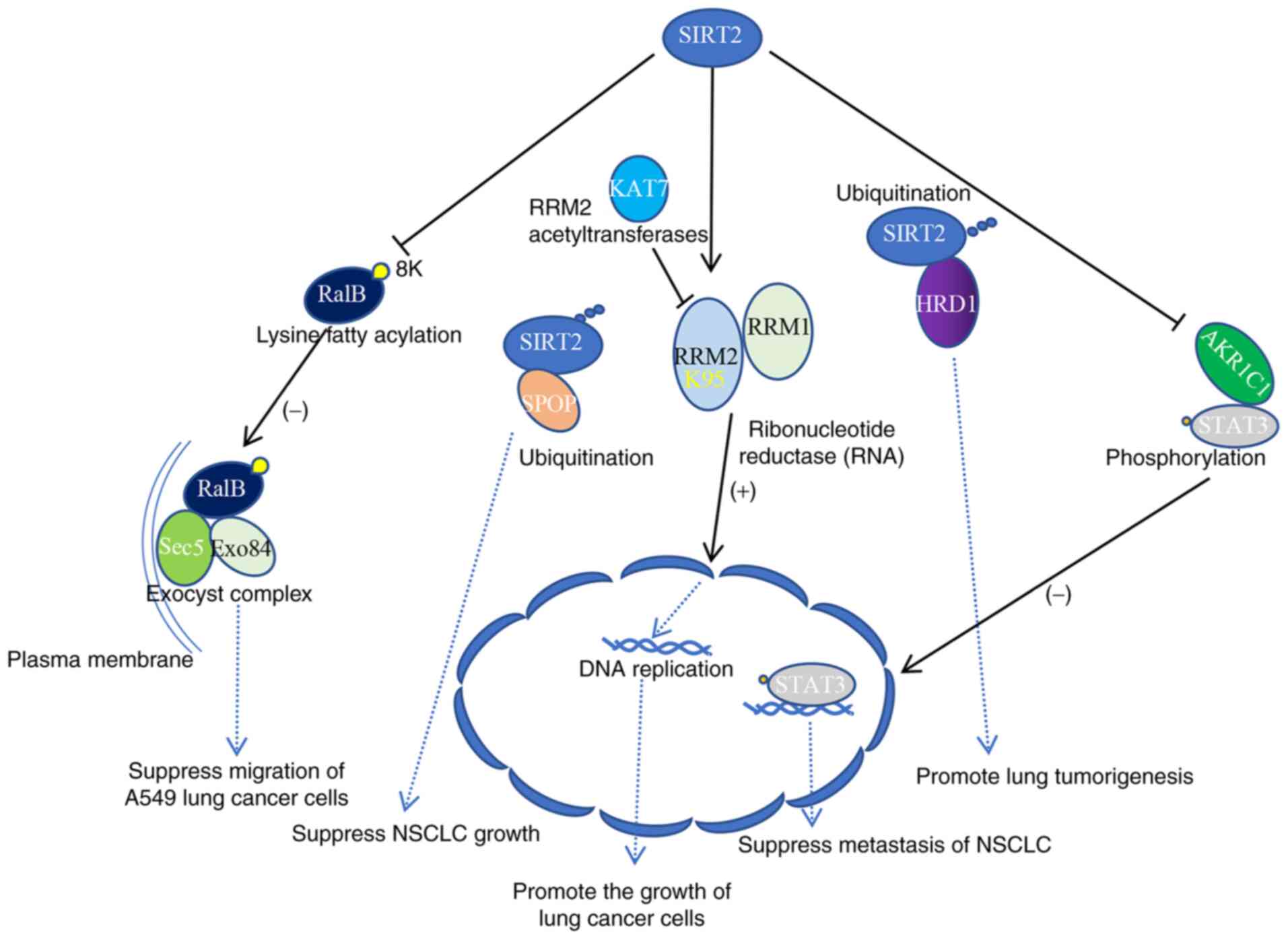
Taken together, the data demonstrate that SIRT2 can
participate in the occurrence and development of various cancer
types by regulating a variety of physiological processes; it has
different mechanisms of action in various cancer types, and a
schematic diagram of its mechanism of action in NSCLC is shown in
Fig. 2.
EGFR-activating mutations are noted in ~20% patients
with NSCLC. EGFR tyrosine kinase inhibitors are currently the
standard treatment for patients with NSCLC and EGFR mutations.
