Introduction
As the most common type of malignancy originating
from the gynecological system in developed countries, endometrial
cancer (EC) primarily affects women >55 years of age (1). EC accounted for ~4.8% of all cases of
cancer in women between 1999 and 2006 (2). With advances in the treatment of EC,
>95% of patients with localized tumors survive for 5 years
following diagnosis (3–5). However, effective treatment for
metastatic EC remains poor (6). In
addition, distant tumor metastasis, such as rectal and bladder
metastases, is common in patients with EC (6). Once tumors have spread to distant
sites, <17% of patients with EC survive for 5 years (3–5).
Therefore, the development of novel approaches for EC treatment is
required.
The main risk factors for EC include obesity, aging,
diet, type 2 diabetes and a history of breast or ovarian cancer
(7). However, the exact mechanisms
underlying EC remain unclear. EC growth and metastasis, as well as
the development of drug resistance in EC cells, involve molecular
components (8–10). Progress in the elucidation of the
molecular pathways involved in EC has resulted in the development
of novel therapies that aim to treat EC by regulating the mRNA
expression level of EC-related genes (8–10).
However, effective targets for EC-targeted therapy are lacking.
Circular RNAs (circRNAs) have either no or limited protein-coding
capacity, but participate in human cancer by regulating
transcription and translation (11,12).
Therefore, circRNAs may serve as potential targets for EC
treatment. A previous study described a novel circRNA, circ_POLA2,
with congenic functions in lung and cervical cancers (13,14). Our
previous study, using deep sequencing analysis revealed altered
circ_POLA2 expression level in EC, with an inverse correlation with
microRNA (miR)-31, which has also been associated with cancer
biology (15). The present study
aimed to investigate the mechanism of regulation in EC cell
proliferation via miR-31.
Materials and methods
Tissue acquisition
Between April 2018 and April 2020, 60 patients with
EC (all female; 26 cases at stage I or II and 34 cases at stage III
or IV) were admitted to Beilun District People's Hospital
(Zhejiang, China). Patients with recurrent EC were not enrolled
into the present study. Patients receiving current treatment
therapies were also excluded from the study, as were patients with
other severe clinical complications, such as chronic renal or
hepatic disease, chronic obstructive pulmonary disease, asthma and
congenital diseases. The age range of the patients was 52–68 years,
with a mean age of 60.7 ± 5.5 years. The present study was approved
by the Ethics Committee of Beilun District People's Hospital
(Zhejiang, China) and written informed consent was obtained from
all patients. Before treatment, specimens of both EC and paired
adjacent normal (within 5 cm around tumors) tissues were collected
from each patient using fine-needle aspiration. Histopathological
analysis was performed on all tissue specimens to confirm that the
correct specimens were collected, according to the guidelines of
the German Working Group on Gynecological Oncology (16). The tissue specimens were stored at
−80°C or in liquid nitrogen before the subsequent experiments.
Association between miR-31 expression level and the
clinicopathological parameters (age, sex, histopathological grade,
depth of myometrial invasion, lymphatic metastasis and distant
metastasis) in patients with EC are shown in Table I.
 | Table I.Association between miR-31 expression
level and the clinicopathological parameters in patients with
endometrial carcinoma. |
Table I.
Association between miR-31 expression
level and the clinicopathological parameters in patients with
endometrial carcinoma.
| Clinicopathological
parameter | Number | miR-31 expression
levela | P-value |
|---|
| Age, years |
|
| 0.059 |
|
<60 | 37 | 0.013
(0.005–0.042) |
|
| ≥60 | 23 | 0.156 (0.
068–0.450) |
|
| Stage |
|
| 0.0002b |
| I | 16 | 0.123
(0.051–0.279) |
|
| II | 10 | 0.008
(0.006–0.021) |
|
|
III/IV | 34 | 0.004
(0.003–0.005) |
|
| Histopathological
grade |
|
| 0.011b |
| G1 and
G2 | 30 | 0.120
(0.045–0.251) |
|
| G3 | 20 | 0.116
(0.006–0.743) |
|
| Depth of myometrial
invasion |
|
| 0.0001b |
| No
infiltration | 20 | 0.342
(0.130–0.632) |
|
| ≤1/2 | 34 | 0.014
(0.005–0.023) |
|
|
>1/2 | 6 | 0.004
(0.002–0.022) |
|
| Lymphatic
metastasis |
|
| 0.431 |
| Yes | 6 | 0.370
(0.006–0.900) |
|
| No | 54 | 0.028
(0.004–0.127) |
|
| Distant
metastasis |
|
| 0.016b |
| Yes | 2 | 0.178
(0.126–0.262) |
|
| No | 58 | 0.025
(0.005–0.226) |
|
EC transfection
A total of two human EC cell lines, HEC-1-A and
RL95-2 (both from American Type Culture Collection) were used as
the EC cell models. Cell culture was cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% FBS
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified incubator with 5% CO2 according to the
manufacturer's instructions. Subsequent assays were performed using
cells at ~85% confluence. The cells were transfected with miR-31
mimics and/or circ_POLA2 overexpression plasmid using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The cells were cultured
as aforementioned for 48 h after transfection before subsequent
experiments.
