Introduction
Human osteosarcoma is the most frequent malignant
bone tumor, and its characteristics of malignant proliferation and
invasion have made it the leading cause of cancer-associated
mortality among teenagers and children (1–3). The
incidence of osteosarcoma is approximately 4–5 cases per million,
which accounts for >60% of all bone malignancies and ~20% of all
primary osteosarcomas in children >5 years old (4). Surgery and chemotherapy are the most
common treatment methods for osteosarcoma (5). With improvements in therapeutic
technologies, the 5-year survival rate of newly diagnosed patients
with osteosarcoma without metastasis has improved to ~60-70%
(6). However, the 5-year survival
rate of patients with osteosarcoma is only 20% because of the
frequency of recurrent or metastatic disease (7). Thus, it is important to understand the
molecular mechanisms underlying the initiation, progression,
invasion and recurrence of osteosarcoma to identify novel
therapeutic targets and develop rational strategies for clinical
treatment of osteosarcoma.
MicroRNAs (miRNAs/miRs) are small (17–25
nucleotides) non-coding RNAs (8).
These molecules are highly conserved endogenous RNAs that have an
important role in post-transcriptional regulation of mRNAs for
translation, and influence diverse biological activities (9). Although only the biological functions
of a few miRNAs have been elucidated, miRNAs have been associated
with osteosarcoma initiation, progression and invasion (10,11).
Increasing evidence suggest that miRNAs may perform gene regulation
in different types of cancer, such as direct regulation of target
genes, including oncogenes or tumor suppressor genes (12). For example, miR-142-3p has been
demonstrated to inhibit osteosarcoma cell proliferation by
targeting Rac1 (13). In addition,
miR-142-3p has been reported to be highly overexpressed in
osteosarcoma cells, to have pivotal roles in cellular invasion,
proliferation, migration and apoptosis, and to induce E-cadherin
expression and decrease expression of matrix metalloproteinase
(MMP)2 and MMP9 (13). Thus, miRNAs
are considered potential biomarkers and therapeutic targets for
osteosarcoma.
Increasing evidence suggest that miR-382-5p is
enriched in the serum of patients with ischemic stroke (14). Notably, serum miR-382-5p may be used
as a non-invasive biomarker for potential diagnosis of ischemic
stroke (15). In addition, aberrant
miR-382-5p expression is frequently observed in different types of
human cancer, including oral, breast cancer, ovarian, prostate,
colorectal, lung and glioblastoma (16–19).
However, the underlying molecular mechanisms and functional role of
miR-382-5p in osteosarcoma tissues and cell lines remain largely
unclear.
The present study aimed to investigate the
biological effects of miR-382-5p on osteosarcoma cell migration,
invasion, proliferation and apoptosis, both in vitro and
in vivo. The results demonstrated that miR-382-5p expression
was markedly downregulated in human osteosarcoma tumor tissues and
cell lines, including MG63 and U2OS cells. In addition,
overexpression of miR-382-5p inhibited malignant biological
behaviors, including colony formation, proliferation, invasion and
migration of osteosarcoma cell lines. Further studies demonstrated
that miR-382-5p suppressed osteosarcoma development and progression
by targeting VEZF1, and its aberrant expression remarkably reversed
the antitumor effects of miR-382-5p overexpression in human
osteosarcoma cell lines in vitro. Both the in vivo
and in vitro experiments provide novel insights into the
molecular functions of miR-382-5p in human osteosarcoma, which may
be used as a potential therapeutic target for patients with
osteosarcoma.
Materials and methods
Clinical samples and cell culture
A total of 20 paired (13 men and seven women; age
range, 22–48 years; mean age, 33.8 years) adjacent normal tissues
(within 5 mm from the tumor boundary) and osteosarcoma tumor
tissues were collected from patients who underwent surgical
resection at Nanchong Central Hospital (Nanchong, China) between
January 2018 and December 2019. Resected tissues were immediately
frozen in liquid nitrogen and stored at −80°C until subsequent
experimentation. The present study was approved by the
Institutional Center Ethics Review Committee of Nanchong Central
Hospital (Nanchong, China; approval no. 2020-002) and written
informed consent was provided by all patients or their guardians
prior to the study start.
Human osteosarcoma cell lines, including HFOB, HOBC,
143B and U2OS were purchased from The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences, while MG63 cells
were purchased from the American Type Culture Collection (ATCC,
cat. no. ATCC® CRL-1690™) and U2 osteosarcoma
(ATCC® CRL-2611™) were purchased from the indicated
vendors and maintained in Dulbecco's Modified Eagle's Medium (DMEM,
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; AccuRef Scientific, http://www.accurefsci.com), 100 IU/ml penicillin and
100 µg/ml streptomycin (all from Thermo Fisher Scientific, Inc.),
at 37°C with 5% CO2.
Transfection of miRNA mimics and
plasmids
Human miR-382-5p mimics (50 nM,
5′-GAAGUUGUUCGUGGUGGAUUGG-3′), antisense or negative controls (50
nM, NC-mimic; 5′-CTCGCTTCGGCAGCACA-3′) were purchased from Shanghai
GenePharma Co., Ltd.. The DNA coding sequence for VEZF1 was cloned
into the pcDNA™3.1 (+) expression vector (Thermo Fisher Scientific,
Inc.), and the empty vector pcDNA3.1 was used as the NC.
Osteosarcoma cell lines (MG63 and U2OS) in logarithmic growth phase
were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS. Cells were incubated
at 37°C with 5% CO2 in a sterility incubator and
cultured for 24 h. Cells were transfected with miR-382-5p mimics
using the Lipofectamine® 3000 kit (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Cells
were collected for subsequent experimentation 48 h
post-transfection.
Lentivirus packaging and
infection
The 2nd lentiviral system purchased from Shanghai
Genechem Co., Ltd., was used for lentivirus generation. Lentiviral
particles were produced in Lenti-X™ 293T cells (Takara Bio Inc.;
cat. no. 632180) in 10-cm dish by transiently co-transfecting
control lentiviral vector (4.5 µg) or miR-382-5p overexpressing
lentiviral vector (4.5 µg) together with helper plasmids pHelper
1.0 (Gag and Pol; 2.5 µg) and pHelper 2.0 (VSVG; 2.0 µg) using the
Lipofectamine® 3000 kit (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The vector
constructions, verification by sequencing, virus packaging and
collection of the corresponding viral supernatants were performed
by Shanghai Genechem Co., Ltd. The lentivirus in medium of 293T
cells were harvested by centrifuging at 75,000 × g for 1.5 h at 4°C
48 h post-transfection (Abcam), according to the manufacturer's
protocol. Cells in the logarithmic growth phase were seeded into
96-well microplates (Costar; Corning, Inc.) at a density of
2×104 cells/well. Subsequently, the medium was replaced
with DMEM supplemented with 10% FBS after overnight incubation at
37°C. Cells were incubated for an additional 24, 48, 72, and 96 h,
respectively. At each time point, 100 µl CCK-8 reagent was added to
each well and cells were incubated for an additional 2 h at 37°C.
Against a background control, the sample absorbance was measured at
a wavelength of 490, using a microplate reader (Bio-Rad
Laboratories, Inc.).
Apoptosis assay
MG63 and U2OS cells seeded into 6-well plates were
transfected with the indicated miRNA mimics or the indicated
plasmids and incubated at 37°C for an additional 24 h. Cells were
digested with 0.25% trypsin-EDTA solution (Sigma-Aldrich; Merck
KGaA), terminated with cell medium, and harvested via centrifuged
at 300 × g for 5 min at 4°C. After washing with PBS, the Annexin
V-fluorescein Isothiocyanate (FITC)/PI Apoptosis kit (BD
Biosciences) was used to detect cell apoptosis according to the
manufacturer's protocols. Cells were washed three times with PBS
and the binding buffer, and stained with Annexin V-FITC/PI for
15–20 min at room temperature in the dark. Cells were re-washed and
the labeled cells were detected using a flow cytometer (CytoFLEX;
Beckman Coulter, Inc.). Flow cytometry data were analyzed using
Expo32 software (version 1.2; Beckman Coulter, Inc.).
Cell migration and invasion
assays
Transwell® Boyden chambers (BD
Biosciences) were used for in vitro cell migration assays,
and chambers coated with or without Matrigel (BD Biosciences) were
used to assess the migratory and invasive abilities of MG63 and
U2OS cells in vitro. For pre-coating, Matrigel was
maintained at 4°C overnight and the diluted with serum-free DMEM at
dilution of 1:4 on ice. Subsequently, Matrigel was added to each
upper chamber for coverage. Following incubation at 37°C for 1 h,
the chamber was rinsed with serum free DMEM followed by 50 µl of
DMEM supplemented with 10 g/l BSA at 37°C. Cells were resuspended
in FBS-free fresh medium 48 h post-transfection. Cells were plated
at a density of 1×105 cells/well into the upper chamber,
and 200 µl complete medium was loaded into the lower chamber.
