Introduction
The World Health Organization estimated that ~10
million individuals succumbed to cancer in 2020, of which colon
cancer exhibited the third highest rate of incidence (1). There are several methods for detecting
colon cancer, such as colonoscopy, fecal occult blood tests (FOBT)
and fecal DNA tests (2). However, it
should be emphasized that FOBT has low specificity and sensitivity.
A promising technique for the diagnosis of colon cancer is the
detection of DNA in stool. However, there are certain limitations
for the use of this technique, since both DNA from the patient and
intestinal bacteria can be detected, the latter of which is more
highly prevalent in the stool (3).
Colonoscopy is the most reliable method for the
early detection of colon cancer and premalignant lesions. However,
its invasive nature and high cost restrict its widespread
application (4). Timely diagnosis of
colon cancer and early treatment are two fundamental factors for
the successful approach of patients with colon cancer (5). Furthermore, it is essential to evaluate
the cancer biomarkers of patients during different pharmacological
treatments, in order to identify the most appropriate therapy.
During a retrospective study of a phase I trial, in which patients
with advanced disease received at least two cycles of treatment
with anti-programmed cell death protein 1/programmed cell death 1
ligand 1, a decrease in the neutrophil-to-lymphocyte ratio was
detected, and treatment was associated with prolonged
progression-free survival (6).
MicroRNAs (miRNAs/miRs) are small, conserved,
non-coding RNAs of 17–24 nucleotides, transcribed by RNA polymerase
II (7). miRNAs can trigger
post-transcriptional gene silencing by binding the 3′ untranslated
region of target genes, rendering messenger RNA (mRNA)
inaccessible, and thus inhibiting translation (8). During altered physiological processes,
such as cancer, different cells can secrete miRNAs into the
extracellular space, which are further transported in systemic
circulation (9). These circulating
miRNAs can be found in different tissues and biological fluids,
including peripheral blood, saliva, follicular fluid, urine, breast
milk and semen (10). The stability
of extracellular miRNAs is the result of packaging into exosomes,
extracellular vesicles released into biological fluids that act as
carriers and prevent the RNase-associated degradation of nucleic
acids (11). Exosomal miRNAs are
known to be potential biomarkers for cancer screening, which may
reduce the need for invasive techniques such as colonoscopy
(12). miRNAs can also be used to
predict chemotherapeutic efficiency; previously, miR-20a, −130 and
−145 have been reported as novel predictive markers for FOLFOX
regimen resistance in colorectal cancer (13). Furthermore, due to its upregulation
in patients, miR-31 has been used for the diagnosis of colon cancer
(14). miRNA detection is primarily
based on reverse transcription-quantitative (RT-qPCR) and/or
microarray analysis. However, since miRNAs are short sequences that
can be difficult to isolate, these techniques may be of limited use
in certain conditions, including paraffin embedded tissue. To
overcome this problem, conventional quantification techniques, such
as nanotechnology-based approaches, could be implemented to
generate the most sensitive and stringent tests for miRNA detection
(15). Metal nanoparticles, such as
silver and gold nanoparticles (AuNPs), are of great interest in the
medical field due to their properties and applications, especially
as diagnostic and therapeutic tools (16). AuNPs are reliable fluorescence
quenchers, and as such, enhance the sensitivity and selectivity of
fluorescence-based approaches, including the detection of miRNAs in
cancer cells, tissues and biological fluids (17). During tumor development, miRNAs can
be secreted into the extracellular environment and encapsulated in
exosomes; since exosomes can be detected by AuNPs coupled to CD81,
a cell surface protein highly expressed in cellular proliferation
and differentiation events, these miRNAs can serve as cancer
biomarkers (18).
Chlorogenic acid (CGA) is a compound found in a
variety of fruits and vegetables, including apples, pears and
carrots. CGA is highly abundant in nature, and due to its high
content in coffee beans, is the primary polyphenol consumed in
various populations (19). CGA is
also active against a wide range of microorganisms, including
bacteria, viruses, yeasts, molds and amoebas (20). Likewise, the primary metabolite of
CGA (dihydrocaffeic acid) is produced by the gut microbiota,
possesses anticancer activity against colon cancer (21,22).
Considering both the advantages of miRNAs for the
diagnosis of several types of human cancer, and the inhibitory
effects of CGA on oncogene expression, could highlight novel
mechanisms of non-invasive molecular cancer diagnosis, resulting in
the identification of adjuvant treatments for patients with colon
cancer (23). The aim of the present
study was to evaluate the antitumor potential of CGA, quantifying
the suppression of miR-31 expression in an in vitro model of
colon cancer. Furthermore, AuNPs were used to evaluate whether
their use increased the detection of circulating miRNAs in the
culture medium.
