Introduction
Lung cancer is one of the most common malignant
tumors (1–3). Owing to the high incidence and
mortality rates of lung cancer, it has become one of the most
notable causes of cancer-related death worldwide (4). According to pathological
classification, lung cancer primarily includes small cell and
non-small cell lung cancer (NSCLC) (5). NSCLC accounts for ~66% of lung cancer
cases (6,7), among which lung squamous cell carcinoma
(LUSC) is an important subtype (8).
LUSC and its associated complications are reportedly responsible
for >400,000 worldwide deaths annually (9). In addition to a lack of effective
prognostic biomarkers, as LUSC frequently results in local
infiltration and metastasis, the 5-year survival rate of patients
with advanced disease is <17% (10,11).
Improving the prognosis of patients is likely to increase the
survival rate (12,13); therefore, it is necessary to identify
reliable prognostic markers of LUSC for developing effective early
therapeutic strategies.
Noncoding RNAs play an important regulatory role in
the initiation of gene expression (12,14,15).
They are primarily divided into two subtypes, namely small
noncoding RNAs [microRNAs (miRNAs/miRs)] with a transcript size of
<200 nucleotides (nt), and long non-coding RNAs (lncRNAs) with a
transcript size between 200 nt and 100 kb (16–18).
previous research on noncoding RNAs has mainly focused on miRNAs,
and lncRNAs were often regarded as ‘junk RNAs’. However, current
literature demonstrates that the abnormal expression of lncRNAs
directly influences the occurrence and development of different
diseases, including various type of cancer (13,17,19). Cui
et al (20) identified that
PSMG3-antisense (AS) 1 serves as an oncogenic lncRNA in breast
cancer, which has also been revealed to play an important role in
lung adenocarcinoma (LUAD) (13).
Investigating whether PSMG3-AS1 can associate with, or influence
the biological functions of, cancer cells by interacting with
miRNAs has become a major topic of interest. Numerous studies have
suggested that a negative correlation exists between PSMG3-AS1 and
miR-143-3p (13,20). As reported, miR-143-3p targets
PSMG3-AS1 in liver cancer, the expression levels of which were
closely and inversely correlated (13); however, their association with LUSC
remains to be elucidated. Therefore, the focus of the present study
was to investigate the potential therapeutic significance of
PSMG3-AS1 in LUSC.
Materials and methods
Patients and clinical specimens
A total of 130 patients, who were diagnosed with
LUSC for the first time at The Forth People's Hospital (Shenyang,
China) between Februry 2012 and December 2015, were selected. The
inclusion criteria were: i) Diagnosed with LUSC by
histopathological examination; ii) no history of tumor treatment;
and iii) full clinical characteristics and 5-year follow-up
information available. Paired LUSC and matched adjacent lung
epithelial tissue specimens were collected during surgery, and
confirmed by two pathologists. All specimens were stored in liquid
nitrogen for further experimentation. The present study complied
with the Ethics Committee of the Fourth People's Hospital of
Shenyang [approval no. k(2012)11], and writtern informed consent
was obtained from patients before the initiation of any
study-related procedure. Patient clinicopathological
characteristics were recorded (Table
I), and information regarding 5-year survival was obtained via
telephone.
 | Table I.Correlation between PSMG3-AS1
expression and the clinical characteristics of patients with lung
squamous cell carcinoma. |
Table I.
Correlation between PSMG3-AS1
expression and the clinical characteristics of patients with lung
squamous cell carcinoma.
|
|
| PSMG3-AS1
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Cases (n130) | Low (n=60) | High (n=70) | P-value |
|---|
| Age |
|
|
| 0.663 |
| ≤60 | 58 | 28 | 30 |
|
|
>60 | 72 | 32 | 40 |
|
| Sex |
|
|
| 0.828 |
| Male | 68 | 32 | 36 |
|
|
Female | 62 | 28 | 34 |
|
| Tumor size |
|
|
| 0.745 |
| ≤5
cm | 67 | 30 | 37 |
|
| >5
cm | 63 | 30 | 33 |
|
| Smoking status |
|
|
|
|
|
Non-smoker | 65 | 34 | 31 | 0.159 |
|
Smoker | 65 | 26 | 39 |
|
|
Differentiation |
|
|
| 0.172 |
| Well,
Moderate | 74 | 38 | 36 |
|
|
Poor | 56 | 22 | 34 |
|
| Lymph node
metastasis |
|
|
| 0.001 |
|
Negative | 80 | 46 | 34 |
|
|
Positive | 50 | 14 | 36 |
|
| TNM stage |
|
|
| 0.006 |
| I,
II | 81 | 45 | 36 |
|
| III,
IV | 49 | 15 | 34 |
|
Cell lines and transfection
Human LUSC cell lines (H2170, H520, HCC95 and
SK-MES-1) and the human lung epithelial BEAS-2B cell line were
obtained from the Shanghai Cell Bank of the Chinese Academy of
Sciences (Shanghai, China). All cells were cultured in DMEM
supplemented with 10% FBS (both Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a 5% CO2 incubator.