A backbone vector expressing circ_POLA2 was
constructed using the pcDNA3.1(+) circRNA mini vector (Addgene,
Inc.). The following primer sequences were used to construct
circ_POLA2: forward, 5′-GGAATTCATGTCCGCATCCGCC-3′ and reverse
5′-ATAAGAATTCAGATCCTGACGACC-3′. miR-31 mimics were synthesized by
Shanghai GenePharma Co., Ltd., and negative control (NC; cat. no.
miR1N0000001-1-5) miRNAs were purchased from Guangzhou RiboBio Co.,
Ltd. The sequence of the miR-31 mimics and NC miRNA are as follows:
5′-UAGCAGCACAGAAAUAUUGGC-3′ and 5′-UUGUACUACACAAAAGUACUG-3′,
respectively. To overexpress circ_POLA2 and miR-31, the HEC-1-A and
RL95-2 cells (1×108) were transfected with circ_POLA2
expression vector (1 µg) or miR-31 mimics (40 nM) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The same number of cells were transfected with
empty vector or NC miR mimics using the same method as the
overexpression vector and miR-31 mimics. Empty vector or untreated
cells were used as the control (C) group. Subsequent assays were
performed 48 h later.
Methylation-specific PCR (MSP)
DNA was extracted from the HEC-1-A and RL95-2 cells
transfected with circ_POLA2 overexpression plasmid using a
Monarch® Genomic DNA Purification kit (New England
Biolabs, Inc.). An EZ DNA Methylation kit (cat. no. D5001; Zymo
Research Corp.) was used to convert the genomic DNA into bisulfite
modified DNA. Firstly, MSP was performed to analyze miR-31
methylation, in which the DNA was modified by sodium bisulfite
treatment to convert unmethylated cytosine to uracil, while
methylated cytosine remained intact. Secondly, following the
removal of bisulfite and completion of the chemical conversion, the
modified DNA was used as the template for PCR using Taq DNA
polymerase (Takara Bio, Inc.) The following primer sequences were
used: Methylation forward, 5′-TTGTGTATAATTTGGGGCGTC-3′ and reverse,
5′-CCAACTTACCTACGAATCCGA-3′, and unmethylation forward,
5′-TTGTGTATAATTTGGGGTGTTGT-3′ and reverse,
5′-CTCCCAACTTACCTACAAATCCA-3′. The following thermocycling
conditions were used: Initial denaturation at 95°C for 30 sec, then
55°C for 30 sec, 72°C 30 sec. A total of 2 PCRs were performed for
each DNA sample, one specific for originally methylated DNA without
methylation for the gene of interest and one specific for
originally unmethylated DNA with methylation. The PCR products were
separated using a 6–8% non-denaturing polyacrylamide gels and the
bands were visualized by staining with ethidium bromide. The
presence of a band of the appropriate molecular weight indicated
the presence of unmethylated and/or methylated alleles in the
original sample.
RNA extraction
Total RNA was isolated from the HEC-1-A and RL95-2
cells transfected with circ_POLA2 overexpression plasmid or miR-31
mimics, as well as paired tissue specimens from 60 patients with EC
using RNAzol (Sigma-Aldrich; Merck KGaA). DNase I (Invitrogen;
Thermo Fisher Scientific, Inc.) was used to remove the genomic DNA.
RNA integrity was analyzed using agarose gel electrophoresis.
Reverse transcription-quantitative PCR
(RT-qPCR)
The cDNA samples were prepared using a Bio-Rad cDNA
Supermix kit (Bio-Rad Laboratories, Inc.). The following
temperature protocol was used for RT: Incubation at 50°C for 15 min
then 75°C for 5 min. The following housekeeping genes were used as
the internal controls: GAPDH and U6. The expression level of mature
miR-31 was determined using the All-in-One™ miRNA RT-qPCR Detection
kit using SYBRGreen (GeneCopoeia, Inc.) following the
manufacturer's instructions. For circ_POLA2, qPCR was performed
using a LightCycler® 480 SYBR Green I Master (Roche
Diagnostics). The following thermocycling conditions were used for
both miRNA and cir_POLA2: Initial denaturation at 95°C for 30 sec,
then 95°C for 5 sec, 60°C 34 sec for 40 cycles; then 95°C for 15
sec, 60°C for l min and 95°C for 15 sec. The threshold cycle (Cq)
values were analyzed using the 2−ΔΔCq method. Analysis
of relative gene expression was performed using the
2−ΔΔCq method (17).