Materials and methods
Cell culture and CGA treatment
RKO human colon cancer cells (ATCC; CRL-2577) were
cultured in Advanced Dulbecco's Modified Eagle's Medium (ADMEM)
supplemented with 10% fetal bovine serum (FBS), 1%
penicillin-streptomycin (all Gibco; Thermo Fisher Scientific, Inc.)
and 1% L-glutamine (Sigma-Aldrich; Merck KGaA), in a humidified
incubator at 37°C (5% CO2). For passage, the cells were
washed with sterile 1X phosphate-buffered saline (PBS), and
digested with trypsin (0.25%)-EDTA solution (Gibco; Thermo Fisher
Scientific, Inc.) for 5–10 min at 37°C. To inactivate the enzyme,
an equal volume of medium supplemented with FBS was added to the
trypsin/cell mixture, which was subsequently centrifuged at 300 × g
for 5 min at room temperature. The supernatant was removed, and the
cell pellet was resuspended in 2–4 ml culture medium. The cells
were subcultured at a ratio of 1:3 to 1:12 and incubated at 37°C
with 5% CO2 until 80% confluent.
CGA treatment and viability
assessment
To determine the effects of CGA on survival, cell
cultures underwent a cell viability assay (Sigma-Aldrich; Merck
KGaA) following treatment with 250, 500, 750 and 1,000 µM CGA for
24, 48 and 72 h. The viability assay was performed in triplicate
using Alamar Blue (Invitrogen; Thermo Fisher Scientific, Inc.) with
4×104 RKO cells/well in a 96-well plate, with each well
initially containing 200 µl ADMEM; three cell-free wells served as
the negative control. Following CGA treatment at the specified time
and concentration, the medium was removed, and 200 µl Alamar Blue
was added to each well. The plate was incubated until a color
change was observed in cells without treatment, and the optical
density (OD) was determined using a Multiskan Go microplate reader
(Thermo Fisher Scientific, Inc.) at 570/600 nm.
We selected 1,000 µM CGA after the results of the
viability assay with alamar blue. Subsequently, 1.2×105
RKO cells were transferred to a 24-well plate and cultured in the
presence of 1,000 µM CGA. The cells were incubated for 24, 48 and
72 h, after which 300 µl supernatant was recovered from each well
and stored at −20°C until RNA isolation.
RNA isolation with AuNPs + CD81
Exosomal miR-31 quantification was conducted from a
cell culture sample treated with 1,000 µM CGA; prior to the
extraction of miR-31, the aliquot of culture medium was treated
with AuNPs coupled to CD81 (AuNPs + CD81). Briefly, miRNA was
extracted from the supernatant (culture medium) of RKO cells. This
medium was treated with different combinations of factors,
following a 3×2×2 mixed factorial design (in triplicate).
Specifically, the three analyzed factors were as follows:
incubation time (24, 48 and 72 h), CGA (treated with 1,000 µM or
untreated) and AuNPs + CD81 (presence or absence).
For treatment with AuNPs + CD81, the Gold
Conjugation Kit (40 nm, 20 OD; Abcam) was used according to the
manufacturer's instructions. Firstly, 50 µl of a mixture containing
Gold Reaction Buffer, Gold Quencher, CD81 antibody (0.02 mg/ml) and
lyophilized AuNPs was added to 300 µl culture medium, the mix was
incubated 10 min at room temperature.
For miRNA isolation, the mirVana™ miRNA Isolation
kit (Ambion; Thermo Fisher Scientific, Inc.) was used, per the
manufacturer's protocol. To homogenize the solution, miRNA
Homogenate Additive was added (incubation 10 min at 4°C), followed
by acid-phenol-chloroform (5:1), pH 4.5. The samples were later
centrifuged at 25°C for 5 min at 10,000 × g. The aqueous phase
containing miRNAs was recovered and transferred to a new tube,
leaving behind lipids, proteins, and salts. Subsequently, 100%
ethanol was mixed with the aqueous solution, and filtered by
passing the samples through filter cartridges (containing a
glass-fiber filter which immobilizes the RNA; Ambion; Thermo Fisher
Scientific, Inc.) and spinning at 10,000 × g for 15 sec at room
temperature. Washing solutions were added to the filter cartridges,
and finally, miRNA was eluted with 100 µl preheated elution
solution (95°C), and stored at −20°C.
RT-qPCR
RNA was reverse transcribed from the total RNA
extracted using TaqMan Advanced miRNA Assays (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and a MyCycler™ 1709713 (Bio-Rad
Laboratories, Inc.). The reaction conditions were as follows: 42°C
for 15 min and 85°C for 5 min. The amount of total cDNA obtained
after RT was measured using a NanoDrop 2000c (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). Then, qPCR was
performed to quantify miR-31. Expression levels were normalized to
that of miR-191, which is constitutively expressed. The primers
used in both cases were 20X miR-Amp Primer Mix (A25576; Applied
Biosystems; Thermo Fisher Scientific, Inc.) TaqMan fluorescent
probes for miR-31 (ref. 477827_miR-31; Applied Biosystems) and
miR191 (ref. 477952_mir191; Applied Biosystems) were used, as well
as specific primers for both miR-31 and miR-191. The reaction was
carried out using the StepOnePlus Real-Time PCR System (Applied
Biosystems) with the following thermocycling conditions: 95°C for
20 sec, then 40 cycles of 95°C for 1 sec and 60°C for 20 sec. The
results were analyzed using the 2−ΔΔCq method (24). Each experiment was performed in
triplicate and using untreated cells as a negative control.