Following incubation at 37°C for 24 h, the migratory and invasive
cells on the lower surface of the membrane were fixed with 4%
paraformaldehyde at 4°C for 1 h and stained with 0.1% crystal
violet (Sigma-Aldrich; Merck KGaA) at room temperature for 15 min,
followed by a light microscopy (Nikon E600; Nikon Instrument Inc.)
for visualization and photography. Stained cells were counted in
five randomly selected fields at low-power (×100)
magnification.
Dual-luciferase reporter assay
Given that miRNAs potentially interact with VEZF1,
the binding sites between VEZF1 and miR-382-5p were predicted using
lncBase version 2.0 (20). Candidate
genes were targeted by miR-382-5p, and the binding sites of
miR-382-5p were predicted using the online tools STarMirDB
(http://sfold.wadsworth.org/starmirDB.php) and miRWalk
(http://mirwalk.umm.uni-heidelberg.de). The predicted
binding sites within the 3′-untranslated region (UTR) of VEZF1 mRNA
(5′-AGGACAUUAAAUUGUACAACUUU-3′) and the corresponding mutated
binding region (5′-AGGACAUUAAAUUGUCCUACCGU-3′) were cloned into a
luciferase-expressing vector pcDNA3.1. For the luciferase reporter
assay, MG63 cells were transfected with different combinations of
miR-382-5p mimics, control mimics, and pcDNA3.1-VEZF1 3′-UTR
wild-type or mutant using the Lipofectamine® 3000 kit
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Following transfection for 48 h, cells were collected and
luciferase activities were detected using the dual-luciferase
reporter assay system (Promega Corporation) according to the
manufacturer's instructions. Firefly luciferase activity was
normalized to Renilla luciferase activity.
Reverse transcription-quantitative
(RT-q)PCR
According to the manufacturer's protocols, total RNA
and miRNA were extracted from osteosarcoma tissues and cell lines
using TRIzol® reagent (AccuRef Scientific) and the
miRNeasy mini kit (Qiagen GmbH), respectively. The
PrimeScript® RT Master Mix Perfect Real-Time Reagent kit
(Takara Bio, Inc.) was used to synthesize cDNA from total RNA,
according to the manufacturer's instructions. For miRNA RT, the
corresponding cDNA was synthesized using a universal tag (miScript
II RT kit; Qiagen GmbH). qPCR for miRNA and mRNA was subsequently
performed using a standard protocol from the SYBR Green PCR kit
(AccuRef Scientific) on an AB7500 RT-PCR instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.) The following
thermocycling conditions were used for qPCR: 95°C for 10 min, and
40 cycles of 95°C for 10 sec, 60°C for 30 sec and 72°C for 10 sec.
Relative quantification was determined by normalization to U6 or
GAPDH. The following primer sequences were used for qPCR: VEZF1
forward, 5′-GCCTCAACTGACAGAGGAGAAG-3′ and reverse,
5′-TCCAAAGTACTCCTTGGCTCC−3′; AKIRIN1 forward,
5′-GCACCTAGCTCTCCAGAACA-3′ and reverse, 5′-CTTGTCCCATACCGTCGCAT-3′;
SLAIN1 forward, 5′-TTTTCCCCAGTGTTTTCGGAG−3′ and reverse,
5′-CCCTGGAAGTGTAACCTTGCT−3′; LMTK3 forward,
5′-GTGTAATGTCTGCGTAACCGC−3′ and reverse,
5′-CCCCCAGTTGACAGAACTCC-3′; FOXN2 forward,
5′-GGGCTGGAATCTGCTGTTAGGG-3′ and reverse,
5′-TCCATGGCTTCAACCAACTGT−3′; TOP1 forward,
5′-GTTCAAGCCACAGACCGAGA-3′ and reverse, 5′-GCCTGGTAGAACGCTGACAA-3′;
AMOTL2 forward, 5′-GGGTGATTCAGTTGGGTGCT-3′ and reverse,
5′-GTACCGTGGGTCAGTGACAG-3′; ARIH2 forward,
5′-ACTTGGGTGACATCTGCCTG-3′ and reverse, 5′-GTCCTCTATGTCCCCAGGGT-3′;
DDX3X forward, 5′-GTAGCAGTCGTGGACGTTCT-3′ and reverse,
5′-ACCTGTGTGCCAAGGTTTGA-3′; GAPDH forward,
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3′; miR-382-5p forward,
5′-ATCCGTGAAGTTGTTCGTGG-3′ and reverse,
5′-TATGGTTGTAGAGGACTCCTTGAC−3′; and U6 forward,
5′-CGCTTCACGAATTTGCGT−3′ and reverse, 5′-CTCGCTTCGGCAGCACA-3′. All
experiments were performed in triplicate. PCR product specificity
was confirmed via melting curve analysis. Relative expression
levels were calculated using the 2−ΔΔCq method (21).
Western blotting
Protein samples from osteosarcoma tumor tissues and
cell lines were isolated using RIPA lysis buffer (Sigma-Aldrich;
Merck KGaA) and subsequently centrifuged at 12,000 × g for 30 min
at 4°C. Total protein was quantified using the BCA method (Thermo
Fisher Scientific, lnc.). Equal amounts of protein samples (20
µg/lane) were separated via 10% SDS-PAGE, transferred onto PVDF
membranes and blocked with 5% skimmed milk at room temperature for
1 h. The membranes were incubated with primary antibodies against
VEZF1 (1:1,000; cat. no. SAB2102675; Sigma-Aldrich; Merck KGaA) and
β-actin (1:10,000; cat. no. MFCD00164531; Sigma-Aldrich; Merck
KGaA) at 4°C overnight. All primary antibodies were diluted in a
Primary Dilution Solution (AccuRef Scientific) prior to incubation.
Following the primary incubation, membranes were incubated with
secondary HRP-conjugated polyclonal antibody anti-rabbit IgG
(1:10,000; cat. no. MFCD00163923; Sigma-Aldrich; Merck KGaA) at
room temperature for 1 h. Protein bands were visualized using the
Enhanced Chemiluminescence kit (Millipore, Sigma) and qualified
using ImageJ software (version 1.49, National Institutes of
Health).
Colony formation assay
Following trypsinization, MG63 and U2OS cells were
suspended in medium that included 0.3% agar (low melt, Bio-Rad
Laboratories, Inc.) and 10% FBS (AccuRef Scientific), seeded into
12-well plates and subsequently transfected with the indicated
miRNA mimics or the indicated plasmids followed by culturing for an
additional 24 h. Subsequently, 500 cells of each group were seeded
into 35-mm dishes, and the medium was replaced with fresh complete
DMEM every 2 days. Following culturing for 14 days at 37°C in the
incubator and washing three times with PBS, the cells were fixed
with 4% paraformaldehyde for 10 min at room temperature. Cells were
subsequently stained with 0.1% crystal violet (Sigma-Aldrich; Merck
KGaA) for 20 min at room temperature and washed with tap water.
Cells were observed under a light microscope (Nikon E600; Nikon
Instrument Inc.) with ×200 magnification. All experiments were
performed in triplicate.
Human osteosarcoma xenograft
model
A total of 15 female BALB/c nude mice (4–6 weeks old
and weighed 18–20 g) were obtained from Taconic Biosciences Company
and acclimated in a specific-pathogen-free facility at the
Experimental Animal Center of Nanchong Central Hospital, with a
12/12 h light/dark cycle, 40–60% humidity, 24–26°C temperature
condition and free access to food and water for ≥1 week. Stable NC
lentivirus and stable miR-382-5p-expressing lentivirus-infected
MG63 cells were resuspended in normal saline and mixed with an
isopycnic of Matrigel® (BD Biosciences) at a final
concentration of 5×106 cells/ml, respectively. Cells
were injected into the left flank of nude mice subcutaneously at a
density of 1×106 cells/mouse. Tumor size was measured
every 7 days using vernier calipers. Following tumor cell
inoculation, the mice were euthanatized at day 21 using
CO2 inhalation with a flow rate of 3 l/min and
euthanasia was confirmed via cardiac and respiration arrest. The
tumors were subsequently isolated, weighed, imaged and subjected to
subsequent analyses. All animal experiments were performed in
accordance with the Guide for the Care and Use of Laboratory
Animals (National Academies Press, 2011; Washington, USA). The
animal experimental protocol was approved by the Ethical Review
Committee for Animal Experiments of Nanchong Central Hospital
(Nanchong, China; approval no. NSMC-2020-42), and all experiments
were performed according to the AVMA guidelines.
Immunohistochemistry
While the tumors were pictured and weighed, partial
tumor tissues were collected and cut into 5 mm3 size.