Before transfection, LUSC cells (2×105
cells/well) were cultured in 6-well plates. Small interfering RNA
(siRNA) targeting PSMG3-AS1 (si-PSMG3-AS1 sense,
5′-GGACGUCUCCCAUUCUGAATT-3′ and antisense,
5′-UUCAGAAUGGGAGACGUCCTT-3′) and the siRNA negative control (si-NC
sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′) were synthesized by Shanghai
GenePharma Co., Ltd. The miR-143-3p inhibitor
(5′-GAGCUACAGUGCUUCAUCUCA-3′), inhibitor NC
(5′-CAGUACUUUUGUGUAGUACAA-3′), miR-143-3p mimic
(5′-UGAGAUGAAGCACUGUAGCUC-3′) and mimic NC
(5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from Guangzhou RiboBio
Co., Ltd. All transfections were conducted at a final concentration
of 50 nM using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) for 48 h at 37°C (according to the
manufacturer's instructions), with untreated cells as the control.
Follow-up experiments were carried out within 24 h.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated from LUSC cells and tumor
tissues using RNAzol (Sigma-Aldrich; Merck KGaA). To harvest
miRNAs, RNA precipitation and washing were performed using 85%
ethanol, with each centrifugation step at 13,840 × g (4°C) for 10
min. A Precision nanoScript2 Reverse Transcription Kit
(Primerdesign Ltd.) and miRNA 1st Strand cDNA Synthesis Kit (Vazyme
Biotech Co., Ltd.) were used for reverse transcription of lncRNAs
and miRNAs, respectively (A260/A280 ratio, 1.8–2.0). Subsequently,
qPCR was conducted using a 7500 Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with ChamQ SYBR qPCR
Green Master Mix (Vazyme Biotech Co., Ltd.) and miRNA Universal
SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.). The qPCR condition
were as follows: 95°C for 30 sec, followed by 40 cycles at 95°C for
15 sec, 60°C for 30 sec, 72°C for 15 sec, and then final extension
at 72°C for 5 min. GAPDH and U6 were used as the endogenous
controls for PSMG3-AS1 and miR-143-3p, respectively. The expression
levels of PSMG3-AS1 and miR-143-3p were calculated using the
2−ΔΔCq method (21), and
were normalized to those of GAPDH and U6, respectively. The primer
sequences were as follows: PSMG3-AS1 forward,
5′-AAATGTGGGAGGGATGGCAG-3′ and reverse, 5′-AATGGTGCCTTCCCCATCAG-3′;
GAPDH forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse,
5′-AAGTGGTCGTTGAGGGCAATG-3′; miR-143-3p forward,
5′-CTGGCGTTGAGATGAAGCAC-3′ and reverse, 5′-CAGAGCAGGGTCCGAGGTA-3′;
and U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′ and reverse,
5′-TTCACGAATTTGCGTGTCATC-3′.
Cell Counting Kit 8 (CCK-8)
analysis
The CCK-8 kit (Dojindo Molecular Technologies, Inc.)
was used to assess H520 and SK-MES-1 cell proliferative capacity.
Transfected cells were seeded into 96-well culture plates
(4×103 cells/well) and cultured in a humidified
incubator with 5% CO2 at 37°C for 0, 24, 48 and 72 h.
Subsequently, 10 µl CCK-8 solution was added. After a further 2-h
incubation, the absorbance value of each well was measured at 450
nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Transwell migration and invasion
assays
A 24-well Transwell plate (8 µm pore; Corning, Inc.)
was used for Transwell migration and invasion assays. The
experimental procedure was similar in both the migration and
invasion assays, except that the upper chamber was precoated with
Matrigel (BD Biosciences) for 6 h at 37°C in the invasion assay.
Briefly, transfected cells (5×104 cells/well) were
seeded into the upper chamber in serum-free DMEM medium, and DMEM
with 10% FBS was added to the lower chamber as a chemoattractant.
After incubation for 24 h at 37°C, the migrated or invaded cells
were fixed with 4% paraformaldehyde for 15 min at room temperature,
and stained with 0.1% crystal violet for 20 min at room
temperature. The number of cells was observed under a light
microscope in five random fields of view.
Bioinformatisc analysis
The interaction between PSMG-AS1 and miR-143-3p was
predicted using starBase v2.0 database (http://starbase.sysu.edu.cn/index.php).
Dual-luciferase reporter assay
The PSMG3-AS1 and corresponding mutant sequences
(without PSMG3-AS1 binding sites) were synthesized and subcloned
into luciferase reporter vectors (Promega Corporation), which were
subsequently named WT-PSMG3-AS1 and MUT-PSMG3-AS1, respectively.
SK-MES-1 cells were then seeded into 24-well plates and
co-transfected with miR-143-3p mimics, miR-143-3p inhibitor,
mimic-NC and inhibitor-NC (as aforementioned) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, the firefly luciferase activity was
determined using a dual-luciferase reporter assay kit (Promega
Corporation) in accordance with the manufacturer's protocol and
normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using SPSS 20.0
(IBM Corp) and GraphPad Prism 5.0 software (GraphPad Software,
Inc.). All experiments were repeated at least three times, and the
data are presented as the mean ± standard deviation. The
χ2 test was used to determine the association between
PSMG3-AS1 expression and patient clinical data. One-way ANOVA
followed by Tukey's post hoc test was used to determine significant
differences among multiple groups, and paired Student's t-test was
used for comparing data between two groups. Kaplan-Meier (and the
log-rank test) and Cox regression analysis were used to evaluate
the prognostic significance of PSMG3-AS1, and Pearson's test was
used for correlation analysis. P<0.05 was considered to indicate
a statistically significant difference.