However, drug resistance is a major factor affecting the efficacy
of anticancer therapy. Using large-scale screening, Bajpe et
al (72) demonstrated that loss
of SIRT2 conferred resistance to EGFR inhibitors in NSCLC and colon
cancer. SIRT2 deacetylates MEK1 and inhibits its activation. Since
loss of SIRT2 results in increased levels of MEK1 acetylation and
phosphorylation, the increase in MEK1 activation and downstream ERK
phosphorylation may lead to cancer recurrence. Similarly, SIRT2
loss conferred resistance to the effects of BRAF and MEK inhibitors
in BRAF-mutant melanoma and Kras-mutant colon cancer, respectively
(72). In contrast to these
findings, an increase in SIRT2 expression levels may cause
multidrug resistance in acute myelogenous leukemia by activating
the ERK1/2 signaling pathway (73).
In addition, SIRT2 exhibited protective effects against
chemotherapy-induced peripheral neuropathy in a subcutaneous lung
cancer mouse model. This condition is one of the most common causes
of chemotherapy dose reduction and discontinuation. Cisplatin
induces SIRT2 nuclear accumulation in dorsal root ganglia neurons,
allowing SIRT2 to participate in the repair of cisplatin-generated
DNA damage (74). Multiple studies
have shown that SIRT2 contributes to the stemness of cancer stem
cells (CSCs), which provides a further link between SIRT2 and
chemoresistance in NSCLC (75,76).
CSCs are considered the ‘seeds’ of cancer cells,
with self-renewal ability and multilineage differentiation
potential. CSCs are resistant to radiotherapy and chemotherapy, and
are closely associated with tumor recurrence and metastasis
(77). It has been shown that SIRT2
is involved in regulating the stemness of embryonic stem cells
(ESCs) and CSCs. SIRT2 expression is significantly downregulated in
both human ESCs and human pluripotent stem cells, while its
upregulation is noted during mouse ESC differentiation. In
addition, a study showed that depletion of SIRT2 prominently
increased iPSC generation, while overexpression of SIRT2
significantly reduced this process (19). These effects are mainly attributed to
the deacetylation and inactivation of glycolytic enzymes by SIRT2;
Cha et al (19) reported that
downregulation of SIRT2 by miR-200c promotes glycolysis and
acetylation of glycolytic enzymes, which contributes to cellular
reprogramming of human PSCs. Aldehyde dehydrogenase 1A1 (ALDH1A1)
serves as a marker of CSCs in NSCLC and participates in their
maintenance (78). ALDH1A1 activity
is inhibited by K353 acetylation, which can be further deacetylated
by SIRT2. Activation of the NOTCH signaling pathway induces
deacetylation of ALDH1A1, which is catalyzed by SIRT2. This leads
to activation of ALDH1A1 and increased self-renewal properties of
CSCs (75). Similar pro-self-renewal
effects of SIRT2 have been demonstrated in renal cell carcinoma
CSCs; Wei et al (76)
reported that SIRT2 may be highly expressed in the RCC stem-like
cells and that it contributes to cancer metastasis. In addition, it
was reported that the SIRT2-selective inhibitor
2-cyano-3-(5-(2,5-dichlorophenyl)-2-furanyl)-N-5-quinolinyl-2-propenamide
(AGK2) showed the most potent antiproliferative effect in
glioblastoma multiforme CSCs (79).
Tumor infiltrating lymphocyte (TIL) therapy is
considered a promising option for treating patients with metastatic
NSCLC, whereas loss of T-cell effector functions within the tumor
microenvironment can limit the clinical efficacy of this
therapeutic method. SIRT2 has been suggested as a master regulator
of T-cell metabolism and an immune checkpoint in TILs (38). By utilizing sirt2-knockout mice and
SIRT2 inhibitors, one study concluded that SIRT2 inhibited T-cell
metabolism and impaired T-cell effector functions by deacetylating
a number of metabolic enzymes. This conclusion was supported by the
fact that SIRT2 was only increased in TILs from patients with NSCLC
that responded partially to TIL therapy (38).
Recent studies have shown that inhibition of SIRT2
exhibits broad anticancer activity (59,80).
