Introduction
Melanoma is the most common malignancy of the skin,
with increasing incidence rate (1).
Melanoma has a high risk of metastasis. Current treatment options
for melanoma primarily consist of surgical resection, radiotherapy
and chemotherapy; however, these methods are only effective for
tumors at early stages (2). Once
metastasis occurs, the median survival time is only 6–9 months
(3).
Ubiquitin C-terminal hydrolase-L3 (UCHL3) is a
member of the ubiquitin carboxyl terminal hydrolase (UCH) family
(4). Our previous studies have
demonstrated that neural precursor cell-expressed developmentally
downregulated protein (NEDD) is upregulated in melanoma tissues and
cells, with associated changes in several regulatory enzymes, among
which UCHL3 exhibits the most significant alterations (5,6).
Recently, studies have been focusing on the role of the UCH family
in cancer (7,8). Unlike other members of this family,
UCHL3 not only functions as a ubiquitin hydrolase, but also
regulates NEDD8 hydrolase (9).
Furthermore, it acts as a cleavage enzyme of the NEDD8 precursor to
promote the maturation of NEDD8 (10).
NEDD8 is a ubiquitin-like protein, and neddylation
plays a key role in regulating ubiquitin ligase E3 and promoting
the function of the ubiquitin pathway (11). The process of neddylation is similar
to ubiquitination, which is maintained by a series of regulatory
enzymes. Following transcription, NEDD8 is present in the form of
its precursor, which is subsequently converted to mature NEDD8
following hydrolysis by UCHL3 hydrolase (12). Mature NEDD8 is activated by NEDD8
activase and subsequently transferred to NEDD8 ligase (13). NEDD8 carried by E2 is transferred to
its substrate and associated with the cullin protein (14). However, NEDD8 hydrolase can cause an
isomeric change of NEDD8, dissociating NEDD8 from cullin, which
plays a negative regulatory role, a process referred to as
de-neddylation (8). Thus,
neddylation and de-neddylation maintain the balance by
simultaneously acting on the NEDD8 protein (15). UCHL3 not only acts on the NEDD8
precursor to convert it to its mature form, but also hydrolyzes the
bound NEDD8 (16). Thus, the present
study aimed to investigate the effect of UCHL3 knockdown on the
biological activities of melanoma cells, and determine its role in
downstream NEDD8 signaling via in vitro cell
experiments.
Materials and methods
Cell culture
Normal human epidermal cells, HaCat, were purchased
from CLS Cell Lines Service GmbH, while the human melanoma cell
lines, SK-MEL-2, MV3, A375 and MuM-2B were purchased from the
American Type Culture Collection. Cells were maintained in DMEM
(HyClone; Cytiva) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin
(Gibco; Thermo Fisher Scientific, Inc.), at 37°C with 5%
CO2.
Small interfering (si)RNA
transfection
One day prior to transfection, SK-MEL-2 and A375
cells (1×105) were seeded into the 6-well cell culture
plate and cultured until they reached a confluence of 70–80%.
Serum-free Opti-MEM (Sigma-Aldrich; Merck KGaA, 125 µl) was used to
dilute 5 µl 20 µM siRNA and negative control (NC), which were
gently mixed and incubated at room temperature for 5 min.
Subsequently, serum-free Opti-MEM (125 µl) was used to dilute 5 µl
Lipofectamine® (lipo)3,000 (cat. no. L300001;
Invitrogen; Thermo Fisher Scientific, Inc.), which was gently mixed
and incubated for 5 min at room temperature. The solutions were
incubated for an additional for 20 min at room temperature. The
plasmid-lipo3,000 mixture was added into culture wells of the 750
µl complete culture medium containing 1×105 cells for
complete mixing, and the culture plates were incubated at 37°C for
48 h with 5% CO2. The following sequences were used for
transfection: NC forward, 5′-UGACCUACAACUUCUAUGGTT−3′ and reverse,
5′-UUCUCCGAACGUGUCACGUTT−3′; UCHL3 (human) siRNA-1 forward,
5′-GAACAGAAGAGGAAGAAAATT−3′ and reverse,
5′-UUUUCUUCCUCUUCUGUUCTT−3′; UCHL3 (human) siRNA-2 forward,
5′-CUGAAGAACGAGCCAGAUATT−3′ and reverse,
5′-UAUCUGGCUCGUUCUUCAGTT−3′; UCHL3 (human) siRNA-3 forward,
5′-UGGAACAAUUGGACUGAUUTT−3′ and reverse,
5′-AAUCAGUCCAAUUGUUCCATT−3′; NEDD8 (human) siRNA-1 forward,
5′-CAGACAAGGUGGAGCGAAUTT−3′ and reverse,
5′-AUUCGCUCCACCUUGUCUGTT−3′; NEDD8 (human) siRNA-2 forward,
5′-CGGAAAGGAGAUUGAGAUUTT−3′ and reverse,
5′-AAUCUCAAUCUCCUUUCCGTT−3′; and NEDD8 (human) siRNA-3 forward,
5′-UGGAGGAGAAAGAGGGAAUTT−3′ and reverse,
5′-AUUCCCUCUUUCUCCUCCATT−3′.
MTT assay
Cells in the NC group were treated with normal
medium; cells in the si-NC groups were transfected with si-NC;
cells in the si-UCHL3 group were transfected with si-UCHL3, which
inhibited UCHL3 expression; cells in the si-NEDD8 group were
transfected with si-NEDD8, which inhibited NEDD8 expression; and
cells in the si-UCHL3+ si-NEDD8 group were transfected with
si-UCHL3 and si-NEDD8.
Following treatment for 48 h, 20 µl MTT solution (5
g/l; Amresco, LLC) was added and cells were incubated for an
additional 4 h at 37°C. The excessive culture medium was discarded
and 150 µl DMSO was added for 10 min of oscillatory reaction.
Absorbance (OD value) was detected at a wavelength of 490 nm, using
a microplate reader (BioTek ELx800). The cell proliferation rate
was calculated in each group.
Apoptosis analysis
Cells in each group were digested with EDTA-free
trypsin (Sigma-Aldrich; Merck KGaA) centrifuged at 8,000 × g for 30
sec at 4°C, collected, rinsed twice with PBS and resuspended in
binding buffer (Sigma-Aldrich; Merck KGaA) (1×105
cells/l). Cells were subsequently stained with Annexin V-FITC and
PI for 10 min at 4°C in the dark, according to the manufacturer's
instructions (Nanjing KeyGen Biotech Co., Ltd.). Fluorescence
intensity was measured at an excitation wavelength of 488 nm and
emission wavelength of 530 nm, using a flow cytometer
(Becton-Dickinson and Company). The experiment was performed in
triplicate.
Reverse transcription-quantitative
(RT-q)PCR
Following the corresponding treatments in each group
for 48 h, total RNA was extracted from cells of different groups
using TRIzol® reagent (Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. The purity and
concentration of RNA were detected using microNucleic Acid Analyzer
(Thermo Fisher Scientific, Inc.). RNA was reverse transcribed into
cDNA using the Takara reverse transcription kit (Takara
Biotechnology Co., Ltd.), at 30°C for 10 min, 42°C for 30 min, 99°C
for 5 min and 4°C for 5 min. qPCR was subsequently performed using
the Takara fluorescent quantitative kit (Takara Biotechnology Co.,
Ltd.), according to the manufacturer's instructions. GAPDH was used
as the internal reference for PCR amplification. A DNA Engine with
a Chromo 4 detector (MJ Research, Inc.; Bio-Rad Laboratories, Inc.)
was used for qPCR. The following thermocycling conditions were used
for qPCR: 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec for
a total of 40 cycles, followed by extension at 60°C for 5 min.
Relative expression levels were calculated using the
2−ΔΔCq method (12). The
primer sequences used for qPCR were designed and synthesized by
Shanghai Shenggong Biology Engineering Technology Service, Ltd. The
following primer sequences were used for qPCR: GAPDH forward,
5′-GGTGAAGGTCGGTGTGAACG-3′ and reverse,
5′-GCTCCTGGAAGATGGTGATGG−3′; UCHL3 forward,
5′-AGCCCTGAAGAACGAGCCAGAT-3′ and reverse,
5′-ACTTGGTGCCTCAGTCTGACCT-3′; NEDD8 forward,
5′-AATCAAGGAGCGTGTGGAGGAG-3′ and reverse,
5′-AGAGCCAACACCAGGTGAAGGA-3′; and LC3B forward,
5′-TCAGCGTCTCCACACCAATCTCA-3′ and reverse,
5′-CGAACGTCTCCTGGGAGGCATA-3′. The amplification conditions were as
follows: Initial denaturation at 50°C for 2 min, 95°C for 5 min,
followed by 50 cycles of 95°C for 15 sec and 60°C for 30 sec. The
experiment was repeated in triplicate.
TUNEL assay
SK-MEL-2 and A375 cells in each group were treated
with the corresponding treatments for 48 h. Cells were subsequently
fixed in 4% paraformaldehyde at room temperature for 25 min and
washed twice with PBS. Cells were permeabilized with 0.2% Triton
X-100 for 5 min, washed twice with PBS, treated with fresh 3%
H2O2 at room temperature for 5 min, followed
by another two washes with PBS. After the slides were dried, 50 µl
TUNEL reaction mixture (TdT and dUTP mixed at a ratio of 1:9) was
added to the cells at 37°C for 1 h. Only 50 µl dUTP solution was
added to the NC group, and the reaction time was 60 min at 37°C in
a wet box in the dark. After washing three times with PBS, 50 µl
converter-POD was added to the cells after drying the slides for 30
min at 37°C in a wet box in the dark. Subsequently, 100 µl DAB
developer was added for 10 min at room temperature after washing
three time with PBS. After another three washes with PBS, cells
were re-stained with hematoxylin, immediately washed with tap water
after 30 sec of reaction, dehydrated with gradient alcohol,
transparentized with xylene and sealed with neutral gum. The ratio
of positive cells was observed under a fluorescence microscope
(Olympus Corporation BX43; magnification, ×200).
