Introduction
Colorectal cancer (CRC) is the third most frequently
occurring cancer type worldwide, and with a high mortality rate,
accounted for ~930,000 deaths in 2020 (1). Surgery remains the principal CRC
treatment method to achieve complete resection of the primary tumor
and metastatic lesions (2). However,
in a large number of cases, complete resection is difficult. As
such, minimizing tumor size and inhibiting further growth and
proliferation are the primary aims for patients whose tumors cannot
be completely removed, or those who are unable to tolerate surgery,
for which chemotherapy and radiotherapy are the key treatment
options (3). In addition, adjuvant
chemotherapy has been used to extend the lifespan of patients with
CRC (4).
5-Fluorouracil (5-FU) is a thymidylate synthase
inhibitor that prevents the methylation of deoxyuridine acid to
deoxythymidine acid, and with notable anticancer properties, was
one of the first therapeutic drugs developed for clinical cancer
treatment (5,6). Since 5-FU is widely utilized as a
first-line treatment, the incidence of CRC resistance to 5-FU is
gradually increasing (5). Therefore,
strategies for enhancing the chemosensitivity of CRC cells to 5-FU
are urgently required.
N-myc downstream-regulated gene 4 (NDRG4) belongs to
the NDRG family, the members of which are expressed in a variety of
human organs, and are associated with a wide range of biological
processes, such as organ development, tumor inhibition,
angiogenesis and growth regulation (7). NDRG4 plays a tumor-suppressive role in
various cancer types, including pancreatic ductal and esophageal
adenocarcinoma (8,9). In addition, hypermethylation of the
NDRG4 promoter is associated with gastric cancer tumorigenesis, and
is a predictor of poor prognosis in patients with the disease
(10). NDRG4 also plays an important
role in CRC, as molecular analysis of the NDRG4 promoter region in
stool samples can be used to screen for CRC (11).
The DNA damage inducible transcript 3 (DDIT3) gene
encodes a member of the CCAAT/enhancer binding protein
transcription factor family. DDIT3 is primarily involved in
apoptosis associated with endoplasmic reticulum (ER) stress, as it
enhances the biological function of the BH3-only protein BCL2
interacting mediator of cell death and inhibits the antiapoptotic
function of BCL2 (12). In addition,
DDIT3 can inhibit CRC by promoting the cell apoptosis (13). In terms of chemotherapeutic
resistance, low expression levels of DDIT3 have been associated
with the chemoresistance of lung cancer cells to cisplatin
(14). The aim of the present study
was to investigate the tumor-suppressive effect of NDRG4 in the
SW480 and SW620 CRC cell lines, as well as whether NDRG4 enhanced
the sensitivity of CRC cells to 5-FU, and the associated molecular
mechanism. The results of the present study may indicate a novel
mechanism that reflects the important role of NDRG4 in CRC
inhibition.
Materials and methods
Cell lines
The SW480 and SW620 cell lines were purchased from
Procell Life Science & Technology Co., Ltd., (cat. no. CL-0223
and CL-0225, respectively) and cultured in Leibovitz's L15 medium
(Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.) at 37°C (100%
air). The 293T cell line was purchased from Procell Life Science
& Technology Co., Ltd. (cat. no. CL-0005) and cultured in DMEM
(Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. All
cell lines were authenticated by STR authentication.
Stable transfection
The NDRG4 lentivirus and its lentiviral vector
GV358, were purchased from Shanghai GeneChem Co., Ltd. SW480 and
SW620 cells were infected with lentivirus at a multiplicity of
infection (MOI) of 10 and 80 for 24 h, respectively. And the stably
transfected cells were selected with 3 µg/ml puromycin (Beyotime
Institute of Biotechnology) for 2 weeks. In subsequent experiments,
puromycin was maintained at 0.5 µg/ml. Short hairpin (sh)RNAs
targeting the DDIT3 gene (shRNA-1,
5′-GATCCCTGCACCAAGCATGAACAATTCTCGAGAATTGTTCATGCTTGGTGCAGTTTTTG-3′;
shRNA-2,
5′-GATCCTGAACGGCTCAAGCAGGAAATCTCGAGATTTCCTGCTTGAGCCGTTCATTTTTG-3′),
and non-targeting shRNA-negative control (shRNA-NC
5′-GATCCCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTTG-3′)
were cloned into the lentiviral vector PLVshRNA-EGFP(2A)Puro by
Inovogen Biotechnology Pvt. Ltd. According to the manufacturer's
protocol, 4 µg lentiviral shRNA plasmids were mixed with packaging
vector (PAX2 plasmid) and envelope vector (PMD2G plasmid) at the
mass ratio of 4:3:1, and subsequently transfected into 293T cells
using 20 µl Lipofectamine 2000 Transfection Reagent (Thermo Fisher
Scientific, Inc.). Following incubation at 37°C with serum-free
DMEM for 12 h, the 293T cells were changed to be cultured in
complete DMEM at 37°C for 48 h. The lentivirus particles were
subsequently collected and purified from the cell supernatants
through a 0.45 µm filter, and meanwhile, the titer of the
lentivirus was determined using the qPCR Lentivirus Titer kit
(Applied Biological Materials, Inc.). Then SW480 cells in the
logarithmic growth phase were added to the lentivirus suspension
(MOI=10) and incubated at 37°C for 48 h, after which the medium was
discarded and the SW480 cells were cultivated with screening
Leibovitz's L15 medium containing 3 µg/ml puromycin for 2 weeks to
select the positive infected cells. In subsequent experiments, they
were maintained with 0.5 µg/ml puromycin.
