Introduction
Colorectal cancer (CRC) is the development of cancer
from the colon or the rectum. It is the third most common cancer
type in males and the second most common cancer type in females
(1). CRC also accounts for ~10% of
annual global cancer incidence, which is increasing with economic
development (2). Furthermore, the
5-year survival rate decreases at lower levels of income, with
rates reaching 60% in high-income countries but falling to 30% or
less in low-income countries (3,4). The low
survival rate is due to metastasis of primary CRC to the liver,
lungs and other organs (1,3). Despite breakthroughs in clinical
research that have identified numerous genetic and protein
abnormalities in CRC, it remains the second leading cause of
cancer-related mortality (1). The
molecular mechanisms underlying CRC tumorigenesis and progression
have not yet been elucidated.
Collapsin response mediator proteins (CRMPs), namely
CRMP 1–5, are a family of five homologous multifunctional cytosolic
phosphoproteins highly expressed in the developing brain (5,6). They
are involved in regulating cell migration through interactions with
the cytoskeleton and altered expression levels of CRMPs have been
detected in several cancer types, including breast, prostate and
lung carcinoma (7–10). The most recently identified CRMP,
CRMP5, also known as DPYSL5, is widely expressed in regions of
neurogenesis in the adult brain (11,12).
CRMP5 was identified as a specific marker of high-grade lung
neuroendocrine carcinoma, including small cell lung carcinomas and
large cell neuroendocrine carcinomas (13). In glioblastoma, CRMP5 was found to
control cancer cell proliferation and survival via Notch-dependent
signaling (9). However, the role of
CRMP5 in CRC is unclear.
The mitogen-activated protein kinase (MAPK) family
consists primarily of extracellular-signal-regulated kinase (ERK),
c-jun N-terminal kinase (JNK) and p38MAPK (14). Each MAPK signaling pathway contains
at least three major components, including mitogen-activated
protein 3 kinase (MAPKKK), mitogen-activated protein 2 kinase
(MAPKK) and MAPK (14,15). Various extracellular and
intracellular stimuli, including genotoxic agents, cytokines,
hormones, oxidative stress and cell adhesion, can promote
sequential activation of these kinase components which
phosphorylate various substrates, including transcription factors
and enzymes, to regulate multiple cellular physiological or
pathological processes, including inflammation, cell proliferation,
differentiation, metabolism, survival and death (15–17).
However, MAPK serves a pleiotropic role in cancer, and the
associated mechanisms of the MAPK pathway in regulating tumors
remain elusive.
The present study demonstrated that high CRMP5
expression is associated with poor survival in 367 patients with
CRC. CRMP5 regulates CRC cell proliferation and development via
MAPK signaling. Additionally, an unexpected role of
CRMP5-overexpression in promoting CRC chemotherapy resistance and
tumor recurrence was identified. The results of the present study
elucidated novel mechanisms of CRMP5-induced CRC cell proliferation
and development and provided a potential novel target for CRC
treatments.
Materials and methods
Cell culture and transfection
Human CRC HCT116 and HT29 cell lines were purchased
from the Cell Resource Center of Shanghai Institutes for Biological
Sciences, Chinese Academy of Sciences. Cell lines were
authenticated by STR analysis (Shanghai Biotechnology Co., Ltd.,
http://www.cellresource.cn/index.aspx). Cells were
grown in Dulbecco's minimal essential medium (DMEM, Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin
streptomycin-glutamine (Gibco; Thermo Fisher Scientific, Inc.) at
37°C with 5% CO2.
Human CRMP5-overexpression or -knockdown plasmids
were constructed using pCDH and pLKO.1 vectors (Genomeditech),
respectively. Recombinant plasmids and packaging plasmids,
including VSVG (0.75 µg, Addgene) and psPAX2 (1.72 µg, Addgene),
were transiently transfected into 293 cells (Shanghai Zhongqiao
Xinzhou Biotechnology Co., Ltd.) to produce lentivirus using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Cultured CRC cells were infected with the appropriate lentivirus in
the presence of polybrene (5 µg/ml; Sigma-Aldrich; Merck KGaA) at
37°C for 12 h, prior to being selected using puromycin (2 µg/ml;
Sigma-Aldrich; Merck KGaA). An empty vector or a nonsense scrambled
oligonucleotide was used as a negative control. Following selection
with puromycin (2 µg/ml; Sigma-Aldrich; Merck KGaA) for 2 week at
37°C, transfected cells were harvested for subsequent
experimentation. CRMP5 gene expression of the transfected cells was
evaluated via reverse transcription-quantitative (RT-q)PCR and
western blot analyses.
Western blotting
Cultured CRC cells treated without or with
selumetinib (500 nM) at 37°C for 24 h were collected and lysed with
1X RIPA Lysis Buffer (Sigma-Aldrich; Merck KGaA) containing 1X
Protease Inhibitor Cocktail (Sigma-Aldrich; Merck KGaA). The
concentration of total protein was measured with a BCA protein
assay kit (Beyotime Institute of Biotechnology). Total protein (30
µg/lane) was separated using 6–12.5% SDS-PAGE, transferred to a
polyvinylidene fluoride membrane, and blocked with 5% skimmed milk
at room temperature for 1 h. Next, membranes were incubated with
primary antibodies against CRMP5 (Abcam; cat. no. ab36203;
1:2,000), Ki67 (Abcam; cat. no. ab15580; 1:1,000), Caspase-3 (CST;
cat. no. 96659665; 1:1,000), E-cadherin (CST; cat. no. 3195;
1:1,000), MMP-2 (CST; cat. no. 4022; 1:1,000), MMP-9 (CST; cat. no.