Observation of cellular ultrastructure
under a transmission electron microscope (TEM)
Following the corresponding treatments in each group
for 48 h, cells were collected via centrifugation at 1,000 × g for
10 min at room temperature. After washing three times with PBS,
cells were immediately fixed with 25 g/l glutaraldehyde overnight
at 4°C. Subsequently, cells were fixed with 10 g osmium tetroxide
for 2 h at 4°C, dehydrated with gradient ethanol, soaked and
embedded with Epon812 epoxy resin at 4°C for 2 h, followed by
semi-ultrathin slicing (80 nm), as well as double staining with
uranium acetate and lead citrate at 4°C for 2 h. Cells were
observed and photographed under a Hitachi H-600 TEM (Hitachi,
Ltd.).
Detection of LC3B protein expression
in cells under a laser confocal microscope
Following the corresponding treatments in each group
for 48 h, cells were fixed with 4% paraformaldehyde at room
temperature for 10 min. After washing three times with PBS (5 min
each), cells were lysed using 0.1% Triton-100 (Nanjing KeyGen
Biotech Co., Ltd.) for 20 min. After re-washing three times with
PBS (5 min each), 5% BSA (Sigma-Aldrich; Merck KGaA) was added for
sealing at room temperature for 20 min. Subsequently, rabbit
anti-mouse LC3B monoclonal antibody (cat. no. ab205718; Abcam;
1:100) was added and incubated overnight at 4°C, followed by
washing three times with PBS (5 min each). Following the primary
incubation, samples were incubated with HRP-labeled goat
anti-rabbit IgG (cat. no. ab192890; Abcam; 1:500) at 37°C for 45
min, in the dark. Cells were washed with PBS, and Hochest 33342 dye
(1:200; Nanjing KeyGen Biotech Co., Ltd.) was used to stain the
nuclei for 5 min at room temperature, and the treated cells were
subsequently washed with PBS. Cells were observed and photographed
under a laser confocal microscope (magnification, ×200). Mean
fluorescence intensity was detected using ImageJ software
v1.8.0.112 (National Institutes of Health). The experiment was
performed in triplicate.
Western blotting
Total protein from each group was extracted from
cells using the total protein extraction kit (cat. no. KGP250;
Nanjing KeyGen Biotech Co., Ltd.), according to the manufacturer's
instructions. The BCA protein concentration assay kit was used to
detect total protein concentration and calculate the loading
amount. Following denaturation by heating at 100°C, 30 µg of total
protein was loaded per lane. The proteins were separated by 10%
SDS-PAGE, transferred onto PVDF membranes and blocked with 5%
skimmed milk at room temperature for 1 h, followed by three washes
in 20% TBST (5 min each). The membranes were incubated with primary
antibodies against NEDD8 (1:2,000; cat. no. ab81264; Abcam), LC3
(1:2,000; cat. no. ab192890; Abcam) and UCHL3 (cat. no. ab126621;
Abcam; 1:2,000) overnight at 4°C. Membranes were washed three times
with TBS-0.5% Tween-20 (TBS-T) and subsequently incubated with
diluted goat anti-Rabbit IgG-HRP secondary antibody (1:500; cat.
no. KGAA35; Nanjing KeyGen Biotech Co., Ltd.) for 2 h at room
temperature. Membranes were re-washed, placed into a clean culture
dish and evenly smeared with ECL development solution (cat. no.
KGP116; Nanjing KeyGen Biotech Co., Ltd.) prepared at a ratio of
1:1. Membranes were exposed in the Bio-Rad exposure system (cat.
no. 170-4150; Bio-Rad Laboratories, Inc.), using ImageJ software
(version 1.52r; National Institutes of Health) for image
acquisition.
Statistical analysis
Statistical analysis was performed using SPSS 19.0
software (IBM Corp.). Measurement data are consistent with normal
distribution. All experiments were performed in triplicate and data
are presented as the mean ± SD. One-way ANOVA followed by Tukey's
post hoc test were used to compare differences among multiple
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
UCHL3 gene expression in cells of each
group
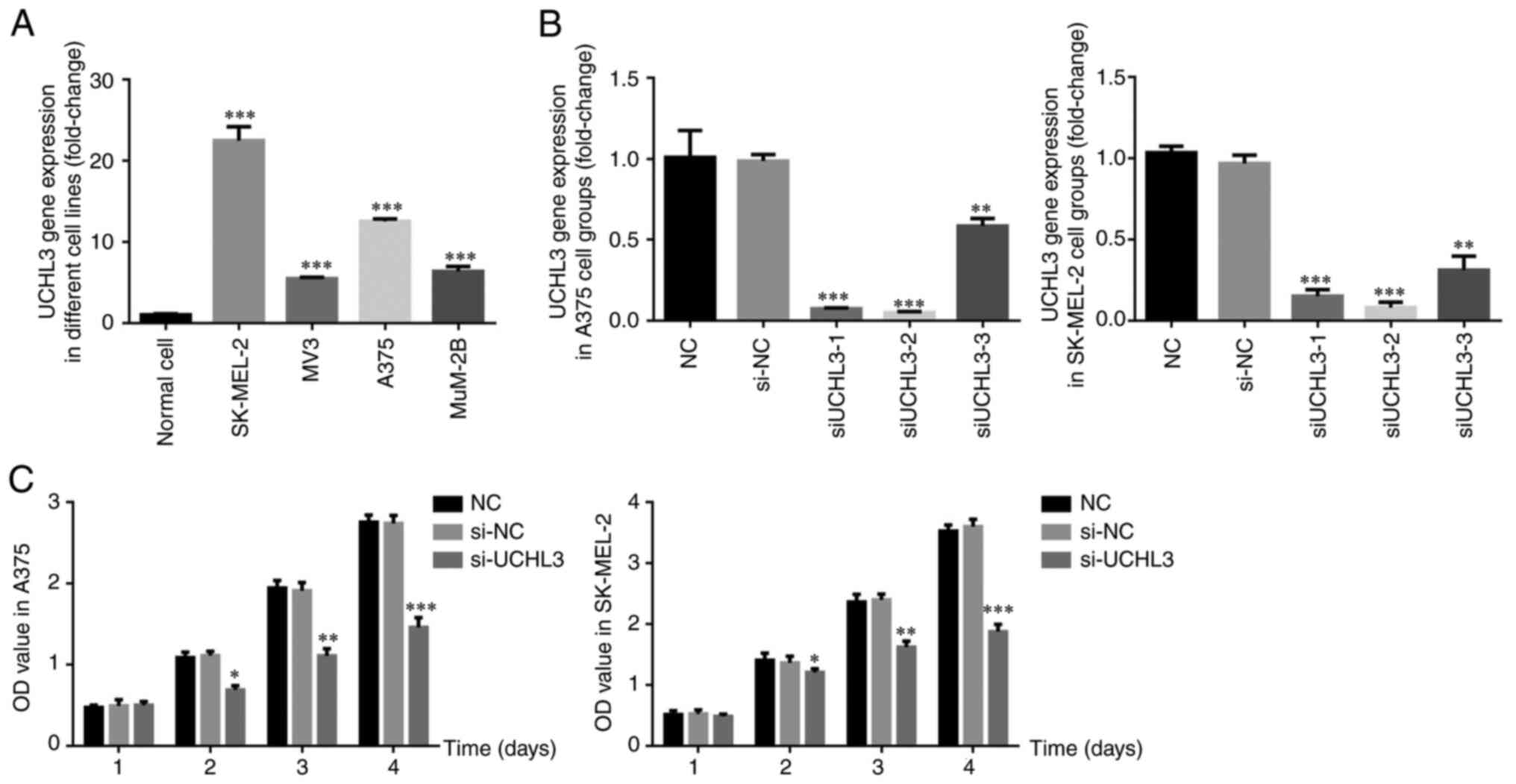
As presented in Fig.
1A, UCHL3 expression was significantly higher in the melanoma
cell lines, SK-MEL-2, MV-3, A375 and MUN2B (all P<0.001)
compared with normal HaCat cells. Among these, SK-MEL-2 and A375
cells exhibited the highest expression of UCHL3, and thus were
selected for subsequent experimentation. To assess the effect of
UCHL3 inhibition, si-UCHL3-1, si-UCHL3-2 and si-UCHL3-3 were
transfected into SK-MEL-2 and A375 cells. The results demonstrated
that UCHL3 expression significantly decreased in all groups
compared with the si-NC group (all P<0.01; Fig. 1B), particularly in the si-UCHL3-2
group. Thus, si-UCHL3-2 was selected for UCHL-3 knockdown
experiments.
Effect of UCHL3 knockdown on the
proliferation of melanoma cells
The results of the MTT assay demonstrated no
significant differences in the proliferation rate of A375 and
SK-MEL-2 cells between the si-NC and NC groups (P>0.05), while
the proliferation rate significantly decreased in the si-UCHL3
group on days 2, 3 and 4 (all P<0.05; Fig. 1C).