MTT assay
Cells were seeded into a 96-well plate with five
biological replicates per group. Before detection, 20 µl MTT
solution (Beyotime Institute of Biotechnology) was added to each
well. The medium was replaced with 150 µl DMSO (Amresco, LLC) after
4 h at 37°C, and the plates were then shaken for 10 min. The OD
values were determined at 570 nm using a microplate reader. To
investigate the effect of NDRG4 on the proliferative ability of CRC
cell lines, activity was detected at 0, 1, 2, 3 and 4 days of cell
culture. To determine the effects of 5-FU on cell viability,
different concentrations of 5-FU (5, 10, 20, 40 and 50 µg/ml) were
added to cells in the logarithmic growth phase, and the absorbance
was measured 48 h after treatment.
Western blotting
Cells were lysed using RIPA lysis buffer (Beyotime
Institute of Biotechnology), and the protein concentration was
determined using a BCA protein assay kit (Thermo Fisher Scientific,
Inc.). A total of 40 µg protein per lane was separated by 10% or
15% SDS-PAGE and transferred onto PVDF membranes. The PVDF membrane
was then blocked with TBST (Tween-20 at 0.1%) containing 5% skim
milk at room temperature for 1.5 h. The following primary
antibodies were diluted 1:1,000 in Primary Antibody Dilution Buffer
(Beyotime Institute of Biotechnology): anti-NDRG4 (monoclonal,
rabbit anti-human; cat. no. 9039; Cell Signaling Technology, Inc.),
anti-DDIT3 (monoclonal, mouse anti-human; cat. no. 2895; Cell
Signaling Technology, Inc.), anti-p-AKT (monoclonal, rabbit
anti-human; cat. no. 4060; Cell Signaling Technology, Inc.),
anti-AKT (monoclonal, rabbit anti-human; cat. no. 4685; Cell
Signaling Technology, Inc.), anti-p-ERK (polyclonal, rabbit
anti-human; cat. no. ab4819; Abcam), anti-ERK (polyclonal, rabbit
anti-human; cat. no. ab17942; Abcam), anti-cleaved caspase-3
(polyclonal, rabbit anti-human; cat. no. 9661; Cell Signaling
Technology, Inc.), anti-PARP (monoclonal, rabbit anti-human; cat.
no. 9532; Cell Signaling Technology, Inc.), and anti-β-actin
(monoclonal, mouse anti-human; cat. no. D191047; Sangon Biotech,
Co., Ltd.). After incubated by the primary antibody overnight at
4°C, the membrane was washed with TBST (Tween-20 at 0.1%) at room
temperature three times for 10 min each. Appropriate secondary
antibodies derived from the same species as the primary antibodies
(anti-rabbit IgG, HRP-linked; cat. no. 7074; Cell Signaling
Technology, Inc.; and anti-mouse IgG, HRP-linked; cat. no. 7076;
Cell Signaling Technology, Inc.) were diluted at 1:5,000 and added
to the membranes, incubated at room temperature for 1 h. After
that, the membrane was washed with TBST three times for 10 min
each, and then FDbio-dura ECL Kit (Hangzhou Fude Biological
Technology Co., Ltd.) was added for visualization. The blots were
detected using the Tanon 550 Imaging System (Tanon Science and
Technology Co., Ltd.).
Reverse transcription-quantitative
(RT-q) PCR and PCR array
The RNeasy Mini kit (Qiagen, Inc.) was used to
extract total cellular RNA, from which cDNA was then synthesized
using the PrimeScript RT-PCR kit (Takara Bio, Inc.) according to
the manufacturers' protocols. qPCR was performed using qPCR
SYBR-Green Master Mix (Shanghai Yeasen Biotechnology Co., Ltd.) per
the manufacturer's protocol. The thermocycling conditions were as
follows: Pre-denaturation at 95°C for 5 min, then 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec and
extension at 72°C for 20 sec. Relative mRNA expression levels were
determined using the 2−∆∆Cq method (15). The primer sequences were as follows:
DDIT3 forward, 5′-GGAAACAGAGTGGTCATTCCC-3′ and reverse,
5′-CTGCTTGAGCCGTTCATTCTC-3′; CASP7 forward,
5′-AGTGACAGGTATGGGCGTTC-3′ and reverse,
5′-CGGCATTTGTATGGTCCTCTT-3′; and β-actin forward,
5′-CCTGGGCATGGAGTCCTGTG-3′ and reverse, 5′-TCTTCATTGTGCTGGGTGCC-3′.
For PCR array analysis, the extracted cDNA was used for with the
real-time RT2 Profiler PCR Array (Qiagen, Inc.)
according to the manufacturer's protocol.
TdT-UTP nick end labeling (TUNEL)
assay
TUNEL assays were performed using the One Step TUNEL
Apoptosis Assay Kit (Beyotime Institute of Biotechnology) according
to the manufacturer's protocol. TUNEL regent was added to cells
after 48 h of treatment with PBS or 5-FU. Each sample was observed
by microscopy in five visual fields. Images of the cells were
acquired using a fluorescence microscope.
Flow cytometry
Apoptosis assays were performed with cell lines
using an Annexin V-FITC/PI apoptosis detection kit (Shanghai Yeasen
Biotechnology Co., Ltd.) according to the manufacturer's protocol.
The experiments were conducted 48 h after treatment with PBS or
5-FU. Flow cytometry was performed using a FC500 Flow Cytometer
(Beckman Coulter Co., Ltd.), and the data were analyzed using
FlowJo 10 software (FlowJo LLC).