High expression of SIRT2 in NSCLC samples can be used to ensure the
efficacy of SIRT2 inhibitors in NSCLC treatment. The existing SIRT2
inhibitors (AEM1 and AEM2) have shown p53-dependent proapoptotic
activity in NSCLC (81). In
addition, combination chemotherapy is a potentially promising
approach used to enhance anticancer activity. The combination of
SIRT2 inhibitors (AGK2 and Sirtinol) and the pyruvate dehydrogenase
kinase inhibitor (dichloroacetic acid) was highly effective for
inhibiting the proliferation of NSCLC cells (82). Bisnaphthalimidopropyl
diaminodicyclohexylmethane is a polyamine derivative that inhibits
growth and induces the apoptosis of NSCLC cancer cells. This
anticancer effect is possibly mediated by SIRT2 inhibition
(83). Moreover, inhibition of SIRT1
and SIRT2 by salermide, a reverse amide compound, leads to the
upregulation of death receptor 5 through the activating
transcription factor (ATF)4/ATF3/DNA damage inducible transcript 3
pathway in NSCLC, resulting in the apoptosis of human lung cancer
cells (84). The anticancer effect
of SIRT2 inhibitors in NSCLC provides a rational therapeutic
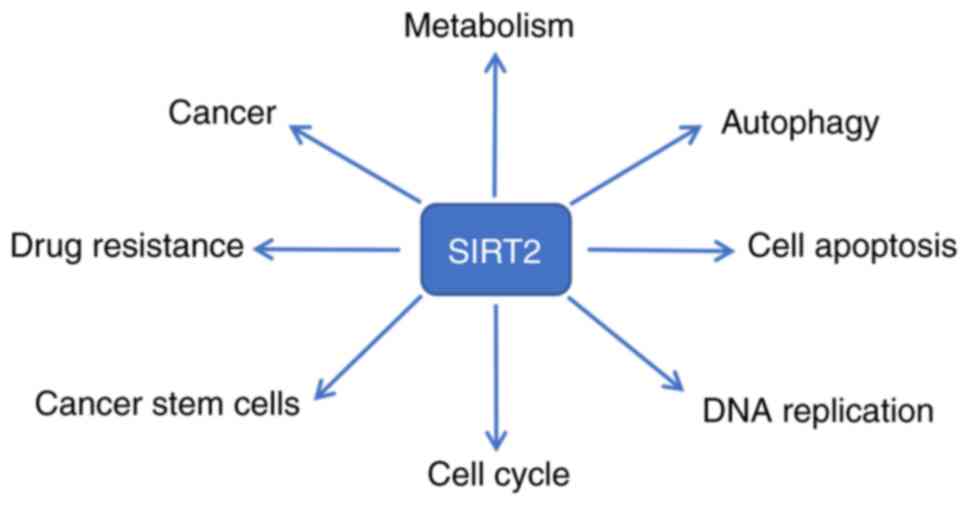
strategy for NSCLC tumors with high expression of SIRT2. A
schematic diagram of the biological functions of SIRT2 is shown in
Fig. 3.
As aforementioned, NSCLC is a malignant cancer with
a complicated etiology and poor prognosis, accounting for ~80% of
the total incidence of lung cancer. SIRT2 functions either as a
tumor suppressor or as an oncogene in NSCLC, depending on the
experimental conditions. It is notable that SIRT2 inhibitors
exhibit a protective effect in patients with lung cancer and high
SIRT2 expression, providing a theoretical basis for successful
cancer therapy.
SIRT2 is mainly localized in the cytoplasm. However,
it is also found in the nucleus and mitochondria. A secretome study
performed in mouse macrophages revealed 775 proteins including
SIRT2, which were reproducibly detected in the culture medium
following lipopolysaccharide stimulation (85), suggesting a potential role of SIRT2
in the extracellular compartment. The applications of
high-throughput liquid chromatography tandem mass spectrometry
analysis have resulted in the identification of a large number of
extracellular matrix and peripheral proteins that are acetylated in
both healthy and diseased tissues (86–88).
However, additional studies are required to assess whether the
SIRT2 deacetylase activity in the extracellular tumor
microenvironment can regulate tumorigenesis.
In conclusion, SIRT2 is implicated in a wide range
of physiological and pathological processes via deacetylation or
fatty deacylation of specific substrates. Dysregulation of SIRT2 is
closely associated with NSCLC progression. SIRT2 exhibits both
oncogenic and tumor suppressive functions depending on the cancer
stage, cell molecular characteristics and experimental conditions.
Therefore, despite the broad anticancer activity of SIRT2
inhibitors, extensive research is still required to validate the
potential of SIRT2 as a target for NSCLC treatment.
Not applicable.
