Medulloblastoma (MB) is a malignant tumor, which
exhibits the highest incidence and mortality amongst central
nervous system tumors (1). This
disease predominately affects children. According to the 2016 World
Health Organization's redefined classification of central nervous
system tumors, MB is divided into four types as follows:
Wnt-activated MB, Sonic hedgehog (SHH)-activated MB, group-3 MB and
group-4 MB (2), of which, the latter
two are the most common types. The prognosis of different subtypes
varies considerably. The 5-year overall survival (OS) rate of
Wnt-activated MB can reach 95%, whereas group-3 exhibits the worst
prognosis (45–60%), and group-4 and SHH-activated MB have an
intermediate OS (75–80%) (3,4). Since MB is commonly located in the
cerebellum, the main symptoms and signs are caused by intracranial
hypertension and hydrocephaly, which may be secondary to direct
tumor compression or obstruction of cerebrospinal fluid circulation
(5). At present, the treatment of MB
is primarily based on surgery, radiotherapy and chemotherapy.
Although MB is sensitive to radiotherapy and chemotherapy,
excessive treatment usually gives rise to serious secondary side
effects such as infection, peripheral neuropathy, ototoxicity and
myelosuppression in children, who are typically in the
developmental stage (6,7). Tumor cells can spread to the spinal
cord along the cerebrospinal fluid circulation pathway and ~30% of
patients will develop early tumor spread (8). Concomitantly, since MB exhibits a high
degree of malignancy, it commonly relapses after surgery. The
efficacy of traditional treatment remains poor. Therefore, the
investigation of the molecular mechanism responsible for MB
development and the identification of novel therapeutic targets are
crucial for the treatment and prevention of this disease.
Approximately 80% of the genes found in the human
genome possess transcriptional activity, whereas only 2% of the RNA
produced by transcription encodes proteins (9,10). The
remaining RNA that does not encode proteins is referred to as
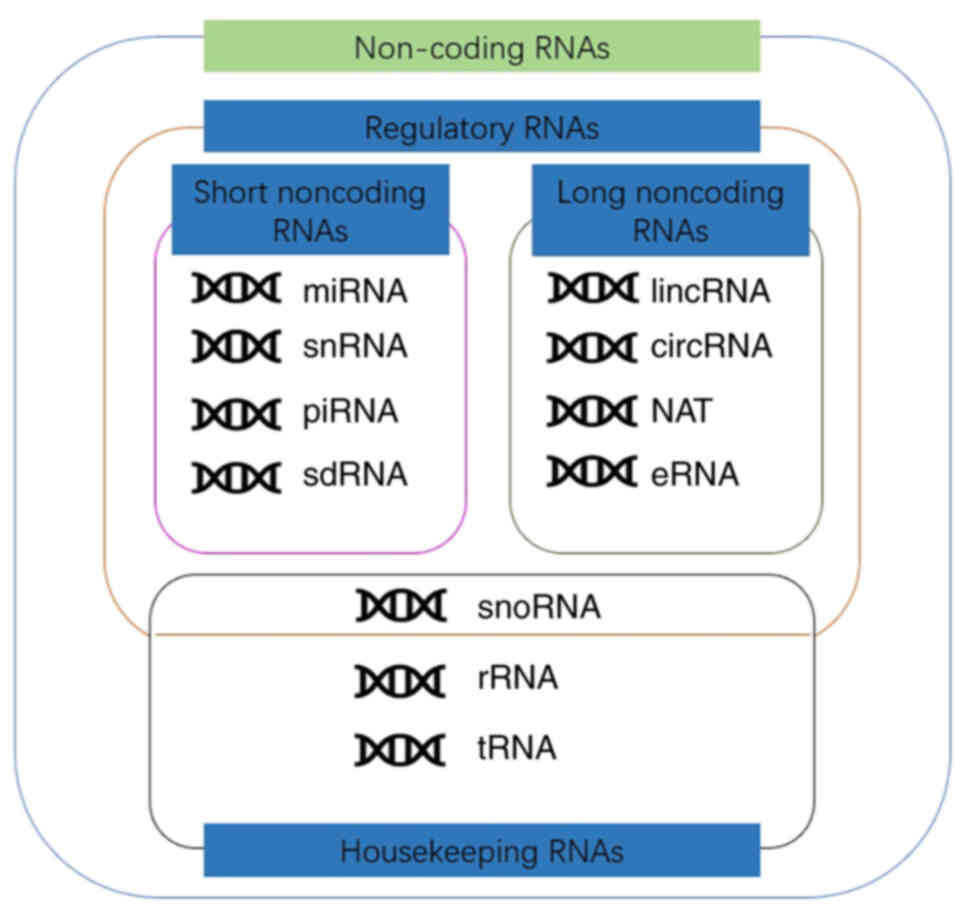
non-coding RNA (ncRNA) (9,10). Based on their functions, ncRNA
molecules are divided into housekeeping RNAs and regulatory RNAs
(11). According to the length of
the gene, they are divided into short ncRNAs (sncRNAs) and long
ncRNAs (lncRNAs). By using 200 nt as the cut-off, sncRNAs may be
further subdivided into PIWI-interacting RNAs (piRAN), small
nuclear RNAs (snRNA), small nucleolar RNAs (snoRNA) and microRNAs
(miRs/miRNAs). Similarly, lncRNAs can be divided into long
intergenic non-coding RNAs, natural antisense transcripts, enhancer
RNAs (eRNAs), partially unparalleled lncRNAs and circular RNAs
(circRNAs), which possess specific structural motifs (Fig. 1) (12,13). At
present, the involvement of ncRNAs in tumor cell regulation remains
unclear. However, numerous studies have investigated various
applications based on their functions. For example, miRNAs regulate
the stability of transcripts in order to silence genes at the
post-transcriptional level (14).