Colony formation assay
Single cells (~200 per dish) were seeded into cell
culture dishes with a diameter of 6 cm. After 18 days of culture at
37°C, visible colonies (>50 cells per colony) had formed and the
culture was terminated. The medium was replaced every 3 days during
the culture period. The colonies were then washed, fixed with 4%
paraformaldehyde (Beyotime Institute of Biotechnology) at room
temperature for 15 min and stained with crystal violet
(Sigma-Aldrich; Merck KGaA) at room temperature for 20 min.
Finally, images of the stained colonies were captured, and the
colonies were manually counted.
EdU staining
EdU staining was performed using the BeyoClick EdU
Cell Proliferation Kit with Alexa Fluor 594 (cat. no. C0078S;
Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Cells in the logarithmic growth phase
(~24–48 h in culture) were used for detection. Images of the cells
were captured using a fluorescence microscope.
Statistical analysis
All data are expressed as the mean ± SD. Statistical
analyses were performed using GraphPad Prism 8 (GraphPad Software,
Inc.). Student's t-test was used to analyze two independent groups.
For comparisons between multiple groups, ANOVA was applied; Sidak's
multiple comparisons test was used following two-way ANOVA, and
Dunnett's multiple comparisons test was used following one-way
ANOVA. P<0.05 was considered to indicate a statistically
significant difference.
Results
NDRG4 inhibits the proliferation of
CRC cells
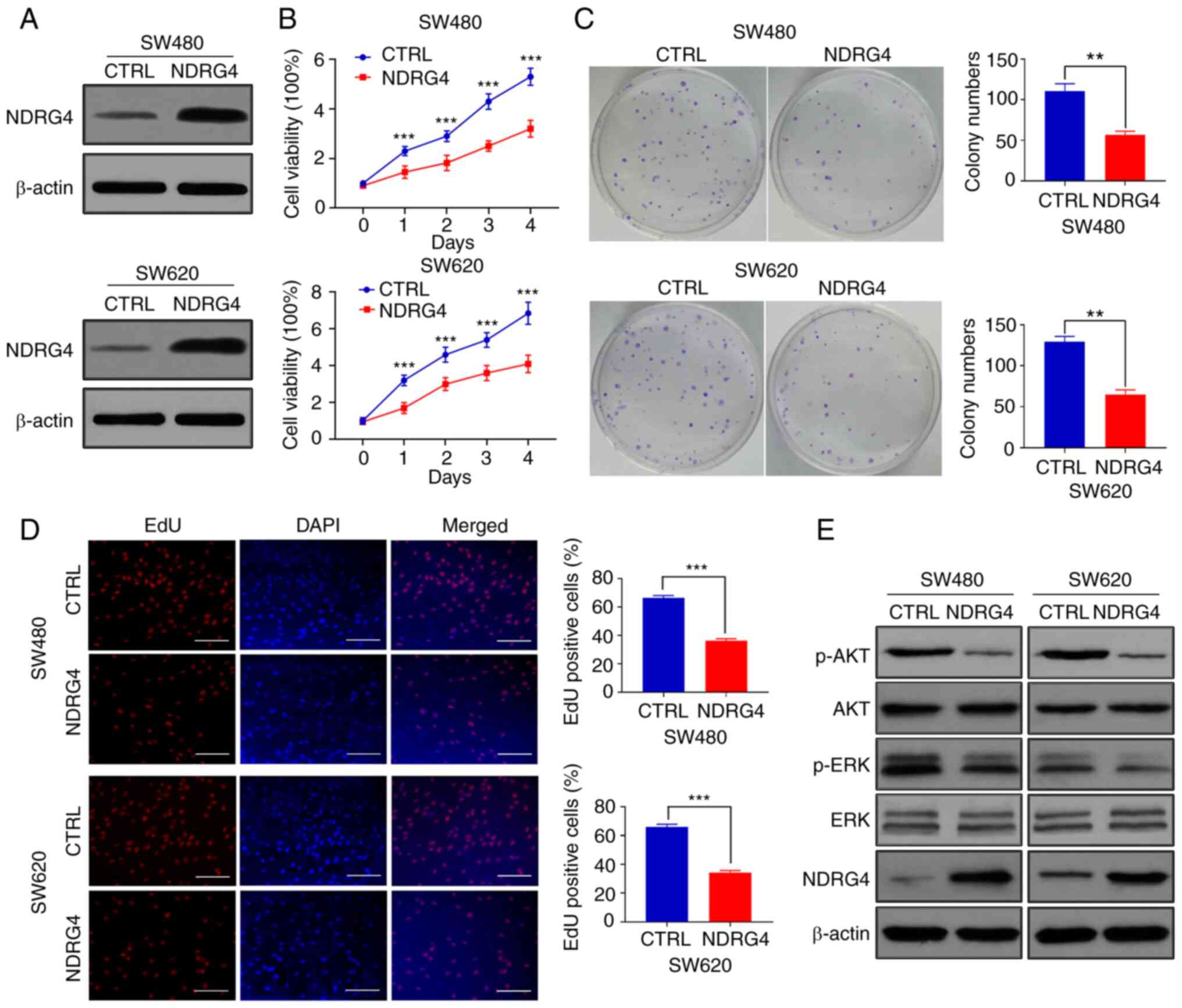
SW480 and SW620 CRC cells overexpressing NDRG4 were
successfully constructed through lentiviral infection. The NDRG4
protein levels of the overexpression cells were notably higher than
those of their control counterparts (Fig. 1A). The MTT assay results indicated
that the proliferative capacity of NDRG4-overexpressing cells was
significantly lower than that of the control cells on days 1–4 of
cell culture (Fig. 1B). The colony
formation assay revealed that the NDRG4-overexpressing cells formed
fewer visible clones than the control cells (Fig. 1C). Furthermore, EdU analysis indicted
that the proliferation of NDRG4-overexpressing cells was inhibited
relative to that of the control cells (Fig. 1D). These results indicate that NDRG4
inhibited CRC cell proliferation.