Effect of UCHL3 knockdown on the
apoptosis of melanoma cells
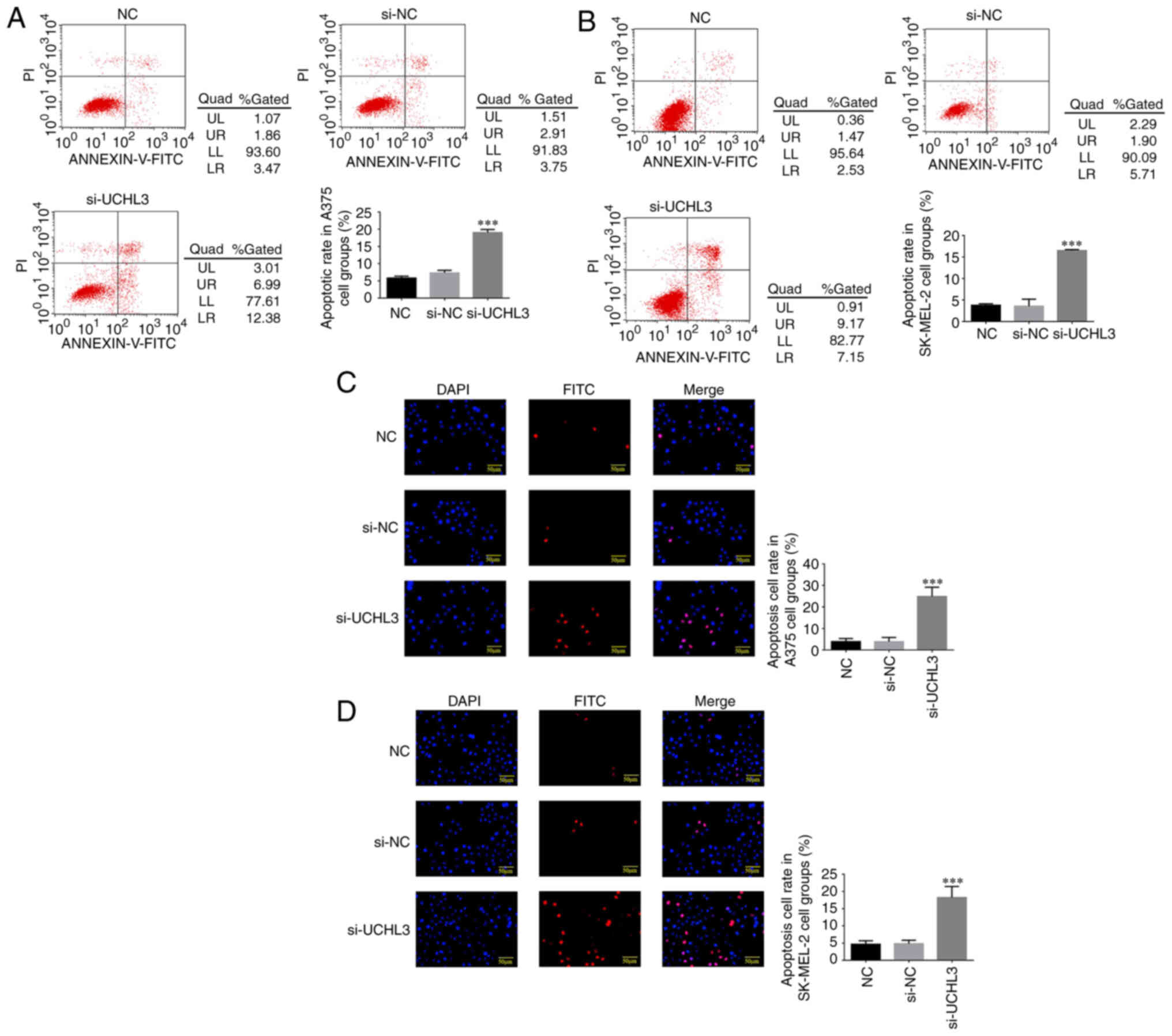
Flow cytometric analysis demonstrated no significant
differences in the apoptotic rates of A375 and SK-MEL-2 cells
between the si-NC and NC groups (P>0.05). However, the apoptotic
rates significantly increased in the si-UCHL3 group of both cell
lines compared with NC group (P<0.001; Fig. 2A and B).
TUNEL staining analysis exhibited no significant
differences in the number of apoptotic A375 and SK-MEL-2 cells
between the NC and si-NC groups (P>0.05), while a significant
increase in the number of apoptotic cells was observed in the
si-UCHL3 group compared with the NC group (P<0.001; Fig. 2C and D).
Effect of UCHL3 knockdown on the
ultrastructure and LC3B protein expression of melanoma cells
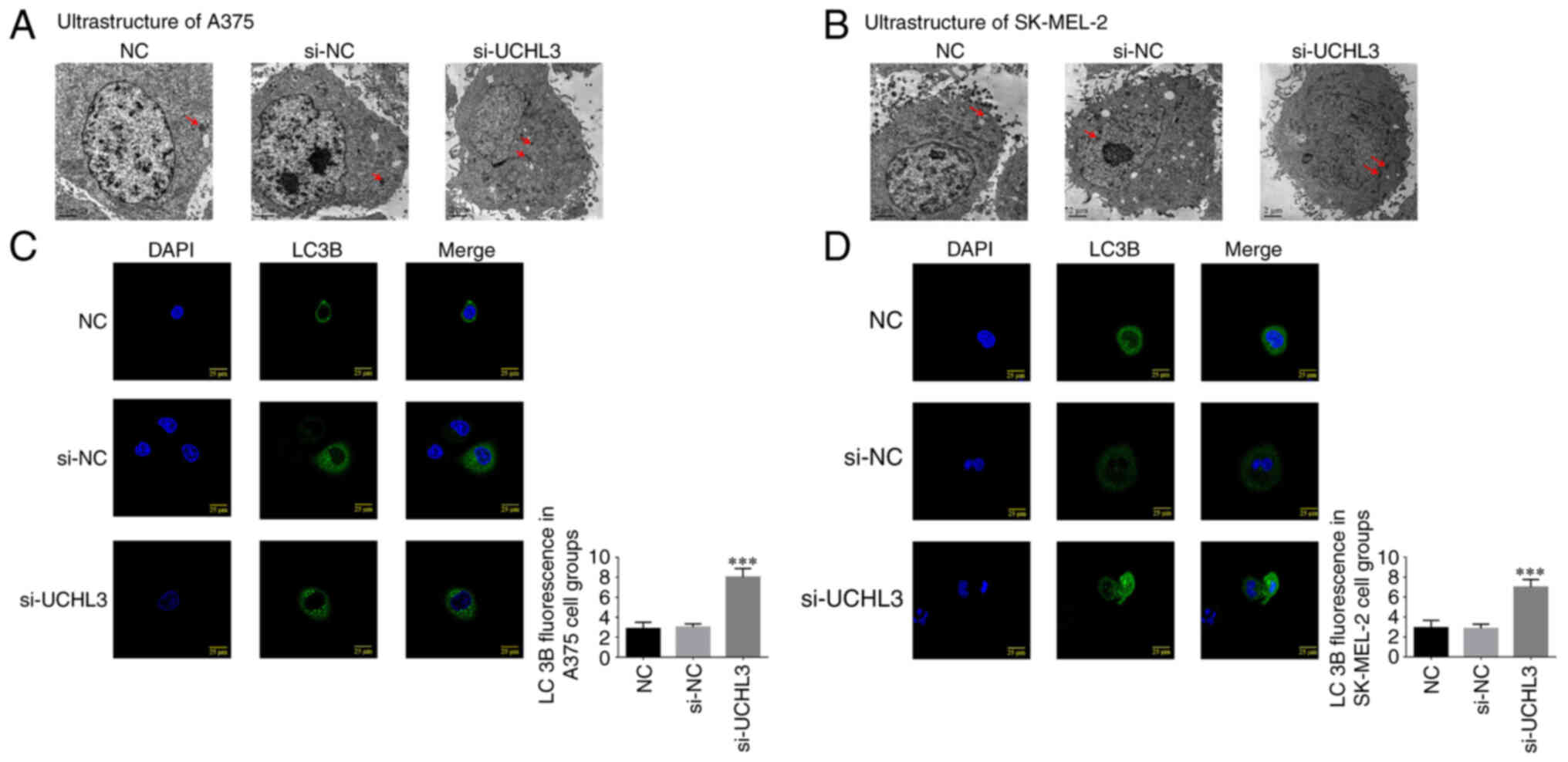
No significant changes in the ultrastructure of A375
and SK-MEL-2 cells were observed between the NC and si-NC groups,
while the number of autophagosomes increased in A375 and SK-MEL-2
cells in the si-UCHL3 group (Fig. 3A and
B), suggesting that UCHL3 knockdown may enhance autophagy.
Immunofluorescence analysis demonstrated no
significant differences in LC3B protein expression in A375 and
SK-MEL-2 cells between the NC and si-NC groups (P>0.05);
however, UCHL3 knockdown significantly increased LC3B protein
expression in the si-UCHL3 group compared with the NC group
(P<0.001; Fig. 3C and D).