The present study was funded by the Priority
Academic Program Development of Jiangsu Higher Education
Institutions, as well as by a grant from the China Natural Science
Foundation (grant no. 31801058).
Not applicable.
MZ and MW searched the literature and wrote the
manuscript. YEC and CH searched the literature and revised the
manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
No applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Klar AJ, Fogel S and Macleod K: MAR1-a
Regulator of the HMa and HMalpha Loci in Saccharomyces Cerevisiae.
Genetics. 93:37–50. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Loo S and Rine J: Silencing and heritable
domains of gene expression. Annu Rev Cell Dev Biol. 11:519–548.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gottlieb S and Esposito RE: A new role for
a yeast transcriptional silencer gene, SIR2, in regulation of
recombination in ribosomal DNA. Cell. 56:771–776. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guarente L: Diverse and dynamic functions
of the Sir silencing complex. Nat Genet. 23:281–285. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tanny JC, Dowd GJ, Huang J, Hilz H and
Moazed D: An enzymatic activity in the yeast Sir2 protein that is
essential for gene silencing. Cell. 99:735–745. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tanner KG, Landry J, Sternglanz R and Denu
JM: Silent information regulator 2 family of NAD-dependent
histone/protein deacetylases generates a unique product,
1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA. 97:14178–14182.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hirschey MD: Old enzymes, new tricks:
Sirtuins are NAD(+)-dependent de-acylases. Cell Metab. 14:718–719.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Haigis MC, Mostoslavsky R, Haigis KM,
Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos
GD, Karow M, Blander G, et al: SIRT4 inhibits glutamate
dehydrogenase and opposes the effects of calorie restriction in
pancreatic beta cells. Cell. 126:941–954. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Ruijter AJ, van Gennip AH, Caron HN,
Kemp S and van Kuilenburg AB: Histone deacetylases (HDACs):
Characterization of the classical HDAC family. Biochem J. 370((Pt
3)): 737–749. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Feldman JL, Dittenhafer-Reed KE, Kudo N,
Thelen JN, Ito A, Yoshida M and Denu JM: Kinetic and structural
basis for acyl-group selectivity and NAD(+) dependence in
sirtuin-catalyzed deacylation. Biochemistry. 54:3037–3050. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maxwell MM, Tomkinson EM, Nobles J,
Wizeman JW, Amore AM, Quinti L, Chopra V, Hersch SM and Kazantsev
AG: The Sirtuin 2 microtubule deacetylase is an abundant neuronal
protein that accumulates in the aging CNS. Hum Mol Genet.
20:3986–3996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rack JG, VanLinden MR, Lutter T, Aasland R
and Ziegler M: Constitutive nuclear localization of an
alternatively spliced sirtuin-2 isoform. J Mol Biol. 426:1677–1691.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kudo N, Ito A, Arata M, Nakata A and
Yoshida M: Identification of a novel small molecule that inhibits
deacetylase but not defatty-acylase reaction catalysed by SIRT2.
Philos Trans R Soc Lond B Biol Sci. 373:201700702018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mei Z, Zhang X, Yi J, Huang J, He J and
Tao Y: Sirtuins in metabolism, DNA repair and cancer. J Exp Clin
Cancer Res. 35:1822016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
North BJ and Verdin E: Interphase
nucleo-cytoplasmic shuttling and localization of SIRT2 during
mitosis. PLoS One. 2:e7842007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mathias RA, Greco TM, Oberstein A,
Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T and Cristea
IM: Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase
complex activity. Cell. 159:1615–1625. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tong Z, Wang M, Wang Y, Kim DD, Grenier
JK, Cao J, Sadhukhan S, Hao Q and Lin H: SIRT7 Is an RNA-activated
protein lysine deacylase. ACS Chem Biol. 12:300–310. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Eskandarian HA, Impens F, Nahori MA,
Soubigou G, Coppée JY, Cossart P and Hamon MA: A role for
SIRT2-dependent histone H3K18 deacetylation in bacterial infection.