Results
Expression of PSMG3-AS1 in LUSC
tissues and cell lines
PSMG3-AS1 expression levels in LUSC and
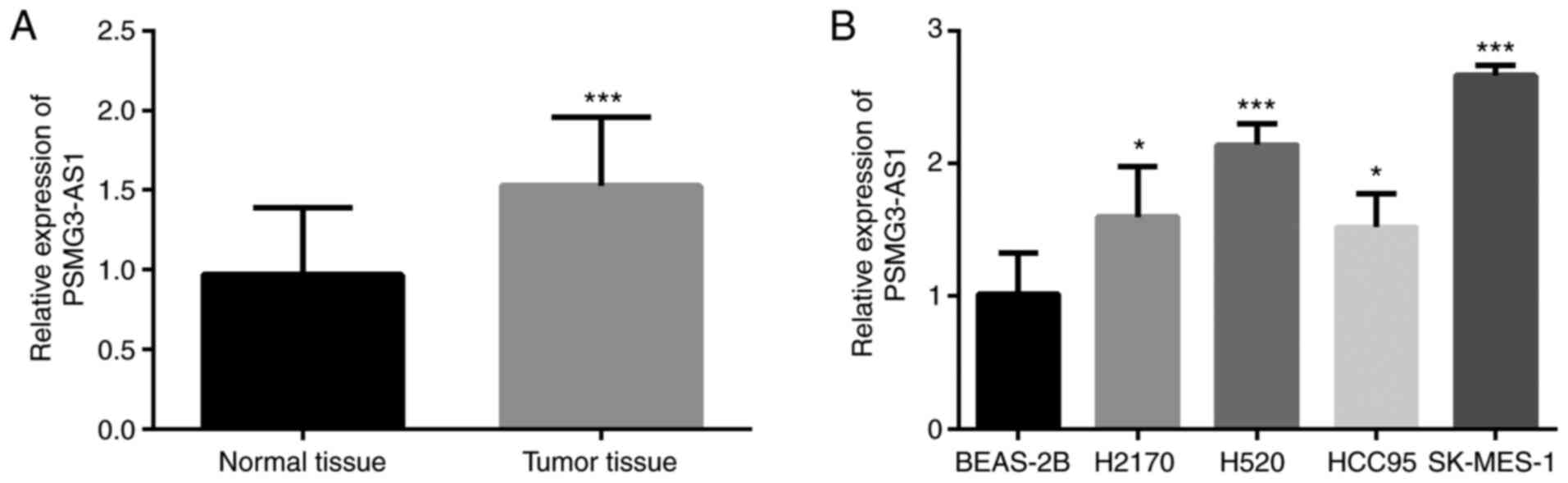
adjacent-normal tissues were determined using RT-qPCR analysis.
Compared with adjacent-normal tissues, the expression levels of
PSMG3-AS1 were higher in tumor tissues (Fig. 1A; P<0.001). In addition, the
expression of PSMG3-AS1 in four different LUSC cell lines and one
normal cell line was assessed. As illustrated in Fig. 1B, the expression of PSMG3-AS1 was
higher in all LUSC cell lines than in the normal cell line (all
P<0.05). The two cell lines exhibiting the highest relative
expression of PSMG3-AS1 (SK-MES-1 and H520) were selected for
subsequent experimentation. These results indicated that PSMG3-AS1
may play an oncogenic role in LUSC.
Association between PSMG3-AS1 and
clinicopathological characteristics of patients with LUSC
The association between PSMG3-AS1 expression and the
clinical parameters of patients with LUSC was evaluated to
determine whether PSMG3-AS1 expression was involved in LUSC tumor
progression. Patients were divided into low (60 patients) and high
(70 patients) PSMG3-AS1 expression groups, based on the mean
PSMG3-AS1 expression value. The χ2 test revealed that
upregulated PSMG3-AS1 expression was associated with TNM stage
(P=0.006) and lymph node metastasis (P=0.001), suggesting that
PSMG3-AS1 expression may be involved in the progression of LUSC.
However, PSMG3-AS1 expression was not significantly associated with
any of the other investigated characteristics, including age, sex,
tumor size, smoking status, and tumor differentiation (P>0.05;
Table I).