circRNAs can be used as miR molecular sponges that participate in
adjustment of cell biological function by affecting the functions
of miRNAs (15). eRNAs and binding
transcription factors form complexes to promote interactions
between gene enhancers and promoters (16). In recent years, certain studies have
provided significant evidence on the structure and function of
ncRNAs. It has been demonstrated that some short ncRNAs, including
circRNA, can not only regulate cell function through the
aforementioned pathways, but also directly encode various
regulatory proteins (17–19). Mounting evidence has shown that
ncRNAs can regulate the growth of central nervous system malignant
tumors, including gliomas (11,20,21). In
addition, it has been shown that the expression levels of various
ncRNAs are significantly different between MB and normal cerebellar
cells (22). Therefore, it was
hypothesized that deregulated expression of ncRNAs may serve as a
marker and/or therapeutic target for MB. Currently, research in
this field focuses on the mechanisms of miRNAs has revealed the
functions of a large number of miRNAs. However, there are
relatively fewer studies on the mechanisms of circRNA and other
ncRNAs, which are equally important. This article focuses on the
research progress of the role of ncRNAs in MB. In addition, this
paper also reviews some of the studies that are expected to be
translated into clinical therapeutic and diagnostic targets. The
aim of the present article was to review the above content to show
the potential of ncRNA in the clinical application of MB.
miRNAs belong to the ncRNA family, which usually
includes classes of RNA molecules of 18–25 nucleotides in length.
These motifs are highly conserved across different species
(11). miRNAs were initially
identified in Caenorhabditis elegans in 1993 (23). Since then, research on this topic has
been gaining increasing popularity, with a wealth of studies
assessing the roles of numerous miRNAs in almost all types of
diseases. The current point of view suggests that the main function
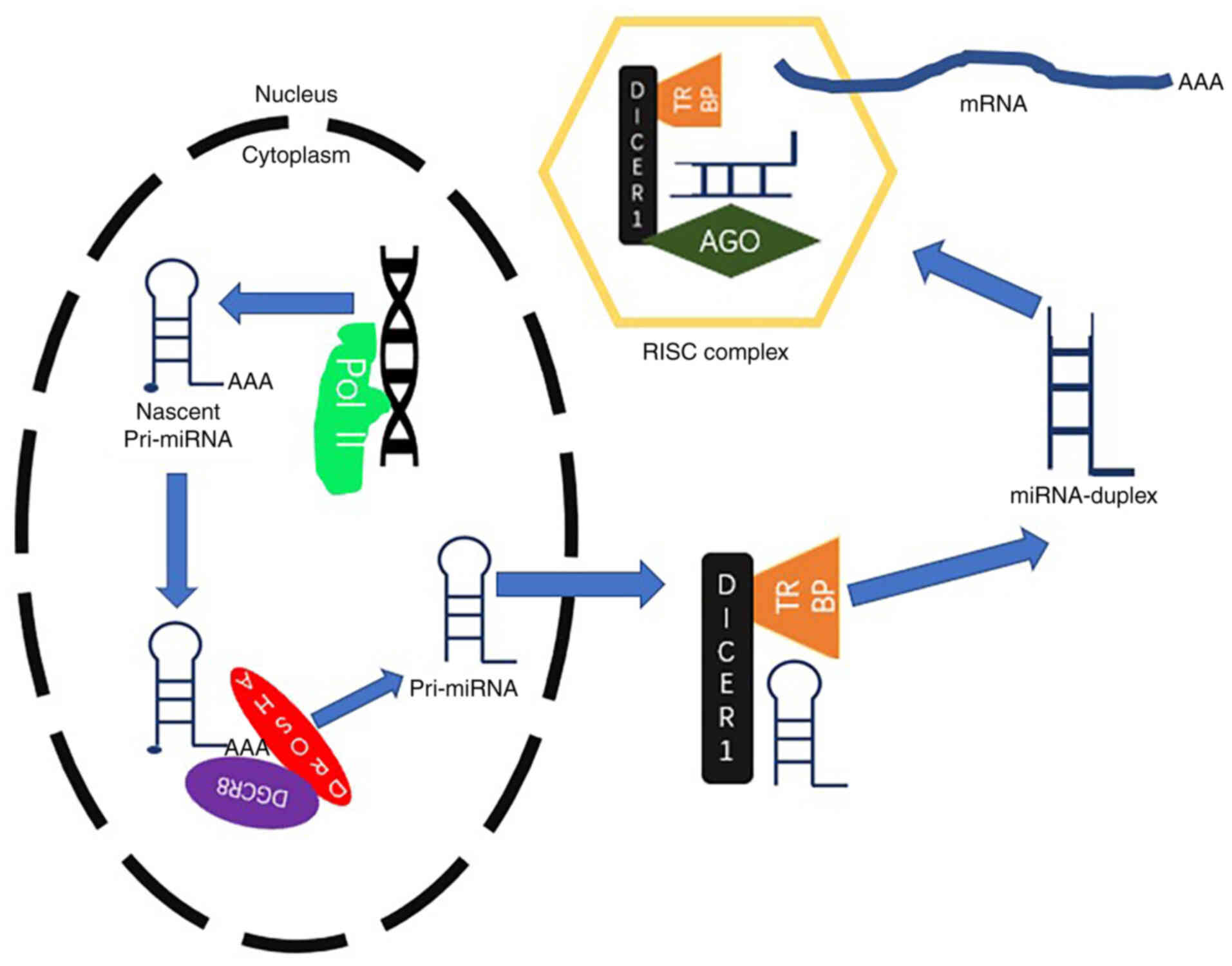
of miRNAs is to form RNA-induced silencing complexes (RISCs), which
include the Argonaute family protein in the cytoplasm (14). RISCs act on the downstream encoding
RNAs or ncRNAs to affect cell metabolism at the
post-transcriptional level (Fig. 2)
(14). It has been shown that the
expression levels of specific miRNAs differ on the type and stage
of the tumor (24–26). The specific expression patterns of
miRNAs determines the development of tumors. Therefore, miRNAs have
a wide range of applications for early diagnosis, targeted
treatment and prognostic assessment of the tumors.
The analysis of the miRNA expression levels in
tissues derived from normal cerebellum and MB has shown
similarities with regard to deregulated miRNA expression between MB
and other malignancies (22). The
first report that examined the expression of specific miRNAs in MB
was performed in 2008 by Pierson et al (27), in which miR-124 expression was
predicted to participate in the regulation of the MB prognostic
marker cyclin-dependent kinase 6 (CDK6). Subsequent experiments
confirmed that miR-124 expression was decreased in MB and that it
affected the expression levels of CKD6 in tumor cells (27). Ferretti et al (28) compared the expression levels of
specific miRNAs in normal brain and MB tissues. It was found that
miR-18a, miR-19a, miR-21 and miR-25 expression levels in MB were