Subsequently, tissues were fixed with 4% formaldehyde at 4°C more
than 12 h. Then, tissues were paraffin-embedded at 60°C overnight
and cut into 5-µm thick slices. Tumor tissues from xenograft mice
were deparaffinized and rehydrated in an ethanol series. Antigen
retrieval was performed according to previous experimental
protocols (22). After washing with
PBS and subsequent blocking with 3% hydrogen peroxide at room
temperature for 10 min, the sections were incubated with
rabbit-anti-Ki-67 polyclonal antibody (1:400; cat. no. PA5-19462;
Thermo Fisher Scientific, Inc.) overnight at 4°C. Sections were
washed three times with PBS, and the samples were incubated with
HRP-conjugated secondary donkey-anti-rabbit monoclonal antibody
(1:200; cat. no. ab6802; Abcam) at room temperature for 1 h.
Samples were re-washed five times with PBS and subsequently stained
with 0.5% diaminobenzidine and counterstained with Mayer's
hematoxylin at room temperature for 2–3 min. The mounted slices
were observed under a light microscope (Nikon E600; Nikon
Instrument Inc., magnification, ×200).
Wound healing assay
U2OS and MG63 cells were seeded into 6-well plates
at a density of 2×105 cells/well and allowed to adhere
for 24 h. Mimics were introduced into the cells and monolayer cells
were scratched using a 200 µl pipette tip after 24 h. The medium
was replaced with fresh serum-free RPMI-1640 medium. Wounded areas
were observed at 0 and 48 h after culturing under a light
microscope (Nikon E600; Nikon Instrument Inc., magnification,
×100). The wound-healing rate (%) was calculated using the
following equation: Wound-healing rate (%) = (width at 0 h-width at
48 h)/width at 0 h) ×100. All experiments were performed in
triplicate.
Statistical analysis
All quantitative data are presented as the mean ±
standard deviation. Normal distribution of all data were confirmed
using the Kolmogorov-Smirnov D test, and differences between
unpaired two groups were compared using unpaired two-tailed
Student's t-test. Paired two-tailed Student's t-test was used to
compare differences between adjacent tissues and tumor tissues.
Comparisons among multiple groups were performed using ANOVA
analysis. All experiments were performed in triplicate. SPSS v.19.0
software (IBM Corp.) was used to assess the correlation between
miR-382-5p and VEZF1 mRNA expression, using Pearson's correlation
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
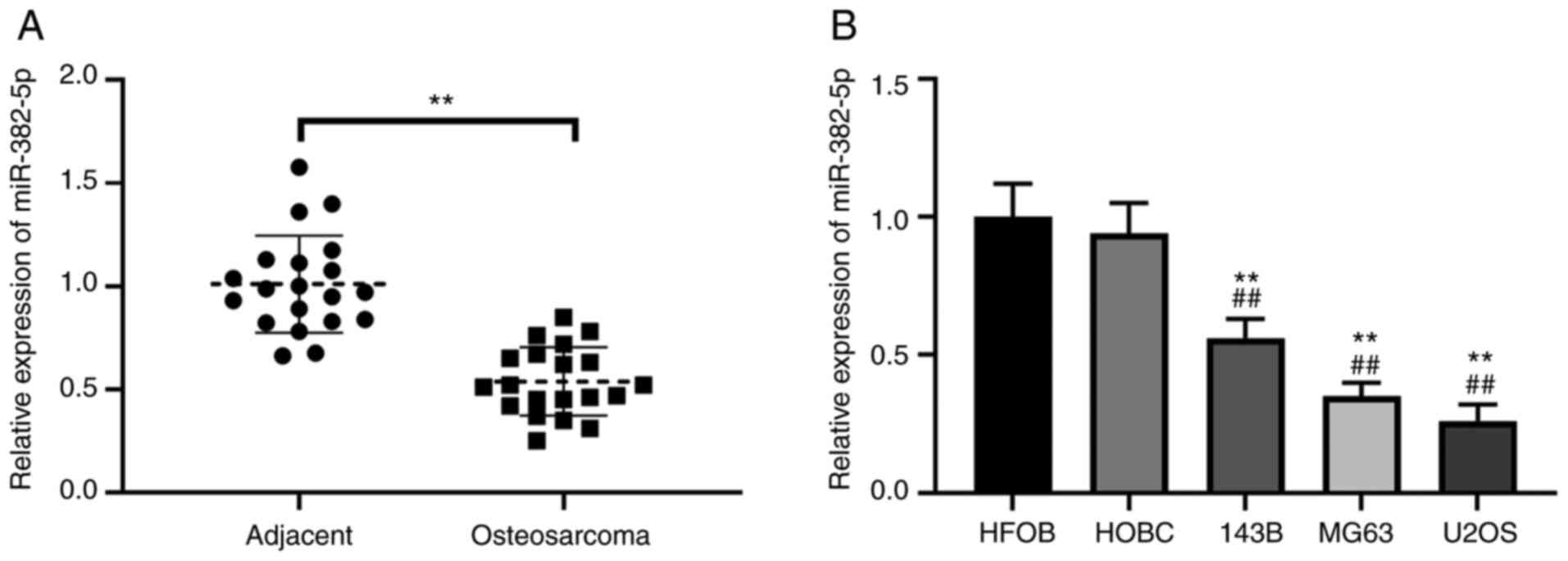
miR-382-5p expression is downregulated
in osteosarcoma tumor tissues and cell lines
To investigate the role of miR-382-5p in
osteosarcoma, RT-qPCR analysis was performed to detect miR-382-5p
expression in 20 paired human osteosarcoma tissues and adjacent
normal tissues. The results demonstrated that miR-382-5p expression
was significantly downregulated in osteosarcoma tumor tissues
compared with adjacent normal tissues (P<0.01; Fig. 1A). In addition, miR-382-5p expression
was detected in human osteosarcoma cell lines. The results
demonstrated that miR-382-5p expression was significantly
downregulated in human osteosarcoma cell lines (143B, MG63 and
U2OS) compared with the normal HFOB and HOBC cell lines (P<0.01;
Fig. 1B). Notably, MG63 and U2OS
cells exhibited ~80% reduction of miR-382-5p expression (Fig. 1B). Thus, these cell lines were
selected for subsequent experimentation.
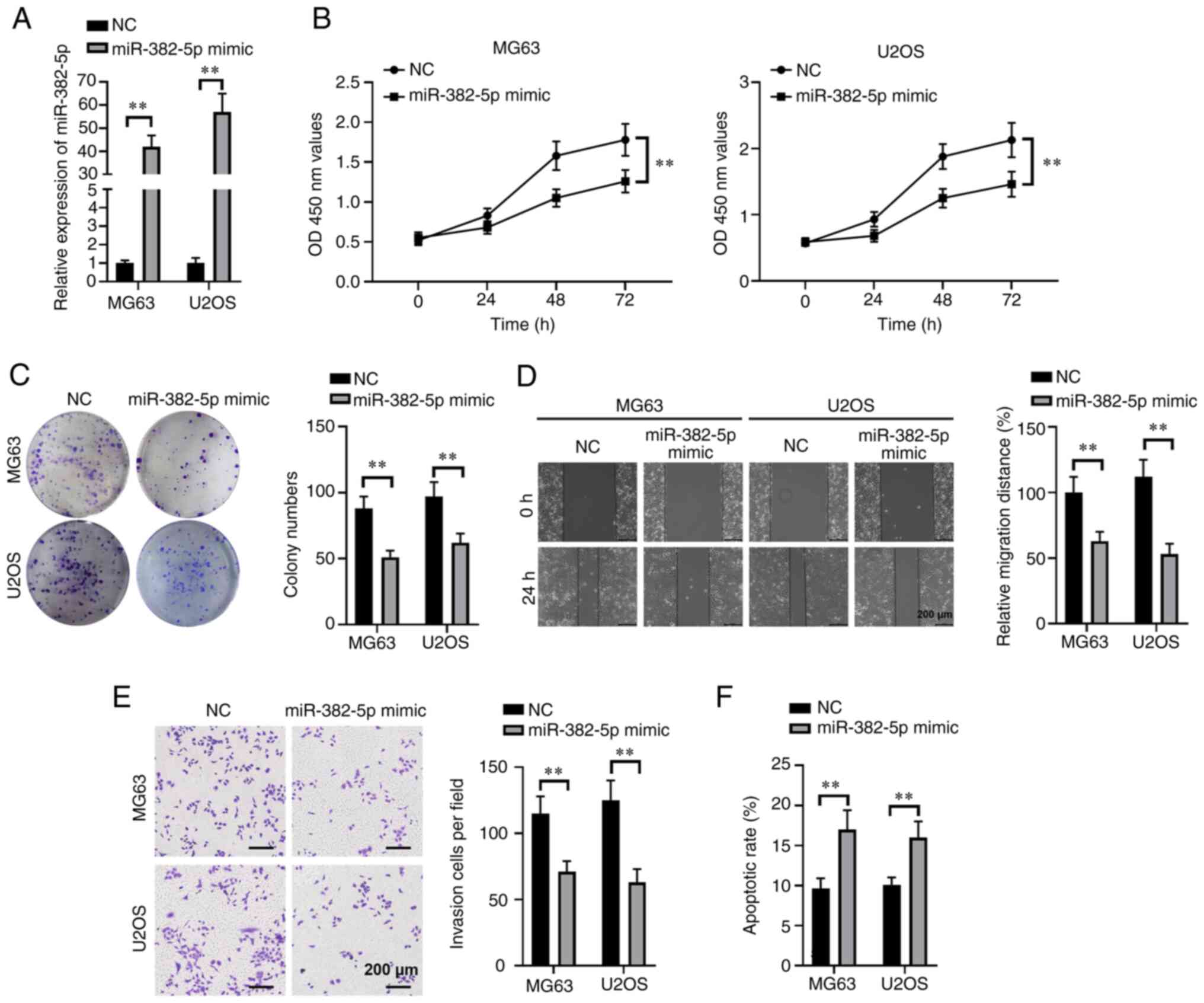
Overexpression of miR-382-5p inhibits
proliferation, invasion and migration of osteosarcoma cells, and
promotes apoptosis
According to the identifications in Fig. 1, downregulated miR-382-5p expression
in human osteosarcoma tissues and osteosarcoma cell lines suggests
that miR-382-5p may have important antitumor functions in the
initiation and progression of osteosarcoma. Thus, the effects of
miR-382-5p overexpression on the malignant behaviors of human
osteosarcoma cells were investigated, including colony formation,
proliferation, invasion and migration. MG63 and U2OS cells were
transfected with miR-382-5p mimic or a NC miRNA mimic, and a
40-fold increase in miR-382-5p expression was observed at 48 h in
miR-82-5p mimic-transfected MG63 and U2OS cells (P<0.01;
Fig. 2A). We then compared the
proliferation of these cells by performing CCK-8 assays. In
addition, overexpression of miR-382-5p significantly impeded the
proliferation of both MG63 and U2OS cells (both P<0.01; Fig. 2B). A nearly 50% decrease in
OD450nm absorbance values was observed in miR-382-5p
mimic-transfected MG63 and U2OS cells at 72 h after culturing.