NDRG4 inhibits the activation of
PI3K/AKT and ERK signaling
Since both the PI3K/AKT and ERK signaling pathways
are associated with cellular proliferation (16), the levels of AKT and ERK
phosphorylation can be measured to reflect their degrees of
activation. Western blotting revealed decreased levels of p-AKT and
p-ERK in NDRG4-overexpressing cells (Fig. 1E), indicating that NDRG4 inhibited
the activation of the PI3K/AKT and ERK signaling pathways.
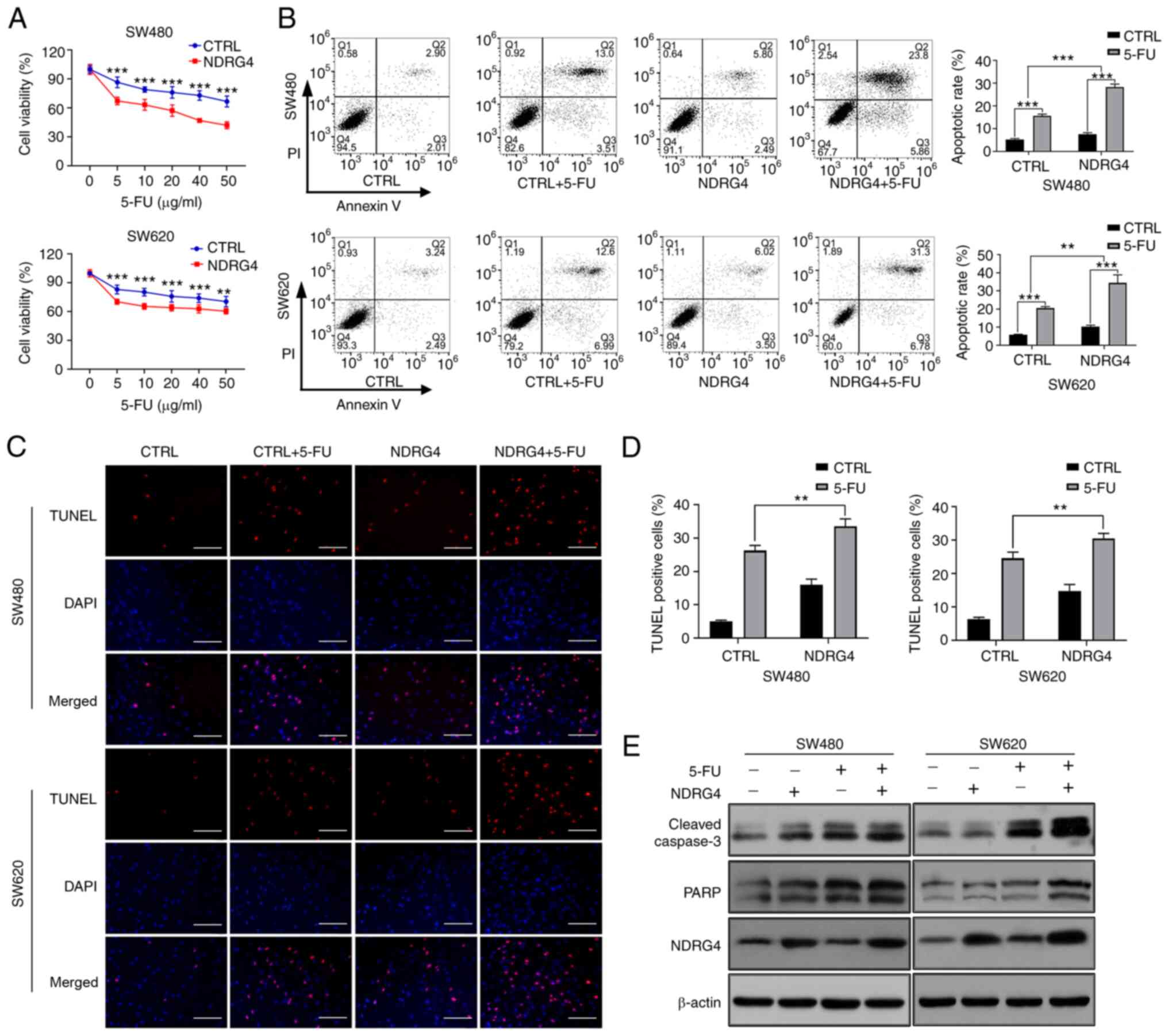
NDRG4 promotes 5-FU-induced CRC cell
apoptosis
MTT assays were used to assess cell viability after
48 h of culture with different concentrations of 5-FU. The
overexpression of NDRG4 significantly decreased the viability of
cells treated with 5-FU at five different concentrations (Fig. 2A). Moreover, the survival rates of
NDRG4-overexpressing SW480 and SW620 cells were decreased most
significantly following treatment with 40 and 10 µg/ml 5-FU,
respectively. Therefore, subsequent experiments were carried out
using these two concentrations of 5-FU. Flow cytometry was
performed to detect apoptosis, which showed that NDRG4
overexpression increased the rates of SW480 and SW620 apoptosis
induced by 5-FU, compared with those of the control cells;
measurements were based on the percentage of annexin-V-positive
cells (Q2 + Q3), and two-way ANOVA revealed that the interaction
between NDRG4 and 5-FU was statistically significant (Fig. 2B). Subsequently, a TUNEL apoptosis
assay was conducted, and the experimental results confirmed that
NDRG4 overexpression increased the apoptotic rate induced by 5-FU,
compared with that of control cells (Fig. 2C and D). Furthermore, expression of
the apoptosis-associated molecules cleaved caspase-3 (C-caspase-3)
and poly-ADP-ribose polymerase (PARP, the cleaved substrate of
caspase) was also examined. The expression level of C-caspase-3 was
increased in NDRG4-overexpressing cells compared with the control
cells, indicating an increase in cellular apoptosis. After adding
5-FU, the expression of C-caspase-3 was notably increased, and the
difference between NDRG4-overexpressing cells and control cells was
still apparent. Meanwhile, NDRG4 overexpression and 5-FU treatment
also increased PARP expression (Fig.