Effect of UCHL3 knockdown on relevant
gene expression in melanoma cells
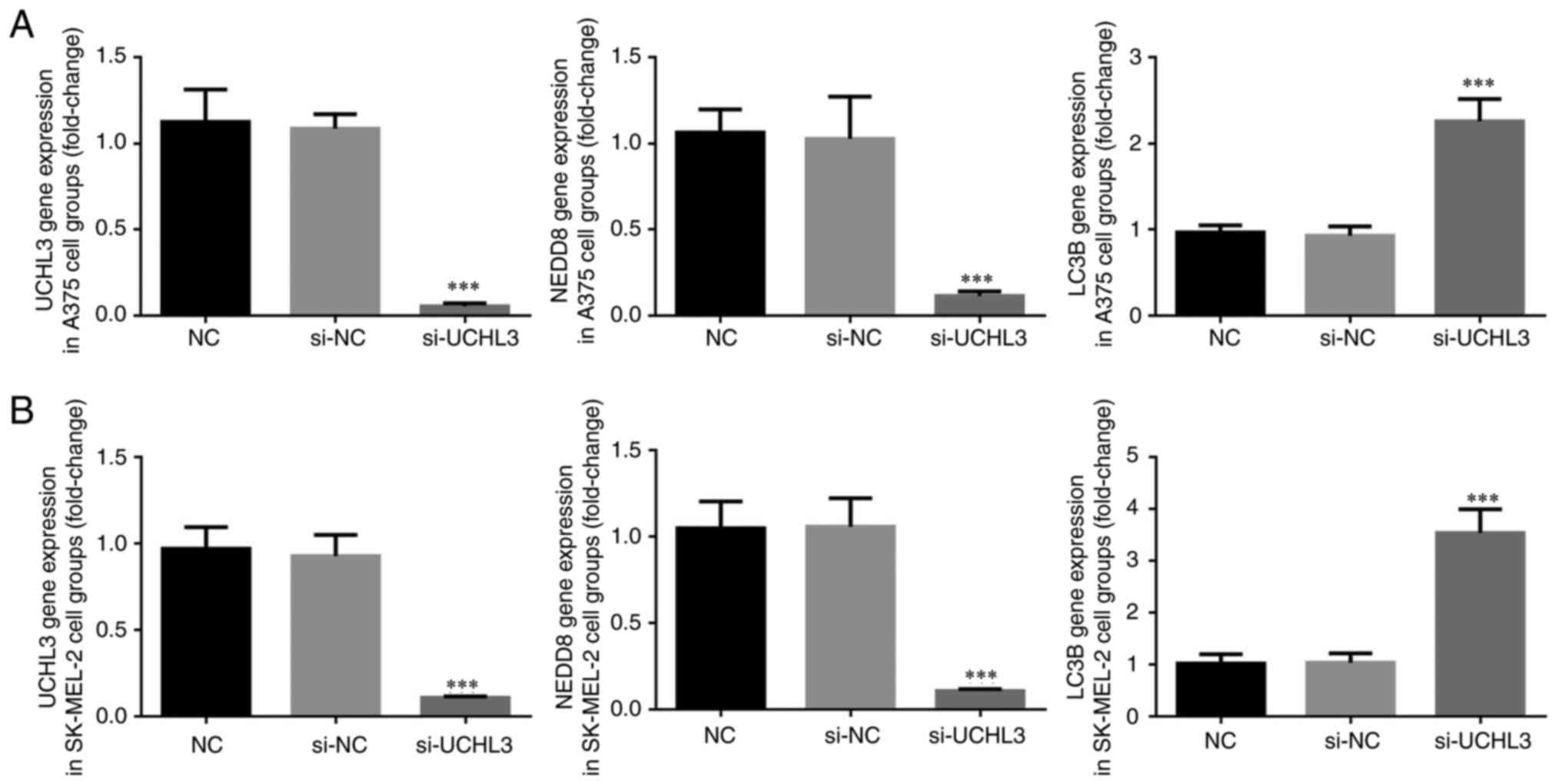
RT-qPCR analysis demonstrated no significant
differences in UCHL3, NEDD8 and LC3B expression levels between the
NC and si-NC groups (P>0.05; Fig. 4A
and B); however, UCHL3 knockdown significantly decreased the
expression levels of UCHL3 and NEDD8, and significantly increased
LC3B expression in the si-UCHL3 group compared with the NC group
(all P<0.001; Fig. 4A and B).
Effect of UCHL3 knockdown on relevant
protein expression and NEDD8 ubiquitination
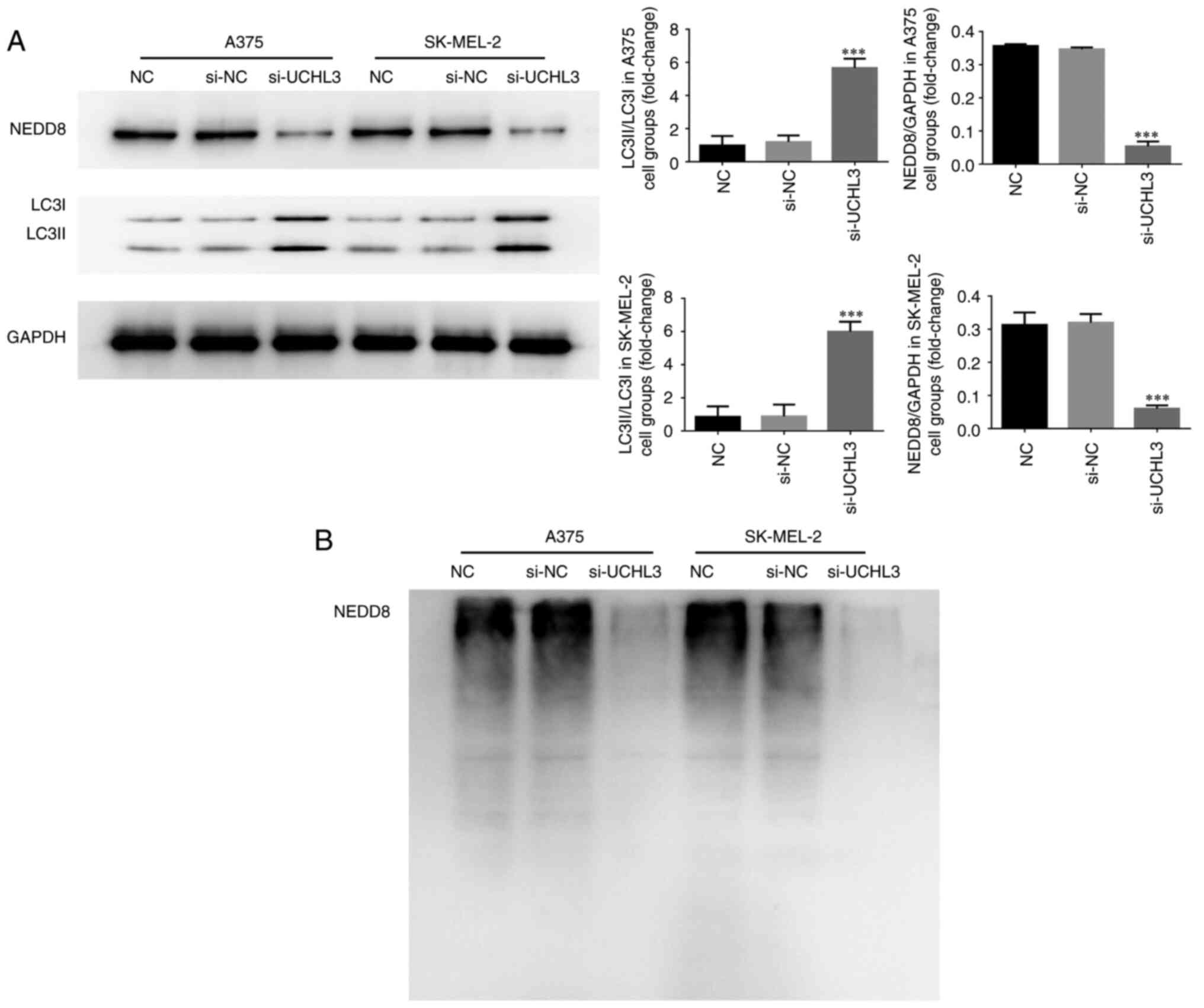
Western blot analysis demonstrated no significant
differences in NEDD8 protein expression, the LC3II/LC3I ratio and
NEDD8 ubiquitination in A375 and SK-MEL-2 cells between the NC and
si-NC groups (all P>0.05; Fig. 5A and
B). However, UCHL3 knockdown significantly decreased NEDD8
protein expression and markedly increased the LC3II/LC3I ratio in
the si-UCHL3 group compared with the NC group (all P<0.001;
Fig. 5A), accompanied by notably
decreased NEDD8 ubiquitination (Fig.
5B).
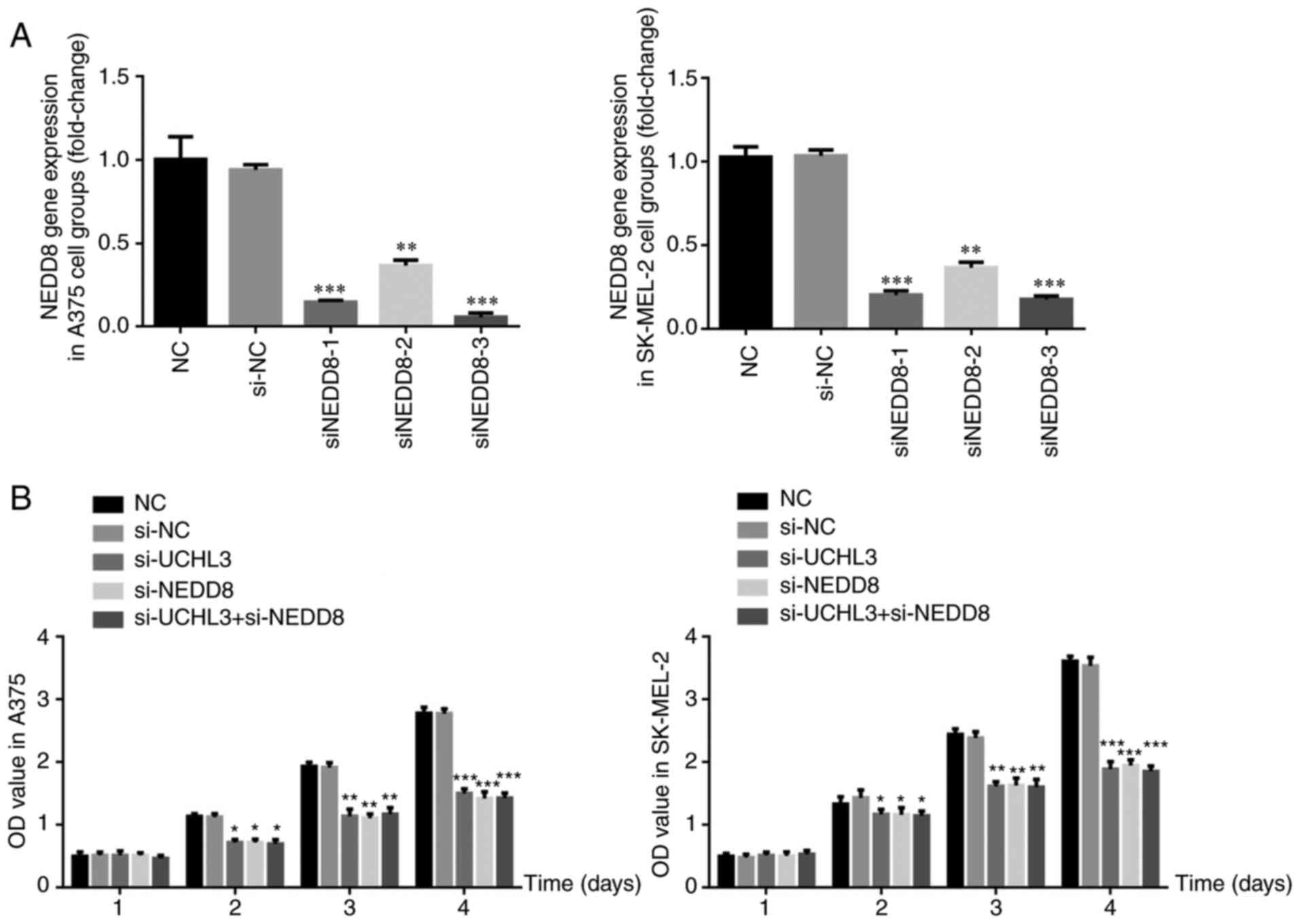
Sequence screening for NEDD8 knockdown
and corresponding expression in each group
RT-qPCR analysis demonstrated no significant
differences in NEDD8 gene expression between the si-NC and NC
groups (P>0.05; Fig. 6A).