Association between PSMG3-AS1 and the
prognosis of LUSC patients
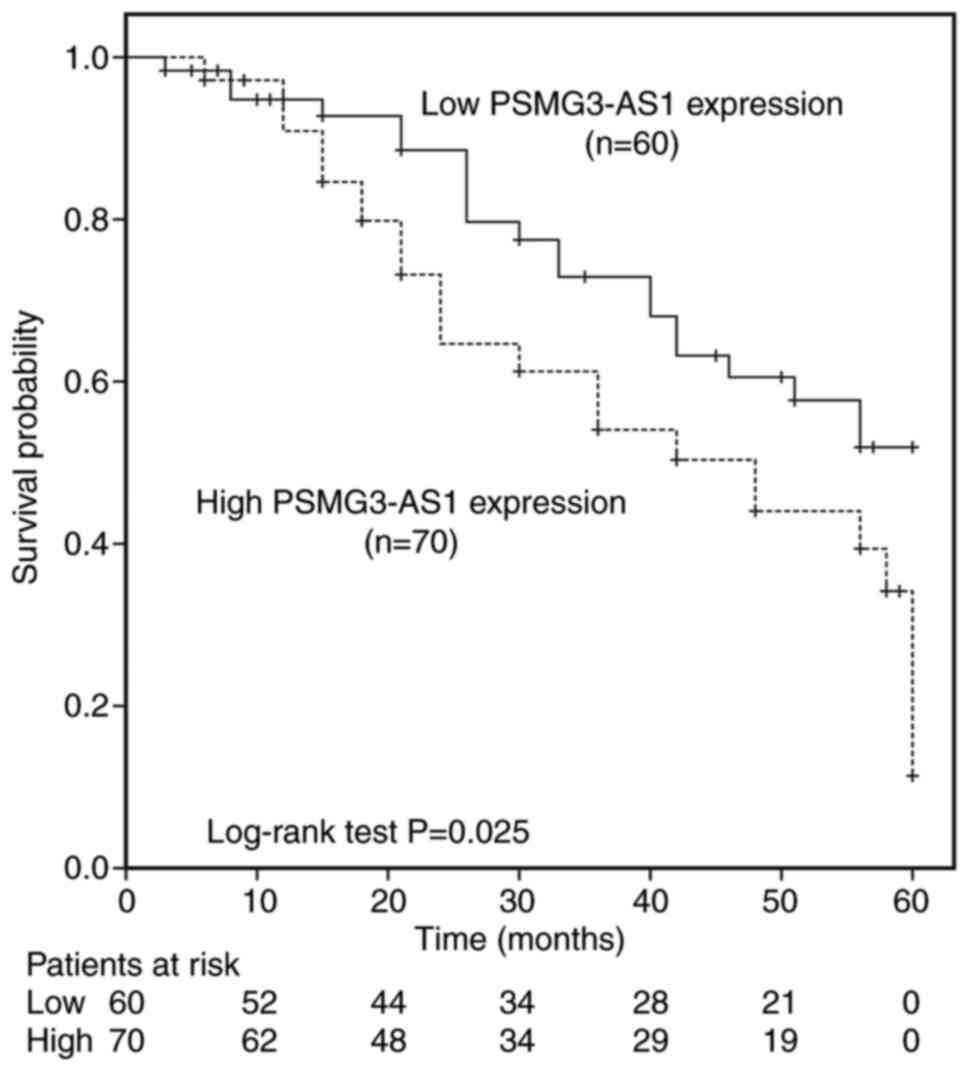
Kaplan-Meier curve analysis and the log-rank test
were used to assess the prognostic value of PSMG3-AS1 in LUSC.
Based on the PSMG3-AS1 expression and overall survival status of
patients with LUSC, the results demonstrated that the 5-year
overall survival rate of patients in the high PSMG3-AS1 expression
group was lower than that of those in the low expression group
(P=0.025; Fig. 2). Moreover,
multivariate Cox regression analysis indicated that PSMG3-AS1
expression level (hazard ratio, 2.068; 95% confidence interval,
1.142–3.744; P=0.016) was an independent prognostic factor for
assessing the 5-year overall survival of patients with LUSC
(Table II). These results suggested
that PSMG3-AS1 may be a prognostic marker for LUSC.
 | Table II.Multivariate Cox analysis of the
clinical characteristics of patients with lung squamous cell
carcinoma in relation to overall survival. |
Table II.
Multivariate Cox analysis of the
clinical characteristics of patients with lung squamous cell
carcinoma in relation to overall survival.