Statistical analysis
The RT-qPCR results were evaluated using one-way
ANOVA followed by Dunnett's post hoc test using SPSS for Windows,
v.20 (IBM Corp.). Data are presented as the mean ± SD. P<0.05
was considered to indicate a statistically significant
difference.
Results
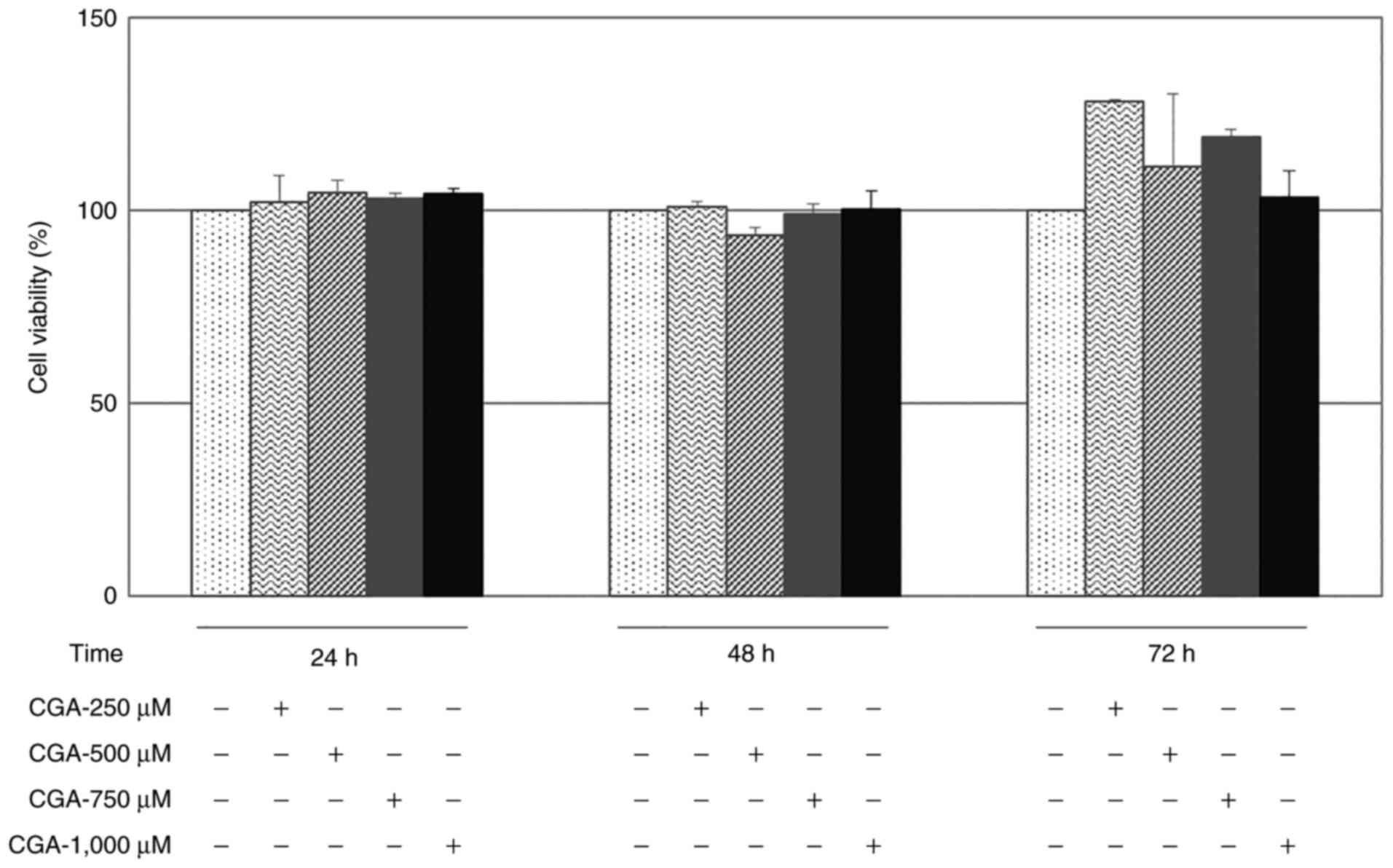
Viability of RKO cells treated with
CGA
To assess viability, RKO cells were incubated for up
to 72 h with 250, 500, 750 and 1,000 µM CGA. There were no
cytotoxic effects of CGA on RKO cells at the evaluated
concentrations at 24, 48 or 72 h post-treatment (Fig. 1). Nevertheless, the highest
concentration (1,000 µM) was selected for subsequent experiments to
obtain the most significant inhibitory effect, and following the
information found in a previous study (25). The CGA did not show a cytotoxic
effect on RKO at any of the assessed concentrations.