Following transfection with three si-NEDD8 sequences, NEDD8 gene
expression significantly decreased in the si-NEDD8 groups compared
with the NC group (all P<0.01; Fig.
6A), with the most significant decrease observed in the
si-NEDD8-3 group. Thus, the si-NEDD8-3 sequence was used for
subsequent experimentation.
Notably, the proliferation of A375 and SK-MEL-2
cells on days 2, 3 and 4 significantly decreased in the si-UCHL3,
si-NEDD8 and si-UCHL3+ si-NEDD8 groups compared with the NC group
(all P<0.05; Fig. 6B).
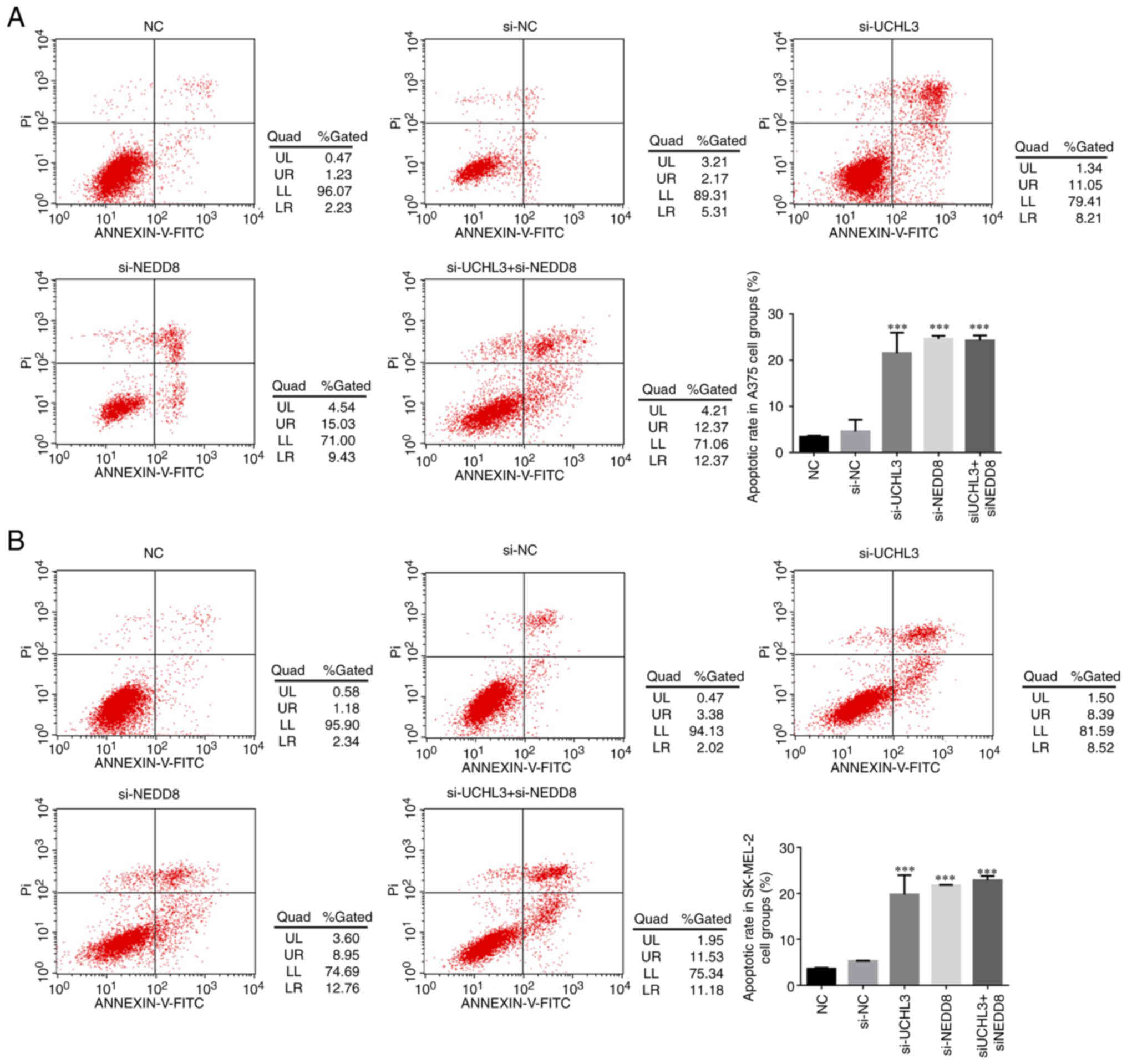
Detection of cell apoptosis in each
group
Flow cytometric analysis demonstrated that the
apoptotic rates of A375 and SK-MEL-2 cells significantly increased
in the si-UCHL3, si-NEDD8 and si-UCHL3+ si-NEDD8 groups compared
with the NC group (all P<0.001; Fig.
7A and B), while no significance differences were observed
between the si-UCHL3, si-NEDD8 and si-UCHL3+ si-NEDD8 groups
(P>0.05).
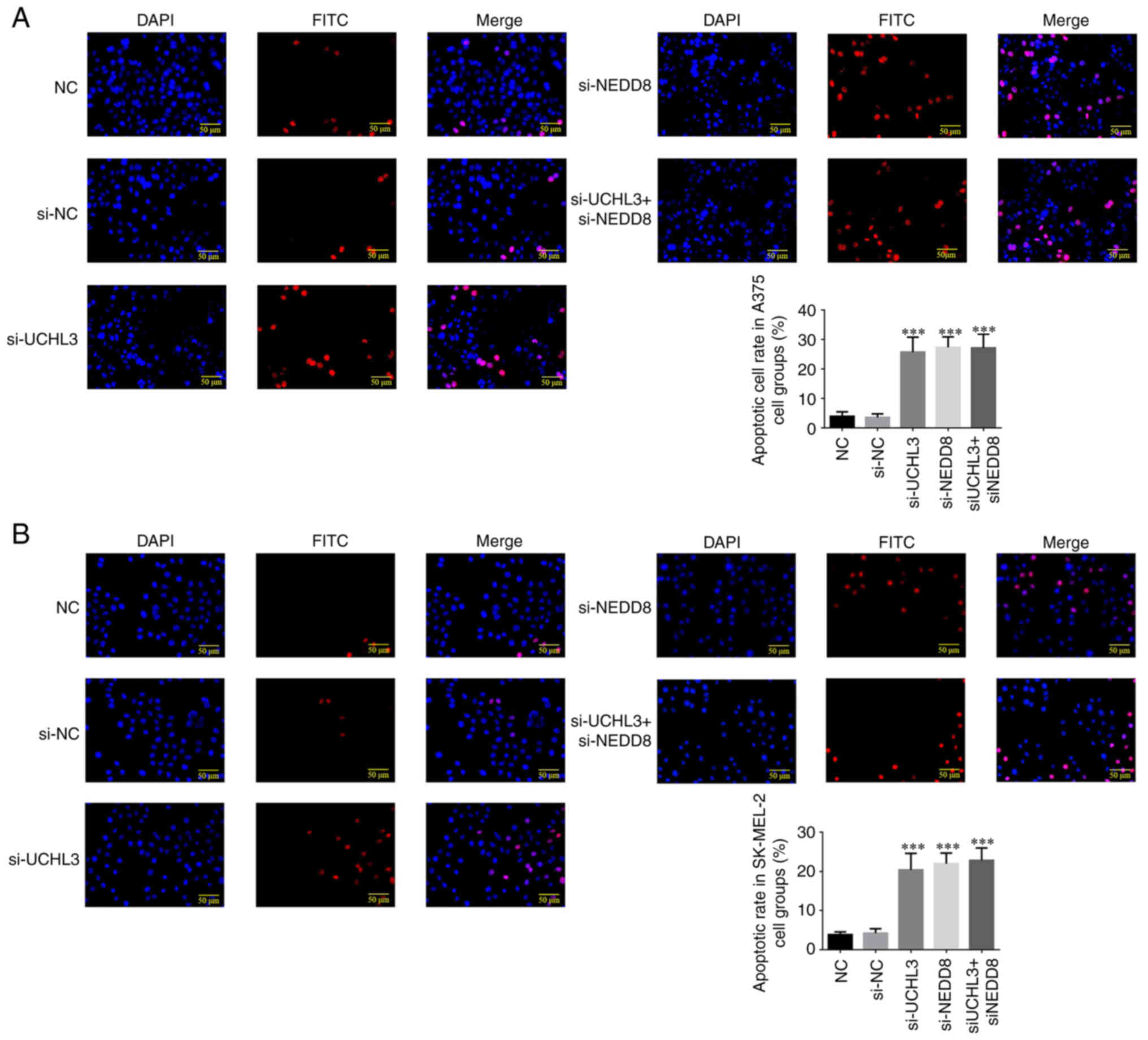
Detection of apoptotic cell count in
each group
The results of the TUNEL assay demonstrated that the
apoptotic rates of A375 and SK-MEL-2 cells significantly increased
in the si-UCHL3, si-NEDD8 and si-UCHL3+ si-NEDD8 groups compared
with the NC group (all P<0.001; Fig.