The following primer sequences were used: Circ_POLA2
forward, 5′-ATGTCCGCATCCGCC-3′ and reverse 5′-TCAGATCCTGACGACC-3′;
GAPDH forward, 5′-GCACCGTCAAGCTGAGAAC-3′ and reverse
5′-GGTGAAGACGCCAGTGGA-3′; miR-31 forward,
5′-AGGCAAGAUGCUGGCAUAGCU-3′ and reverse
5′-AAAGGCAAGAUGCUGGCAUAG-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse
5′-ACGCTTCACGAATTTGCGTGTC-3′.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assays were performed using a kit from Abcam
(cat. no. ab228554). In a 96-well cell culture plate, 4,000 cells
in 0.1 ml fresh RPMI-1640 cell culture medium, supplemented with
10% FBS were added. A total of 3 wells were used for each
experiment. The cells were cultured at 37°C. Cell proliferation was
assessed by measuring the optical density at 450 nm at different
time points (0.5, 1 and 2 h). The CCK-8 (Abcam) solution was added
to each well to a final concentration of 10%, 2 h before the
samples were measured.
Statistical analysis
The data were analyzed using GraphPad Prism v6
(GraphPad Software, Inc.). A total of 3 replicates were included in
each experiment and the data in all figures are presented as the
mean ± SD, while the data in Table I
is presented as the median + IQR. Unpaired t-tests were used to
compare 2 groups, while comparisons among multiple groups were
performed using ANOVA followed by Tukey's post hoc test.
Correlations were analyzed by linear regression. The mRNA
expression levels of circ_POLA2 and miR-31 in EC and paired
non-tumor tissues were compared using paired t-tests. The
association between the relative mRNA expression level of miR-31
and the clinicopathological data was analyzed using either a Mann
Whitney U test or Kruskall-Wallis test for 2 or >3 groups,
respectively. P<0.05 was considered to indicate a statistically
significant difference.
Results
circ_POLA2 and miR-31 expression level
in EC tissues is altered
EC and paired adjacent normal tissue specimens were
collected from 60 patients with EC. RT-qPCR was then performed to
determine the mRNA expression levels of circ_POLA2 and miR-31.
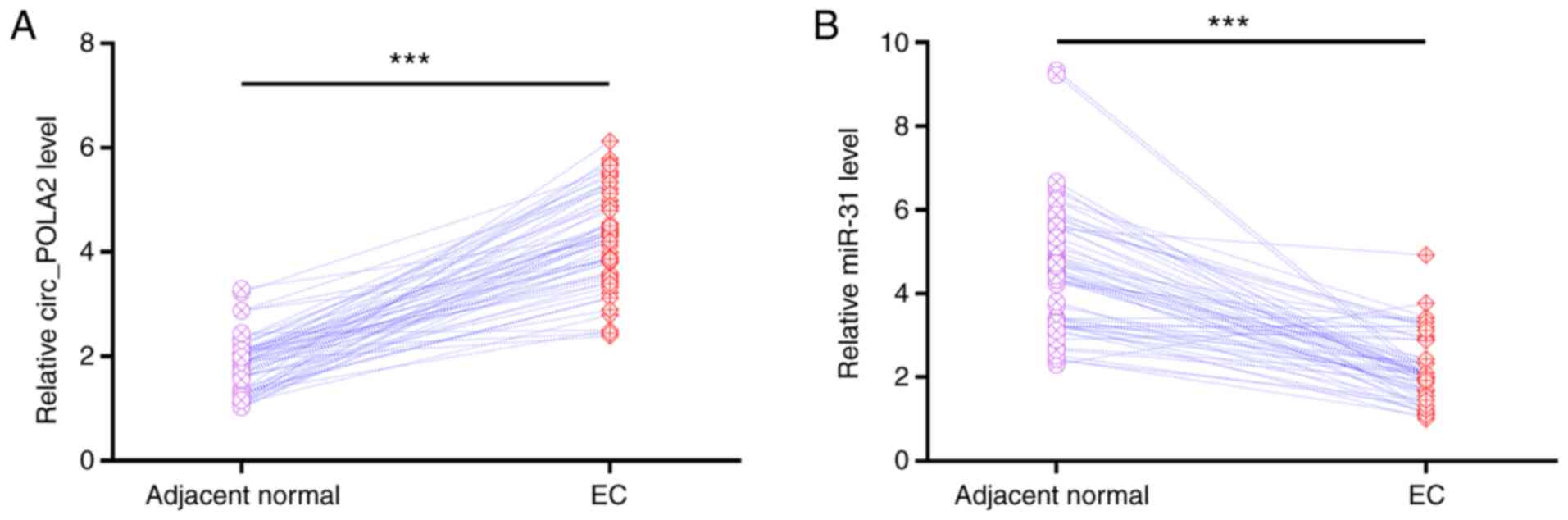
Compared with that in the adjacent normal tissues, circ_POLA2 was
significantly upregulated in EC tissues (Fig. 1A). In addition, miR-31 was
significantly downregulated in EC tissues compared with that in
adjacent normal tissues (Fig. 1B).
Therefore, circ_POLA2 and miR-31 may serve roles in EC.