13667; 1:1,000) and vimentin (CST; cat. no. 5741; 1:1,000)
overnight at 4°C, followed by incubation with
fluorescence-conjugated goat ant-mouse IgG H&L (Abcam; cat. no.
ab216772) or goat an-rabbit IgG H&L (Abcam; cat. no. ab216773)
secondary antibodies (dilution, 1:1,000) at room temperature for 1
h. Finally, bands were detected using a two-color infrared laser
imaging system (Odyssey; LI-COR Biosciences).
Cell proliferation assays
For colony formation assay, cells (1,000/well) were
seeded into 12-well plates and cultured in fresh DMEM medium
(Gibco; Thermo Fisher Scientific, Inc.) with or without selumetinib
(500 nM) for approximately two weeks to allow colony formation.
Cells were then fixed with 4% paraformaldehyde at room temperature
for 10 min, and stained with 1% crystal violet at room temperature
for 20 min. The number of colonies was counted using ImageJ
software (v1.51; National Institutes of Health). Cell viability was
measured using the Cell Counting kit-8 (CCK-8) assay. Cultured CRC
cells (2,000 cells/well) were seeded into 96-well plates for 24 h.
The next day, cells were treated with increasing doses (0, 1, 5,
10, 50, 100 and 200 µM) of 5-FU (Sigma-Aldrich; Merck KGaA) at 37°C
for 48 h. Next, CCK-8 solution (10 µl/well; Dojindo Molecular
Technologies, Inc.) was added to each well and absorbance was
measured at 450 nm using a microplate reader.
Transwell migration assay
CRC cells (2×105 cells/well) in 200 µl
complete DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) were
seeded into the upper Transwell chamber (12-µM pore size; BD
Biosciences) and DMEM medium (Gibco; Thermo Fisher Scientific,
Inc.) with 15% FBS was plated into the lower chamber. After 48 h at
37°C in culture, cells adhering to the underside of the
transmembrane were fixed with 20% methanol for 10 min at room
temperature, and then stained with 0.1% crystal violet for 1 h at
room temperature. Finally, the images were captured using an
inverted microscope (Carl Zeiss AG; magnification, ×100).
RT-qPCR
Total RNA was extracted from cells using the RNeasy
Mini kit (Qiagen GmbH) and reverse transcribed into cDNA with
specific oligo (dT) primers using the Prime Script RT Reagent kit
(Takara Bio, Inc.). The temperature and duration of RT were: 37°C
for 15 min, followed by 85°C for 5 sec. RT-qPCR was performed in
96-well plates using SYBR Premix Ex Taq II kit (Takara
Biotechnology Co., Ltd.), on an Applied Biosystems 7300 Real-Time
PCR System (Thermo Fisher Scientific, Inc.) to measure mRNA levels.
The thermocycling conditions used were as follows: 95°C for 30 sec,
followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. The
2−ΔΔCq method (18) was
used to calculate relative mRNA levels and GAPDH was used for
normalization of specific mRNA expression. Primers used in RT-qPCR
are listed in Table SI.
The Cancer Genome Atlas (TCGA) data
analysis
The Colorectal adenocarcinoma dataset (dataset no.
TCGA-COADREAD) from TCGA (https://portal.gdc.cancer.gov) was downloaded to
determine the correlation between selected gene (DPYSL5) and
patient survival (19,20). A total of 376 patients with
transcriptional profiles and basic clinical information, including
age, sex, stage, overall survival and survival status were enrolled
in the present study. RNaseq v2 mRNA expression data and clinical
information were retrieved from the cBioportal (21). Survival analysis was performed using
the R package survival (v3.5.3; http://www.bioconductor.org/packages), and survival
curves were fit using the survfit function. The difference between
high- and low-expression groups was tested by survdiff using the
median as the cut-off.
RNA-sequencing
RNA was extracted using Direct-zol™ RNA kit (Zymo
Research Corp.), purified with RNeasy Plus Mini kit (Qiagen GmbH).
One hundred nanograms of mRNA was used as input to the TruSeq mRNA
Sample Prep v2 kit (Illumina, Inc.), with the poly-A pulldown step.
Sample preparation was performed according to the manufacturer's
protocols. The libraries were sequenced on HiSeq X ten (Illumina,
Inc.) using 150 bp paired-end reads. The sequencing data termed raw
data (raw reads) were subjected to quality control (QC). Following
QC, raw data were filtered into clean data, and clean reads were
aligned to the reference genome (February 2009, GRCh37/hg19) using
BWA (v0.7.12). Genes with an adjusted P-value of <0.05
identified by DESeq2 (v1.12.3) were defined as differentially
expressed. The statistical enrichment of differentially expressed
genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
analysis was determined by Cluster Profiler (v3.0.4).
Statistical analysis
Data are presented as the mean ± standard deviation
of three independent experiments. Student's t-test or one-way
analysis of variance, followed by the Bonferroni post hoc
test, was used to compare the treated and corresponding control
groups. Survival curves were estimated using the Kaplan-Meier
method and analyzed by the log-rank test. P<0.05 was considered
to indicate a statistically significant difference.
Results
High CRMP5 expression is associated
with poor survival in TCGA patients with colorectal cancer
To evaluate the clinical significance of CRMP5 in
CRC, survival analysis was conducted using TCGA COADREAD data.