|
| Multivariate
analysis |
|---|
|
|
|
|---|
| Characteristic | HR | 95% CI | P-value |
|---|
| PSMG3-AS1 | 2.068 | 1.142–3.744 | 0.016 |
| Age | 0.784 | 0.467–1.317 | 0.358 |
| Sex | 0.908 | 0.535–1.540 | 0.720 |
| Tumor size | 1.127 | 0.669–1.898 | 0.654 |
| Smoking status | 1.477 | 0.878–2.486 | 0.142 |
|
Differentiation | 1.466 | 0.863–2.490 | 0.157 |
| Lymph node
metastasis | 1.728 | 1.004–2.975 | 0.048 |
| TNM stage | 1.730 | 1.014–2.953 | 0.044 |
PSMG3-AS1 promotes LUSC cellular
characteristics
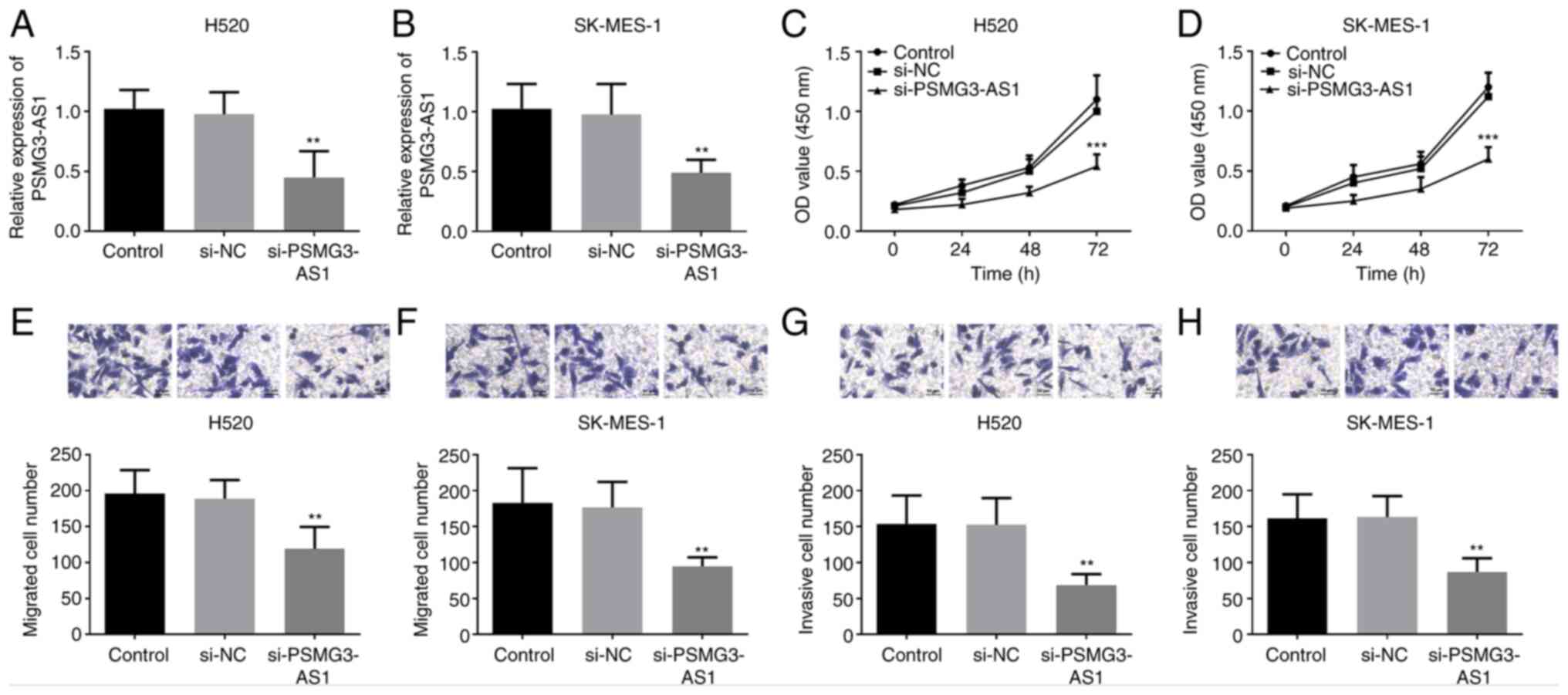
To determine its functional role in LUSC, PSMG3-AS1
expression in H520 and SK-MES-1 cells was quantified using RT-qPCR
following si-PSMG3-AS1 transfection. As illustrated in Fig. 3A and B, PSMG3-AS1 expression was
significantly decreased by si-PSMG3-AS1 in H520 and SK-MES-1 cells
(P<0.01). Subsequently, a CCK-8 assay was performed to assess
cellular proliferative capacity. The results indicated that
PSMG3-AS1-knockdown suppressed cellular proliferation (P<0.001;
Fig. 3C and D). Moreover, the
Transwell assay results demonstrated that si-PSMG3-AS1 suppressed
the migratory and invasive abilities of H520 and SK-MES-1 cells
(P<0.01; Fig. 3E-H). Of note, no
marked differences were observed between the H520 and SK-MES-1 cell
lines in characterizing LUSC. Ultimately, these findings confirmed
the oncogenic role of PSMG3-AS1 in LUSC cells.