Furthermore, the association between the mRNA expression levels of
miR-31 and certain clinicopathological parameters were also
analyzed. The median tumor/normal tissue miR-31 expression ratio in
EC was 0.034 in patients with a depth of myometrial invasion
(18) of ≤1/2 and 0.004 in patients
with tumor invasion >1/2. The results also showed that the
miR-31 expression level was associated with the clinical stage. The
median tumor/normal tissue miR-31 expression ratio was 0.123 in 16
patients with stage I cancer and 0.004 in 34 patients with stage
III/IV cancer. These results indicated an association between lower
miR-31 mRNA expression levels and increasing stage and depth of
tumor invasion. In addition, the median tumor/normal tissue miR-31
expression ratio was 0.120 in 30 patients with G1 and G2
histopathological grade and 0.116 in 20 patients with G3
histopathological grade. The median tumor/normal tissue miR-31
expression ratio was 0.178 in 2 patients with distant metastasis
and 0.025 in 58 patients without distant metastasis. These results
indicated an association between lower miR-31 mRNA expression
levels and increasing histopathological grade and no distant
metastasis. miR-31 expression was not associated with other
clinicopathological factors, such as patient age and lymphatic
metastasis (Table I).
circ_POLA2 and miR-31 expression
levels are correlated
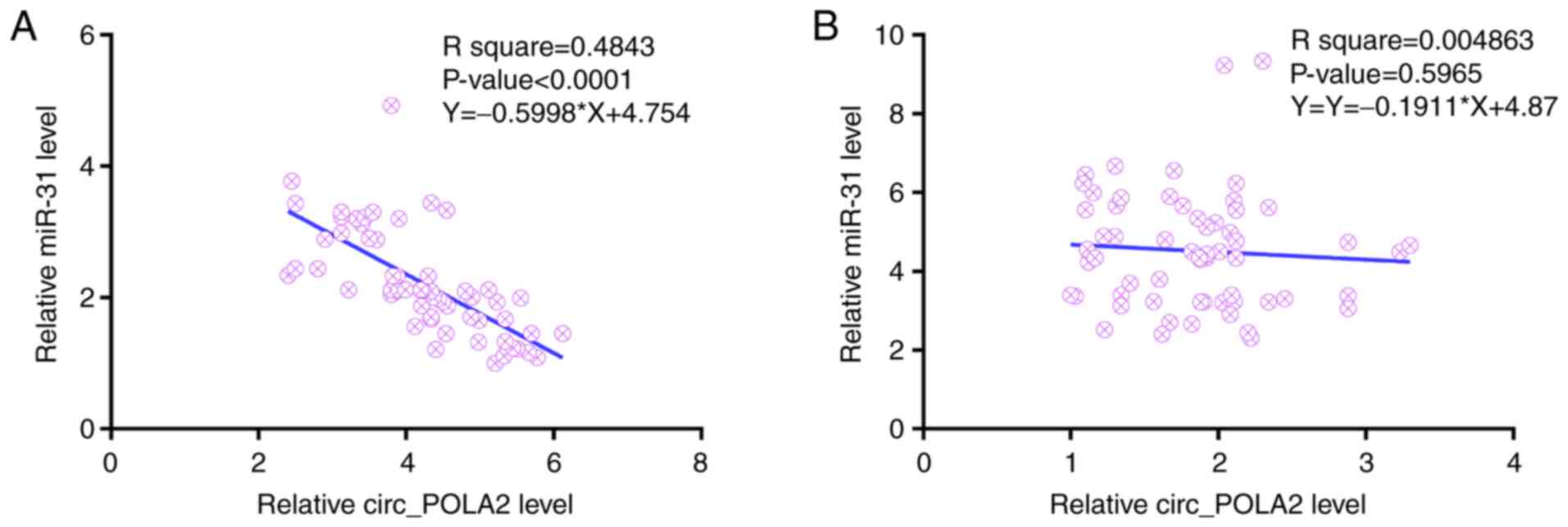
Linear regression was performed to determine the
correlation between circ_POLA2 and miR-31 expression levels in both
EC (Fig. 2A) and adjacent normal
(Fig. 2B) tissues. In EC tissue,
circ_POLA2 mRNA expression level was inversely correlated with
miR-31 mRNA expression level. However, their expression levels were
not correlated in adjacent normal tissues. Therefore, circ_POLA2
and miR-31 may interact in EC.