Evaluation of miR-31 levels following
treatment with CGA
To evaluate whether 1,000 µM CGA could modulate
miR-31 expression in RKO cells, the miR-31 level was quantified
from cell supernatants by RT-qPCR. miR-31 expression was
downregulated at 24 (~97%; P=1.7×10−5), 48 (~75%;
P=0.002) and 72 h (~85%; P=3.08×10−4) post-treatment
with CGA (Fig. 2), compared with the
untreated controls at the same timepoints. Therefore, 1,000 µM CGA
can reduce the expression levels of the oncogene, miR-31.
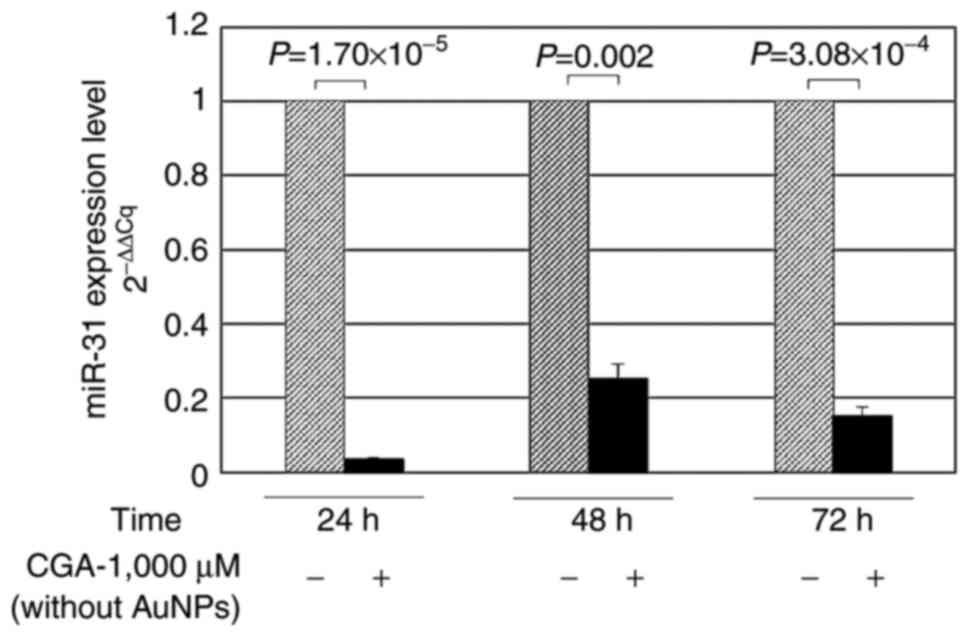 | Figure 2.Quantification of miR-31 isolated from
the supernatants of RKO colon cancer cells treated with CGA,
without AuNPs. Cells were cultured in 24-well plates and treated
with 1,000 µM CGA. Total RNA was isolated, and miR-31 was
quantified at 24, 48 and 72 h by reverse transcription-quantitative
PCR. miR-31 quantification was significantly higher in cells that
were not treated with CGA, demonstrating the antitumoral properties
of CGA by the reduction of the tumoral marker miR-31. (−),
Untreated cells; (+), cells treated with 1,000 µM CGA. miR,
microRNA; CGA, chlorogenic acid; AuNPs, gold nanoparticles. |
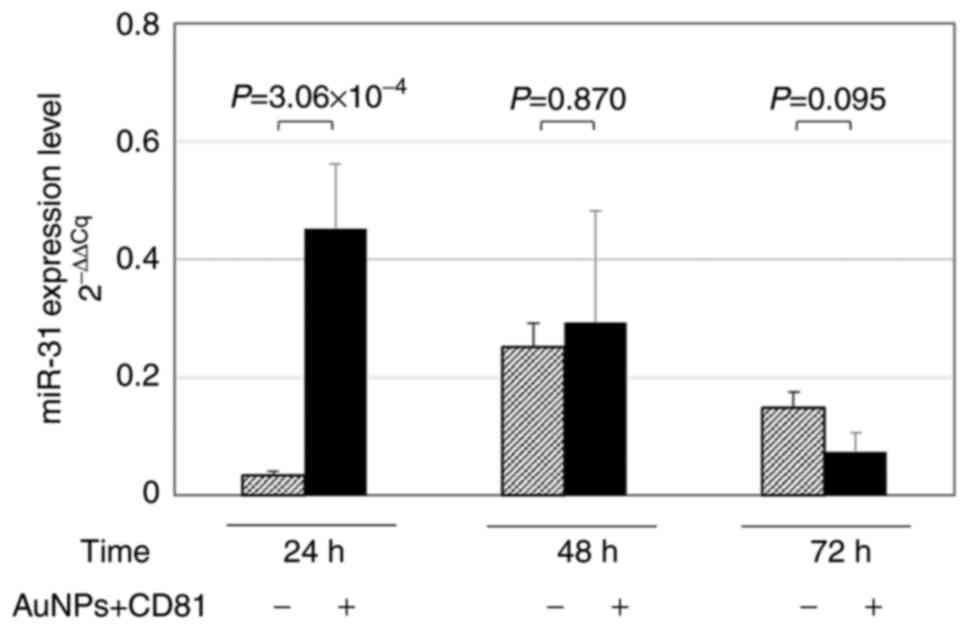
miR-31 levels following treatment with
CGA and isolation with AuNPs + CD81
RKO cells were harvested at 24, 48 or 72 h for miRNA
isolation in the presence or absence of AuNPs + CD81. This
experiment aimed to evaluate whether the use of nanoparticles
allowed the detection of higher miR-31 levels after treatment with
CGA, compared with the isolation of miRNAs without AuNPs + CD81.