8A and B). However, no significant differences were observed
between the si-UCHL3, si-NEDD8 and si-UCHL3+ si-NEDD8 groups
(P>0.05).
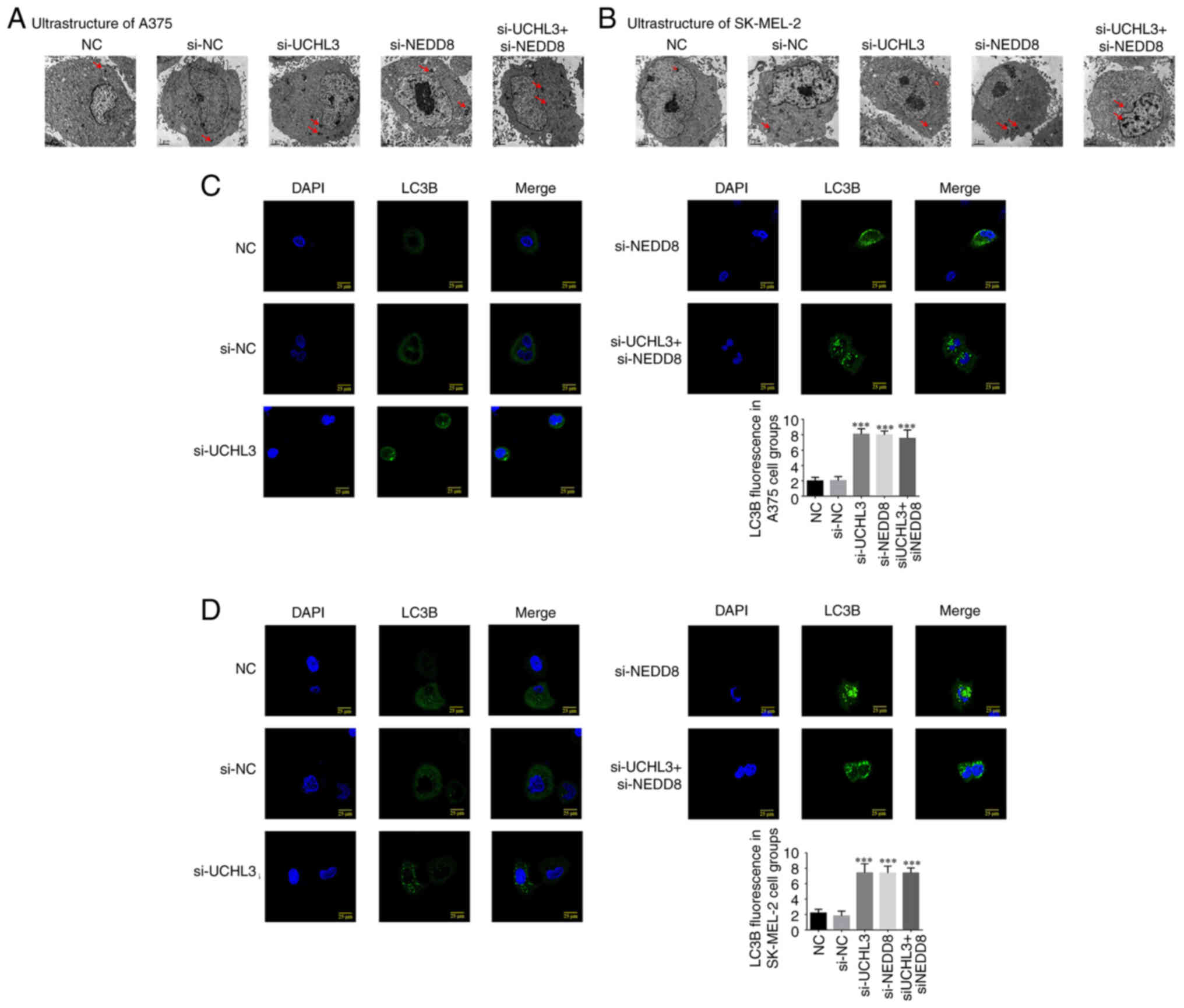
Ultrastructure of melanoma cells and
LC3B protein expression
TEM analysis demonstrated no nuclear damage or
autophagosome formation in A375 and SK-MEL-2 cells in the NC and
si-NC groups, while autophagosomes were observed in cells in the
si-UCHL3, si-NEDD8 and si-UCHL3+ si-NEDD8 groups (Fig. 9A and B).
Immunofluorescence analysis demonstrated that LC3B
protein expression in A375 and SK-MEL-2 cells was relatively lower
in the NC and si-NC groups. Furthermore, LC3B protein expression
was significantly higher in the si-UCHL3, si-NEDD8 and si-UCHL3+
si-NEDD8 groups compared with the NC group (all P<0.001;
Fig. 9C and D), while no significant
differences were observed among the three groups (P>0.05).
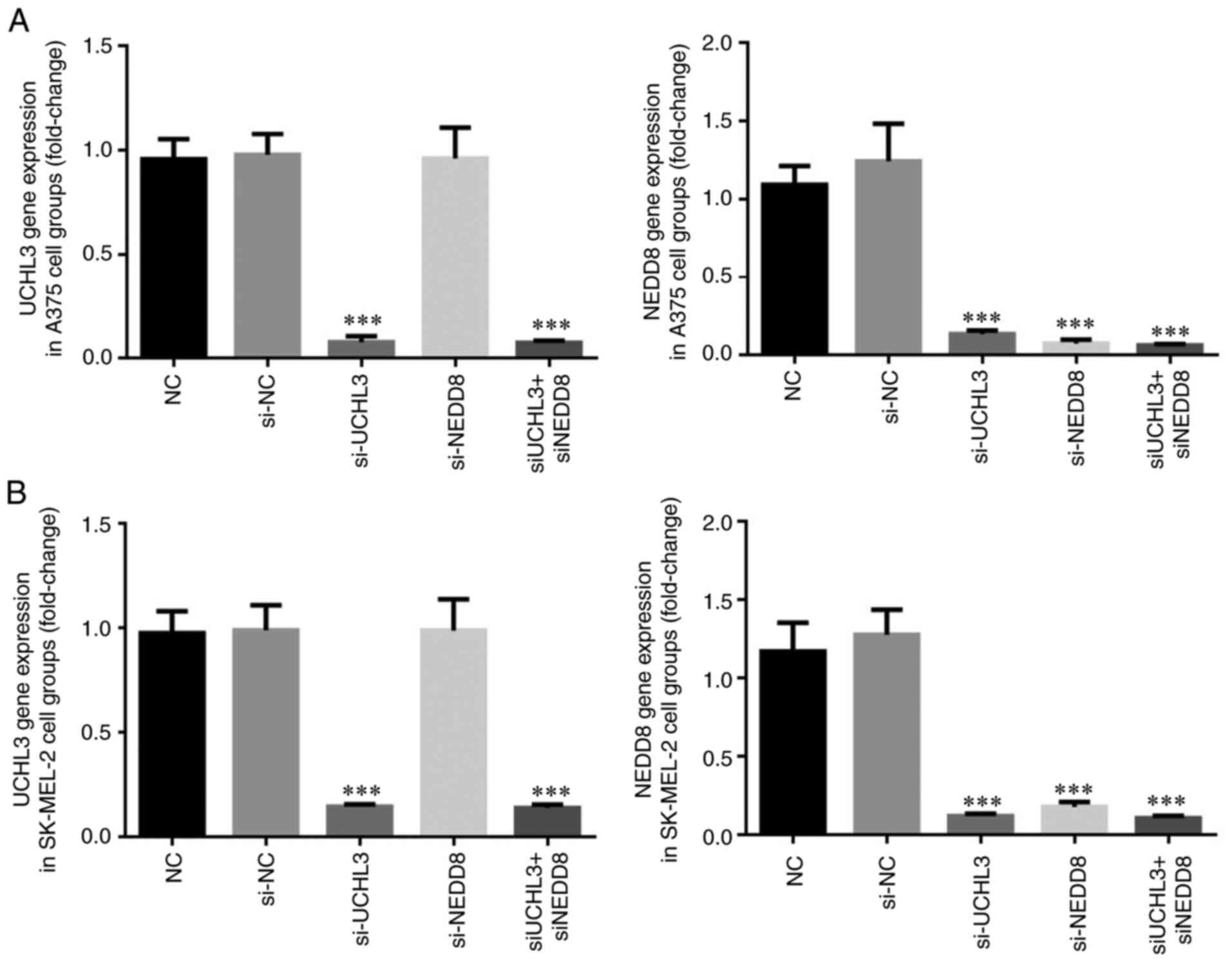
UCHL3 and NEDD8 gene expression
levels
RT-qPCR analysis demonstrated no significant
differences in UCHL3 and NEDD8 gene expression levels in A375 and
SK-MEL-2 cells between the si-NC and NC groups (P>0.05; Fig. 10). However, UCHL3 expression
significantly decreased in the si-UCHL3 and si-UHL3+ si-NEDD8
groups (all P<0.001; Fig. 10A and
B), while NEDD8 gene expression significantly decreased in the
si-UCHL3, si-NEDD8 and si-UCHL3+ si-NEDD8 groups (all P<0.001;
Fig. 10A and B).