Furthermore, similar reductions in colony numbers were also
observed in MG63 (P<0.01; Fig.
2C) and U2OS (P<0.01) cells with ectopic miR-382-5p
expression, which suggests that miR-382-5p notably decreases the
colony formation ability of human osteosarcoma cells. Notable
reductions in the migratory (P<0.01; Fig. 2D) and invasive (P<0.01; Fig. 2E) abilities of both MG63 and U2OS
cells were also observed following transfection with miR-382-5p
mimics. In addition, overexpression of miR-382-5p significantly
elevated the apoptotic rates of MG63 and U2OS cells (both
P<0.01; Fig. 2F). Taken together,
these results suggest that miR-382-5p exhibits tumor-suppressive
functions in human osteosarcoma cells.
miR-382-5p directly targets VEZF1 in
osteosarcoma cells
To further investigate the molecular mechanisms
underlying the tumor-suppressive function of miR-382-5p in
osteosarcoma, bioinformatics analysis was performed, using
STarMirDB and miRWalk (23,24), to predict the target genes based on
the consensus binding sites of miR-382-5p. In the present study,
the potential targets of miR-385-5p were predicted using ENCORI and
TargetScan databases and the overlapped targets were screened using
a Venn analysis (Fig. 3A). A total
of nine overlapped targets of miR-382-5p were identified, and
overexpression of miR-382-5p significantly inhibited the expression
levels of VEZF1, FOXN2, and AMOTL2, particularly VEZF1 (P<0.01;
Fig. 3B). The dual-luciferase
reporter assay was performed to determine whether VEZF1 is a direct
target of miR-382-5p in osteosarcoma. The results demonstrated that
luciferase activity significantly deceased following
co-transfection of wild-type VEZF1 3′-UTR and miR-382-5p mimics but
remained unchanged following co-transfection with mutated VEZF1
3′-UTR and miR-382-5p mimics or NC in MG63 cells (P<0.01;
Fig. 3C). These results suggest
direct binding of miR-382-5p to the predicted 3′-UTR of VEZF1.
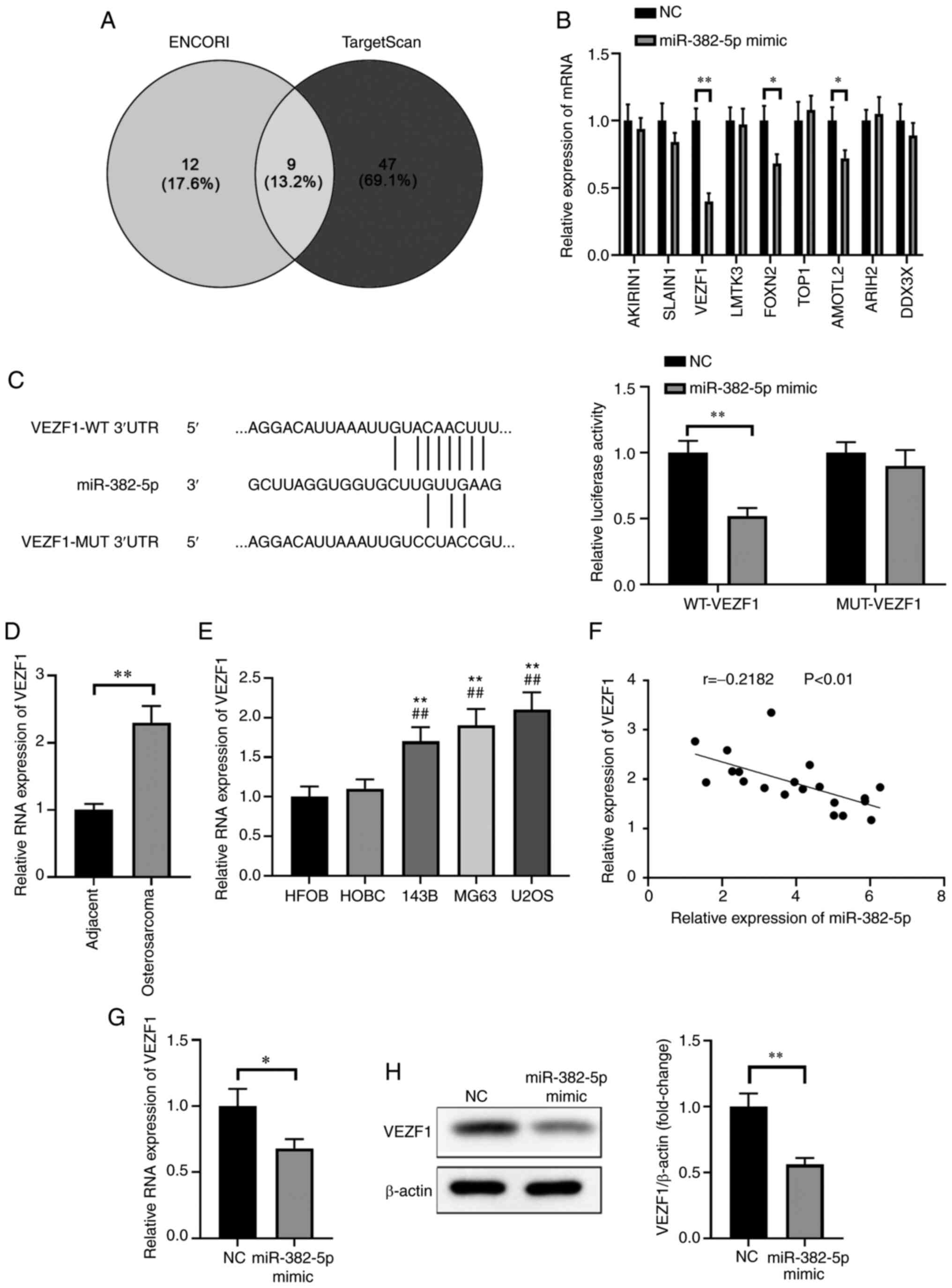 | Figure 3.VEZF1 is a direct target of
miR-382-5p in osteosarcoma. (A) Bioinformatics analysis was
performed to identify the potential targets of miR-382-5p, among
which nine mRNAs were selected for subsequent experimentations. (B)
Relative expression of potential targets of miR-382-5p in MG63
cells following transfection with miR-382-5p mimic. (C) The mature
miR-382-5p sequence, the putative binding sites of miR-382-5p and
the corresponding wild-type and mutant sites of VEZF1 mRNA 3′-UTR
(left) and the luciferase activity of the reporter vectors
demonstrated that VEZF1 mRNA expression significantly decreased in
MG63 cells following transfection with miR-382-5p mimics (right).