significantly higher than those in normal brain tissues.
Concomitantly, the expression levels of the majority of miRNAs,
such as miR-9 and miR-125a, were downregulated in tumor samples.
These downregulated miRNAs may possess tumor suppressive functions
(28). The same is true for cancer
and neural stem cells (29). miRNA
microarray data analysis from the Gene Expression Omnibus database
demonstrated that the expression levels of 22 miRNAs were
upregulated in MB, and 26 miRNAs were downregulated (30). It has also been shown that the
expression levels of certain miRNAs in MB do not differ from those
in normal cerebellum cells. Differential expression of miRNAs has
been used as an identification marker of MB molecular subtypes. For
example, it was shown that miR-148a expression was specifically
enriched in the Wnt-activated MB subtype (31). High expression of the miR-17-92
cluster was observed in SHH-activated MB (32). Zhu et al (33) demonstrated that the expression levels
of miR-181a-5p and miR-125b-5p were increased in group-3 MB.
However, 12 miRNAs, including miR-18a, miR-135b and miR-660, were
overexpressed in group-4 MB (34).
These differentially expressed miRNAs were expected to be key
indicators for the identification of specific MB-associated
markers. Visani et al (35)
indicated that there were differences in miR-196B-5P and
miR-200B-3P expression between adults and children. Their study
suggested that miRNAs may be involved in the development of MB, but
not in the differences noted in the biological responses of adults
and children with MB. An increasing number of studies have
confirmed that the differences in the expression of these miRNAs
can affect specific biological processes of MB cells, such as
proliferation, migration, invasion and apoptosis. For example,
miR-10b is specifically overexpressed in MB cells and it can
promote proliferation by mediating the downregulation of the
expression of the apoptotic protein Bcl-2 (36). Therefore, miR-10b overexpression can
induce apoptosis and inhibit colony formation (36). In SHH-activated MB, the expression of
the miR-17-92 cluster regulates N-myc proto-oncogene overexpression
(32). Northcott et al
(32) confirmed that miR-17-92
clusters can promote proliferation of tumor cells and enhance the
invasion of MB cells in vitro. Grunder et al
(37) demonstrated that miR-21
overexpression negatively regulated the expression of the transfer
inhibitory factor programmed cell death protein 4 in MB compared
with that observed in normal tissues, thereby increasing the
expression levels of the downstream invasion medium proteins
mitogen-activated protein kinase kinase and JNK, which promote
tumor cell invasion. Yang et al (38) analyzed the miRNA expression profiles
in 29 patients with MB and screened this group for miR-192
expression, which was downregulated in tumor cells. The results of
their study confirmed that miR-192 inhibited the proliferation and
anchorage capability of MB cell lines by regulating the downstream
target genes dihydrofolate, integrin α-V precursor, integrin β-1/3
precursor and cluster of differentiation 47. This finding was also
confirmed in a nude mouse xenograft model, suggesting that miR-192
is a type of metastasis inhibitory factor (38). In MB, the natural antisense
transcript HOTAIR of lncRNA HOX competitively binds to miR-1 and
miR-206 and causes upregulation of Yin Yang 1 protein expression.