The greatest effect was observed at 24 h (~55%,
P=7.3×10−5) without AuNPs + CD81 (Fig. 3). In Fig.
1 and 2, the inhibitory effect
of CGA on miR-31 can be observed; however, the use of AuNPs
optimized the capacity of the quantification method. Consequently,
when compared with isolation without AuNPs, a higher level of
miR-31 was quantified at 24 h when AuNPs were used to optimize
RT-qPCR detection, (in both cases the cells were treated with CGA
1,000 µM) (P=3.06 ×10−4; Fig
4). According to the results of the present study, this
strategy could be implemented in patients with colon cancer to
isolate exosomes using AuNPs + CD81 and optimize the quantification
of miRNAs derived from serum/plasma.
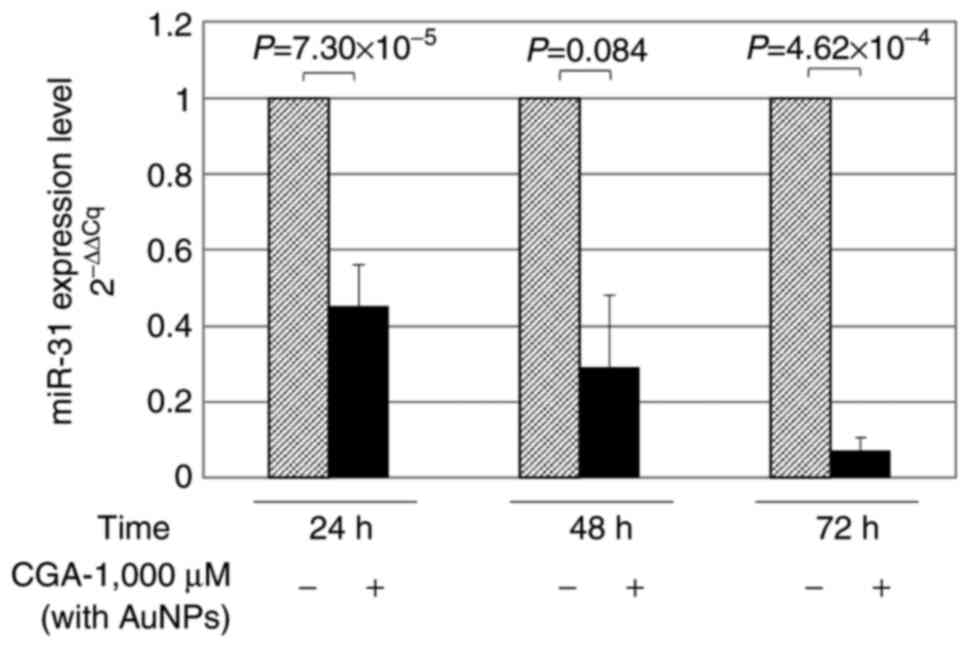 | Figure 3.Quantification of miR-31 isolated from
the supernatants of RKO colon cancer cells treated with CGA, with
AuNPs. Cells were cultured in 24-well plates and treated with 1,000
µM CGA. Total RNA, and miRNAs encapsulated in exosomes coupled with
AuNPs + CD81, were isolated. miR-31 was quantified at 24, 48, and
72 h by reverse transcription-quantitative PCR. Decreased
concentrations of miR-31 were found when treated with CGA for
different times of exposure. (−), Untreated cells; (+) cells
treated with CGA 1,000 µM and AuNPs. miR, microRNA; CGA,
chlorogenic acid; AuNPs, gold nanoparticles. |
Discussion
The aim of the present study was to evaluate the
antitumor potential of CGA by quantifying the suppression of the
miR-31 in an in vitro model of colon cancer. Furthermore,
the potential use of AuNPs to increase the levels of circulating
miRNAs in the culture medium (and thus enhance their detection) was
also assessed. AuNPs + CD81 were used to isolate miRNAs
encapsulated in exosomes from the culture medium of RKO colon
cancer cells, which were treated with 1,000 µM CGA for 24, 48 and
72 h. The RT-qPCR results showed that CGA significantly inhibited
miR-31 expression at 24 (~97%), 48 (~75%,) and 72 h (~85%) compared
with negative controls. Numerous studies have reported that CGA
serves important therapeutic roles, including as an antioxidant,
antibacterial, hepatoprotective, cardioprotective,
anti-inflammatory, antipyretic, neuroprotective, anti-obesity,
antiviral, anti-microbial and anti-hypertensive agent, as well as a
free radical scavenger, central nervous system stimulator and
oncogene inhibitor (20–22,26–28). CGA
was also found to regulate the expression of apoptosis-related
genes in lung and colon cancer cell lines (25,29), the
latter of which appear to be especially affected by CGA. For
example, Hou et al (30)
showed that CGA has several effects on colon cancer cells (HCT116
and HT29), including S-phase arrest, extracellular signal-related
kinase inactivation, induced reactive oxygen species and a decrease
in cell viability. These results suggest CGA as a potential
treatment for colorectal cancer (30).