(D) RT-qPCR analysis was performed to detect VEZF1 mRNA expression
in osteosarcoma tissues and adjacent normal tissues. *P<0.05,
**P<0.01. (E) RT-qPCR analysis was performed to detect VEZF1
mRNA expression in osteosarcoma cell lines (143B, MG63 and U2OS)
and normal cell lines (HFOB and HOBC). **P<0.01 vs. HFOB cells;
##P<0.01 vs. HOBC cells. (F) Pearson's correlation
analysis was performed to assess the correlation between miR-382-5p
and VEZF1 expression in osteosarcoma. (G) RT-qPCR analysis was
performed to detect VEZF1 mRNA expression in MG63 cells transfected
with miR-382-5p mimic or NC mimic. *P<0.05. (H) Western blot
analysis was performed to detect VEZF1 protein expression in cells
transfected with miR-382-5p mimic or NC mimic. **P<0.01. VEZF1,
vascular endothelial zinc finger 1; miR, microRNA; UTR,
untranslated; RT-qPCR, reverse transcription-quantitative PCR; NC,
negative control; WT, wild-type; MUT, mutant. |
RT-qPCR and western blot analyses were performed to
further verify the association between miR-382-5p and VEZF1
expression in osteosarcoma tissues and adjacent normal tissues, and
the respective cell lines. The results demonstrated that the mRNA
and protein expression levels of VEZF1 were significantly lower in
adjacent normal tissues compared with osteosarcoma tissues
(P<0.01; Fig. 3D). Similarly, the
mRNA and protein expression levels of VEZF1 were significantly
lower in normal cells compared with osteosarcoma cells (P<0.01;
Fig. 3E). Furthermore, mRNA VEZF1
expression was inversely correlated with miR-382-5p expression in
osteosarcoma tissues (r=−0.2182; P<0.01; Fig. 3F).
VEZF1 mRNA expression significantly decreased in
MG63 cells transfected with miR-382-5p mimic compared with the
control group (P<0.05; Fig. 3G).
Furthermore, western blot analysis demonstrated that VEZF1 protein
expression significantly decreased in MG63 cells transfected with
miR-382-5p mimic compared with the control group (Fig. 3H). Collectively, these results
suggested that VEZF1 is a direct downstream target of miR-382-5p in
human osteosarcoma.
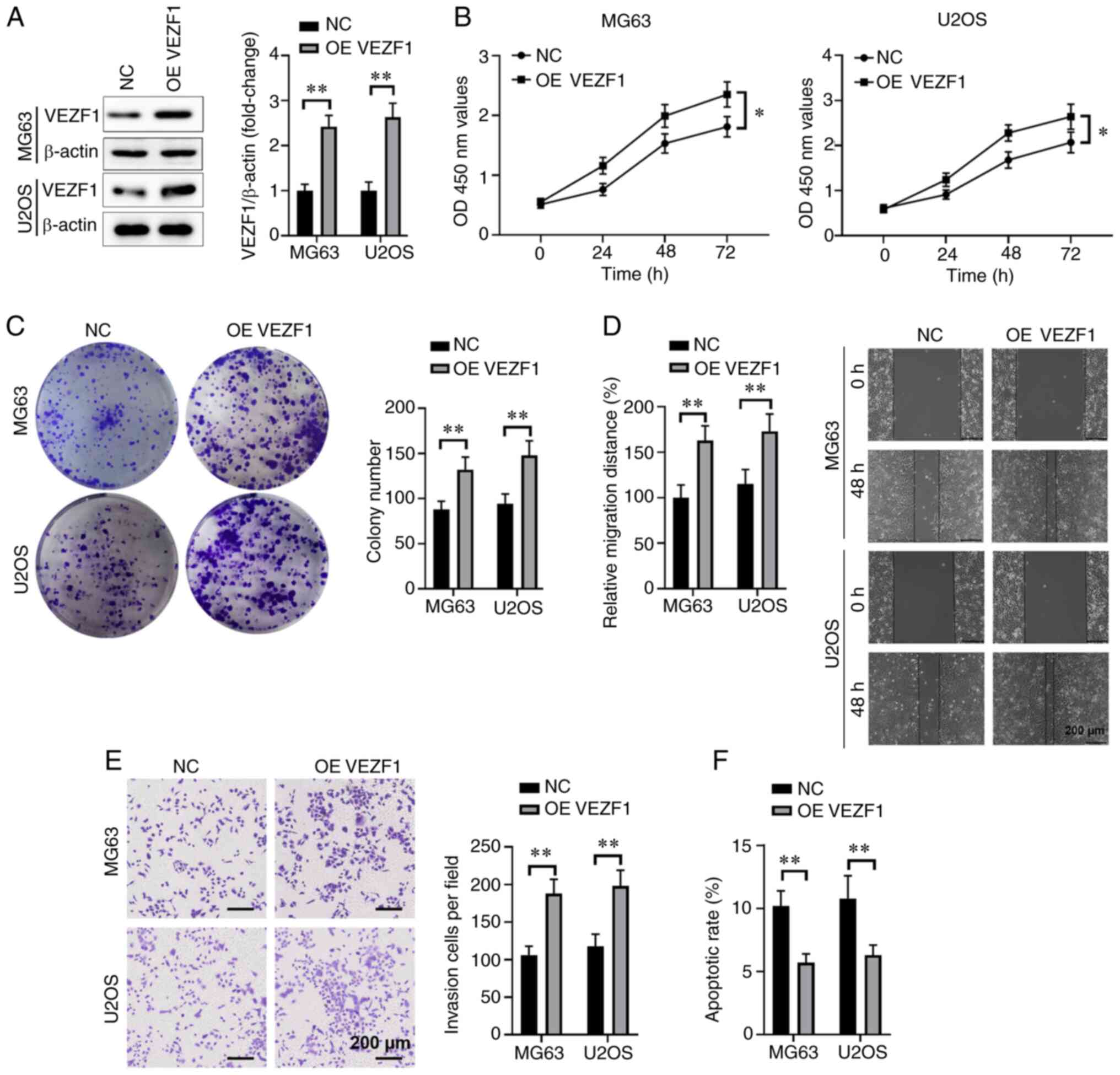
Overexpression of VEZF1 enhances cell
proliferation, invasion and migration, while inhibiting apoptosis
in osteosarcoma cells
To determine whether VEZF1 promotes the malignant
behaviors of osteosarcoma cell, including invasion, proliferation
and migration in MG63 or U2OS cells, MG63 and U2OS cells were
transfected with VEZF1-expressing plasmid to elevate VEZF1 protein
expression (P<0.01; Fig. 4A). The
results of the CCK-8 assay demonstrated that overexpression of
VEZF1 significantly enhanced the proliferation of MG63 and U2OS
cells (P<0.05; Fig. 4B).
Furthermore, overexpression of VEZF1 significantly increased the
colony formation ability of MG63 and U2OS cells (P<0.01;
Fig. 4C). The results of the wound
healing assay demonstrated that overexpression of VEZF1 promoted
osteosarcoma cell migration in MG63 or U2OS cells compared with the
NC group (P<0.01; Fig. 4D).
Similarly, the results of the Transwell assay demonstrated that
overexpression of VEZF1 promoted the invasive ability of MG63 and
U2OS cells compared with the NC group (P<0.01; Fig. 4E). Flow cytometric analysis
demonstrated that overexpression of VEZF1 significantly decreased
cell apoptosis compared with the control group (P<0.01; Fig. 4F). Taken together, these results
suggest that VEZF1 promotes malignant behaviors of MG63 and U2OS
cells.