Science. 341:12388582013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cha Y, Han MJ, Cha HJ, Zoldan J, Burkart
A, Jung JH, Jang Y, Kim CH, Jeong HC, Kim BG, et al: Metabolic
control of primed human pluripotent stem cell fate and function by
the miR-200c-SIRT2 axis. Nat Cell Biol. 19:445–456. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fiskus W, Coothankandaswamy V, Chen J, Ma
H, Ha K, Saenz DT, Krieger SS, Mill CP, Sun B, Huang P, et al:
SIRT2 deacetylates and inhibits the peroxidase activity of
peroxiredoxin-1 to sensitize breast cancer cells to oxidant
stress-inducing agents. Cancer Res. 76:5467–5478. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rothgiesser KM, Erener S, Waibel S,
Lüscher B and Hottiger MO: SIRT2 regulates NF-kappaB dependent gene
expression through deacetylation of p65 Lys310. J Cell Sci.
(123)((Pt 24)): 4251–4258. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsusaka T, Guo T, Yagura T, Inoue T,
Yokode M, Inagaki N and Kondoh H: Deacetylation of phosphoglycerate
mutase in its distinct central region by SIRT2 down-regulates its
enzymatic activity. Genes Cells. 19:766–777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sarikhani M, Mishra S, Desingu PA, Kotyada
C, Wolfgeher D, Gupta MP, Singh M and Sundaresan NR: SIRT2
regulates oxidative stress-induced cell death through deacetylation
of c-Jun NH2-terminal kinase. Cell Death Differ.
25:1638–1656. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fabrizio P, Gattazzo C, Battistella L, Wei
M, Cheng C, McGrew K and Longo VD: Sir2 blocks extreme life-span
extension. Cell. 123:655–667. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Horio Y, Hayashi T, Kuno A and Kunimoto R:
Cellular and molecular effects of sirtuins in health and disease.
Clin Sci (Lond). 121:191–203. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Michan S and Sinclair D: Sirtuins in
mammals: Insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
North BJ, Rosenberg MA, Jeganathan KB,
Hafner AV, Michan S, Dai J, Baker DJ, Cen Y, Wu LE, Sauve AA, et
al: SIRT2 induces the checkpoint kinase BubR1 to increase lifespan.
EMBO J. 33:1438–1453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dryden SC, Nahhas FA, Nowak JE, Goustin AS
and Tainsky MA: Role for human SIRT2 NAD-dependent deacetylase
activity in control of mitotic exit in the cell cycle. Mol Cell
Biol. 23:3173–3185. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vaquero A, Scher MB, Lee DH, Sutton A,
Cheng HL, Alt FW, Serrano L, Sternglanz R and Reinberg D: SirT2 is
a histone deacetylase with preference for histone H4 Lys 16 during
mitosis. Genes Dev. 20:1256–1261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Serrano L, Martínez-Redondo P,
Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB,
Kane-Goldsmith N, Tong Q, Rabanal RM, et al: The tumor suppressor
SirT2 regulates cell cycle progression and genome stability by
modulating the mitotic deposition of H4K20 methylation. Genes Dev.
27:639–653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim HS, Vassilopoulos A, Wang RH, Lahusen
T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, et al: SIRT2
maintains genome integrity and suppresses tumorigenesis through
regulating APC/C activity. Cancer Cell. 20:487–499. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen G, Luo Y, Warncke K, Sun Y, Yu DS, Fu
H, Behera M, Ramalingam SS, Doetsch PW, Duong DM, et al:
Acetylation regulates ribonucleotide reductase activity and cancer
cell growth. Nat Commun. 10:32132019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gonfloni S, Iannizzotto V, Maiani E,
Bellusci G, Ciccone S and Diederich M: P53 and Sirt1: Routes of
metabolism and genome stability. Biochem Pharmacol. 92:149–156.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cha YI and Kim HS: Emerging role of
sirtuins on tumorigenesis: Possible link between aging and cancer.
BMB Rep. 46:429–438. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Y, Zhang M, Dorfman RG, Pan Y, Tang D,
Xu L, Zhao Z, Zhou Q, Zhou L, Wang Y, et al: SIRT2 promotes the
migration and invasion of gastric cancer through RAS/ERK/JNK/MMP-9
pathway by increasing PEPCK1-Related metabolism. Neoplasia.
20:745–756. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park SH, Ozden O, Liu G, Song HY, Zhu Y,
Yan Y, Zou X, Kang HJ, Jiang H, Principe DR, et al: SIRT2-Mediated
deacetylation and tetramerization of pyruvate kinase directs
glycolysis and tumor growth. Cancer Res. 76:3802–3812. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang YP, Zhou LS, Zhao YZ, Wang SW, Chen
LL, Liu LX, Ling ZQ, Hu FJ, Sun YP, Zhang JY, et al: Regulation of
G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and
cell survival during oxidative stress. EMBO J. 33:1304–1320.