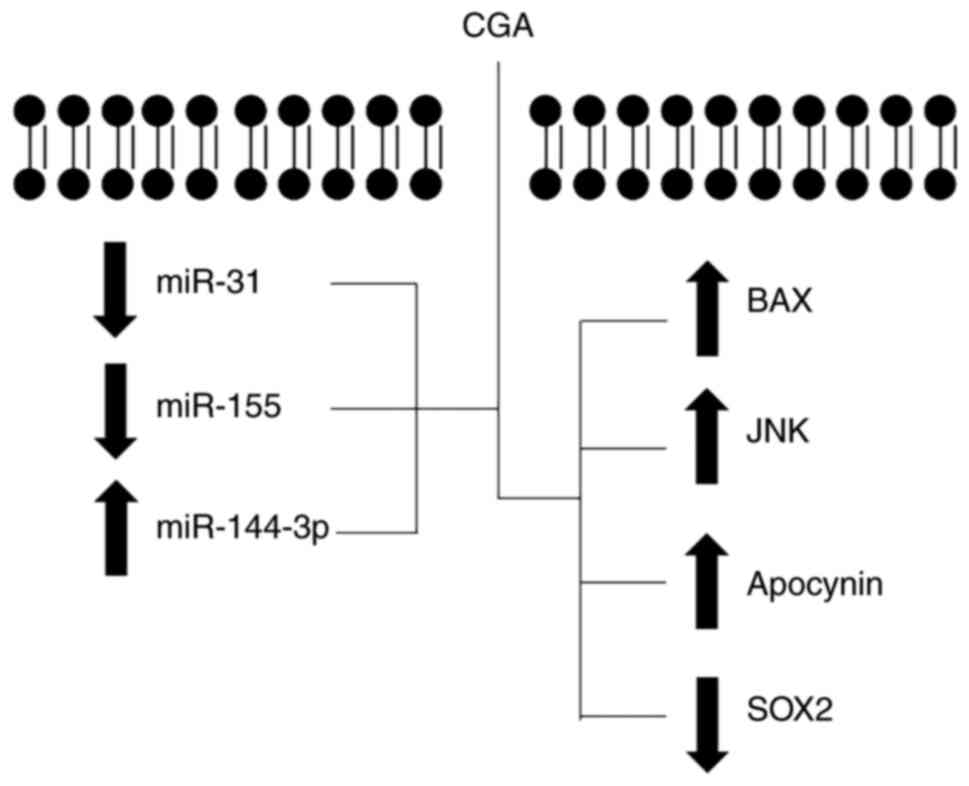
miRNAs regulate ~30% of human genes; they act as
oncogenic and tumor suppressor genes, and as such, are frequently
dysregulated in cancer (31). The
oncomiR-31 inhibits the translation of RAS P21 protein activator 1,
an inhibitor of KRAS protooncogene, thus stimulating the
transcription of the KRAS oncogene, which promotes cell
cycle progression in colon cancer cells (32). According to previous research,
miR-31, and other miRNAs such as miR155 (33) and miR144-3p (34), are modulated by CGA (Fig. 5). Hence, they have great potential as
biomarkers for colon cancer diagnosis, and in disease monitoring
during treatment. However, as short-sequence molecules, miRNAs are
difficult to isolate from some biological samples (such as feces).
Therefore, in conventional quantification techniques,
nanotechnology-based approaches may be used to develop the most
sensitive and stringent methods for miRNA detection (16).