circ_POLA2 overexpression decreases
miR-31 expression levels in EC cell lines
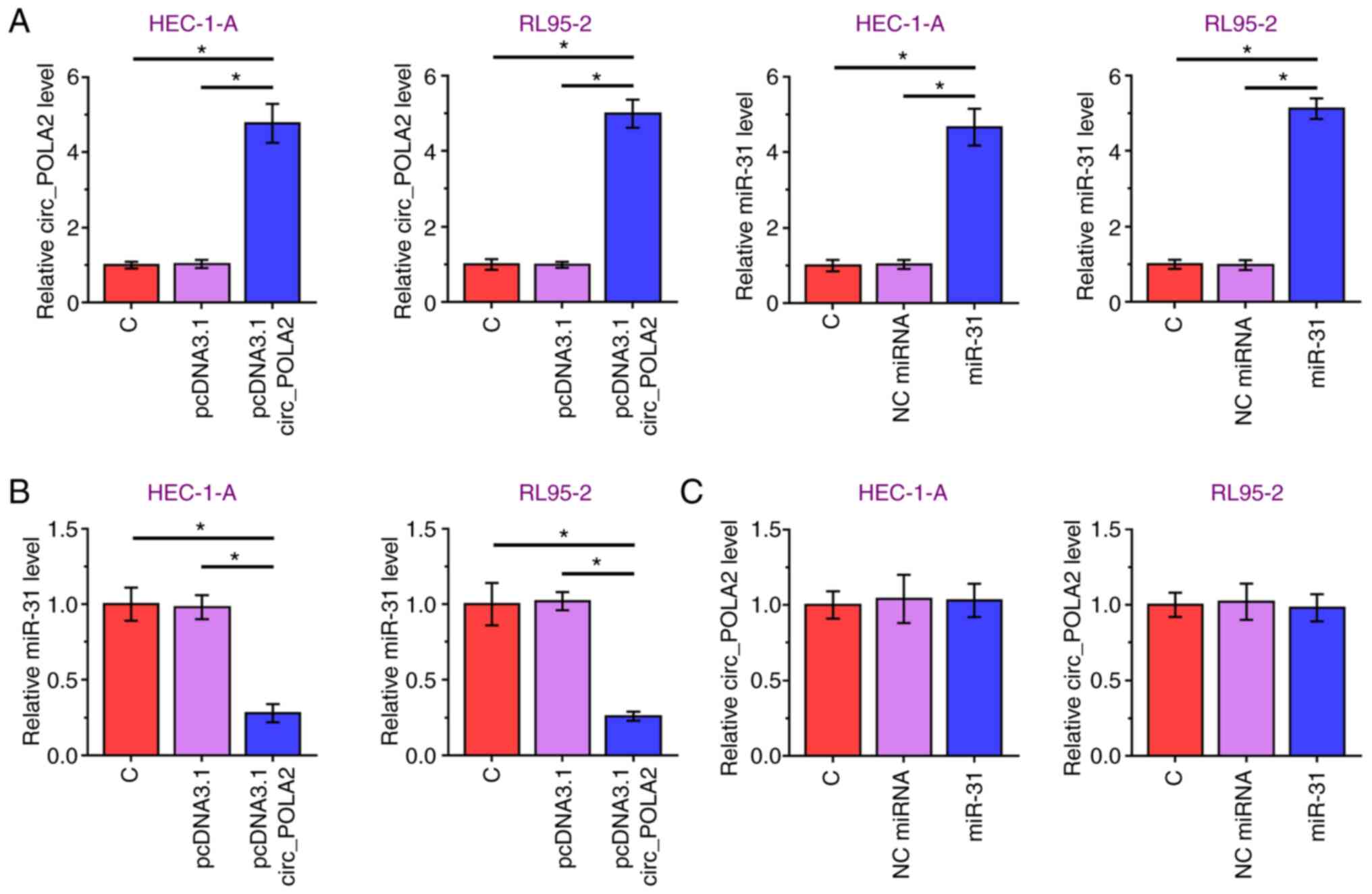
circ_POLA2 and miR-31 were overexpressed in the
HEC-1-A and RL95-2 cell lines (Fig.
3A). Overexpression of circ-POLA2 significantly decreased
miR-31 mRNA expression level, compared with that in the empty
vector group (Fig. 3B). However,
miR-31 overexpression did not affect circ_POLA2 mRNA expression
level, compared with that in the NC miRNA group (Fig. 3C). Therefore, circ_POLA2 may
downregulate miR-31 expression level in the EC cell lines
analyzed.