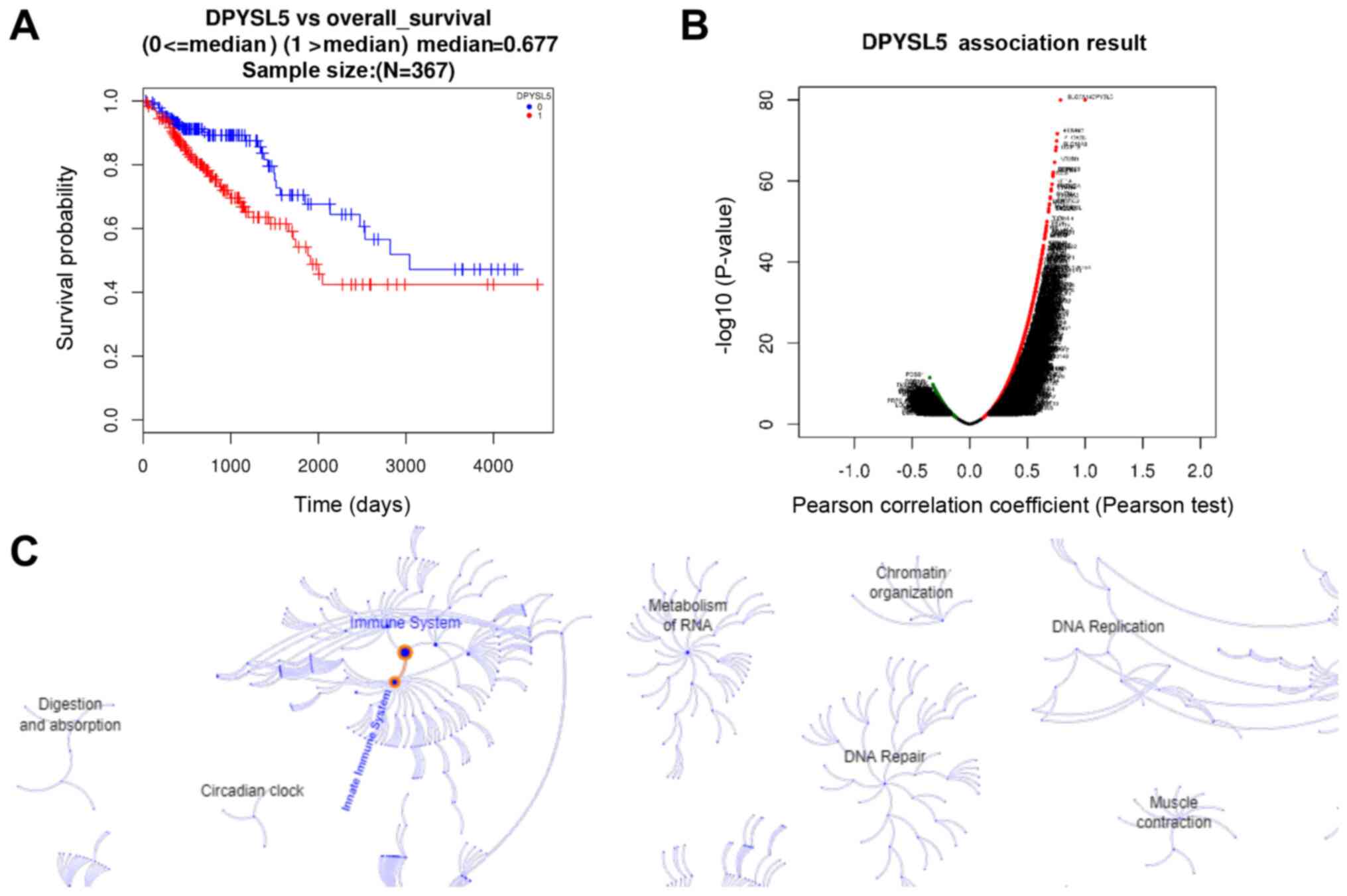
Notably, high expression of CRMP5 was significantly associated with
poor survival in 367 patients from COADREAD (Fig. 1A). Furthermore, co-expression
analysis by Pearson correlation demonstrated that the majority of
genes co-expressed with CRMP5 were mainly associated with the
immune response (Fig. 1B and C).
These data suggested that CRMP5 may serve an important role in
tumor inflammation, thereby promoting cancer development and
subsequently causing poor survival.
CRMP5-overexpression promotes cell
proliferation and development of CRC
To investigate the potential role of CRMP5 in CRC,
CRMP5 was overexpressed in HT29 cells using lentivirus-mediated
human CRMP5 cDNA. CRMP5-overexpression was confirmed by RT-qPCR and
Western blotting (Fig. 2A). HT29
cells overexpressing CRMP5 had an increase in cell number compared
with the wild-type (WT) cells and cells transfected with empty
plasmid (NC), indicating that high levels of CRMP5 promoted CRC
cell proliferation (Fig. 2B). This
result was further confirmed by a colony formation assay which
demonstrated that CRMP5-overexpression significantly increased the
colony forming ability of HT29 cells (Fig. 2C). Additionally, Transwell migration
assays demonstrated that CRMP5-overexpression promoted HT29 cell
migration (Fig. 2D). Additionally,
the expression levels of common markers for cell proliferation,
migration and invasion were measured. As expected, an increase in
the expression of Ki67, a common marker for cellular proliferation,
was observed and commonly used markers of migration and invasion,
including E-cadherin, vimentin, MMP-2 and MMP-9, while expression
of caspase-3, an apoptosis-related protein, was decreased in
CRMP5-OE HT29 cells compared with WT or NC as detected by RT-qPCR
and Western blotting (Fig. 2E and
F).