Interaction between PSMG3-AS1 and
miR-143-3p
The SK-MES-1 cell line (in which the expression of
PSMG3-AS1 was the highest) was used to determine the association
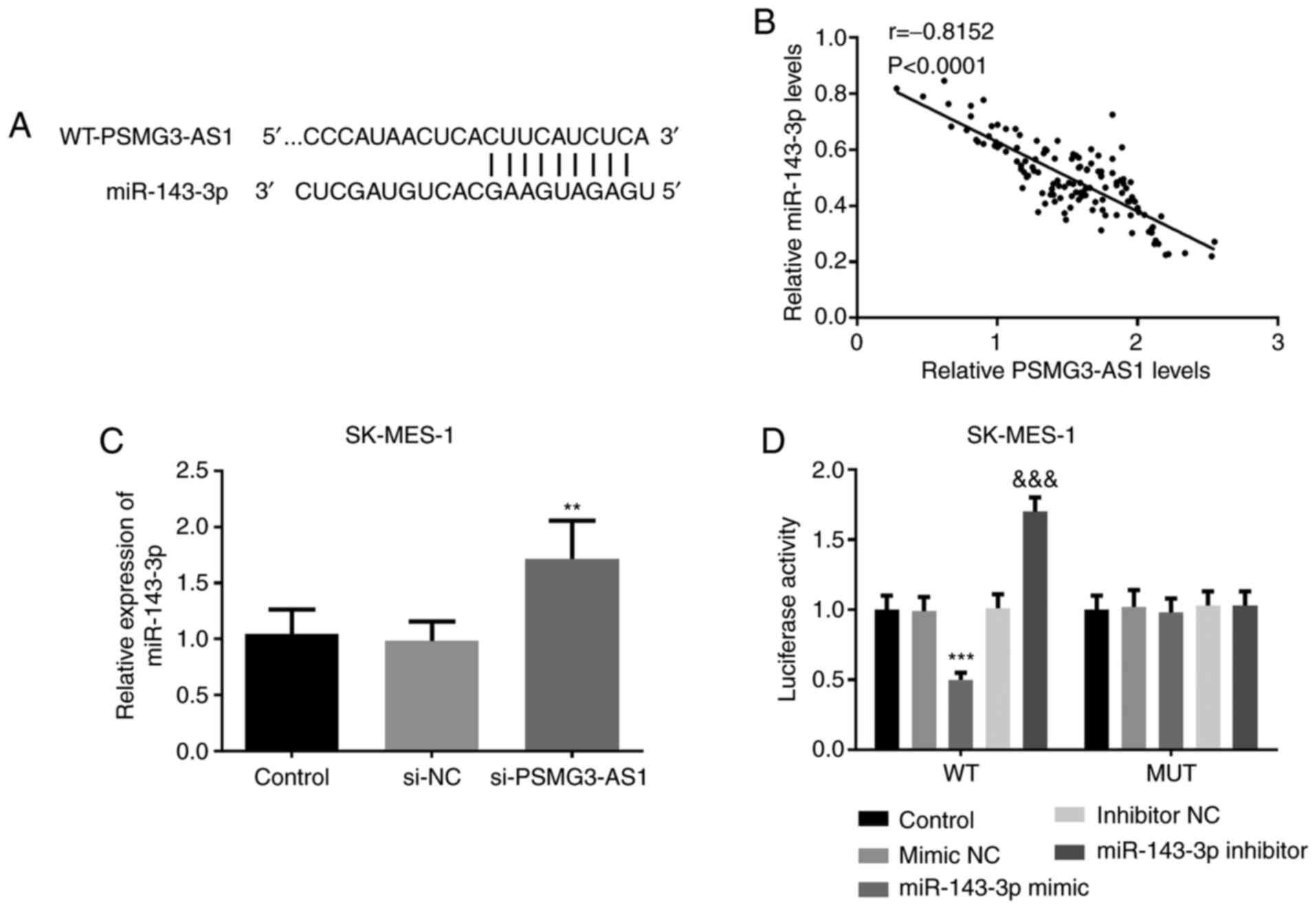
between miR-143-3p and PSMG3-AS1. According to the bioinformatics
analysis, miR-143-3p and PSMG3-AS1 formed multiple base pairings
(Fig. 4A). Spearman's rank
correlation coefficient analysis was used to evaluate the
correlation between PSMG3-AS1 and miR-143-3p. The findings
indicated that the expression of PSMG3-AS1 was inversely correlated
with that of miR-143-3p (r=−0.8152; P<0.0001; Fig. 4B). The silencing of PSMG3-AS1 in
SK-MES-1 cells was confirmed by RT-qPCR, and the results revealed
that miR-143-3p expression was successfully increased (P<0.01;
Fig. 4C). Subsequently, a mutated
PSMG3-AS1 construct was obtained by mutating the miR-143-3p binding
sites within the PSMG3-AS1 3′untranslated region. The luciferase
activity of the MUT-PSMG3-AS1-transfected cells was not affected by
the expression of miR-143-3p, whereas that of the
WT-PSMG3-AS1-transfected cells was significantly decreased by
miR-143-3p overexpression, and increased by miR-143-3p inhibition
(P<0.001; Fig. 4D). This
suggested that miR-143-3p may bind to the predicted sites and
subsequently inhibit the expression of PSMG3-AS1.
Discussion
The aim of the present study was to investigate the
clinical role of PSMG3-AS1, and the interaction between miR-143-3p
and PSMG3-AS1, in LUSC. The findings revealed that the expression
of miR-143-3p and PSMG3-AS1 was altered in LUSC. Moreover,
PSMG3-AS1 may target miR-143-3p to promote cancer cell
proliferation.
As noncoding RNAs, lncRNAs and miRNAs are commonly
perceived as epigenetic markers for diagnosis and/or prognosis
(22–24). However, as a type of lncRNA,
PSMG3-AS1 plays a key role in various physiological processes of
cancer cells. Therefore, in the present study, the role of
PSMG3-AS1 in LUSC was investigated. Firstly, the expression level
of PSMG3-AS1 was determined in LUSC tissue samples and four LUSC
cell lines. The data demonstrated that PSMG3-AS1 was upregulated in
LUSC tissue specimens and all of the cell lines investigated, which
suggests that PSMG3-AS1 may serve as an oncogene in LUSC. This is
in accordance with PSMG3-AS1 upregulation in glioblastomas and
breast cancer cells (20,25). However, a previous study also
reported that PSMG3-AS1 was downregulated in pancreatic cancer
(23), suggesting that its role may
differ between cancer types. Subsequently, further experiments were
conducted to verify the role of PSMG3-AS1 in LUSC. PSMG3-AS1
expression, lymph node metastasis and TNM stage were revealed to be
associated with the overall survival of patients with LUSC.