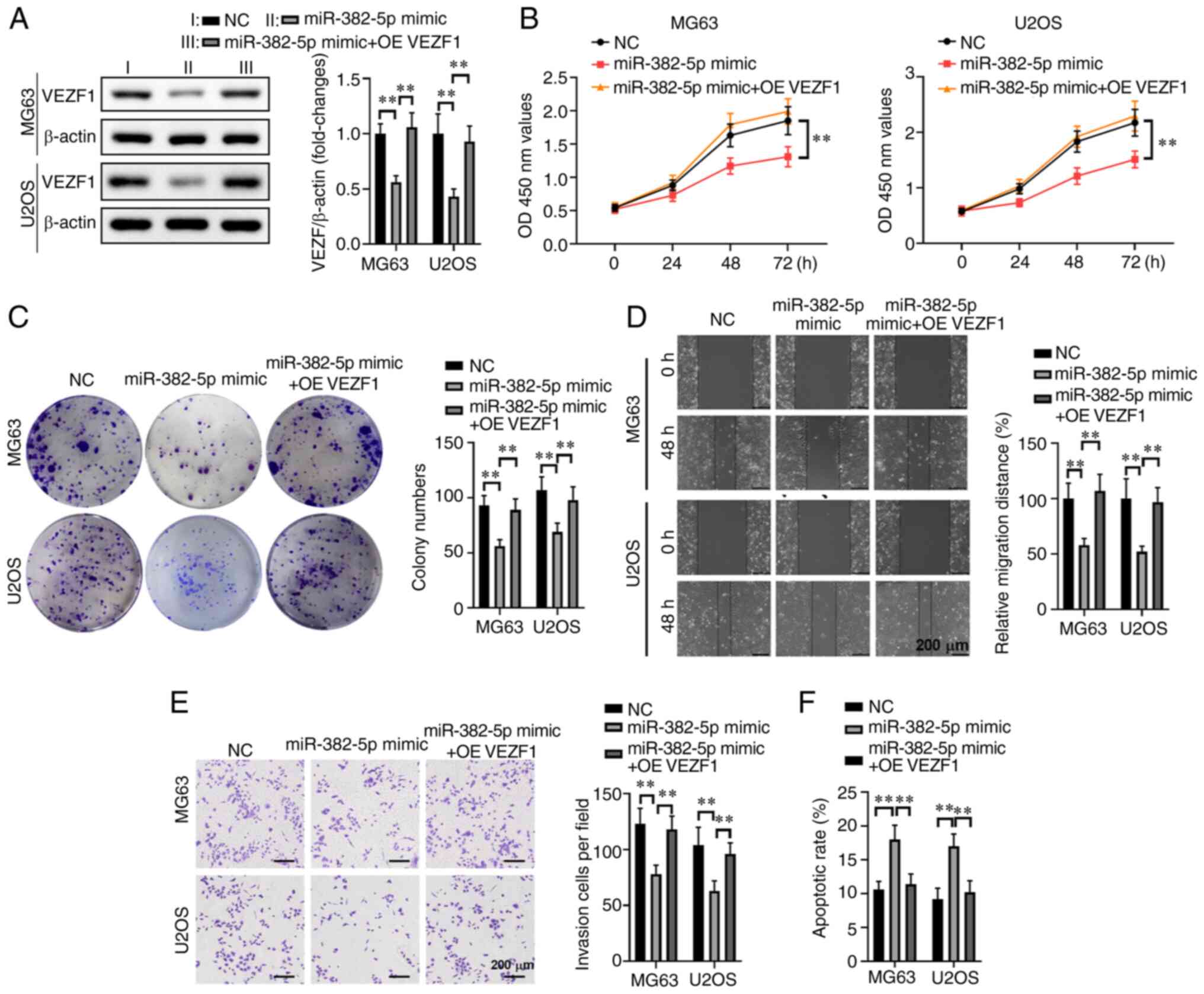
Overexpression of VEZF1 abrogates the
effect of miR-382-5p on cell proliferation, invasion, migration and
cell apoptosis
To further investigate the association between VEZF1
and miR-382-5p in human osteosarcoma cells, the effects of
overexpressing VEZF1 on malignant behaviors of miR-382-5p
mimic-transfected MG63 or U2OS cells were assessed. Western blot
analysis demonstrated that overexpression of VEZF1 significantly
increased VEZF1 expression in MG63 and U2OS cells compared with the
miR-382-5p mimics group (P<0.01; Fig.
5A). Similarly, the results of the CCK-8 assay demonstrated
that overexpression of VEZF1 significantly enhanced the
proliferation of MG63 or U2OS cells compared with the miR-382-5p
mimics group (P<0.01; Fig. 5B).
Furthermore, simultaneous overexpression of VEZF1 in
miR-382-5p-transfected MG63 and U2OS cells significantly increased
the number of cell colonies (P<0.01; Fig. 5C), and significantly induced the cell
migratory (P<0.01; Fig. 5D) and
invasive (P<0.01; Fig. 5E)
abilities, while decreasing apoptosis (P<0.01; Fig. 5F). Taken together, these results
suggested that simultaneous overexpression of VEZF1 reverses the
effects of miR-382-5p overexpression on malignant behaviors of
human osteosarcoma cells.
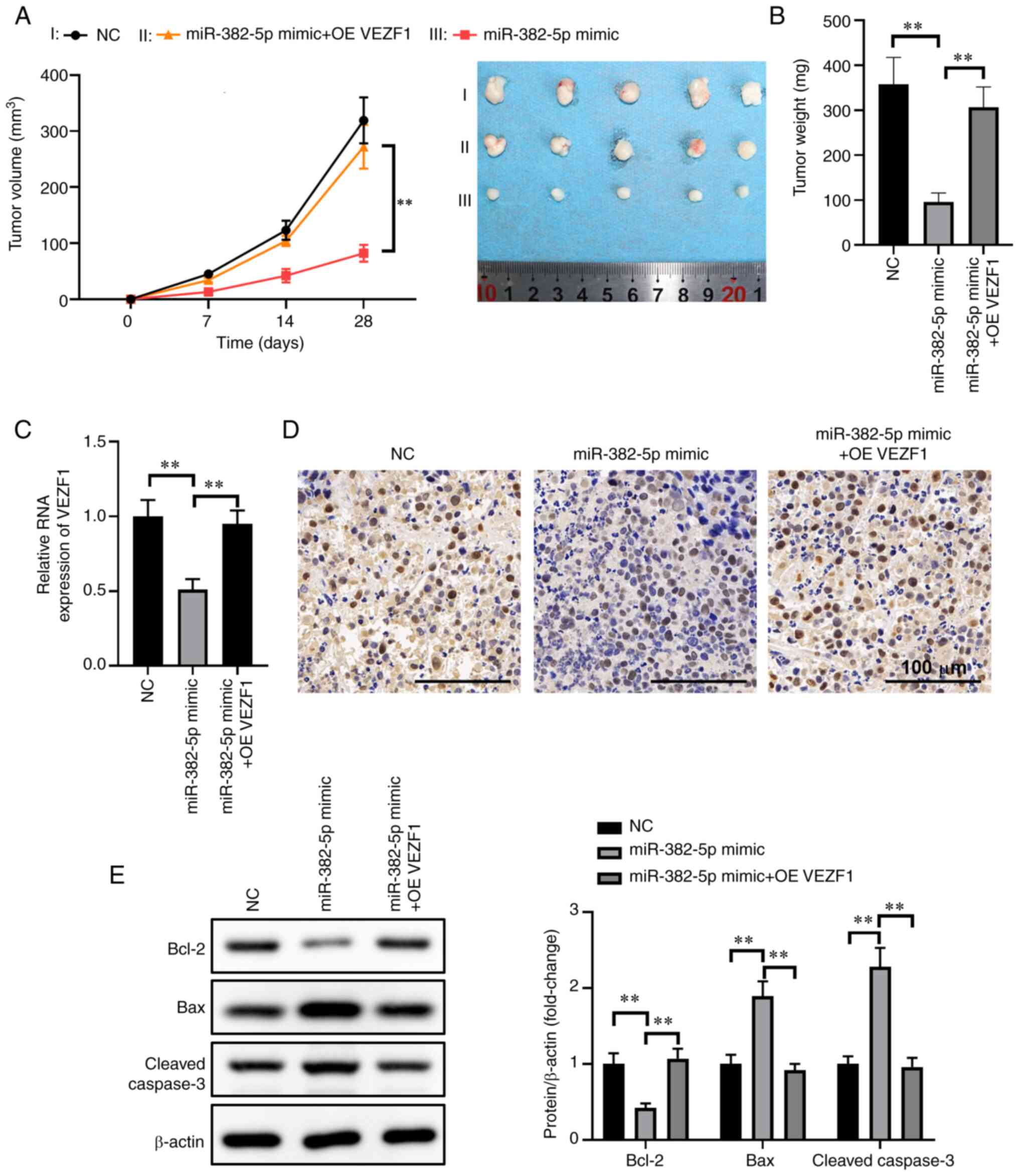
Overexpression of VEZF1 attenuates the
inhibitory effect of miR-382-5p in an animal model
To further investigate the tumor-inhibiting function
of miR-382-5p via VEZF1 in human osteosarcoma in vivo, empty
lentivirus-infected control stable U2OS cells (LV),
miR-382-5p-expressing lentivirus-infected stable U2OS cells
(LV-miR-382-5p) and VEZF1 overexpression vectors (VEZF1) were
inoculated and co-transfected into LV-miR-382-5p cells on the left
flank of model mice via subcutaneous injection. Some of the mice
were sacrificed at day 18 following tumor inoculation. The results
demonstrated that 21 days after xenografting, the tumor volume
significantly decreased in U2OS cells stably transfected with
LV-miR-382-5p significantly. In addition, the weight of the mice
decreased by 70% compared with the miR-382-5p mimics and VEZF1
co-transfection or miR-NC groups (Fig.
6A and B). RT-qPCR analysis demonstrated a notable increase in
miR-382-5p expression following transduction with miR-382-5p
lentivirus but had no obvious change after overexpressing VEZF1,
whereas VEZF1 mRNA expression significantly decreased following
transfection with miR-382-5p mimics but significantly increased
after overexpressing VEZ1 (P<0.01; Fig. 6C), which confirms the association
between miR-382-5p and VEZF1 in human osteosarcoma.