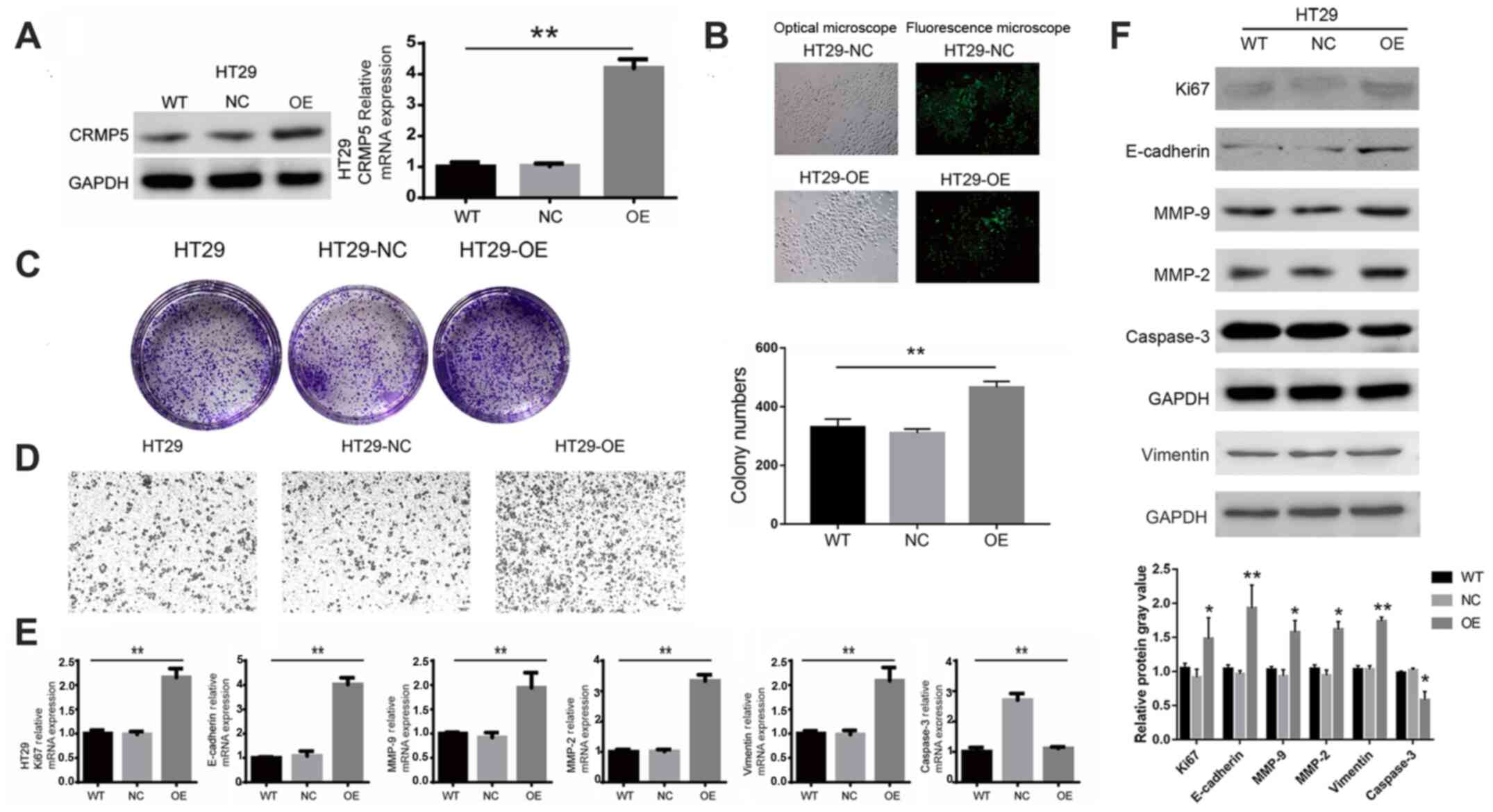 | Figure 2.CRMP5-overexpression promotes
proliferation of HT29 cells. (A) Western blotting and qPCR analysis
of CRMP5 expression levels. (B) Optical and fluorescence microscopy
of HT29 cells. (C) Colony number and (D) cell migration of HT29 WT,
NC and CRMP5 OE cells. (E) qPCR and (F) Western blotting analysis
of Ki67, E-cadherin, vimentin, MMP-2, MMP-9 and caspase-3
expression levels. All experiments were repeated at least three
times. Data are presented as the mean ± standard deviation,
*P<0.05, **P<0.01. CRMP5, Collapsin response mediator protein
5; qPCR, quantitative polymerase chain reaction; WT, wild-type; NC,
negative control; OE, overexpression. |
CRMP5-knockdown inhibits cell
proliferation and development of CRC
CRMP5 expression was downregulated in HCT116 cells
using shRNA-encoding lentiviruses. RT-qPCR and Western blotting
confirmed CRMP5-knockdown in HCT116 cells (Fig. 3A). Additionally, the results of the
present study demonstrated that CRMP5-knockdown markedly inhibited
cell proliferation and migration (Fig.
3B-D). Similarly, CRMP5-knockdown also increased caspase-3
expression and decreased the expression of Ki67, E-cadherin,
vimentin, MMP-2 and MMP-9 (Fig. 3E and
F). Taken together, these data indicated that CRMP5 regulates
CRC cell proliferation and serves a critical role in regulating CRC
development.