circ_POLA2 overexpression increases
miR-31 methylation in the EC cell lines
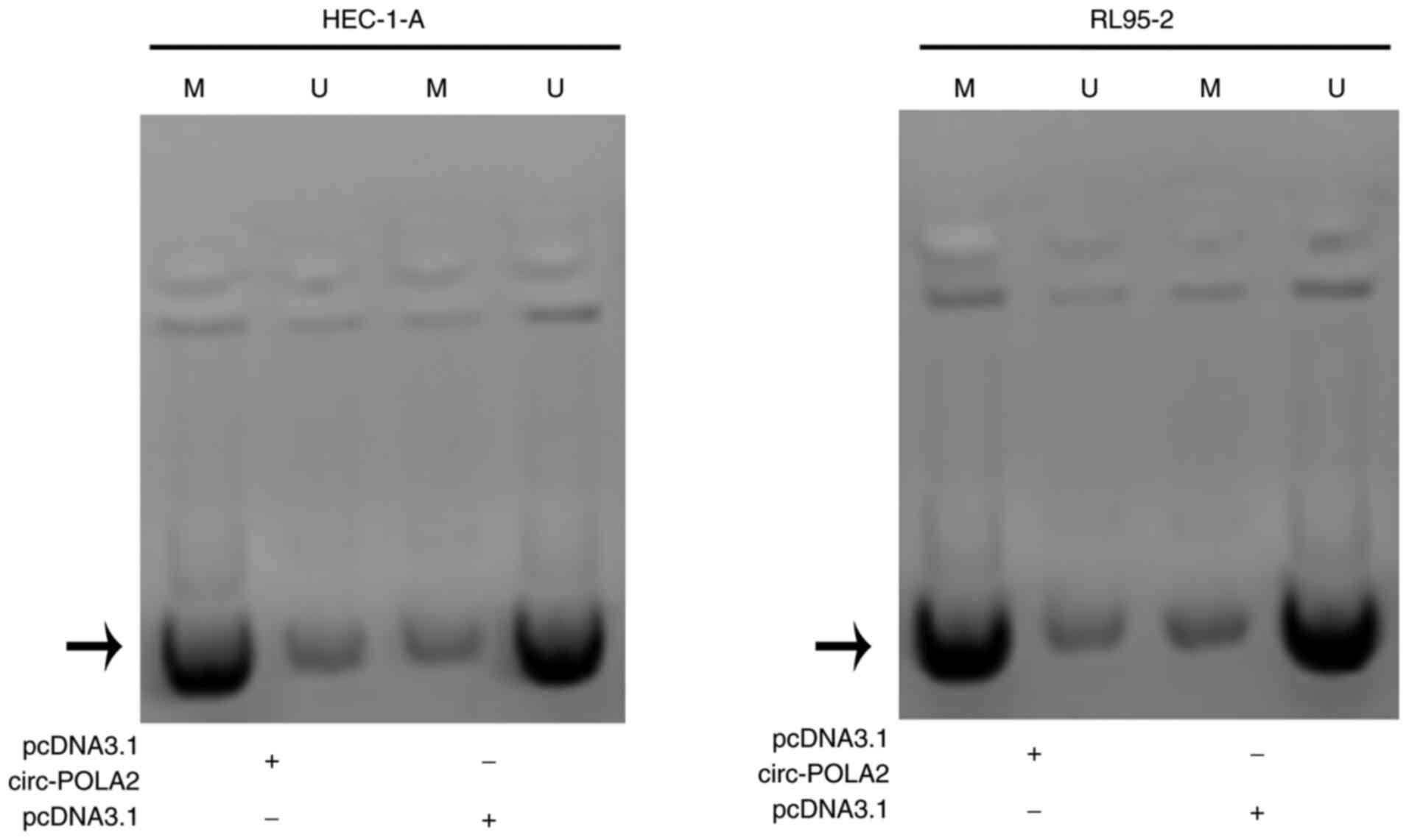
To investigate the mechanism underlying the
interaction between circ_POLA2 and miR-31, MSP was performed. The
results showed increased methylation of the miR-31 gene in cells
transfected with the circ_POLA2 expression vector (Fig. 4). Therefore, circ_POLA2 may
downregulate miR-31 via methylation.
circ_POLA2 overexpression increases
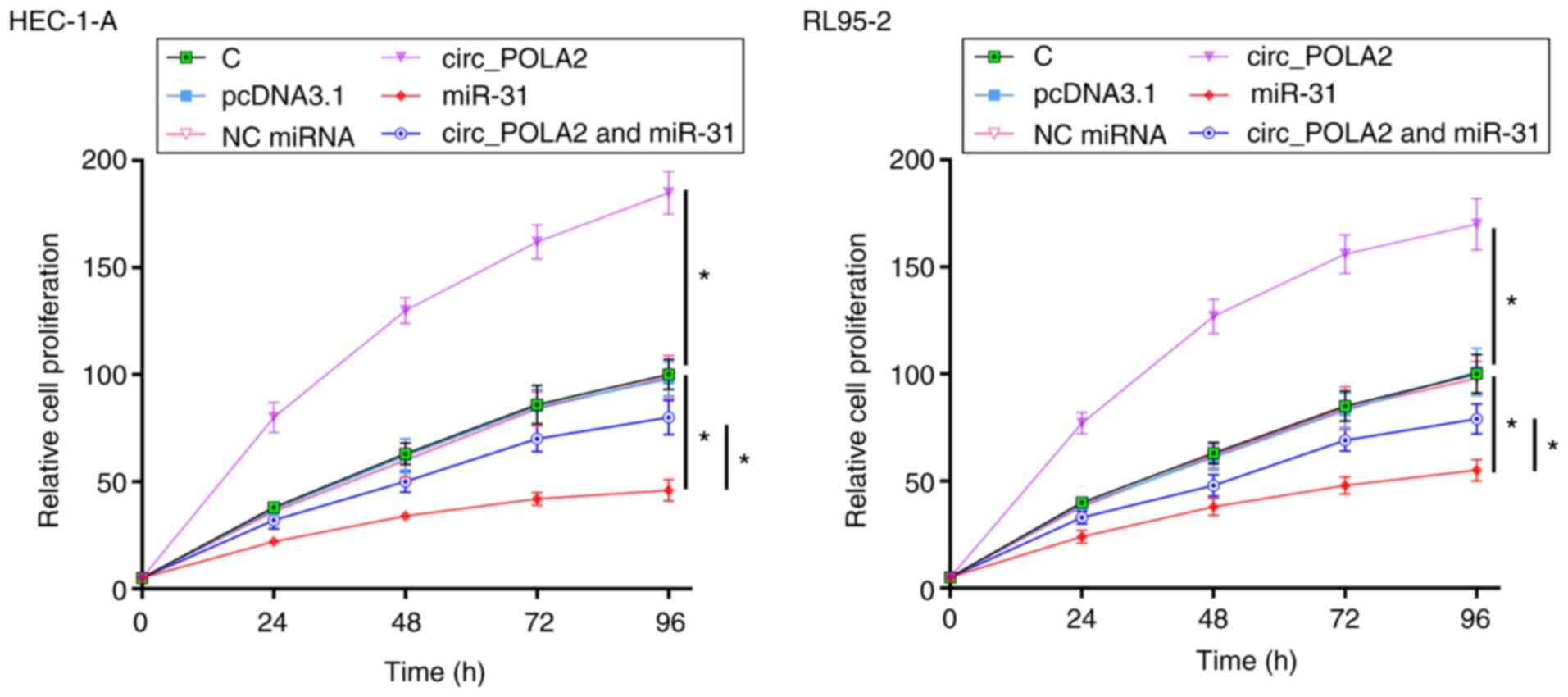
cell proliferation via miR-31
The effects of circ_POLA2 and miR-31 overexpression
on the proliferation of the EC cell lines were evaluated using
CCK-8 assays. circ_POLA2 overexpression increased EC cell
proliferation, whereas miR-31 overexpression decreased cell
proliferation compared with that in the empty vector group and NC
miRNA, respectively. In addition, circ_POLA2 overexpression
suppressed the effect of miR-31, compared with that in the miR-31
overexpression group (Fig. 5).
Discussion
The present study investigated the interaction
between the expression levels of circ_POLA2 and miR-31 in EC. It
was found that circ_POLA2 mRNA expression level was increased in EC
tissues, and following further analysis using miR-31 mimics and
circ_POLA2 overexpression in 2 EC cell lines, circ_POLA2 may
suppress miR-31 expression via methylation to promote EC cell
proliferation.
There have been two recent studies identifying
circ_POLA2 as an oncogenic circRNA in lung and cervical cancers
(13,14). circ_POLA2 was upregulated in lung
cancer and it may interact with the miR-326/GNB1 axis to increase
cancer cell stemness (13).
Moreover, circ_POLA2 upregulation in cervical cancer, as well as
its interaction with the miR-326/GNB1 axis to promote cancer
progression (14). To the best of