This pathway promotes the malignant phenotype of MB (39). Senfter et al (40) demonstrated that the loss of miR-4521
expression led to the activation of the proto-oncogene forkhead box
protein M1 (FOXM1) in MB. Kumar et al (41) indicated that miR-217 promoted tumor
growth by negatively regulating the target genes sirtuin 1,
Roundabout1, FOXO3 and SMAD7. Early diagnosis and prognostic
assessment are important steps in the treatment of MB. Currently,
the detection of miR expression offers an alternative to imaging
data. Li et al (42)
demonstrated that miR-449a expression was downregulated in MB of
all subtypes, with the exception of Wnt-MB. This suggests the
potential applications of this miR in the diagnosis of Wnt-MB
(42). Pezuk et al (43) examined the expression levels of
Polo-like kinase family members and their associated miRNAs
(miR-100, miR-126, miR-219 and miR-593) in 32 clinical samples of
MB in association with disease prognosis. The results indicated
that patients with higher expression of miR-100 and lower
expression of miR-126 and miR-219 had improved OS (43). Although increasing evidence has
suggested that miR-targeted therapy is of considerable value,
miRNA-targeting for the treatment of MB remains under
investigation. Some targeted treatments for specific miRNAs have
been shown to inhibit the development of the malignant phenotype of
MB cells. For example, overexpression of miR-34a can reduce the
expression of transmembrane protein δ-Like1 in MB to inhibit tumor
cell proliferation and induce apoptosis (44). De Antonellis et al (44) demonstrated inhibition of tumor growth
in a nude mouse MB model by administration of adenoviral vectors
carrying miR-34a. The minichromosome maintenance protein (MCM2-7)
complex has the ability to influence DNA transcription and
replication. CDK6, which is part of the MCM2-7 complex, is
overexpressed in one-third of MB cases. Therefore, it may be used
as a specific marker of disease prognosis. Silber et al
(45) established a nude mouse model
of heterotopic transplanted tumor with D425 cells, which were
infected with lentiviruses containing miR-124. The results
indicated that miR-124-targeted CDK6 and effectively inhibited
tumor growth (45). However, the
development of high-efficiency miR-targeted drugs that can be
applied clinically requires further exploration due to the
limitations of the current technologies.
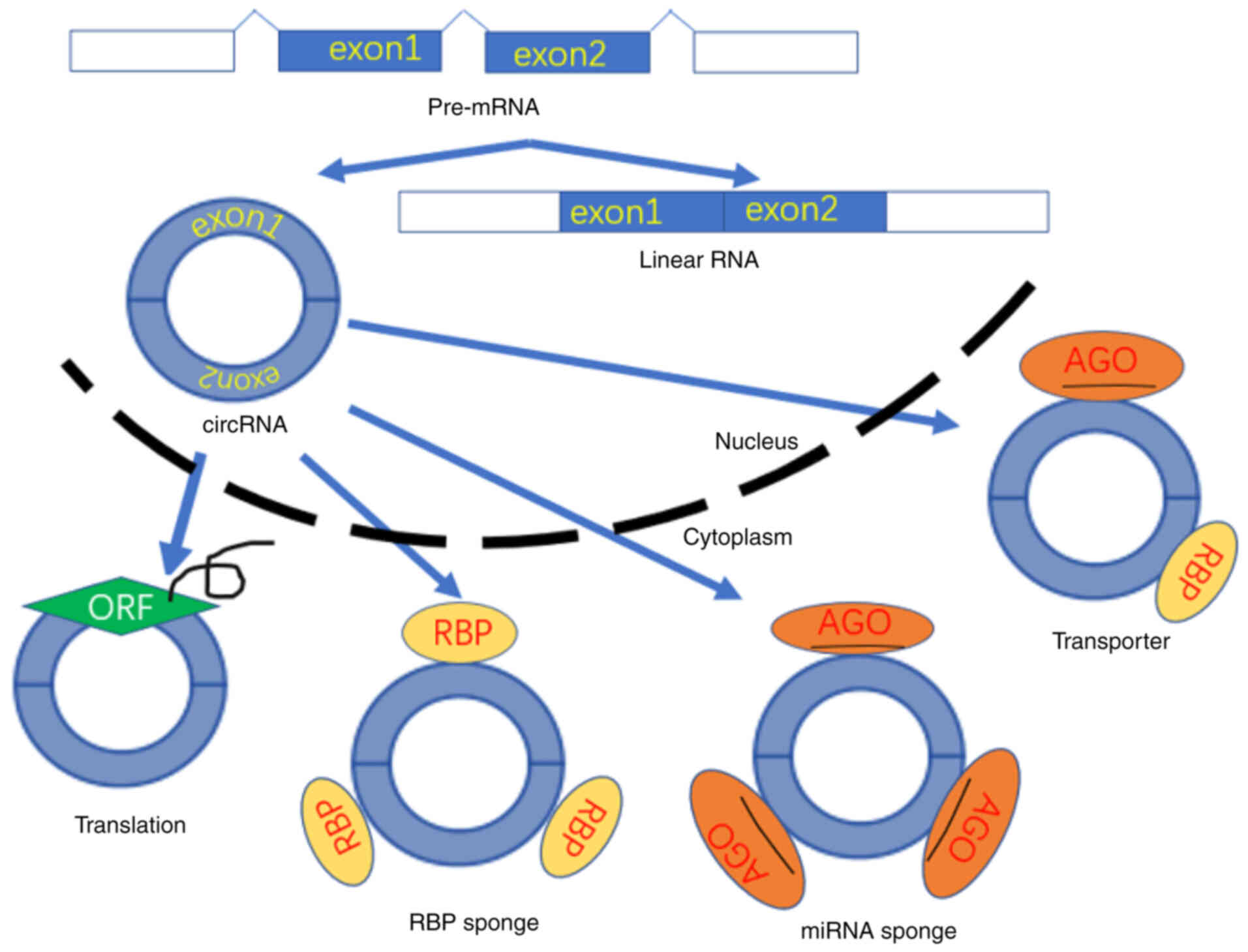
Currently, the prevailing hypothesis is that the
main role of circRNAs is to combine with corresponding miRNAs
through conserved sites. Subsequently, circRNAs act as molecular
sponges of miRNAs and regulate various cellular functions (53–58)
(Fig. 3). circRNAs are often
dysregulated in a variety of malignant tumors, including glioma
(59–61). These molecules are involved in
regulation of tumor growth, suggesting that they may be important
regulators in the development of MB. Lv et al (62) selected 4 pairs of normal cerebellum
and MB tissue samples for gene sequencing and identified 33
differentially expressed circRNAs in MB tissues. The upregulation
of two of these circRNAs, which were identified as circular-spindle
and kinetochore associated complex subunit 3 (circ-SKA3) and
circ-DTL, reverted the malignant phenotype of MB (62). A previous study observed
significantly higher expression levels of circ-SKA3 in MB compared
with the corresponding expression noted in normal tissues. This was