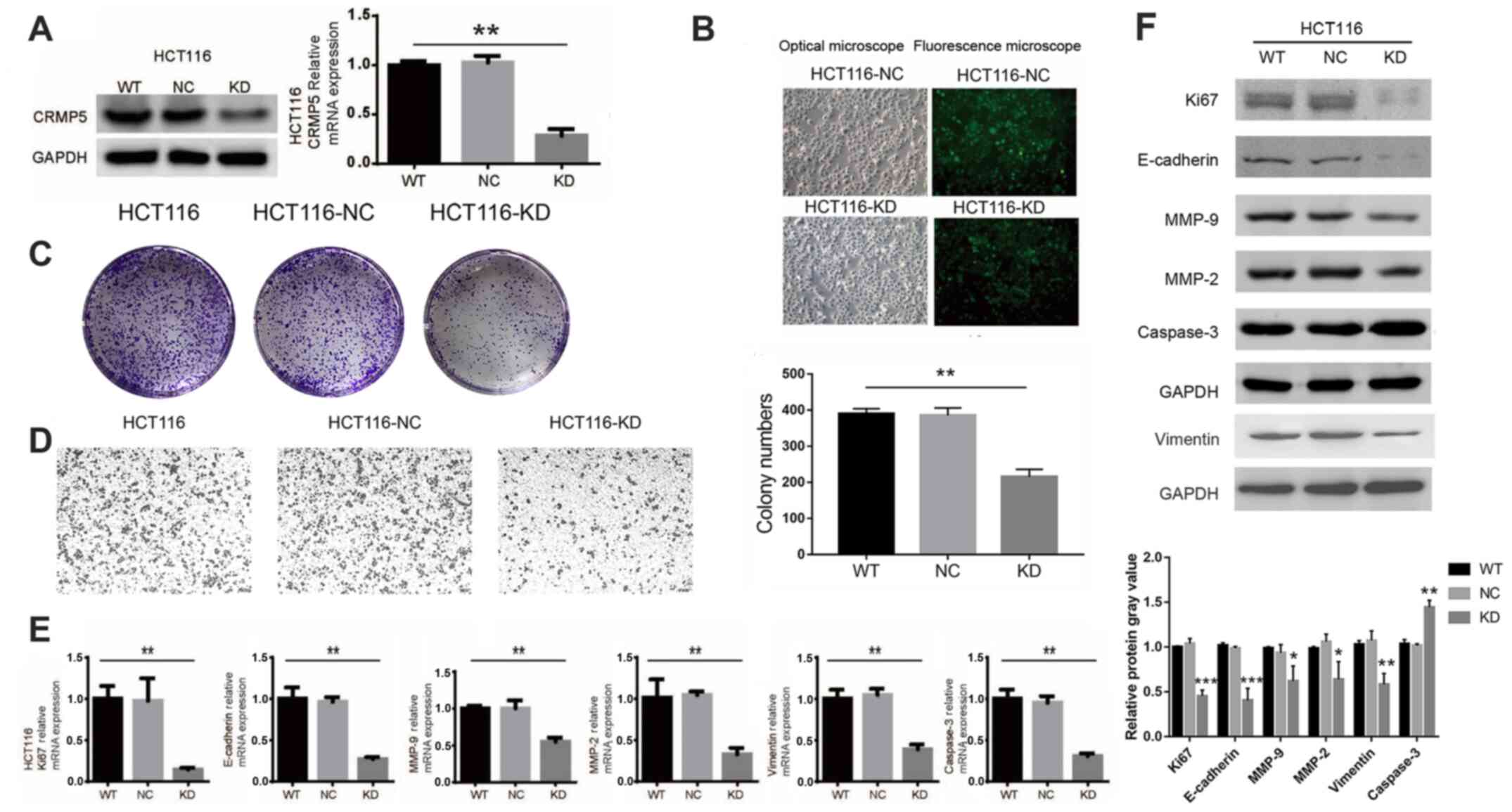 | Figure 3.CRMP5-KD inhibits proliferation of
HCT116 cells. (A) Western blotting and qPCR analysis of CRMP5
expression levels. (B) Optical and fluorescence microscopy of the
HCT116 cells. (C) Colony number and (D) cell migration of HCT116
WT, NC and CRMP5-KD cells. (E) qPCR and (F) Western blot analysis
of Ki67, E-cadherin, vimentin, MMP-2, MMP-9 and caspase-3
expression levels. All experiments were repeated at least three
times. Data are presented as the mean ± standard deviation,
*P<0.05, **P<0.01, ***P<0.001. CRMP5, Collapsin response
mediator protein 5; qPCR, quantitative polymerase chain reaction;
WT, wild-type; NC, negative control; KD, knockdown. |
MAPK signaling is involved in
regulating CRMP5-induced CRC cell proliferation and tumor
recurrence
To further investigate the molecular mechanisms of
CRMP5 in regulating CRC development, RNA-Seq analysis was performed
in CRMP5-overexpressed and -knockdown cell lines. KEGG pathway
enrichment analysis demonstrated that differentially expressed
genes in CRMP5-overexpression or -knockdown cells were
significantly enriched in the MAPK and metabolic signaling pathways
(Fig. 4A and B). To further
investigate the regulatory mechanisms of CRMP5 in CRC, MAPK
signaling was selectively blocked by using Selumetinib (AZD6244), a
potent, selective and ATP-uncompetitive inhibitor of MAPK/ERK1/2 to
determine their effect on cell proliferation. Notably, it was
observed that inhibiting ERK1/2 activity with selumetinib markedly
alleviated the change in colony formation, as well as the changes
in Ki67, E-cadherin, vimentin, MMP-2, MMP-9 and caspase-3
expression levels in HCT116 and HT29 cell lines induced by CRMP5
(Fig. 4C and D). These data
suggested that CRMP5 may regulate CRC cell proliferation and
development through MAPK signaling.