2E). These results suggest that NDRG4 promoted the apoptosis of
CRC cells induced by 5-FU.
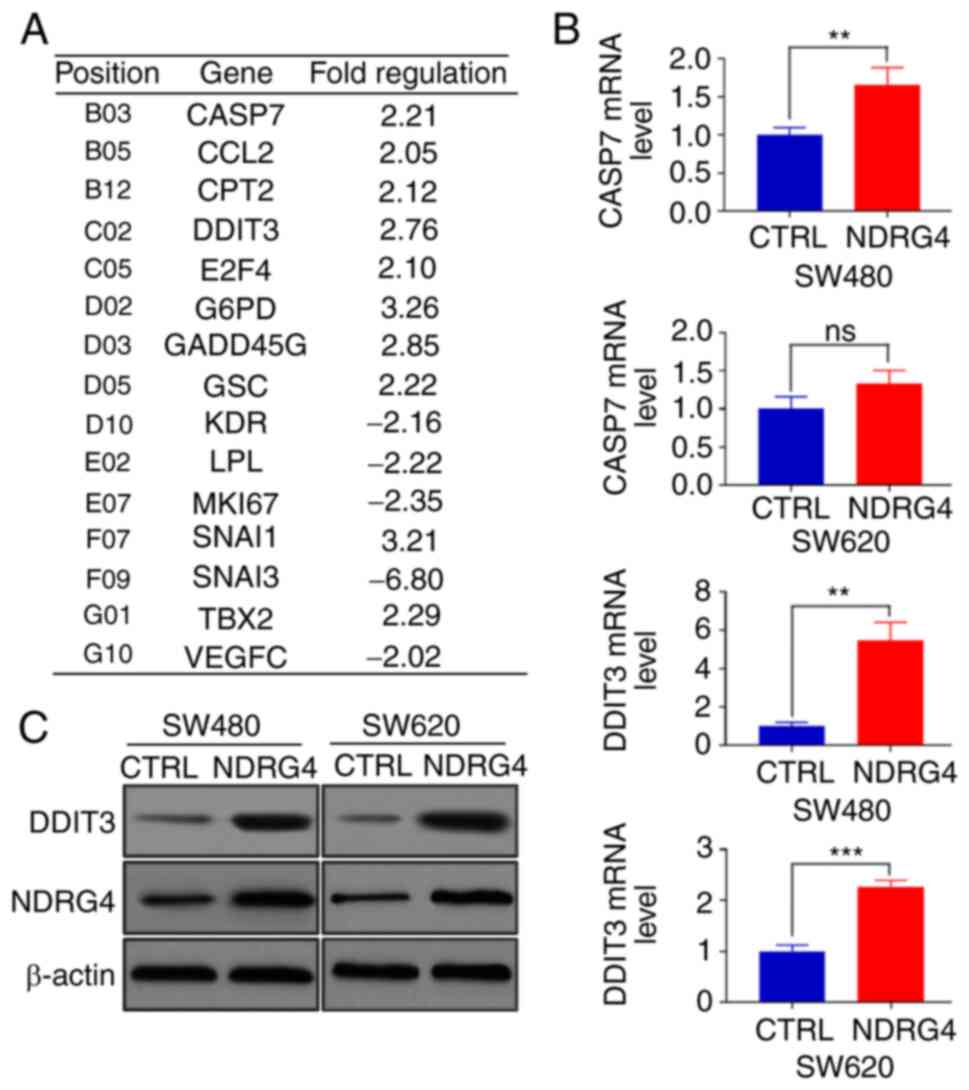
NDRG4 upregulates DDIT3
expression
To investigate the molecular mechanism by which
NDRG4 inhibits CRC cells, a PCR array experiment was conducted
using SW480 cells to identify genes with considerable fold changes
between NDRG4-overexpressing cells and control cells (Fig. 3A). Then, apoptosis-related genes,
such as CASP7 and DDIT3, were selected for qPCR verification. DDIT3
exhibited the greatest differential expression between
NDRG4-overexpressing cells and control cells (Fig. 3B). The western blot results also
confirmed that DDIT3 was expressed at higher levels in
NDRG4-overexpressing SW480 and SW620 cells than in their control
counterparts (Fig. 3C). These
results indicate that NDRG4 upregulated DDIT3 expression in CRC
cells.
Proapoptotic effect of NDRG4 under
5-FU treatment is dependent on DDIT3
To further confirm whether the promotional effect of
NDRG4 on 5-FU-induced CRC cell apoptosis was associated with the
increase in DDIT3 expression, two shRNAs were designed to target
the DDIT3 gene, and transfected into SW480 cells, which showed a
successful decrease in DDIT3 mRNA expression (Fig. S1), and then into
NDRG4-overexpressing SW480 cells to verify the gene silencing
effect (Fig. 4A); further
experiments were performed with shRNA-2, which exhibited the most
prominent gene-silencing effect. The expression levels of the
apoptosis-related proteins C-caspase-3 and PARP were decreased
following DDIT3-knockdown in SW480 NDRG4-overexpressing cells.