achieved by detecting the expression levels of specific circRNAs
that were differentially expressed in MB. The expression levels of
the downstream target miR-383-5p were determined by luciferase
assay. Subsequent experiments indicated that miR-383-5p expression
was influenced by low expression of circ-SKA3 in tumor cells.
Following restoration of miR-383-5p expression by silencing
circ-SKA3, the expression levels of the downstream FOXM1 protein
were also affected and the proliferation, migration and invasion of
the tumors were inhibited, whereas the induction of apoptosis was
enhanced (63). Although current
evidence suggests that circRNAs play an important role in the
development of MB, additional studies are required to assess their
specific mechanisms and potential applications in the clinical
treatment of MB.
At present, the regulatory mechanism of ncRNA
expression in MB has been primarily examined by assessing the
expression levels of miRNAs and circRNAs. Pertinent ncRNA
regulatory functions have not been thoroughly investigated. For
example, eRNAs, which were identified relative more recently, play
a regulatory role by forming a complex with RNA polymerase II DNA
binding transcription factor and the RNA binding transcription
factor that binds to the gene enhancer (64). Lin et al (65) investigated the binding of H3K27AC and
bromodomain-containing protein 4 by chromatin immunoprecipitation
assays in MB tissue matched samples. DNA methylation and
transcription data were also provided to describe 28 cis-regulatory
elements in MB. The results indicated that the differential
regulation of enhancers was heterogeneous between subgroups
(65). That is, eRNAs may play an
important role in the phenotypic changes of MB. SPRY4 intronic
transcript 1 (SPRY4-IT1) is a type of lncRNA with a length of ~706
bp. It has a hairpin structure and is expressed in gliomas. Shi
et al (66) demonstrated that
inhibition of SPRY4-IT1 affected the expression of MMP-2 in MB, and
the migration of the MB cell line Daoy was decreased by this
pathway.
MB is one of the most common types of central
nervous system malignant tumors encountered in children, and it is
characterized by a high incidence and a poor prognosis. Although
significant progress has been made in exploring the development and
pathogenesis of MB, the effects of currently available treatments
are often unsatisfactory due to the propensity of the tumor for
recurrence and metastatic spread, and the occurrence of serious
complications. In recent years, it has been confirmed that several
ncRNAs, such as miRNAs and circRNAs, are associated with the
regulatory mechanisms involved in the occurrence and development of
various tumors. It has also been shown that several ncRNAs,
including miRNAs and circRNAs, regulate tumor metabolism and are
involved in the development of MB. At present, a number of studies
have been conducted on the regulatory mechanism of miRNAs in MB. It
has been confirmed that miRNAs play an important role in the
occurrence and development of MB. However, a limited number of
studies have been performed on the mechanisms associated with the
expression of other ncRNAs, including circRNAs, and the development
of MB. It is considered that wider adoption of high-throughput
microarray and second-generation sequencing technologies will
enable the full investigation of ncRNA-associated mechanisms and
their clinical applications in the diagnosis and treatment of
MB.