2014.PubMed/NCBI
|
|
38
|
Hamaidi I, Zhang L, Kim N, Wang MH,
Iclozan C, Fang B, Liu M, Koomen JM, Berglund AE, Yoder SJ, et al:
Sirt2 inhibition enhances metabolic fitness and effector functions
of Tumor-Reactive T Cells. Cell Metab. 32:420–436.e412. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jing H, Zhang X, Wisner SA, Chen X,
Spiegelman NA, Linder ME and Lin H: SIRT2 and lysine fatty
acylation regulate the transforming activity of K-Ras4a. Elife.
6:e324362017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Spiegelman NA, Zhang X, Jing H, Cao J,
Kotliar IB, Aramsangtienchai P, Wang M, Tong Z, Rosch KM and Lin H:
SIRT2 and Lysine fatty acylation regulate the activity of RalB and
cell migration. ACS Chem Biol. 14:2014–2023. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kosciuk T, Price IR, Zhang X, Zhu C,
Johnson KN, Zhang S, Halaby SL, Komaniecki GP, Yang M, DeHart CJ,
et al: NMT1 and NMT2 are lysine myristoyltransferases regulating
the ARF6 GTPase cycle. Nat Commun. 11:10672020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen G, Huang P and Hu C: The role of
SIRT2 in cancer: A novel therapeutic target. Int J Cancer.
147:3297–3304. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu Y, Zhang Y, Zhu K, Chi S, Wang C and
Xie A: Emerging role of Sirtuin 2 in Parkinson's disease. Front
Aging Neurosci. 11:3722020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Y, Anoopkumar-Dukie S, Arora D and
Davey AK: Review of the anti-inflammatory effect of SIRT1 and SIRT2
modulators on neurodegenerative diseases. Eur J Pharmacol.
867:1728472020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Reifenberger J, Reifenberger G, Liu L,
James CD, Wechsler W and Collins VP: Molecular genetic analysis of
oligodendroglial tumors shows preferential allelic deletions on 19q
and 1p. Am J Pathol. 145:1175–1190. 1994.PubMed/NCBI
|
|
46
|
Head PE, Zhang H, Bastien AJ, Koyen AE,
Withers AE, Daddacha WB, Cheng X and Yu DS: Sirtuin 2 mutations in
human cancers impair its function in genome maintenance. J Biol
Chem. 292:9919–9931. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang B, Ye Y, Yang X, Liu B, Wang Z, Chen
S, Jiang K, Zhang W, Jiang H, Mustonen H, et al: SIRT2-dependent
IDH1 deacetylation inhibits colorectal cancer and liver metastases.
EMBO Rep. 21:e481832020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Carafa V, Altucci L and Nebbioso A: Dual
tumor suppressor and tumor promoter action of sirtuins in
determining malignant phenotype. Front Pharmacol. 10:382019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li Z, Xie QR, Chen Z, Lu S and Xia W:
Regulation of SIRT2 levels for human non-small cell lung cancer
therapy. Lung Cancer. 82:9–15. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Grbesa I, Pajares MJ, Martínez-Terroba E,
Agorreta J, Mikecin AM, Larráyoz M, Idoate MA, Gall-Troselj K, Pio
R and Montuenga LM: Expression of sirtuin 1 and 2 is associated
with poor prognosis in non-small cell lung cancer patients. PLoS
One. 10:e01246702015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao CX, Chen B, Xie HK, Han CN and Luo J:
Immunohistochemistry and clinical value of sirtuin 2 in
non-metastasized non-small cell lung cancer. J Thorac Dis.
11:3973–3979. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
McGlynn LM, Zino S, MacDonald AI, Curle J,
Reilly JE, Mohammed ZM, McMillan DC, Mallon E, Payne AP, Edwards J
and Shiels PG: SIRT2: Tumour suppressor or tumour promoter in
operable breast cancer? Eur J Cancer. 50:290–301. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Gong J, Wang H, Lou W, Wang G, Tao H, Wen
H, Liu Y and Xie Q: Associations of sirtuins with
clinicopathological parameters and prognosis in non-small cell lung
cancer. Cancer Manag Res. 10:3341–3356. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Berggrund M, Enroth S, Lundberg M,
Assarsson E, Stålberg K, Lindquist D, Hallmans G, Grankvist K,
Olovsson M and Gyllensten U: Identification of candidate plasma
protein biomarkers for cervical cancer using the multiplex
proximity extension assay. Mol Cell Proteomics. 18:735–743. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Inoue K, Mallakin A and Frazier DP: Dmp1
and tumor suppression. Oncogene. 26:4329–4335. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Das C, Lucia MS, Hansen KC and Tyler JK:
CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature.