Silencing DDIT3 reduced the increase in C-caspase-3 and PARP
expression induced by 5-FU treatment (Fig. 4B). Subsequently, SW480
NDRG4-overexpressing cells were analyzed by flow cytometry,
revealing that DDIT3-knockdown resulted in a decreased apoptotic
rate compared with that of control cells, and that the decreasing
trend was more apparent after the addition of 5-FU (Fig. 4C and D). These results indicate that
DDIT3 plays an important role in the NDRG4-mediated promotion of
CRC cell apoptosis induced by 5-FU.
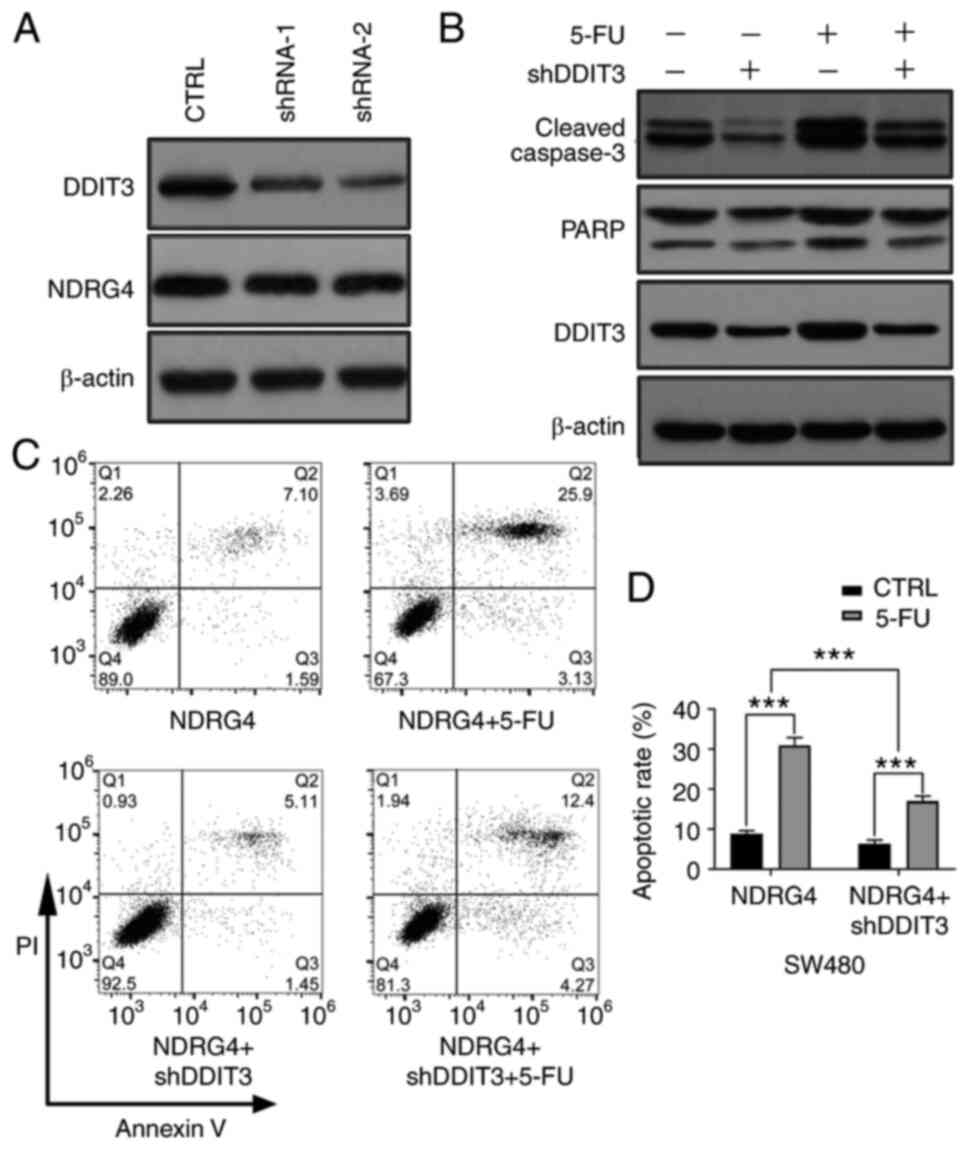 | Figure 4.Proapoptotic effect of NDRG4 under
5-FU treatment is dependent on DDIT3. (A) Western blot analysis of
NDRG4 and DDIT3 in SW480 NDRG4-overexpressing cells treated with or
without DDIT3 shRNA (n=3). (B) Apoptosis-associated protein
expression in SW480 NDRG4-overexpressing cells with or without
DDIT3-knockdown, and with or without 5-FU as determined by western
blotting. Multiple C-caspase3 bands represent the large fragment
(17/19 kDa) of activated caspase-3 resulting from cleavage adjacent
to Asp175. The upper band of PARP represents full-length PARP-1,
and the lower band represents the large fragment produced by
caspase cleavage at Asp214 (n=3). (C) Representative flow
cytometric images of SW480 NDRG4-overexpressing cells with or
without DDIT3-knockdown and with or without 5-FU treatment (n=3).
(D) Statistical analysis of the apoptotic rate of SW480
NDRG4-overexpressing cells with or without DDIT3-knockdown, and
with or without 5-FU. Statistical analysis by two-way ANOVA.