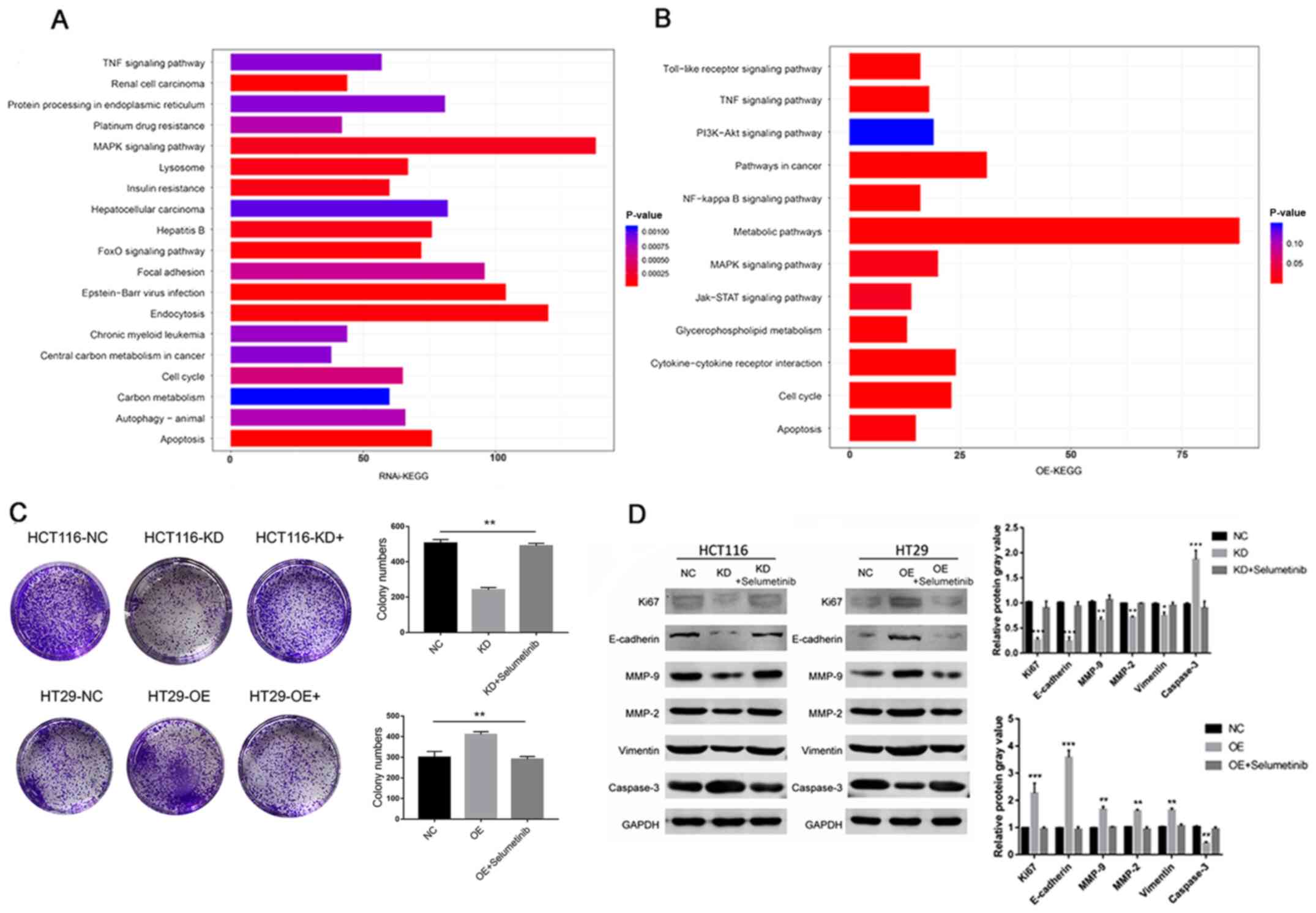 | Figure 4.MAPK signaling is involved in
regulating CRMP5-induced cell proliferation. KEGG pathway
enrichment analysis of CRC cells following (A) CRMP5-KD or (B) -OE.
(C) Colony numbers and (D) Ki67, E-cadherin, vimentin, MMP-2, MMP-9
and caspase-3 expression levels of ERK1/2 signaling inhibiting by
selumetinib in HCT116 cells following KD of CRMP5 or HT29 cells
following OE of CRMP5. All experiments were repeated at least three
times. Data are presented as the mean ± standard deviation,
*P<0.05, **P<0.01, ***P<0.001. CRMP5, Collapsin response
mediator protein 5; KEGG, Kyoto Encyclopedia of Genes and Genomes;
CRC, colorectal cancer; NC, negative control; KD, knockdown; OE,
overexpression. |
Overexpression of CRMP5-induced
chemotherapy resistance and tumor recurrence of CRC
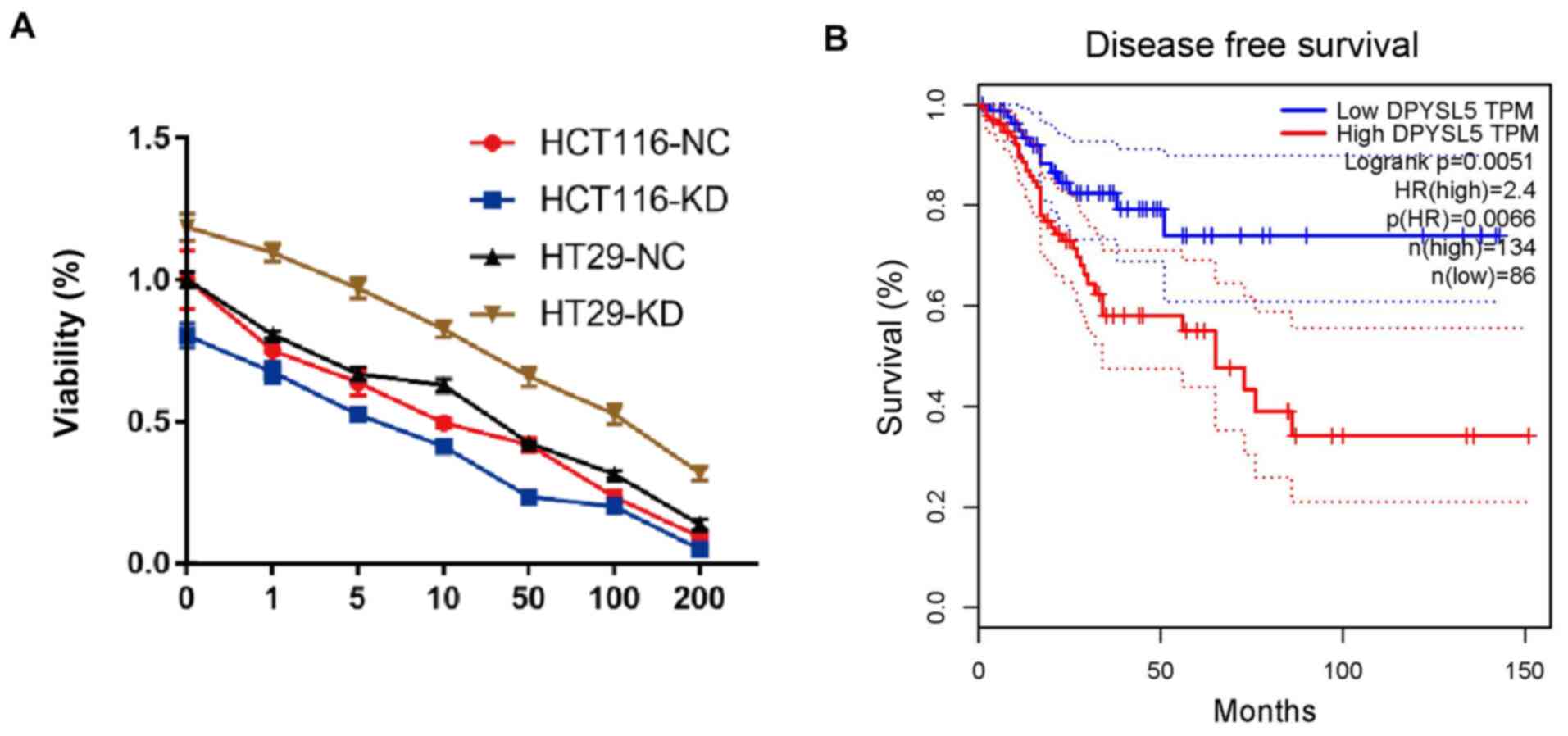
Therefore, the role of CRMP5 in chemotherapeutic
drug resistance was examined. CRMP5-overexpression and control CRC
cells were treated with increasing doses of 5-fluorouracil (5-FU),
a chemotherapeutic drug commonly used in CRC (22). Results of CCK-8 assays demonstrated
that 5-FU treatment significantly decreased the viability of CRC
cells while CRMP5 overexpression significantly reversed the
inhibitory effects induced by the drug (Fig. 5A). Additionally, a retrospective
study was performed using 220 clinical samples from patients with
CRC in a well-established TCGA-COAD cohort to determine the role of
CRMP5 in CRC recurrence. The results of the present study
demonstrated that high expression of CRMP5 significantly indicated
poor disease-free survival of patients with CRC (Fig. 5B). Taken together, these data
suggested that CRMP5-overexpression induces chemotherapy resistance
and tumor recurrence of CRC.
Discussion
CRC is one of the most frequent malignancies
worldwide with incidence and mortality rates still rising rapidly
in many low-income and middle-income countries (3). Combination therapies, including
surgery, local radiotherapy and chemotherapy, are effective against
the early stages of CRC. However, resistance to traditional
chemotherapeutic drugs is frequently observed in patients with
advanced stages of disease, eventually resulting in the development
and relapse of tumors (22,23). Therefore, the identification of novel
biomarkers involved in CRC development and recurrence has potential
applications in anticancer treatment.
Altered CRMP expression levels have been detected in
multiple cancer types, and several CRMP proteins have been reported
to be upregulated in certain cancer types and contribute toward
cancer progression and metastasis (7–10). As a
member of the CRMP family, CRMP5 antibodies have been demonstrated
to be associated with thymoma or small-cell lung cancer, suggesting
that CRMP5 may be involved in these malignancies (24,25). A