459:113–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jin YH, Kim YJ, Kim DW, Baek KH, Kang BY,
Yeo CY and Lee KY: Sirt2 interacts with 14-3-3 beta/gamma and
down-regulates the activity of p53. Biochem Biophys Res Commun.
368:690–695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhao D, Zou SW, Liu Y, Zhou X, Mo Y, Wang
P, Xu YH, Dong B, Xiong Y, Lei QY and Guan KL: Lysine-5 acetylation
negatively regulates lactate dehydrogenase A and is decreased in
pancreatic cancer. Cancer Cell. 23:464–476. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jing H, Hu J, He B, Negrón Abril YL,
Stupinski J, Weiser K, Carbonaro M, Chiang YL, Southard T,
Giannakakou P, et al: A SIRT2-Selective inhibitor promotes c-Myc
oncoprotein degradation and exhibits broad anticancer activity.
Cancer Cell. 29:6072016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang MH, Laurent G, Bause AS, Spang R,
German N, Haigis MC and Haigis KM: HDAC6 and SIRT2 regulate the
acetylation state and oncogenic activity of mutant K-RAS. Mol
Cancer Res. 11:1072–1077. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Spiegelman NA, Hong JY, Hu J, Jing H, Wang
M, Price IR, Cao J, Yang M, Zhang X and Lin H: A Small-Molecule
SIRT2 inhibitor that promotes K-Ras4a lysine fatty-acylation.
ChemMedChem. 14:744–748. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu PY, Xu N, Malyukova A, Scarlett CJ,
Sun YT, Zhang XD, Ling D, Su SP, Nelson C, Chang DK, et al: The
histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death
Differ. 20:503–514. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Luo J, Bao YC, Ji XX, Chen B, Deng QF and
Zhou SW: SPOP promotes SIRT2 degradation and suppresses non-small
cell lung cancer cell growth. Biochem Biophys Res Commun.
483:880–884. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xu Y, Li F, Lv L, Li T, Zhou X, Deng CX,
Guan KL, Lei QY and Xiong Y: Oxidative stress activates SIRT2 to
deacetylate and stimulate phosphoglycerate mutase. Cancer Res.
74:3630–3642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhao Y, Yang J, Liao W, Liu X, Zhang H,
Wang S, Wang D, Feng J, Yu L and Zhu WG: Cytosolic FoxO1 is
essential for the induction of autophagy and tumour suppressor
activity. Nat Cell Biol. 12:665–675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Tang HX, Wang MY, Xiao W and Wen JW:
SIRT2-Reverses Drug-Resistance of HL-60/A through autophagy
mechanism. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 27:409–414. 2019.(In
Chinese). PubMed/NCBI
|
|
67
|
Liu L, Yu L, Zeng C, Long H, Duan G, Yin
G, Dai X and Lin Z: E3 Ubiquitin Ligase HRD1 promotes lung
tumorigenesis by promoting sirtuin 2 ubiquitination and
degradation. Mol Cell Biol. 40:e00257–19. 2020. View Article : Google Scholar
|
|
68
|
Zhu H, Hu Y, Zeng C, Chang L, Ge F, Wang
W, Yan F, Zhao Q, Cao J, Ying M, et al: The SIRT2-mediated
deacetylation of AKR1C1 is required for suppressing its
pro-metastasis function in Non-small cell lung cancer.
Theranostics. 10:2188–2200. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lin R, Tao R, Gao X, Li T, Zhou X, Guan
KL, Xiong Y and Lei QY: Acetylation stabilizes ATP-citrate lyase to
promote lipid biosynthesis and tumor growth. Mol Cell. 51:506–518.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Mu N, Lei Y, Wang Y, Wang Y, Duan Q, Ma G,
Liu X and Su L: Inhibition of SIRT1/2 upregulates HSPA5 acetylation
and induces pro-survival autophagy via ATF4-DDIT4-mTORC1 axis in
human lung cancer cells. Apoptosis. 24:798–811. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang L, Xu P, Xie X, Hu F, Jiang L, Hu R,
Ding F, Xiao H and Zhang H: Down regulation of SIRT2 Reduced ASS
induced NSCLC apoptosis through the release of autophagy components
via exosomes. Front Cell Dev Biol. 8:6019532020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bajpe PK, Prahallad A, Horlings H,
Nagtegaal I, Beijersbergen R and Bernards R: A chromatin modifier
genetic screen identifies SIRT2 as a modulator of response to
targeted therapies through the regulation of MEK kinase activity.