***P<0.001. NDRG4, N-myc downstream-regulated gene 4; 5-FU,
5-fluorouracil; DDIT3, DNA damage inducible transcript 3;
C-caspase, cleaved caspase; CTRL, control; PARP, poly-ADP-ribose
polymerase; C-, cleaved; sh, short hairpin (RNA). |
Discussion
The incidence and morbidity rates of CRC are high
worldwide (1), and surgery remains
the primary and most effective treatment type (17). In addition, adjuvant chemotherapy has
been widely utilized to improve the survival of patients with both
International Union Against Cancer (UICC) stage III CRC and
high-risk UICC stage II CRC (18).
Despite progress in cancer treatment, with the implementation of
novel chemotherapeutic agents such as aflibercept, ramucirumab and
bevacizumab, 5-FU remains one of the most effective and commonly
used therapeutic drugs for CRC (19). Chemoresistance to anticancer agents
is a major obstacle to attaining anticancer therapies with
sufficient benefits (5), and the
resistance of CRC to 5-FU is becoming increasingly prevalent.
Chemoresistance to 5-FU may be due to the disruption
of 5-FU metabolic enzymes, drug transporters or crucial cellular
activities, such as apoptosis and the cell cycle (20). For instance, Uppada et al
(21) revealed that MASTL induces
chemoresistance in colon cancer by promoting Wnt/β-catenin
signaling. Correspondingly, 5-FU sensitivity is influenced by a
variety of genes and chemical substances, and CDGSH iron-sulfur
domain-containing protein 2 reportedly augments the
chemosensitivity of gastric cancer by enhancing 5-FU-induced
apoptosis (22). Another study
indicated that dichloroacetate enhanced the chemosensitivity of CRC
to 5-FU through vital metabolic pathways mediated by miRNAs
(23).
The NDRG family contains four members, NDRG1-4,
which share 57–65% identity at the amino acid level, and contain an
α/β hydrolase-fold region (24). The
four members have multiple biological functions (7). NDRG4 was reported to be expressed
primarily in cells of the nervous system, including enteric
neurons, suggesting its involvement in CRC through the enteric
neuron system, and its potential as an early detection marker for
CRC (25). However, the expression
profile of NDRG4 has not been unified. Since human stool contains
exfoliated intestinal epithelial cells (26), detecting the abnormal expression of
certain molecules in stool (including NDRG4) has been used to
screen for CRC, suggesting that NDRG4 may not be exclusively
expressed in the enteric neurons in the colorectum. Although NDRG4
plays a tumor-suppressive role in various cancer types, the
mechanism is rarely studied. Our previous study identified NDRG4 as
a prognostic predictor for patients with CRC, and as a novel
candidate tumor suppressor (27,28). The
present study revealed that NDRG4 inhibited the proliferation of
SW480 and SW620 cells, which further confirmed the
tumor-suppressive effect of NDRG4 in CRC. Interestingly, the effect
of NDRG4 on chemosensitivity to 5-FU has not been previously
reported.
In the present study, a series of experiments was
conducted to determine whether NDRG4 enhanced the sensitivity of
CRC cells to 5-FU. The inhibitory effect of 5-FU on CRC cells has
been reported to be positively correlated with its concentration
(29). In order to determine the
concentration used in subsequent experiments, five different
concentrations of 5-FU were initially evaluated for their effects
on the viability of NDRG4-overexpressing cells and control cells.
The optimal concentration of 5-FU in SW480 and SW620 cells was 40
and 10 µg/ml, respectively. The expression of apoptosis-related
proteins was significantly increased in NDRG4-overexpressing cells
compared with non-overexpressing cells when treated with 5-FU.
However, the levels of NDRG4 were similar in NDRG4-overexpressing
cells in the presence and absence of 5-FU, though the expression of
apoptosis-related proteins was significantly higher in 5-FU treated
NDRG4-overexpressing cells compared with untreated
NDRG4-overexpressing cells (Fig.
2E). This suggests that 5-FU treatment did not affect the
expression of NDRG4, but increased the expression of
apoptosis-related proteins. Lu et al (30) demonstrated that treatment with 5-FU
significantly increased the expression levels of C-caspase-3 and
PARP, which is consistent with the results of the present study.
The reason for the increased expression of apoptotic proteins may
be that 5-FU drives the expression of apoptosis pathway genes by
inducing conformational changes in the chromatin regions containing
binding motifs for activator protein-1 family transcription factors
(31).
To the best of our knowledge, no other studies have
explored the relationship between NDRG4 and DDIT3 in CRC. However,
there are reports supporting the association between DDIT3 and
chemosensitivity. For example, Tan et al (14) noted that increasing the expression of
DDIT3 enhanced the sensitivity of lung cancer cells to cisplatin.
Another study reported that decreased expression of DDIT3 was an
important factor underlying the 5-FU resistance of rectal cancer
resulting from the high expression of rhomboid domain containing 2
(32). These reports are consistent
with the findings of the present study; apoptosis experiments
showed that NDRG4 overexpression enhanced the 5-FU-induced
apoptosis of CRC cells, which was significantly weakened by
DDIT3-knockdown in NDRG4-overexpressing SW480 cells, indicating the
importance of DDIT3 in the apoptosis pathway.
DDIT3 is principally involved in ER stress-related
apoptosis (33), and NDRG4 promotes
the expression of DDIT3, suggesting that ER stress is associated
with the tumor-suppressive effect of NDRG4. In addition, Zhang
et al (34) observed that low
DDIT3 expression was associated with the poor prognosis of patients
with advanced gastric cancer, for whom it was suggested as a
potential prognostic marker. Furthermore, as our previous studies
showed that NDRG4 was associated with the prognosis of CRC
(27,28), DDIT3 may also be a prognostic
biomarker for CRC.