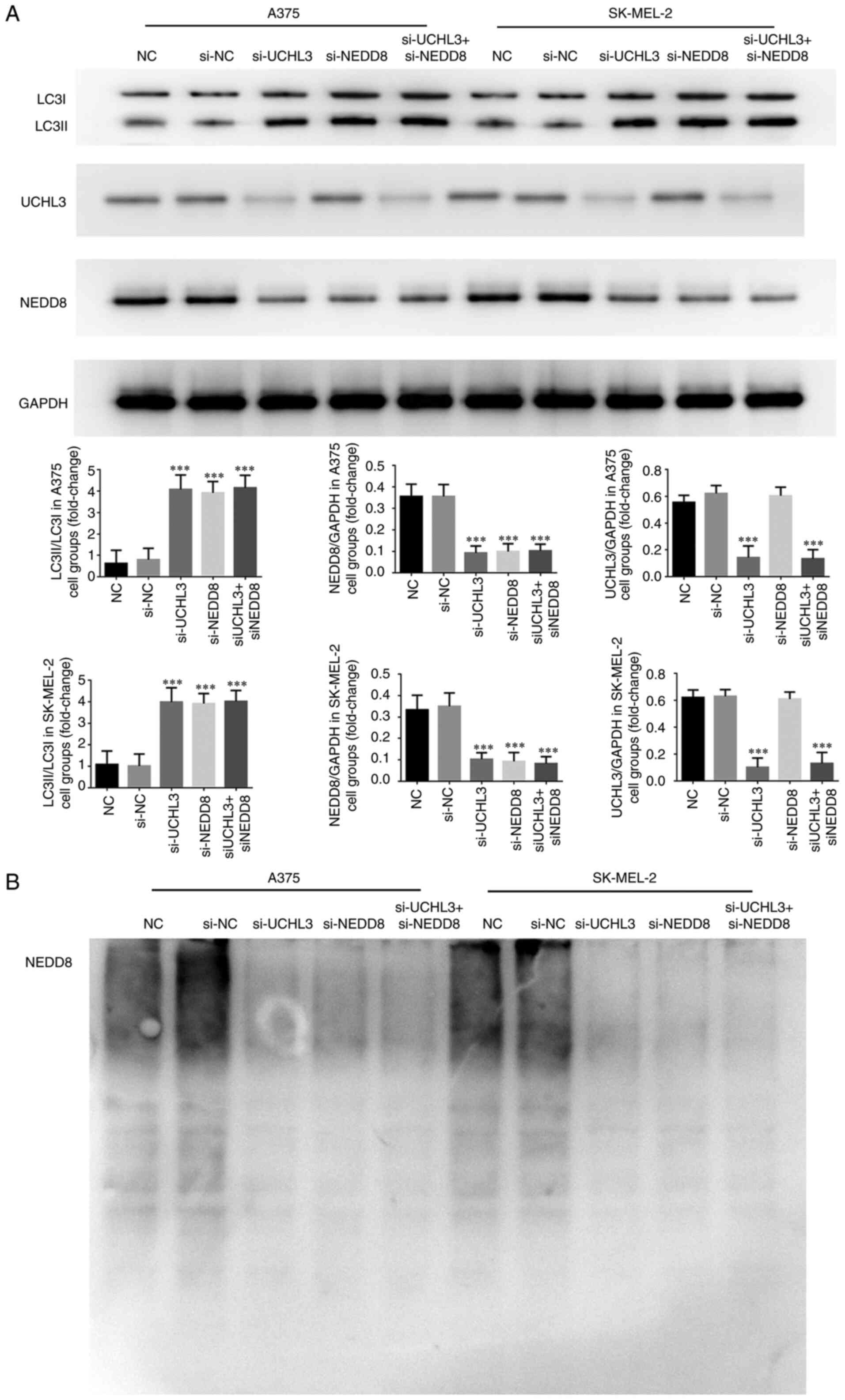
Detection of relevant protein
expression and NEDD8 ubiquitination
Western blot analysis demonstrated no significant
differences in NEDD8 protein expression, the LC3II/LC3I ratio and
NEDD8 ubiquitination in A375 and SK-MEL-2 cells between the NC and
si-NC groups (P>0.05, Fig. 11A and
B). Notably, NEDD8 protein expression significantly decreased
and the LC3II/LC3I ratio increased in the si-UCHL3, si-NEDD8 and
si-UCHL3+ si-NEDD8 groups compared with the NC group (all
P<0.001; Fig. 11A), with no
significant differences among the three groups. In addition, NEDD8
ubiquitination was significantly suppressed in the si-UCHL3,
si-NEDD8 and si-UCHL3+ si-NEDD8 groups (Fig. 11B). Notably, UCHL3 protein
expression significantly decreased in the si-UCHL3 and
si-UCHL3+si-NEDD8 groups compared with the NC group (all
P<0.001; Fig. 11A).
Discussion
UCHL3 is a member of the UCH family, which has
attracted recent interest in tumor research (17). Among the UCH family members, UCHL1 is
the most extensively investigated in the context of several
malignancies, including esophageal cancer (18), gastric cancer (19), renal cancer (20), prostate cancer (21) and ovarian cancer (13). UCHL1 mainly functions as a ubiquitin
hydrolase and ubiquitin ligase, and has limited distribution,
primarily in the ovary, testis and neurons (22). A previous study demonstrated that
UCHL3 can regulate DNA repair, and thus may participate in tumor
development (23). It has been
reported that interference with UCHL3 may cause meiotic arrest of
mature oocytes (24,25). Downregulating UCHL3 expression in
metastatic prostate cancer may interfere with UCHL3 expression in
normal prostate cells and promote their migratory and invasive
abilities (7). UCHL3 expression is
upregulated in breast cancer (26)
and cervical cancer (27), and it
may be involved in the occurrence and malignant behavior of these
two malignancies. Thus, whether UCHL3 acts as an oncogene or tumor
suppressor varies across different tumors.
The results of the present study demonstrated that
UCHL3 was highly expressed in melanoma cell lines (SK-MEL-2, MV3,
A375 and MuM-2B), and UCHL3 mRNA expression was upregulated in
SK-MEL-2 and A375 cell lines. Thus, SK-MEL-2 and A375 cell lines
were selected for subsequent experimentation. The biological
activities (cell proliferation, invasion and migration) of A375 and
SK-MEL-2 melanoma cells were significantly inhibited following
UCHL3 knockdown. Taken together, these results suggest that
abnormally high UCHL3 expression may be an important factor in
melanoma, and its knockdown may effectively inhibit the occurrence
and development of melanoma.
Autophagy is another important pathway for protein
degradation. Autophagy is widely present in eukaryotic cells, where
it plays a key role in degrading longevity-related proteins and
organelles, and is important for maintaining cell stability
(28). In addition, autophagy is a
natural process in cells that enables self-digestion of
intracellular elements (29). Under
conditions of injury, autophagy can provide energy through lysosome
digestion of its own contents to maintain cell survival (30). Under normal physiological conditions,
autophagy is maintained at basic levels; however, it is notably
enhanced under conditions of nutrient deficiency, growth factor
deficiency, hypoxia and other stress conditions (31). Furthermore, autophagy is an important
cell process involved in cell proliferation, differentiation,
senescence and immunity (30).
Disordered autophagy has been reported in multiple diseases, such
as infections (32),
neurodegenerative conditions (33),
cardiovascular diseases (34),
metabolic diseases (35) and tumors
(36,37). Autophagy may play a role in tumors in
an environment-dependent manner. Conversely, it can destroy damaged
organelles, cause cell aging and inhibit the proliferation of
precancerous cells, exhibiting a tumor-inhibiting effect (28). It can also provide nutrients for
tumor cells under conditions of nutritional and oxygen deficiency,
thus promoting tumor progression (38). Autophagy also plays different roles
in the development of melanoma. For example, a previous study
reported a lower level of autophagy in the tumor specimens of 194
patients with early melanoma compared with benign nevus (39). Another study revealed that patients
with melanoma with higher autophagy levels may have a longer
progression-free survival time (40), while nodular melanoma with low
autophagy levels is associated with an increased risk of ulceration
and tumor invasion, accompanied by a low 5-year survival rate
following surgery (41), suggesting
that autophagy may inhibit melanoma growth. Conversely, another
study on superficial spreading melanoma reported that the autophagy
level was higher in tumor tissues compared with normal melanocytes
(39). In addition, the level of
autophagy is higher in metastatic melanoma compared with primary
tumor (42). In a previous study,
UCHL3 knockdown increased the number of autophagosomes on
examination of the ultrastructure of melanoma cells, which was
associated with a notable increase in LC3B levels and the
LC3II/LC3I ratio (43). The results
of the present study suggest that the increase in autophagy level
may be an important factor mediating the inhibitory effects of
UCHL3 knockdown on the biological activity of melanoma cells.
Ubiquitination is closely associated with autophagy
(44,45). The results of the present study
demonstrated that UCHL3 knockdown in melanoma cells not only
inhibited NEDD8 expression, but also decreased the ubiquitination
of NEDD8. Furthermore, autophagy was significantly enhanced along
with the decrease in NEDD8 ubiquitination.