Additionally, multivariate Cox regression analysis revealed that
PSMG3-AS1 expression was an independent prognostic predictor for
LUSC. Furthermore, the Kaplan-Meier analysis results confirmed that
the high PSMG3-AS1 expression group exhibited a significantly
poorer prognosis than the low-expression group in a 5-year
follow-up study. Liu et al (23) also identified that PSMG3-AS1 was
associated with the 5-year overall survival rate, and was involved
in the progression of pancreatic cancer. Yue et al (13) also reported that the increased
expression of PSMG3-AS1 in LUAD predicted poor survival. These
aforementioned studies indicate that PSMG3-AS1 is associated with
poor prognosis in LUSC, in which it may act as a prognostic
biomarker.
To investigate the functional role of PSMG3-AS1 in
LUSC cell function, loss-of-function experiments were performed by
downregulating the expression of PSMG3-AS1 in H520 and SK-MES-1
cells. The results indicated that, compared with untreated cells,
si-PSMG3-AS1 inhibited the proliferative, migratory and invasive
capacities of LUSC cells. A similar finding was observed in a
previous study, whereby transfection with the si-PSMG3-AS1
decreased proliferation in two LUAD cell lines (13). Therefore, we hypothesize that
PSMG3-AS1 may serve as an oncogenic factor and may be involved in
the progression of LUSC.
PSMG3-AS1 is reported to be upregulated in breast
cancer, and may accelerate cancer cell migration and proliferation
by sponging miR-143-3p (20).
However, PSMG3-AS1 was reported to promote tumor progression by
activating the PI3K-Akt pathway (23,26,27).
Another study demonstrated that PSMG3-AS1 promoted cancer cell
characteristics through the lncRNA-PSMG3-AS1/miR-143-3p/COL1A1
signaling axis (20). Furthermore,
the relationship between PSMG3-AS1 and miR-143-3p in LUSC was
analyzed in the present study. We hypothesized that PSMG3-AS1 may
bind miR-143-3p to regulate the biological functions of LUSC cells.
In SK-MES-1 cells, PSMG3-AS1-knockdown was demonstrated to
upregulate miR-143-3p expression. Simultaneously, Spearman's rank
correlation coefficient analysis revealed a negative association
between miR-143-3p and PSMG3-AS1 expression. Moreover, the
luciferase activity of MUT-PSMG3-AS1-transfected cells was not
affected by the expression of miR-143-3p, while that of the
WT-PSMG3-AS1-transfected cells was significantly decreased by
miR-143-3p overexpression, and increased by miR-143-3p inhibition.
Therefore, the present study revealed that PSMG3-AS1 may also drive
LUSC cell proliferation, migration and invasiveness by targeting
miR-143-3p.
The findings regarding LUSC (namely, an upregulation
in PSMG3-AS1) support the recent discussion that PSMG3-AS1 may act
as an oncogenic microenvironmental factor (28,29).
However, there were limitations to the present study. Firstly, LUSC
samples and corresponding normal samples (that were used for
determining the expression of PSMG3-AS1) were limited in number.
Secondly, in vivo experiments were not performed to verify
the effect of PSMG3-AS1 in LUSC.
In conclusion, the results of the current study
indicated that PSMG3-AS1 expression was increased in LUSC tissue
specimens and cancer cells, and that PSMG3-AS1 overexpression
accelerated cancer cell proliferation, migration and invasiveness
by targeting miR-143-3p. Moreover, LUSC patients with high
PSMG3-AS1 expression exhibited a lower overall survival rate than
those with low PSMG3-AS1 expression levels. Therefore, the present
study confirms that PSMG3-AS1 has potential as a prognostic
biomarker and a future therapeutic target.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Liaoning
Municipal Basic Medical and Health Technology Project (grant no.
S20110167).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
EJ designed the study. CH, LZ, XZ and ZR conducted
the experiments, analyzed the data and revised the manuscript. SC
collected the sample tissues and clinical data, and wrote the
manuscript. HF interpreted the data. EJ and HF collected the
samples and confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Fourth People's Hospital of Shenyang [approval no.
k(2012)11]. Informed consent was obtained from all patients before
the initiation of any study-related procedure.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Longo DL, Reck M and Rabe KF: Precision
diagnosis and treatment for advanced non-small-cell lung cancer. N
Engl J Me. 377:849–861. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:4462018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rittmeyer A, Barlesi F, Waterkamp D, Park
K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols
MC, et al: Atezolizumab versus docetaxel in patients with
previously treated non-small-cell lung cancer (OAK): A phase 3,
open-label, multicentre randomised controlled trial. Lancet.