our knowledge, the present study is the first to report circ_POLA2
upregulation in EC tissues. In addition, circ_POLA2 overexpression
increased proliferation in the two EC cell lines. Therefore,
circ_POLA2 may serve an oncogenic role in EC by promoting cancer
cell proliferation.
miR-31 serves different roles in different types of
cancer (15,19). miR-31 was downregulated in gastric
cancer, in which its low expression levels predicted poor patient
overall survival time, suggesting its role as a tumor suppressor
(15). By contrast, miR-31 was
upregulated in cervical cancer and promoted cancer cell invasion,
migration and proliferation (19).
Mitamura et al (20)
previously reported miR-31 upregulation in EC cell lines, in which
it suppressed the Hippo tumor suppressor pathway to promote cancer
development. By contrast, the present study observed miR-31
downregulation in EC tissues and its inhibitory effects on EC cell
proliferation following increased expression in EC cell lines,
suggesting a tumor-suppressing role. These contradictory
observations may be explained by the fact that in the study by
Mitamura et al (20) miR-31
mRNA expression level was only analyzed in EC cell lines, while in
the present study the expression of miR-31 was analyzed in patients
with EC. Additional studies are required to further elucidate the
function of miR-31 in EC.
In addition to regulating gene transcription and
translation, circRNAs may also act as miRNA sponges to exert
functions in cancer biology (11,12). In
lung and cervical cancers, circ_POLA2 sponged miR-326 to promote
cancer development (13,14). The results of the present study
indicated that circ_POLA2 downregulated miR-31 mRNA expression
level in the EC cell lines via methylation. These results improved
understanding of the interactions between circ_POLA2 and miRNAs.