Immunohistochemical analysis of Ki-67 in tumor
tissues indicated pleomorphic poorly differentiated neoplastic
cells, confirming cancerous tissue growth (Fig. 6D). Subsequent experiments focused on
miR-382-5p-mediated regulation of B-cell lymphoma 2 (Bcl2) and Bax,
which are downstream of the vascular endothelial growth factor
(VEGF) signaling pathway (25).
Western blot analysis demonstrated that overexpression of
miR-382-5p promoted Bax expression and suppressed Bcl2 expression,
while simultaneously promoting cleaved caspase-3 expression
(P<0.01; Fig. 6E). In addition,
overexpression of VEZF1 attenuated the suppression of miR-382-5p in
osteosarcoma cells. Taken together, these results suggest that the
size and weight of xenograft tumors in nude mice significantly
decrease following transfection with miR-372-3p mimics.
Discussion
The first miRNAs, which are small non-coding RNA
segments (~22 nucleotides in length) were identified in 1993 (in
Caenorhabditis elegans) (26). These miRNAs have diverse roles in
cellular development, differentiation, apoptosis and metabolism,
including osteoma initiation and normal osteogenesis (9). These molecules have important roles in
various functions, such as those of tumor-suppressor genes or
oncogenes, so when they are disordered, they contribute to
initiation and progression of cancer (27). Recent studies have reported that
aberrant expression of miRNAs has been demonstrated in a range of
human diseases, including various malignancies (28–30).
miR-382-5p has been repeatedly observed in various human
malignancies, including lung, oral, ovarian, breast and prostate
cancers (16–19). Given that miR-382-5p has been
reported to be frequently and significantly reduced in tumor
tissues and tumor-associated cell lines, it was speculated that
miR-382-5p functions as a tumor-suppressor gene or oncogene in
osteosarcoma. However, the highly complex molecular mechanisms
underlying miR-382-5p inhibition of osteosarcoma cells remain
unclear.
The results of the present study demonstrated that
miR-382-5p expression was markedly downregulated in clinical
osteosarcoma tumor tissues and osteosarcoma-associated cell lines
compared with normal tissues and cell lines. In addition,
miR-382-5p exerted antitumor functions in human osteosarcoma cells,
including restraining cell colony formation, proliferation,
migration and invasion, as well as enhancing apoptosis in
vitro and suppressing tumor growth in vivo. These
results are in accordance with previous findings, demonstrating
that miR-382-5p expression is notably downregulated in cancer cells
(16,31,32).
Previous study had demonstrated that miR-382-5p acts as tumor
suppressor to inhibit the metastasis and relapse of osteosarcoma by
targeting YB-1 (33), while
circ_0001658 can significantly promote the proliferation and
metastasis of osteosarcoma by modulating the miR-382-5p/YB-1 axis
(34). A recent study also reported
that LINC00265 can promote the proliferation, migration, invasion
and angiogenesis of osteosarcoma by targeting miR-382-5p mediated
SAT1 and VAV3 (35). In the present
study, bioinformatics analysis was performed to identify a target
gene of miR-382-5p as VEZF1, which was identified as a novel and
functional target of miR-382-5p via the dual-luciferase reporter
assay. Overexpression analyses of VEZF1 in osteosarcoma cells with
ectopic expression of VEZF1 was also performed. The results
demonstrated that overexpression of VEZF1 notably attenuated the
tumor-suppressive role of miR-382-5p mimics on the malignant
behaviors of human osteosarcoma cells. Taken together, these
results suggest that the miR-382-5p-VEZF1 axis may act as a novel
molecular target for developing therapeutics against
osteosarcoma.
Osteosarcoma is the most common primary bone
malignancy that predominantly occurs in children, adolescents and
young adults (36). With
improvements in technology, the 5-year overall survival rate has
increased to ~60-70% (37). However,
the response of osteosarcoma to chemotherapy remains poor, and
there is a risk of recurrence and metastasis following surgical
excision and chemotherapy (38).
Biomarkers are considered one of the most effective tools to
determine the prognosis of a tumor following chemotherapy (39). A previous study demonstrated that
miRNAs are significantly upregulated in serum samples from patients
with breast cancer (40). However,
due to the deficiency of serum samples, miR-382-5p expression was
unable to be detected in serum samples of patients with
osteosarcoma in the present study. miR-382-5p expression was only
detected in tissue samples and the results demonstrated that
miR-382-5p was significantly downregulated in clinical osteosarcoma
tissues and osteosarcoma-associated cell lines. Despite the
limitation in serum samples, the results of the present study and
previous findings suggest that miR-382-5p may serve as a novel
biomarker for the prognosis of patients with osteosarcoma.
Considering the prognostic significance, serum miR-382-5p
expression and its prognostic value in osteosarcoma will be
investigated in prospective studies.
Recent studies have reported that miR-382-5p has
important functions, including a tumor-suppressor or oncogene, in
the context of different types of cancer. For examples, previous
studies have demonstrated that miR-382-5p expression is
downregulated in human ovarian tumor tissues and cancer-associated
cell lines (17,32), whereas miR-382-5p expression is
upregulated in breast and glioma tumor tissues and
cancer-associated cell lines (16,31).
However, the expression and function of miR-382-5p in osteosarcoma
remains unclear. The results of the present study demonstrated that
miR-382-5p expression was markedly downregulated in osteosarcoma
tissues and cell lines, and significantly low expression levels
were detected in tumor tissues. In vivo and in vitro
overexpression of miR-382-5p inhibited proliferation, invasion and
migration of osteosarcoma cells, and enhanced apoptosis. Taken
together, these results suggest that overexpression of miR-382-5p
exerts a tumor-suppressive effect in osteosarcoma.
A previous study reported that miRNAs recognize and
combine specific target mRNAs depending on complete or partial
base-pairing, mostly at the 3′-UTR of the target genes, to regulate
gene expression following transcription, which includes both tumor
suppressors and oncogenes (41).
Bioinformatic analysis was performed in the present study to
predict gene targets of miR-382-5p. The results revealed that VEZF1
has a highly conserved miR-382-5p binding sequence in the mRNA
3′-UTR. The results of the dual-luciferase reporter assay confirmed
the direct association between miR-382-5p and VEZF1 in human
osteosarcoma cells.
VEZF1 is a Krüppel-like zinc finger protein that
plays a vital role in vascular development, and is a member of the
VEGF signaling pathway (42).
Abnormal expression of VEZF1 contributes to the pathogenesis of
different disorders, including cancer and cardiovascular diseases
(43). Recent study has reported
that a small molecule inhibitor of VEZF1, Vec6, prevents wound
healing and angiogenesis (44).
Increasing evidences suggest that VEZF1 plays a major role in the
development and progression of malignancy (43,45,46). The
results of the present study demonstrated that overexpression of
VEZF1 in vitro and in a nude mice model increased
proliferation, invasion and migration of osteosarcoma cells, and
decreased apoptosis. Furthermore, overexpression of VEZF1 notably
attenuated the tumor-suppressive effects of miR-382-5p mimics in
human osteosarcoma cells and the animal model, which suggests that
VEZF1 may act as an oncogene in human osteosarcoma cells. However,
further studies are required to screen potential targets of
miR-382-5p other than VEZF1 to determine the tumor-suppressive
functions of miR-382-5p in human osteosarcoma.
In conclusion, the results of the present study
demonstrated that miR-382-5p acts as a tumor-suppressor gene, whose
ectopic expression inhibits the malignant biological behaviors of
human osteosarcoma cells in vitro and in a nude mice model.
VEZF1 was identified as a direct target gene of miR-382-5p, and its
overexpression remarkably attenuated the inhibitory effect of
miR-382-5p mimics in human osteosarcoma cells. The miR-382-5p/VEZF1
axis provides novel insights into the pathogenesis of osteosarcoma,
and miR-382-5p expression may have effects on the malignancy of
osteosarcoma cells. Thus, miR-382-5p expression may potentially act
as a prognostic biomarker or a therapeutic target in
osteosarcoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Sichuan
Science and Technology Program (grant nos. 2020YFS0529, 2019YJ0707
and 2018SZ0377), the Nanchong Science and Technology Program (grant
nos. 19SXHZ0230, 19SXHZ0451, 19SXHZ0345, 18SXHZ0366 and 18SXHZ0370)
and the Science and Technology Project of the Health Planning
Committee of Sichuan (grant nos. 19PJ057 and 20PJ177).
Availability of data and materials
The datasets used and/or analyzed in this study are
available from the corresponding author upon reasonable
request.
Authors' contributions
KL and GS conceived and designed the experiments.