To determine whether the addition of AuNPs increased
the sensitivity of miRNA detection, the present study aimed to
quantify miR-31 expression after treatment with 1,000 µM CGA (24,
48 and 72 h), and to isolate total RNA using AuNPs + CD81. The most
significant increase in miR-31 quantification was observed at 24 h;
this maximum inhibition may be attributed to the antioxidant effect
of CGA during the earliest exposure time (29). AuNPs have a variety of properties and
uses, making them amenable to a variety of detection modalities and
techniques (35). As supported by
the present study results, a novel strategy can be proposed to
quantify miR-31 expression using AuNPs; miR-31 expression can serve
as an effective, non-invasive molecular diagnostic and monitoring
tool for colon cancer. The efficacy of this strategy as an
alternative non-invasive diagnostic method could be validated in
fecal samples by extracting and quantifying miRNAs encapsulated in
exosomes that were previously isolated with nanoparticles (36). In addition, CGA significantly
decreased miR-31 expression at 24, 48 and 72 h, which may be
explained by one of the antitumor mechanisms of CGA in colon
cancer. Moreover, the present results indicate that AuNPs may
optimize the miRNA isolation method. However, additional studies
are necessary to validate this approach, using biological samples
from patients with colon cancer to establish its diagnostic
potential in disease.
Acknowledgements
To authors of the present study would like to thank
Dr Mauricio Salinas-Santander (School of Medicine, Universidad
Autonoma de Coahuila) for their help formatting the figures.
Funding
The present study was supported by the State Council
of Science and Technology (COECyTJAL, grant no. 4673-2016 to Clara
Patricia Rios-Ibarra).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
CPRI designed the present study. CPRI, ACLB, DARC,
GHT, AMCL, DES and JPCG performed the experiments and acquired the
data. CPRI and MLMF performed data curation. CPRI, MLMF and DAJV
analyzed the results and interpretated the data. CPRI, DAJV and
MLMF provided resources. All authors were involved in the
preparation of the original drafted manuscript, which was reviewed
and edited by CPRI, DAJV and MLMF. CPRI, DAJV and MLMF supervised
the study. CPRI and ACLB confirmed the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cancer. https://www.who.int/news-room/fact-sheets/detail/cancer
|
|
2
|
Hamzehzadeh L, Yousefi M and Ghaffari SH:
Colorectal cancer screening: A comprehensive review to recent
non-invasive methods. Int J Hematol Oncol Stem Cell Res.
11:250–261. 2017.PubMed/NCBI
|
|
3
|
Zarkavelis G, Boussios S, Papadaki A,
Katsanos KH, Christodoulou DK and Pentheroudakis G: Current and
future biomarkers in colorectal cancer. Ann Gastroenterol.
30:613–621. 2017.PubMed/NCBI
|
|
4
|
Lin J, Chuang CC and Zuo L: Potential
roles of microRNAs and ROS in colorectal cancer: Diagnostic
biomarkers and therapeutic targets. Oncotarget. 8:17328–17346.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mohammadi R, Ansari Chaharsoghi M,
Khorvash F, Khorvash F, Kaleidari B, Sanei MH, Ahangarkani F,
Abtahian Z, Meis JF and Badali H: An unusual case of
gastrointestinal basidiobolomycosis mimicking colon cancer;
literature and review. J Mycol Med. 29:75–79. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moschetta M, Uccello M, Kasenda B, Mak G,
McClelland A, Boussios S, Forster M and Arkenau HT: Dynamics of
neutrophils-to-lymphocyte ratio predict outcomes of PD-1/PD-L1
blockade. Biomed Res Int. 2017:15068242017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yao P, Wu J, Lindner D and Fox PL:
Interplay between miR-574-3p and hnRNP L regulates VEGFA mRNA
translation and tumorigenesis. Nucleic Acids Res. 45:7950–7964.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang YH, Jin M, Li J and Kong X:
Identifying circulating miRNA biomarkers for early diagnosis and
monitoring of lung cancer. Biochim Biophys Acta Mol Basis Dis.
1866:1658472020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Peng R, Wang J, Qin Z and Xue L:
Circulating microRNAs as potential cancer biomarkers: The advantage
and disadvantage. Clinical Epigenetics. 10:592018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ingenito F, Roscigno G, Affinito A, Nuzzo
S, Scognamiglio I, Quintavalle C and Condorelli G: The role of
Exo-miRNAs in cancer: A focus on therapeutic and diagnostic
applications. Int J Mol Sci. 20:46872019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tinmouth J, Kennedy EB, Baron D, Burke M,
Feinberg S, Gould M, Baxter N and Lewis N: Colonoscopy quality
assurance in ontario: Systematic review and clinical practice
guideline. Can J Gastroenterol Hepatol. 28:251–274. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boussios S, Ozturk MA, Moschetta M,
Karathanasi A, Zakynthinakis-Kyriakou N, Katsanos KH, Christodoulou
DK and Pavlidis N: The developing story of predictive biomarkers in
colorectal cancer. J Pers Med. 9:122019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Chen Z and Chen W: Novel
circulating microRNAs expression profile in colon cancer: A pilot
study. Eur J Med Res. 22:512017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aldewachi H, Chalati T, Woodroofe MN,
Bricklebank N, Sharrack B and Gardiner P: Gold nanoparticle-based
colorimetric biosensors. Nanoscale. 10:18–33. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chaudhary V, Jangra S and Yadav NR:
Nanotechnology based approaches for detection and delivery of
microRNA in healthcare and crop protection. J Nanobiotechnol.