However, the underlying mechanisms remain unclear. It was found
that circ_POLA2 and miR-31 were not correlated in adjacent normal
tissues, suggesting an indirect interaction between them. Future
studies will utilize new experimental approaches, for example,
using circRNA sequencing or microarrays to investigate the function
and mechanism of circ-POLA and miRNAs in EC, and its expression
distribution in EC cell lines (21).
In conclusion, the present study demonstrated that
circ_POLA2 was upregulated in EC tissues and may downregulate
miR-31 via methylation to promote cancer cell proliferation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XF and XZ designed the study. XF and JW performed
the experiments. LC analyzed the data. XF wrote the manuscript. All
authors read and approved the final version of manuscript. XF, XZ,
JW and XF confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Beilun District People's Hospital and written informed
consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCK-8
|
Cell Counting Kit-8
|
|
circRNA
|
circular RNA
|
|
Cq
|
threshold cycle
|
|
EC
|
endometrial cancer
|
|
MSP
|
methylation-specific PCR
|
|
NC
|
negative control
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
References
|
1
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duong LM, Wilson RJ, Ajani UA, Singh SD
and Eheman CR: Trends in endometrial cancer incidence rates in the
United States, 1999–2006. J Womens Health (Larchmt). 20:1157–1163.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Donkers H, Bekkers R, Massuger L and
Galaal K: Systematic review on socioeconomic deprivation and
survival in endometrial cancer. Cancer Causes Control.
30:1013–1022. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sanni OB, Mc Menamin ÚC, Cardwell CR,
Sharp L, Murray LJ and Coleman HG: Commonly used medications and
endometrial cancer survival: A population-based cohort study. Br J
Cancer. 117:432–438. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morrison J: Endometrial cancer follow up;
finding the confidence to let go? BJOG. 125:17152018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Makker V, Green AK, Wenham RM, Mutch D,
Davidson B and Miller DS: New therapies for advanced, recurrent,
and metastatic endometrial cancers. Gynecol Oncol Res Pract.
4:192017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Busch EL, Crous-Bou M, Prescott J, Chen
MM, Downing MJ, Rosner BA, Mutter GL and De Vivo I: Endometrial
cancer risk factors, hormone receptors, and mortality prediction.
Cancer Epidemiol Biomarkers Prev. 26:727–735. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitamura T, Dong P, Ihira K, Kudo M and
Watari H: Molecular-targeted therapies and precision medicine for
endometrial cancer. Jpn J Clin Oncol. 49:108–120. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Remmerie M and Janssens V: PP2A: A
promising biomarker and therapeutic target in endometrial cancer.
Front Oncol. 9:4622019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kunitomi H, Kobayashi Y, Wu RC, Takeda T,
Tominaga E, Banno K and Aoki D: LAMC1 is a prognostic factor and a
potential therapeutic target in endometrial cancer. J Gynecol
Oncol. 31:e112020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shang Q, Yang Z, Jia R and Ge S: The novel
roles of circRNAs in human cancer. Mol Cancer. 18:62019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fan Z, Bai Y, Zhang Q and Qian P: CircRNA
circ_POLA2 promotes lung cancer cell stemness via regulating the
miR-326/GNB1 axis. Environ Toxicol. 35:1146–1156. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cao Y, Li J, Jia Y, Zhang R and Shi H:
CircRNA circ_POLA2 promotes cervical squamous cell carcinoma
progression via regulating miR-326/GNB1. Front Oncol. 10:9592020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Guo J, Li D, Xiao B, Miao Y,
Jiang Z and Zhuo H: Down-regulation of miR-31 expression in gastric
cancer tissues and its clinical significance. Med Oncol.
27:685–689. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Raffone A, Travaglino A, Mascolo M,
Carotenuto C, Guida M, Mollo A, Insabato L and Zullo F:
Histopathological characterization of ProMisE molecular groups of
endometrial cancer. Gynecol Oncol. 157:252–259. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takeuchi M, Matsuzaki K and Harada M:
Evaluating myometrial invasion in endometrial cancer: Comparison of
reduced field-of-view diffusion-weighted imaging and dynamic
contrast-enhanced MR imaging. Magn Reson Med Sci. 17:28–34. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng W, Liu Z, Zhang W and Hu X: MiR-31
functions as an oncogene in cervical cancer. Arch Gynecol Obstet.
292:1083–1089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mitamura T, Watari H, Wang L, Kanno H,
Kitagawa M, Hassan MK, Kimura T, Tanino M, Nishihara H, Tanaka S
and Sakuragi N: MicroRNA 31 functions as an endometrial cancer
oncogene by suppressing Hippo tumor suppressor pathway. Mol Cancer.
13:972014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ye F, Tang QL, Ma F, Cai L, Chen M, Ran
XX, Wang XY and Jiang XF: Analysis of the circular RNA
transcriptome in the grade 3 endometrial cancer. Cancer Manag Res.
11:6215–6227. 2019. View Article : Google Scholar : PubMed/NCBI
|