389:255–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pless M, Stupp R, Ris HB, Stahel RA, Weder
W, Thierstein S, Gerard MA, Xyrafas A, Früh M, Cathomas R, et al:
Induction chemoradiation in stage IIIA/N2 non-small-cell lung
cancer: A phase 3 randomised trial. Lancet. 1049–1056. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song Y, Zhou B, Du X, Wang Y, Zhang J, Ai
Y, Xia Z and Zhao G: Folic acid (FA)-conjugated mesoporous silica
nanoparticles combined with MRP-1 siRNA improves the suppressive
effects of myricetin on non-small cell lung cancer (NSCLC). Biomed
Pharmacother. 125:1095612020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gandara DR, Hammerman PS, Sos ML, Lara PN
Jr and Hirsch FR: Squamous cell lung cancer: From tumor genomics to
cancer therapeutics. Clin Cancer Res. 21:2236–2243. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mentzelopoulos A, Gkiatis K, Karanasiou I,
Karavasilis E, Papathanasiou M, Efstathopoulos E, Kelekis N,
Kouloulias V and Matsopoulos GK: Chemotherapy-induced brain effects
in small-cell lung cancer patients: A multimodal MRI study. Brain
Topogr. 34:167–181. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lo YL, Hsiao CF, Chang GC, Tsai YH, Huang
MS, Su WC, Chen YM, Hsin CW, Chang CH, Yang PC, et al: Risk factors
for primary lung cancer among never smokers by gender in a matched
case-control study. Cancer Causes Control. 24:567–576. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kabir Z, Bennett K and Clancy L: Lung
cancer and urban air-pollution in Dublin: A temporal association?
Ir Med J. 100:367–369. 2007.PubMed/NCBI
|
|
11
|
Loomis D, Grosse Y, Lauby-Secretan B, El
Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Baan R, Mattock
H, Straif K, et al: The carcinogenicity of outdoor air pollution.
Lancet Oncology. 14:1262–1263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yue N, Ye M, Zhang R and Guo Y:
miR-449b-5p targets lncRNA PSMG3-AS1 to suppress cancer cell
proliferation in lung adenocarcinoma. BMC Pulm Med. 20:1522020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao
T, Liu Y, Ou J, Wang D, Yao L, et al: lncRNA-HOST2 regulates cell
biological behaviors in epithelial ovarian cancer through a
mechanism involving microRNA let-7b. Hum Mol Genet. 24:841–852.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Katsushima K, Natsume A, Ohka F, Shinjo K,
Hatanaka A, Ichimura N, Sato S, Takahashi S, Kimura H, Totoki Y, et
al: Targeting the Notch-regulated non-coding RNA TUG1 for glioma
treatment. Nat Commun. 7:136162016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schaukowitch K and Kim TK: Emerging
epigenetic mechanisms of long non-coding RNAs. Neuroscience.
264:25–38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Monroig Pdel C, Chen L, Zhang S and Calin
GA: Small molecule compounds targeting miRNAs for cancer therapy.
Adv Drug Deliv Rev. 81:104–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng G, Liu D, Liang H, Yang H, Chen K
and Zhang X: A cluster of long non-coding RNAs exhibit diagnostic
and prognostic values in renal cell carcinoma. Aging (Albany NY).
11:9597–9615. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Simpson RJ, Lim JW, Moritz RL and
Mathivanan S: Exosomes: Proteomic insights and diagnostic
potential. Expert Rev Proteomics. 6:267–283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui Y, Fan Y, Zhao G, Zhang Q, Bao Y, Cui
Y, Ye Z, Chen G, Piao X, Guo F, et al: Novel lncRNA PSMG3AS1
functions as a miR1433p sponge to increase the proliferation and
migration of breast cancer cells. Oncol Rep. 43:229–239.
2020.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang W and Wang M: New insights into the
immunomodulatory role of exosomes in cardiovascular disease. Rev
Cardiovasc Med. 20:153–160. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Y, Huo Z, Wang W, Zhai S, Wang Y, Weng
Y, Deng X and Lu X: Construction and integrated analysis of a
lncRNA-associated competing endogenous RNA network reveal
functional lncRNAs in pancreatic cancer. Transl Cancer Res.
9:3643–3657. 2020. View Article : Google Scholar
|
|
24
|
Liu QY, Miao Y, Wang XH, Wang P, Cheng ZC
and Qian TM: Increased levels of miR-3099 induced by peripheral
nerve injury promote Schwann cell proliferation and migration.
Neural Regen Res. 14:525–531. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen L, Wang G, Xu Z, Lin K, Mu S, Pan Y
and Shan M: Overexpression of lncRNA PSMG3-AS1 distinguishes
glioblastomas from sarcoidosis. J Mol Neurosci. 70:2015–2019. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okkenhaug K and Vanhaesebroeck B: PI3K in
lymphocyte development, differentiation and activation. Nat Rev
Immunol. 3:317–330. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meo AD, Bartlett J, Cheng Y, Pasic MD and
Yousef GM: Liquid biopsy: A step forward towards precision medicine
in urologic malignancies. Mol Cancer. 16:802017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kalwa M, Hänzelmann S, Otto S, Kuo CC,
Franzen J, Joussen S, Fernandez-Rebollo E, Rath B, Koch C, Hofmann
A, et al: The lncRNA HOTAIR impacts on mesenchymal stem cells via
triple helix formation. Nucleic Acids Res. 44:10631–10643. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Z, Chen Z, Fan R, Jiang B, Chen X,
Chen Q, Nie F, Lu K and Sun M: Over-expressed long noncoding RNA
HOXA11-AS promotes cell cycle progression and metastasis in gastric
cancer. Mol Cancer. 16:822017. View Article : Google Scholar : PubMed/NCBI
|