Not applicable.
The present study was supported by grants from the
Science and Technology Research Project of Gansu Province (grant
nos. 145RTSA012 and 17JR5RA307), the Project of Healty and Family
Planing Commission of Gansu (grant no.
GSWSKY-2014-31/GSWSKY-2015-58/GSWSKY2018-01), the Lanzhou Science
and Technology Bureau Project (grant no. 2018-1-109), the Cuiying
Science and Technology fund (grant no.
CY2017-MS12/CY2017-MS15/CY2017-BJ15/CYXZ-01), Cuiying Graduate
Supervisor Applicant Training Program (grant no. 201803) and the
Special fund project for doctoral training (grant no. YJS-BD-13) of
Lanzhou University Second Hospital.
Not applicable.
YNZ completed the primary writing and proofreading
of the manuscript. KL made partial corrections to the original
anuscript and made the figures. XSH performed the literature
search. YWP directed the writing of the article and made partial
revisions. All authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Ostrom QT, Gino C, Gittleman H, Patil N,
Waite K, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical
report: Primary brain and other central nervous system tumors
diagnosed in the United States in 2012–2016. Neuro Oncol. 21 (Suppl
5):v1–v100. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis D, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the Central Nervous System: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Forbes K, Osborn, Salzman, Barkovich, et
al: Diagnostic Imaging. Brain (2nd edition). Neuroradiology.
54:2692012.
|
|
4
|
Gajjar A and Robinson G:
Medulloblastoma-translating discoveries from the bench to the
bedside. Nat Rev Clin Oncol. 11:714–722. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pollack IF and Jakacki RI: Childhood brain
tumors: Epidemiology, current management and future directions. Nat
Rev Neurol. 7:495–506. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Merchant TE, Mulhern RK, Krasin MJ, Kun
LE, Williams T, Li C, Xiong X, Khan RB, Lustig RH, Boop FA and
Sanford RA: Preliminary results from a phase II trial of conformal
radiation therapy and evaluation of radiation-related CNS effects
for pediatric patients with localized ependymoma. J Clin Oncol.
22:3156–3162. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kortmann RD, Kühl J, Timmermann B, Mittler
U, Urban C, Budach V, Richter E, Willich N, Flentje M, Berthold F,
et al: Postoperative neoadjuvant chemotherapy before radiotherapy
as compared to immediate radiotherapy followed by maintenance
chemotherapy in the treatment of medulloblastoma in childhood:
Results of the german prospective randomized trial hit '91. Int J
Radiat Oncol Biol Phys. 46:269–279. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gerber NU, Mynarek M, Von Hoff K,
Friedrich C, Resch A and Rutkowski S: Recent developments and
current concepts in medulloblastoma. Cancer Treat Rev. 40:356–365.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
ENCODE Project Consortium, . An Integrated
Encyclopedia of DNA Elements in the Human Genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pennisi E: ENCODE project writes eulogy
for junk DNA. Science. 337:1159–1161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ramasamy P, Malhotra M and Massoud TF: The
protean world of non-coding RNAs in glioblastoma. J Mol Med (Berl).
97:909–925. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM,
Li CW, Wang Y, Hsu JL and Hung MC: Long non-coding RNAs: Versatile
master regulators of gene expression and crucial players in cancer.
Am J Transl Res. 4:127–150. 2012.PubMed/NCBI
|
|
13
|
Barrett SP and Salzman J: Circular RNAs:
Analysis, expression and potential functions. Development.
143:1838–1847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trimarchi T, Bilal E, Ntziachristos P,
Fabbri G, Dalla-Favera R, Tsirigos A and Aifantis I: Genome-wide
mapping and characterization of Notch-regulated long noncoding RNAs
in acute leukemia. Cell. 158:593–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H,
Huang N, Yang X, Xiao F, Liu D, et al: A novel tumor suppressor
protein encoded by circular AKT3 RNA inhibits glioblastoma
tumorigenicity by competing with active phosphoinositide-dependent
Kinase-1. Mol Cancer. 18:1312019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X,
Zhong J, Zhao Z, Zhao K, Liu D, et al: Circular RNA-encoded
oncogenic E-cadherin variant promotes glioblastoma tumorigenicity
through activation of EGFR-STAT3 signalling. Nat Cell Biol.
23:278–291. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morlando M, Di Timoteo G, Rossi F,
Morlando M, Briganti F, Sthandier O, Fatica A, Santini T,
Andronache A, Wade M, et al: Circ-ZNF609 is a circular RNA that Can
Be translated and functions in myogenesis. Mol Cell. 66:22–37.e9.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Diederichs S: Non-coding RNA in malignant
tumors. A new world of tumor biomarkers and target structures in
cancer cells. Pathologe. 31 (Suppl 2):S258–S262. 2010.(In German).