In conclusion, the results of the present study
demonstrated that UCHL3 knockdown decreased melanoma cell
proliferation by increasing cell autophagy through regulating NEDD8
expression and autophagosome numbers in vitro. However, the
present study is not without limitations as it only focused on the
effect of UCHL3 on melanoma in vitro, whereas its effects
in vivo remain unclear. Thus, this will be the focus of
prospective studies. In addition, future studies will aim to assess
the effect of UCHL3 on other melanoma cell lines.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
RH, YZ, JL and FC conceptualized and designed the
present study. RH, YZ, JL, XCZ, XLZ, LA and ZL performed the
experiments. XCZ, XLZ, LA and ZL analyzed the data. RH, YZ and JL
drafted the initial manuscript. RH and FC confirmed the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lee AY and Brady MS: Neoadjuvant
immunotherapy for melanoma. J Surg Oncol. 123:782–788. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
González-Cruz C, Ferrándiz-Pulido C and
García-Patos Briones V: Melanoma in solid organ transplant
recipients. Actas Dermosifiliogr (Engl Ed). 112:216–224. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balch CM, Balch GC and Sharma RR:
Identifying early melanomas at higher risk for metastases. J Clin
Oncol. 30:1406–1407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Song Z, Tu X, Zhou Q, Huang J, Chen Y, Liu
J, Lee S, Kim W, Nowsheen S, Luo K, et al: A novel UCHL3 inhibitor,
perifosine, enhances PARP inhibitor cytotoxicity through inhibition
of homologous recombination-mediated DNA double strand break
repair. Cell Death Dis. 10:3982019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng F, Chen H, Zhang L, Li RH, Liu Y and
Sun JF: Inhibition of the NEDD8 conjugation pathway by shRNA to
UBA3, the subunit of the NEDD8-activating enzyme, suppresses the
growth of melanoma cells. Asian Pac J Cancer Prev. 13:57–62. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng F, He R, Zhang L, Li H, Zhang W, Ji
X, Kong F, Sun J and Chen S: Expression of neddylation-related
proteins in melanoma cell lines and the effect of neddylation
onmelanoma proliferation. Oncol Lett. 7:1645–1650. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jang MJ, Baek SH and Kim JH: UCH-L1
promotes cancer metastasis in prostate cancer cells through EMT
induction. Cancer Lett. 302:128–135. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li G, Jin X, Zheng J, Jiang N and Shi W:
UCH-L3 promotes non-small cell lung cancer proliferation via
accelerating cell cycle and inhibiting cell apoptosis. Biotechnol
Appl Biochem. 68:165–172. 2021. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song HM, Lee JE and Kim JH: Ubiquitin
C-terminal hydrolase-L3 regulates EMT process and cancer metastasis
in prostate cell lines. Biochem Biophys Res Commun. 452:722–727.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Artavanis-Tsakonas K, Weihofen WA, Antos
JM, Coleman BI, Comeaux CA, Duraisingh MT, Gaudet R and Ploegh HL:
Characterization and structural studies of the Plasmodium
falciparum ubiquitin and Nedd8 hydrolase UCHL3. J Biol Chem.
285:6857–6866. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rabut G and Peter M: Function and
regulation of protein neddylation. ‘Protein modifications: Beyond
the usual suspects’ review series. EMBO Rep. 9:969–976. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Frickel EM, Quesada V, Muething L, Gubbels
MJ, Spooner E, Ploegh H and Artavanis-Tsakonas K: Apicomplexan
UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout
evolution. Cell Microbiol. 9:1601–1610. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo K, Li L, Li Y, Wu C, Yin Y, Chen Y,
Deng M, Nowsheen S, Yuan J and Lou Z: A
phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51
axis in homologous recombination. Genes Dev. 30:2581–2595. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Enchev RI, Schulman BA and Peter M:
Protein neddylation: Beyond cullin-RING ligases. Nat Rev Mol Cell
Biol. 16:30–44. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen JJ, Schmucker LN and Visco DP Jr:
Identifying de-NEDDylation inhibitors: Virtual high-throughput
screens targeting SENP8. Chem Biol Drug Des. 93:590–604. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mandelker DL, Yamashita K, Tokumaru Y,
Mimori K, Howard DL, Tanaka Y, Carvalho AL, Jiang WW, Park HL, Kim
MS, et al: PGP9.5 promoter methylation is an independent prognostic
factor for esophageal squamous cell carcinoma. Cancer Res.
65:4963–4968. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song Z, Li J, Zhang L, Deng J, Fang Z,
Xiang X and Xiong J: UCHL3 promotes pancreatic cancer progression
and chemo-resistance through FOXM1 stabilization. Am J Cancer Res.
9:1970–1981. 2019.PubMed/NCBI
|
|
18
|
Wang G, Zhang W, Zhou B, Jin C, Wang Z,
Yang Y, Wang Z, Chen Y and Feng X: The diagnosis value of promoter
methylation of UCHL1 in the serum for progression of gastric
cancer. Biomed Res Int. 2015:7410302015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Seliger B, Handke D, Schabel E, Bukur J,
Lichtenfels R and Dammann R: Epigenetic control of the ubiquitin
carboxyl terminal hydrolase 1 in renal cell carcinoma. J Transl
Med. 7:902009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mitsui Y, Shiina H, Hiraki M, Arichi N,
Hiraoka T, Sumura M, Honda S, Yasumoto H and Igawa M: Tumor
suppressor function of PGP9.5 is associated with epigenetic
regulation in prostate cancer-novel predictor of biochemical
recurrence after radical surgery. Cancer Epidemiol Biomarkers Prev.
21:487–496. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brait M, Maldonado L, Noordhuis MG, Begum
S, Loyo M, Poeta ML, Barbosa A, Fazio VM, Angioli R, Rabitti C, et
al: Association of promoter methylation of VGF and PGP9.5 with
ovarian cancer progression. PLoS One. 8:e708782013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kwon J, Wang YL, Setsuie R, Sekiguchi S,
Sakurai M, Sato Y, Lee WW, Ishii Y, Kyuwa S, Noda M, et al:
Developmental regulation of ubiquitin C-terminal hydrolase isozyme
expression during spermatogenesis in mice. Biol Reprod. 71:515–521.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mtango NR, Sutovsky M, Vandevoort CA,
Latham KE and Sutovsky P: Essential role of ubiquitin C-terminal
hydrolases UCHL1 and UCHL3 in mammalian oocyte maturation. J Cell
Physiol. 227:2022–2029. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yi YJ, Sutovsky M, Song WH and Sutovsky P:
Protein deubiquitination during oocyte maturation influences sperm
function during fertilisation, antipolyspermy defense and embryo
development. Reprod Fertil Dev. 27:1154–1167. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miyoshi Y, Nakayama S, Torikoshi Y, Tanaka
S, Ishihara H, Taguchi T, Tamaki Y and Noguchi S: High expression
of ubiquitin carboxy-terminal hydrolase-L1 and -L3 mRNA predicts
early recurrence in patients with invasive breast cancer. Cancer
Sci. 97:523–529. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rolén U, Kobzeva V, Gasparjan N, Ovaa H,
Winberg G, Kisseljov F and Masucci MG: Masucci, Activity profiling
of deubiquitinating enzymes in cervical carcinoma biopsies and cell
lines. Mol Carcinog. 45:260–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hayashi-Nishino M, Fujita N, Noda T,
Yamaguchi A, Yoshimori T and Yamamoto A: A subdomain of the
endoplasmic reticulum forms a cradle for autophagosome formation.
Nat Cell Biol. 11:1433–1437. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Onorati AV, Dyczynski M, Ojha R and
Amaravadi RK: Targeting autophagy in cancer. Cancer. 124:3307–3318.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Glick D, Barth S and Macleod KF:
Autophagy: Cellular and molecular mechanisms. J Pathol. 221:3–12.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rubinsztein DC, Codogno P and Levine B:
Autophagy modulation as a potential therapeutic target for diverse
diseases. Nat Rev Drug Discov. 11:709–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kuma A, Komatsu M and Mizushima N:
Autophagy-monitoring and autophagy-deficient mice. Autophagy.
13:1619–1628. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Martinez-Vicente M and Cuervo AM:
Autophagy and neurodegeneration: When the cleaning crew goes on
strike. Lancet Neurol. 6:352–361. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kirshenbaum LA: Regulation of autophagy in
the heart in health and disease. J Cardiovasc Pharmacol.
60:1092012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
He C, Bassik MC, Moresi V, Sun K, Wei Y,
Zou Z, An Z, Loh J, Fisher J, Sun Q, et al: Exercise-induced
BCL2-regulated autophagy is required for muscle glucose
homeostasis. Nature. 481:511–515. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yousefi S and Simon HU: Autophagy in
cancer and chemotherapy. Results Probl Cell Differ. 49:183–190.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
White E: Deconvoluting the
context-dependent role for autophagy in cancer. Nat Rev Cancer.
12:401–410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Choi AM, Ryter SW and Levine B: Autophagy
in human health and disease. N Engl J Med. 368:651–662. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tschan MP and Simon HU: The role of
autophagy in anticancer therapy: Promises and uncertainties. J
Intern Med. 268:410–418. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li S, Song Y, Quach C, Guo H, Jang GB,
Maazi H, Zhao S, Sands NA, Liu Q, In GK, et al: Transcriptional
regulation of autophagy-lysosomal function in BRAF-driven melanoma
progression and chemoresistance. Nat Commun. 10:16932019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu H, He Z and Simon HU: Autophagy
suppresses melanoma tumorigenesis by inducing senescence.
Autophagy. 10:372–373. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sivridis E, Koukourakis MI, Mendrinos SE,
Karpouzis A, Fiska A, Kouskoukis C and Giatromanolaki A: Beclin-1
and LC3A expression in cutaneous malignant melanomas: A biphasic
survival pattern for beclin-1. Melanoma Res. 21:188–195. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lazova R, Klump V and Pawelek J: Autophagy
in cutaneous malignant melanoma. J Cutan Pathol. 37:256–268. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li JX, Yan Q, Liu N, Zheng WJ, Hu M, Yu
ZM, Zhou YD, Wang XW, Liang FX and Chen R: The prognostic value of
autophagy-related markers bclin-1 and LC-3 in colorectal cancers: A
systematic review and meta-analysis. Evid Based Complement Alternat
Med. 2020:84758402020.PubMed/NCBI
|
|
44
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chu Y, Kang Y, Yan C, Yang C, Zhang T, Huo
H and Liu Y: LUBAC and OTULIN regulate autophagy initiation and
maturation by mediating the linear ubiquitination and the
stabilization of ATG13. Autophagy. 17:1684–1699. 2020. View Article : Google Scholar : PubMed/NCBI
|