HW, YXL, MLZ and JB performed the experiments. WH, MY, RP, NXH and
GF analyzed and interpreted the data. WH and MY collected the
clinical samples and performed the experiments involving the
clinical samples. WH and HW drafted the initial manuscript. GS and
KL revised the manuscript for important intellectual content. KL
and GS confirm the authenticity of all the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Center Ethics Review Committee of Nanchong Central Hospital
(Nanchong, China; approval no. 2020-002) and written informed
consent was provided by all patients or their guardians prior to
the study start. All animal experiments were performed in
accordance with the Guide for the Care and Use of Laboratory
Animals (National Academies Press, 2011; Washington, USA). The
animal experimental protocol was approved by the Ethical Review
Committee for Animal Experiments of Nanchong Central Hospital
(Nanchong, China; approval no. NSMC-2020-42) and all experiments
were performed according to the AVMA guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNAs/miRs
|
microRNAs
|
|
UTR
|
untranslated region
|
References
|
1
|
Broadhead ML, Clark JC, Myers DE, Dass CR
and Choong PF: The molecular pathogenesis of osteosarcoma: A
review. Sarcoma. 2011:9592482011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang J and Zhang W: New molecular insights
into osteosarcoma targeted therapy. Curr Opin Oncol. 25:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu CC, Beird HC, Andrew Livingston J,
Advani S, Mitra A, Cao S, Reuben A, Ingram D, Wang WL, Ju Z, et al:
Immuno-genomic landscape of osteosarcoma. Nat Commun. 11:10082020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rytting M, Pearson P, Raymond AK, Ayala A,
Murray J, Yasko AW, Johnson M and Jaffe N: Osteosarcoma in
preadolescent patients. Clin Orthop Relat Res. 39–50. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Petrilli AS, de Camargo B, Filho VO,
Bruniera P, Brunetto AL, Jesus-Garcia R, Camargo OP, Pena W,
Péricles P, Davi A, et al: Results of the Brazilian osteosarcoma
treatment group studies III and IV: Prognostic factors and impact
on survival. J Clin Oncol. 24:1161–1168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hosseinahli N, Aghapour M, Duijf PHG and
Baradaran B: Treating cancer with microRNA replacement therapy: A
literature review. J Cell Physiol. 233:5574–5588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aredia F and Scovassi AI: A new function
for miRNAs as regulators of autophagy. Future Med Chem. 9:25–36.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song B, Wang Y, Xi Y, Kudo K, Bruheim S,
Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al:
Mechanism of chemoresistance mediated by miR-140 in human
osteosarcoma and colon cancer cells. Oncogene. 28:4065–4074. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Naeli P, Yousefi F, Ghasemi Y,
Savardashtaki A and Mirzaei H: The role of MicroRNAs in lung
cancer: Implications for diagnosis and therapy. Curr Mol Med.
20:90–101. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martens-Uzunova ES, Bottcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng Z, Ding M, Ni J, Song D, Huang J and
Wang J: miR-142 acts as a tumor suppressor in osteosarcoma cell
lines by targeting Rac1. Oncol Rep. 33:1291–1299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qu Q, Chu X and Wang P: MicroRNA-195-5p
suppresses osteosarcoma cell proliferation and invasion by
suppressing naked cuticle homolog 1. Cell Biol Int. 41:287–295.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Ma Z, Kan P and Zhang B: The
diagnostic value of serum miRNA-221-3p, miRNA-382-5p, and
miRNA-4271 in ischemia stroke. J Stroke Cerebrovasc Dis.
26:1055–1060. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang X, Zhao H, Zhang Y, Yang X, Zhang J,
Yi M and Zhang C: The MicroRNA-382-5p/MXD1 axis relates to breast
cancer progression and promotes cell malignant phenotypes. J Surg
Res. 246:442–449. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tan H, He Q, Gong G, Wang Y, Li J, Wang J,
Zhu D and Wu X: miR-382 inhibits migration and invasion by
targeting ROR1 through regulating EMT in ovarian cancer. Int J
Oncol. 48:181–190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shan N, Shen L, Wang J, He D and Duan C:
miR-153 inhibits migration and invasion of human non-small-cell
lung cancer by targeting ADAM19. Biochem Biophys Res Commun.
456:385–391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu Z, He B, He J and Mao X: Upregulation
of miR-153 promotes cell proliferation via downregulation of the
PTEN tumor suppressor gene in human prostate cancer. Prostate.
73:596–604. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44(D1): D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee JH, Kim C, Baek SH, Ko JH, Lee SC,
Yang WM, Um JY, Sethi G and Ahn KS: Capsazepine inhibits JAK/STAT3
signaling, tumor growth, and cell survival in prostate cancer.
Oncotarget. 8:17700–17711. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sticht C, De La Torre C, Parveen A and
Gretz N: miRWalk: An online resource for prediction of microRNA
binding sites. PLoS One. 13:e02062392018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rennie W, Kanoria S, Liu C, Mallick B,
Long D, Wolenc A, Carmack CS, Lu J and Ding Y: STarMirDB: A
database of microRNA binding sites. RNA Biol. 13:554–560. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu HF, Wang YH, Liu D, Sun XQ and Wang
FR: Vanadium rutin complex sensitizes breast cancer cells via
modulation of p53/Bax/Bcl2/VEGF correlated with apoptotic events.
Acta Pol Pharm Drug Res. 77:89–98. 2020.
|
|
26
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nugent M: MicroRNA function and
dysregulation in bone tumors: The evidence to date. Cancer Manag
Res. 6:15–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Fu H, Wang Y, Tie Y, Xing R, Zhu J,
Sun Z, Wei L and Zheng X: MicroRNA-101 regulates expression of the
v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene
in human hepatocellular carcinoma. Hepatology. 49:1194–1202. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vishnol A and Rani S: miRNA biogenesis and
regulation of disease: An overview. Methods Mol Biol. 1509:1–10.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Reddy KB: MicroRNA (miRNA) in cancer.
Cancer Cell Int. 15:382015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ho JY, Hsu RJ, Liu JM, Chen SC, Liao GS,
Gao HW and Yu CP: MicroRNA-382-5p aggravates breast cancer
progression by regulating the RERG/Ras/ERK signaling axis.
Oncotarget. 8:22443–22459. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang J, Chen C, Yan X and Wang P: The role
of miR-382-5p in glioma cell proliferation, migration and invasion.
Onco Targets Ther. 12:4993–5002. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu M, Jin H, Xu CX, Sun B, Song ZG, Bi WZ
and Wang Y: miR-382 inhibits osteosarcoma metastasis and relapse by
targeting Y box-binding protein 1. Mol Ther. 23:89–98. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang L, Wang P, Su X and Zhao B:
Circ_0001658 promotes the proliferation and metastasis of
osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell
Biochem Funct. 38:77–86. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiao Y, Li C, Wang H and Liu Y: LINC00265
targets miR-382-5p to regulate SAT1, VAV3 and angiogenesis in
osteosarcoma. Aging (Albany NY). 12:20212–20225. 2020.PubMed/NCBI
|
|
36
|
Kobayashi E, Hornicek FJ and Duan Z:
MicroRNA involvement in osteosarcoma. Sarcoma. 2012:3597392012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nugent M: microRNA and bone cancer. Adv
Exp Med Biol. 889:201–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu L and Qu X: Cancer biomarker detection:
Recent achievements and challegenges. Chem Soc Rev. 44:2963–2997.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang XX, Ye FG, Zhang J, Li JJ, Chen QX,
Lin PY and Song CG: Serum miR-4530 sensitizes breast cancer to
neoadjuvant chemotherapy by suppressing RUNX2. Cancer Manag Res.
10:4393–4400. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sampson VB, Yoo S, Kumar A, Vetter NS and
Kolb EA: MicroRNAs and potential targets in osteosarcoma: Review.
Front Pediatr. 3:692015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
AlAbdi L, He M, Yang Q, Norvil AB and
Gowher H: The transcription factor Vezf1 represses the expression
of the antiangiogenic factor Cited2 in endothelial cells. J Biol
Chem. 293:11109–11118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He M, Yang Q, Norvil AB, Sherris D and
Gowher H: Characterization of small molecules inhibiting the
pro-angiogenic activity of the zinc finger transcription factor
Vezf1. Molecules. 23:16152018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gerald D, Adini I, Shechter S, Prendergast
GC, Klagsbrun M, Stuhlmann H, Rigby AC, Nagy JA and Benjamin LE:
RhoB controls coordination of adult angiogenesis and
lymphangiogenesis following injury by regulating VEZF1-mediated
transcription. Nat Commun. 4:28242013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yin R, Guo L, Gu J, Li C and Zhang W: Over
expressing miR-19b-1 suppress breast cancer growth by inhibiting
tumor microenvironment induced angiogenesis. Int J Biochem Cell
Biol. 97:43–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ahmed M, Lai TH, Zada S, Hwang JS, Pham
TM, Yun M and Kim DR: Functional linkage of RKIP to the epithelial
to mesenchymal transition and autophagy during the development of
prostate cancer. Cancers (Basel). 10:2732018. View Article : Google Scholar : PubMed/NCBI
|