Oncogene. 34:531–536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xu H, Li Y, Chen L, Wang C, Wang Q, Zhang
H, Lin Y, Li Q and Pang T: SIRT2 mediates multidrug resistance in
acute myelogenous leukemia cells via ERK1/2 signaling pathway. Int
J Oncol. 48:613–623. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang M, Du W, Acklin S, Jin S and Xia F:
SIRT2 protects peripheral neurons from cisplatin-induced injury by
enhancing nucleotide excision repair. J Clin Invest. 130:2953–2965.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun
YP, Xiong Y, Guan KL and Lei QY: NOTCH-induced aldehyde
dehydrogenase 1A1 deacetylation promotes breast cancer stem cells.
J Clin Invest. 124:5453–5465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wei R, He D and Zhang X: Role of SIRT2 in
regulation of stemness of cancer stem-like cells in renal cell
carcinoma. Cell Physiol Biochem. 49:2348–2357. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Eramo A, Haas TL and De Maria R: Lung
cancer stem cells: Tools and targets to fight lung cancer.
Oncogene. 29:4625–4635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Jiang F, Qiu Q, Khanna A, Todd NW, Deepak
J, Xing L, Wang H, Liu Z, Su Y, Stass SA and Katz RL: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Rotili D, Tarantino D, Nebbioso A, Paolini
C, Huidobro C, Lara E, Mellini P, Lenoci A, Pezzi R, Botta G, et
al: Discovery of salermide-related sirtuin inhibitors: Binding mode
studies and antiproliferative effects in cancer cells including
cancer stem cells. J Med Chem. 55:10937–10947. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Hu F, Sun X, Li G, Wu Q, Chen Y, Yang X,
Luo X, Hu J and Wang G: Inhibition of SIRT2 limits tumour
angiogenesis via inactivation of the STAT3/VEGFA signalling
pathway. Cell Death Dis. 10:92018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Hoffmann G, Breitenbücher F, Schuler M and
Ehrenhofer-Murray AE: A novel sirtuin 2 (SIRT2) inhibitor with
p53-dependent pro-apoptotic activity in non-small cell lung cancer.
J Biol Chem. 289:5208–5216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ma W, Zhao X, Wang K, Liu J and Huang G:
Dichloroacetic acid (DCA) synergizes with the SIRT2 inhibitor
Sirtinol and AGK2 to enhance anti-tumor efficacy in non-small cell
lung cancer. Cancer Biol Ther. 19:835–846. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lima RT, Barron GA, Grabowska JA, Bermano
G, Kaur S, Roy N, Vasconcelos MH and Lin PK: Cytotoxicity and cell
death mechanisms induced by a novel bisnaphthalimidopropyl
derivative against the NCI-H460 non-small lung cancer cell line.
Anticancer Agents Med Chem. 13:414–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Liu G, Su L, Hao X, Zhong N, Zhong D,
Singhal S and Liu X: Salermide up-regulates death receptor 5
expression through the ATF4-ATF3-CHOP axis and leads to apoptosis
in human cancer cells. J Cell Mol Med. 16:1618–1628. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Meissner F, Scheltema RA, Mollenkopf HJ
and Mann M: Direct proteomic quantification of the secretome of
activated immune cells. Science. 340:475–478. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lundby A, Lage K, Weinert BT,
Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A,
Choudhary C, Lundby C and Olsen JV: Proteomic analysis of lysine
acetylation sites in rat tissues reveals organ specificity and
subcellular patterns. Cell Rep. 2:419–431. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Simon GM, Cheng J and Gordon JI:
Quantitative assessment of the impact of the gut microbiota on
lysine epsilon-acetylation of host proteins using gnotobiotic mice.
Proc Natl Acad Sci USA. 109:11133–11138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Choudhary C, Kumar C, Gnad F, Nielsen ML,
Rehman M, Walther TC, Olsen JV and Mann M: Lysine acetylation
targets protein complexes and co-regulates major cellular
functions. Science. 325:834–840. 2009. View Article : Google Scholar : PubMed/NCBI
|