16:402018. View Article : Google Scholar
|
|
17
|
Singh H, Du J, Singh P and Yi TH:
Ecofriendly synthesis of silver and gold nanoparticles by Euphrasia
officinalis leaf extract and its biomedical applications. Artif
Cells Nanomed Biotechnol. 46:1163–1170. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Klymiuk MC, Balz N, Elashry MI, Heimann M,
Wenisch S and Arnhold S: Exosomes isolation and identification from
equine mesenchymal stem cells. BMC Vet Res. 15:422019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Loader TB, Taylor CG, Zahradka P and Jones
PJH: Chlorogenic acid from coffee beans: Evaluating the evidence
for a blood pressure-regulating health claim. Nutr Rev. 75:114–133.
2017.PubMed/NCBI
|
|
20
|
Santana-Gálvez J, Cisneros-Zevallos L and
Jacobo-Velázquez DA: Chlorogenic acid: Recent advances on its dual
role as a food additive and a nutraceutical against metabolic
syndrome. Molecules. 22:3582017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Santana-Gálvez J, Villela-Castrejón J,
Serna-Saldívar SO, Cisneros-Zevallos L and Jacobo-Velázquez DA:
Synergistic combinations of curcumin, sulforaphane, and
dihydrocaffeic acid against human colon cancer cells. Int J Mol
Sci. 21:31082020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Santana-Gálvez J, Castrejón JV,
Serna-Saldívar SO and Jacobo-Velázquez DA: Anticancer potential of
dihydrocaffeic acid: A chlorogenic acid metabolite. CyTA J Food.
18:245–248. 2020. View Article : Google Scholar
|
|
23
|
Cekaite L, Eide PW, Lind GE, Skotheim RI
and Lothe RA: MicroRNAs as growth regulators, their function and
biomarker status in colorectal cancer. Oncotarget. 7:6476–6505.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sadeghi Ekbatan S, Li XQ, Ghorbani M,
Azadi B and Kubow S: Chlorogenic acid and its microbial metabolites
exert anti-proliferative effects, S-phase cell-cycle arrest and
apoptosis in human colon cancer Caco-2 cells. Int J Mol Sci.
19:7232018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tajik N, Tajik M, Mack I and Enck P: The
potential effects of chlorogenic acid, the main phenolic components
in coffee, on health: A comprehensive review of the literature. Eur
J Nutr. 56:2215–2244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Naveed M, Hejazi V, Abbas M, Kamboh AA,
Khan GJ, Shumzaid M, Ahmad F, Babazadeh D, FangFang X,
Modarresi-Ghazani F, et al: Chlorogenic acid (CGA): A
pharmacological review and call for further research. Biomed
Pharmacother. 97:67–74. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X, Liu J, Xie Z, Rao J, Xu G, Huang
K, Li W and Yin Z: Chlorogenic acid inhibits proliferation and
induces apoptosis in A498 human kidney cancer cells via
inactivating PI3K/Akt/mTOR signalling pathway. J Pharm Pharmacol.
71:1100–1109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yamagata K, Izawa Y, Onodera D and Tagami
M: Chlorogenic acid regulates apoptosis and stem cell
marker-related gene expression in A549 human lung cancer cells. Mol
Cell Biochem. 441:9–19. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hou N, Liu N, Han J, Yan Y and Li J:
Chlorogenic acid induces reactive oxygen species generation and
inhibits the viability of human colon cancer cells. Anticancer
Drugs. 28:59–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Si W, Shen J, Zheng H and Fan W: The role
and mechanisms of action of microRNAs in cancer drug resistance.
Clin Epigenetics. 11:252019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Strubberg AM and Madison BB: MicroRNAs in
the etiology of colorectal cancer: Pathways and clinical
implications. Dis Model Mech. 10:197–214. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zeng J, Zhang D, Wan X, Bai Y, Yuan C,
Wang T, Yuan D, Zhang C and Liu C: Chlorogenic acid suppresses
miR-155 and ameliorates Ulcerative colitis through the NF-κB/NLRP3
inflammasome pathway. Mol Nutr Food Res. 64:e20004522020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Romualdo GR, Prata GB, da Silva TC,
Evangelista AF, Reis RM, Vinken M, Moreno FS, Cogliati B and
Barbisan LF: The combination of coffee compounds attenuates early
fibrosis-associated hepatocarcinogenesis in mice: Involvement of
miRNA profile modulation. J Nutr Biochem. 85:108479. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Coutinho C and Somoza Á: MicroRNA sensors
based on gold nanoparticles. Anal Bioanal Chem. 411:1807–1824.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Duran-Sanchon S, Moreno L, Augé JM,
Serra-Burriel M, Cuatrecasas M, Moreira L, Martín A, Serradesanferm
A, Pozo À, Costa R, et al: Identification and validation of
MicroRNA profiles in fecal samples for detection of colorectal
cancer. Gastroenterology. 158:947–957.e4. 2020. View Article : Google Scholar : PubMed/NCBI
|