previous inter-laboratory study revealed that strong and extensive
CRMP5 expression was observed in 98.6% of high-grade neuroendocrine
lung tumors (13). These results
suggested that CRMP5 may be a useful marker for routine
pathological evaluation of lung tumor surgical samples to
distinguish between highly aggressive neuroendocrine carcinoma and
other lung cancers (13). Another
study demonstrated that CRMP5 was an indicator of poor survival and
promoted cell proliferation of glioblastoma (9). A recent study also reported that CRMP5
promoted osteosarcoma cell growth, and high expression levels of
DRP5 were associated with a poor prognosis in patients with
osteosarcoma (26). To the best of
our knowledge, the present study was the first to demonstrate that
CRMP5-overexpression was significantly associated with poor
survival in patients with CRC. Cell line experiments demonstrated
that CRMP5-overexpression promoted cell proliferation and
migration, while CPMP5-knockdown in CRC cells decreased cell growth
and migration. Additionally, it was found that CRMP5-overexpression
induced CRC recurrence and chemotherapy resistance to 5-FU, a
commonly used chemotherapeutic drug in CRC. The results suggested
that CPMP5 may be used as a potential novel therapeutic target for
CRC. However, further validation and analysis of the effects of
CPMP5 in related in vivo animal models are required.
Members of the MAPK family, including ERK, JNK and
p38MAPK, are involved in regulating cell proliferation, survival
and chemoresistance of multiple cancer types, including CRC
(14,16). The RNA-Seq and KEGG pathway
enrichment analysis revealed that differentially expressed genes
following CRMP5-overexpression or -knockdown were significantly
enriched in the MAPK signaling pathway. In addition, it was found
that inhibiting ERK1/2 markedly alleviated the changes in migration
and invasion induced by CRMP5 in CT116 and HT29 cells. A previous
study on glioblastoma revealed that increased CRMP5 levels promoted
Notch receptor expression and Akt activation in human tumor cell
lines (9). Additionally, CRMP5
functions by hijacking Notch receptors from Itch-dependent
lysosomal degradation (9). Our
findings suggested that MAPK signaling is involved in regulating
CRMP5-induced CRC cell proliferation and development, suggesting
that MAPK signaling inhibition has the potential to be utilized in
CRC treatment.
In summary, CRMP5 was characterized as a novel
biomarker of poor survival in CRC that regulates tumor cell
proliferation and development. The present study uncovered a novel
mechanism by which CRMP5 regulates the MAPK signaling pathway, to
drive CRC cell proliferation and development. The results of the
present study suggested that CRMP5 may be a novel therapeutic
target for CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81503424),
Guangzhou Science and Technology Innovation Commission (grant no.