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Julia L, Grabowska A, Zarębska Ż,
Kuczyński K, Kuczyńska B and Rolle K: Non-coding RNAs in Brain
Tumors, the Contribution of lncRNAs, circRNAs, and snoRNAs to
cancer development-their diagnostic and therapeutic potential. Int
J Mol Scie. 21:70012020. View Article : Google Scholar
|
|
22
|
Joshi P, Katsushima K, Zhou R, Meoded A,
Stapleton S, Jallo G, Raabe E, Eberhart CG and Perera RJ: The
therapeutic and diagnostic potential of regulatory noncoding RNAs
in medulloblastoma. Neurooncol Adv. 1:vdz0232019.PubMed/NCBI
|
|
23
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans Heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Browne BM, Stensland KD, Patel CK,
Sullivan T, Burks EJ, Canes D, Raman JD, Warrick J and
Reiger-Christ KM: MicroRNA expression profiles in upper tract
urothelial carcinoma differentiate tumor grade, stage, and
survival: Implications for clinical decision-making. Urology.
123:93–100. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Magdalena Z, Fendler W, Zakrzewski K,
Sikorska B, Grajkowska W, Dembowska-Bagińska B, Filipek I,
Stefańczyk Ł and Liberski PP: Altered MicroRNA expression is
associated with tumor grade, molecular background and outcome in
childhood infratentorial ependymoma. PLoS One. 11:e01584642016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rothé F, Ignatiadis M, Chaboteaux C,
Haibe-Kains B, Kheddoumi N, Majjaj S, Badran B, Fayyad-Kazan H,
Desmedt C, Harris AL, et al: Global microRNA expression profiling
identifies MiR-210 associated with tumor proliferation, invasion
and poor clinical outcome in breast cancer. PLoS One. 6:e209802011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pierson J, Hostager B, Fan R and Vibhakar
R: Regulation of cyclin dependent kinase 6 by microRNA 124 in
medulloblastoma. J Neurooncol. 90:1–7. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ferretti E, De Smaele E, Po A, Di
Marcotullio L, Tosi E, Espinola MS, Di Rocco C, Riccardi R,
Giangaspero F, Farcomeni A, et al: MicroRNA profiling in human
medulloblastoma. Int J Cancer. 124:568–577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Po A, Abballe L, Sabato C, Gianno F,
Chiacchiarini M, Catanzaro G, De Smaele E, Giangaspero F, Ferretti
E, Miele E and Besharat ZM: Sonic hedgehog medulloblastoma cancer
stem cells mirnome and transcriptome highlight novel Functional
Networks. Int J Mol Sci. 19:23262018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dai J, Li Q, Bing Z, Zhang Y, Niu L, Yin
H, Yuan G and Pan Y: Comprehensive analysis of a microRNA
expression profile in pediatric medulloblastoma. Mol Med Rep.
15:4109–4115. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yogi K, Sridhar E, Goel N, Jalali R, Goel
A, Moiyadi A, Thorat R, Panwalkar P, Khire A, Dasgupta A, et al:
MiR-148a, a microRNA upregulated in the WNT subgroup tumors,
inhibits invasion and tumorigenic potential of medulloblastoma
cells by targeting Neuropilin 1. Oncoscience. 2:334–348. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Northcott PA, Fernandez-L A, Hagan JP,
Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka
JT, Croce CM, et al: The miR-17/92 polycistron is up-regulated in
sonic hedgehog-driven medulloblastomas and induced by N-myc in
sonic hedgehog-treated cerebellar neural precursors. Cancer Res.
69:3249–3255. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu LY, Wu XY, Liu XD, Zheng DF, Li HS,
Yang B, Zhang J and Chang Q: Aggressive medulloblastoma-derived
exosomal miRNAs promote in vitro invasion and migration of tumor
cells via Ras/MAPK pathway. J Neuropathol Exp Neurol. 79:734–745.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gershanov S, Toledano H, Michowiz S,
Barinfeld O, Pinhasov A, Goldenberg-Cohen N and Salmon-Divon M:
MicroRNA-mRNA expression profiles associated with medulloblastoma
subgroup 4. Cancer Manag Res. 10:339–352. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Visani M, Marucci G, Biase D, Giangaspero
F, Buttarelli FR, Brandes AA, Franceschi E, Acquaviva G, Ciarrocchi
A, Rhoden KJ, et al: miR-196B-5P and miR-200B-3P are differentially
expressed in medulloblastomas of adults and children. Diagnostics
(Basel). 10:2652020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pal R and Greene S: microRNA-10b is
overexpressed and critical for cell survival and proliferation in
medulloblastoma. PLoS One. 10:e01378452015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Grunder E, D'ambrosio R, Fiaschetti G,
Abela L, Arcaro A, Zuzak T, Ohgaki H, Lv SQ, Shalaby T and Grotzer
M: MicroRNA-21 suppression impedes medulloblastoma cell migration.
Eur J Cancer. 47:2479–2490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang SY, Choi SA, Lee JY, Park AK, Wang
KC, Phi JH, Koh EJ, Park WY, Park SH, Hwang DW, et al: miR-192
suppresses leptomeningeal dissemination of medulloblastoma by
modulating cell proliferation and anchoring through the regulation
of DHFR, integrins, and CD47. Oncotarget. 6:43712–43730. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Li N, Fu J and Zhou W: Long
noncoding RNA HOTAIR promotes medulloblastoma growth, migration and
invasion by sponging miR-1/miR-206 and targeting YY1. Biomed
Pharmacother. 124:1098872020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Senfter D, Samadaei M, Mader R, Gojo J,
Peyrl A, Krupitza G, Kool M, Sill M, Haberler C, Ricken G, et al:
High impact of miRNA-4521 on FOXM1 expression in medulloblastoma.
Cell death & disease. 10:6962019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kumar V, Kumar V, Chaudhary AK, Coulter
DW, McGuire T and Mahato RI: Impact of miRNA-mRNA profiling and
their correlation on medulloblastoma tumorigenesis. Mol Ther
Nucleic Acids. 12:490–503. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Y, Jiang T, Shao L, Liu Y, Zheng C,
Zhong Y, Zhang J and Chang Q: Mir-449a, a potential diagnostic
biomarker for WNT group of medulloblastoma. J Neurooncol.
129:423–431. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pezuk JA, Brassesco MS, De Oliveira RS,
Machado HR, Neder L, Scrideli CA and Tone LG: PLK1-associated
microRNAs are correlated with pediatric medulloblastoma prognosis.
Childs Nerv Syst. 33:609–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
de Antonellis P, Medaglia C, Cusanelli E,
Andolfo I, Liguori L, De Vita G, Carotenuto M, Bello A, Formiggini
F, Galeone A, et al: MiR-34a targeting of Notch ligand delta-like 1
impairs CD15+/CD133+ tumor-propagating cells
and supports neural differentiation in medulloblastoma. PLoS One.
6:e245842011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Silber J, Hashizume R, Felix T, Hariono S,
Yu M, Berger MS, Huse JT, VandenBerg SR, James CD, Hodgson JG and
Gupta N: Expression of miR-124 inhibits growth of medulloblastoma
cells. Neuro Oncol. 15:83–90. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs Are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nilsen TW and Graveley BR: Expansion of
the eukaryotic proteome by alternative splicing. Nature.
463:457–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Moore MJ and Proudfoot NJ: Pre-mRNA
processing reaches back to transcription and ahead to translation.
Cell. 136:688–700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Belter A, Popenda M, Sajek M, Woźniak T,
Naskręt-Barciszewska MZ, Szachniuk M, Jurga S and Barciszewski J: A
new molecular mechanism of RNA circularization and the microRNA
sponge formation. J Biomol Struct Dyn. Nov 17–2020.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye
X, Zhang H, Yang P, Cui X, Ren Y, et al: A noncoding regulatory
RNAs Network Driven by Circ-CDYL acts specifically in the early
stages hepatocellular carcinoma. Hepatology. 71:130–147. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang
Y and Cui J: CircRNA circ-BANP-mediated miR-503/LARP1 signaling
contributes to lung cancer progression. Biochem Biophys Res Commun.
503:2429–2435. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Gong J, Jiang H, Shu C, Hu MQ, Huang Y,
Liu Q, Li RF and Wei YZ: Integrated analysis of circular
RNA-associated ceRNA network in cervical cancer: Observational
Study. Medicine (Baltimore). 98:e169222019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Huang M, He YR, Liang LC, Huang Q and Zhu
ZQ: Circular RNA hsa_circ_0000745 may serve as a diagnostic marker
for gastric cancer. World J Gastroenterol. 23:6330–6338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li G, Huang M, Cai Y, Yang Y, Sun X and Ke
Y: Circ-U2AF1 promotes human glioma via derepressing
neuro-oncological ventral antigen 2 by sponging hsa-miR-7-5p. J
Cell Physiol. 234:9144–9155. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lv T, Miao Y, Jin K, Han S, Xu TQ, Qiu ZL
and Zhang XH: Dysregulated circular RNAs in medulloblastoma
regulate proliferation and growth of tumor cells via host genes.
Cancer Med. 7:6147–6157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang X, Xu D, Pei X, Zhang Y, Zhang Y, Gu
Y and Li Y: CircSKA3 modulates FOXM1 to facilitate cell
proliferation, migration, and invasion while confine apoptosis in
medulloblastoma via miR-383-5p. Cancer Manag Res. 12:13415–13426.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shibayama Y, Fanucchi S, Magagula L and
Mhlanga MM: lncRNA and gene looping: What's the connection?
Transcription. 5:e286582014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lin CY, Erkek S, Tong Y, Yin L, Federation
AJ, Zapatka M, Haldipur P, Kawauchi D, Risch T, Warnatz HJ, et al:
Active medulloblastoma enhancers reveal subgroup-specific cellular
origins. Nature. 530:57–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shi PF, Ji HL, Luo YK, Mao TM, Chen X and
Zhou KY: Effect of long noncoding RNA SPRY4-IT1 on proliferation
and metastasis of medulloblastoma. Zhongguo Ying Yong Sheng Li Xue
Za Zhi. 33:78–82. 2017.(In Chinese). PubMed/NCBI
|