201704020171), the Medical Key Discipline Planning Project of
Foshan City (grant no. FSZDZK135045), and the Science and
Technology Planning Project of Guangdong Province (grant no.
2019B020208009).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
FHQ and FQ designed the experiments, analyzed the
data and drafted the initial manuscript. FHQ and FQ confirmed the
authenticity of all the raw data. FHQ, LY, DL and YW performed the
experiments. All authors discussed the results and approved the
final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
El-Shami K, Oeffinger KC, Erb NL, Willis
A, Bretsch JK, Pratt-Chapman ML, Cannady RS, Wong SL, Rose J,
Barbour AL, et al: American cancer society colorectal cancer
survivorship care guidelines. CA Cancer J Clin. 65:428–455. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Keum N and Giovannucci E: Global burden of
colorectal cancer: Emerging trends, risk factors and prevention
strategies. Nat Rev Gastroenterol Hepatol. 16:713–732. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Colombet M, Soerjomataram I,
Dyba T, Randi G, Bettio M, Gavin A, Visser O and Bray F: Cancer
incidence and mortality patterns in Europe: Estimates for 40
countries and 25 major cancers in 2018. Eur J Cancer. 103:356–387.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Charrier E, Reibel S, Rogemond V, Aguera
M, Thomasset N and Honnorat J: Collapsin response mediator proteins
(CRMPs): Involvement in nervous system development and adult
neurodegenerative disorders. Mol Neurobiol. 28:51–64. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Quach TT, Honnorat J, Kolattukudy PE,
Khanna R and Duchemin AM: CRMPs: Critical molecules for neurite
morphogenesis and neuropsychiatric diseases. Mol Psychiatry.
20:1037–1045. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tan F, Thiele CJ and Li Z: Collapsin
response mediator proteins: Potential diagnostic and prognostic
biomarkers in cancers (Review). Oncol Lett. 7:1333–1340. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao X, Mao YH, Xiao C, Li K, Liu W, Li LY
and Pang J: Calpain-2 triggers prostate cancer metastasis via
enhancing CRMP4 promoter methylation through NF-kappaB/DNMT1
signaling pathway. Prostate. 78:682–690. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moutal A, Honnorat J, Massoma P,
Désormeaux P, Bertrand C, Malleval C, Watrin C, Chounlamountri N,
Mayeur ME, Besançon R, et al: CRMP5 controls glioblastoma cell
proliferation and survival through notch-dependent signaling.
Cancer Res. 75:3519–3528. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gozzard P, Woodhall M, Chapman C, Nibber
A, Waters P, Vincent A, Lang B and Maddison P: Paraneoplastic
neurologic disorders in small cell lung carcinoma: A prospective
study. Neurology. 85:235–239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Veyrac A, Reibel S, Sacquet J, Mutin M,
Camdessanche JP, Kolattukudy P, Honnorat J and Jourdan F: CRMP5
regulates generation and survival of newborn neurons in olfactory
and hippocampal neurogenic areas of the adult mouse brain. PLoS
One. 6:e237212011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ricard D, Rogemond V, Charrier E, Aguera
M, Bagnard D, Belin MF, Thomasset N and Honnorat J: Isolation and
expression pattern of human Unc-33-like phosphoprotein 6/collapsin
response mediator protein 5 (Ulip6/CRMP5): Coexistence with
Ulip2/CRMP2 in Sema3a- sensitive oligodendrocytes. J Neurosci.
21:7203–7214. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meyronet D, Massoma P, Thivolet F,
Chalabreysse L, Rogemond V, Schlama A, Honnorat J and Thomasset N:
Extensive expression of collapsin response mediator protein 5
(CRMP5) is a specific marker of high-grade lung neuroendocrine
carcinoma. Am J Surg Pathol. 32:1699–1708. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morrison DK: MAP kinase pathways. Cold
Spring Harb Perspect Biol. 4:a0112542012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su B and Karin M: Mitogen-activated
protein kinase cascades and regulation of gene expression. Curr
Opin Immunol. 8:402–411. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dhillon AS, Hagan S, Rath O and Kolch W:
MAP kinase signalling pathways in cancer. Oncogene. 26:3279–3290.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Burotto M, Chiou VL, Lee JM and Kohn EC:
The MAPK pathway across different malignancies: A new perspective.
Cancer. 120:3446–3456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee JS: Exploring cancer genomic data from
the cancer genome atlas project. BMB Rep. 49:607–611. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Seshagiri S, Stawiski EW, Durinck S,
Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman
V, Jaiswal BS, et al: Recurrent R-spondin fusions in colon cancer.
Nature. 488:660–4. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404,
2012.22. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vodenkova S, Buchler T, Cervena K,
Veskrnova V, Vodicka P and Vymetalkova V: 5-fluorouracil and other
fluoropyrimidines in colorectal cancer: Past, present and future.
Pharmacol Ther. 206:1074472019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hammond WA, Swaika A and Mody K:
Pharmacologic resistance in colorectal cancer: A review. Ther Adv
Med Oncol. 8:57–84. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Monstad SE, Nostbakken JK and Vedeler CA:
CRMP5 antibodies found in a patient with limbic encephalitis and
myasthenia gravis. J Neurol Neurosurg Psychiatry. 80:241–242. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Monstad SE, Drivsholm L, Skeie GO, Aarseth
JH and Vedeler CA: CRMP5 antibodies in patients with small-cell
lung cancer or thymoma. Cancer Immunol Immunother. 57:227–232.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang L, Liu W, Tang H, Xie X, Zou C, Wang
Y, Gao Z and Yin J: DRP5 is involved in cancer cell growth and
predicts poor prognosis in human osteosarcoma. Cancer Med.
6:982–993. 2017. View Article : Google Scholar : PubMed/NCBI
|