Since SW480 cells are a classic CRC cell line with
proliferative, invasive, migratory and tumorigenic characteristics,
they are a commonly used model for the study of CRC in
vitro. SW480 cells were primarily used in the present study,
with partial verification studies conducted using the SW620 cell
line. Therefore, PCR-array and DDIT3-knockdown experiments were not
performed in SW620 cells, which is a study limitation. Organoids
can simulate various real-organ characteristics, and are important
models for studying disease (35).
Considering that the enteric nervous system may play an important
role in the function of NDRG4 (25),
the use of intestinal organoids co-cultured with enteric neurons
may be a future research prospect, along with the relevant
molecular biological experiments, so as to further investigate the
upstream and downstream molecular pathways of NDRG4. Of note, cell
function and animal experiments were also not performed, and
additional experiments, such as transwell assays and subcutaneous
tumor-bearing experiments in athymic mice and tumor-specific
patient-derived xenograft (PDX) models, will be a future
consideration to further elucidate how NDRG4/DDIT3 regulates CRC
cell responses to 5-FU treatment in vivo and in
vitro.
In conclusion, the present study revealed that NDRG4
increased the chemosensitivity of CRC cells to 5-FU by increasing
the expression of DDIT3, though the underlying mechanisms require
further study in the future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81572816 and
82072655).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding authors upon reasonable
request.
Authors' contributions
JYZ and JZ conceived and designed the study and
revised the manuscript. RKL and CXH performed most of the
experiments and analyzed the data. RKL drafted the initial
manuscript. LLS, SW and YS performed the MTT and TUNEL assays and
analyzed the data. FF designed part of the study, analyzed the data
and revised the manuscript. JZ and CXH confirmed the authenticity
of all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
DDIT3
|
DNA damage inducible transcript 3
|
|
ER
|
endoplasmic reticulum
|
|
5-FU
|
5-fluorouracil
|
|
NDRG4
|
N-myc downstream-regulated gene 4
|
|
PARP
|
poly-ADP-ribose polymerase
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuipers EJ, Grady WM, Lieberman D,
Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ and Watanabe T:
Colorectal cancer. Nat Rev Dis Primers. 1:150652015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xie YH, Chen YX and Fang JY: Comprehensive
review of targeted therapy for colorectal cancer. Signal Transduct
Target Ther. 5:222020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brown KGM, Solomon MJ, Mahon K and
O'Shannassy S: Management of colorectal cancer. BMJ. 366:l45612019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vodenkova S, Buchler T, Cervena K,
Veskrnova V, Vodicka P and Vymetalkova V: 5-fluorouracil and other
fluoropyrimidines in colorectal cancer: Past, present and future.
Pharmacol Ther. 206:1074472020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xie P, Mo JL, Liu JH, Li X, Tan LM, Zhang
W, Zhou HH and Liu ZQ: Pharmacogenomics of 5-fluorouracil in
colorectal cancer: Review and update. Cell Oncol (Dordr).
43:989–1001. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu C, Hao X, Zhang S, Hu W, Li J, Sun J
and Zheng M: Characterization of the prognostic values of the NDRG
family in gastric cancer. Therap Adv Gastroenterol.
12:17562848198585072019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi HH, Liu HE and Luo XJ:
Hypermethylation-mediated silencing of NDRG4 promotes pancreatic
ductal adenocarcinoma by regulating mitochondrial function. BMB
Rep. 53:658–663. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao L, Hu T, Lu H and Peng D: N-MYC
downstream regulated gene 4 (NDRG4), a frequent downregulated gene
through DNA hypermethylation, plays a tumor suppressive role in
esophageal adenocarcinoma. Cancers (Basel). 12:25732020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen X, Yang Y, Liu J, Li B, Xu Y, Li C,
Xu Q, Liu G, Chen Y, Ying J and Duan S: NDRG4 hypermethylation is a
potential biomarker for diagnosis and prognosis of gastric cancer
in Chinese population. Oncotarget. 8:8105–8119. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kadiyska T and Nossikoff A: Stool DNA
methylation assays in colorectal cancer screening. World J
Gastroenterol. 21:10057–10061. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Puthalakath H, O'Reilly LA, Gunn P, Lee L,
Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin
J, Motoyama N, et al: ER stress triggers apoptosis by activating
BH3-only protein Bim. Cell. 129:1337–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Chen Q, Yu Y, Chen H, Lu M, Huang Y,
Li P and Chang H: RKI-1447 suppresses colorectal carcinoma cell
growth via disrupting cellular bioenergetics and mitochondrial
dynamics. J Cell Physiol. 235:254–266. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan W, Liao Y, Qiu Y, Liu H, Tan D, Wu T,
Tang M, Zhang S and Wang H: MiRNA 146a promotes chemotherapy
resistance in lung cancer cells by targeting DNA damage inducible
transcript 3 (CHOP). Cancer Lett. 428:55–68. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cao Z, Liao Q, Su M, Huang K, Jin J and
Cao D: AKT and ERK dual inhibitors: The way forward? Cancer Lett.
459:30–40. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stintzing S: Management of colorectal
cancer. F1000Prime Rep. 6:1082014. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sargent D, Sobrero A, Grothey A, O'Connell
MJ, Buyse M, Andre T, Zheng Y, Green E, Labianca R, O'Callaghan C,
et al: Evidence for cure by adjuvant therapy in colon cancer:
Observations based on individual patient data from 20,898 patients
on 18 randomized trials. J Clin Oncol. 27:872–877. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blondy S, David V, Verdier M, Mathonnet M,
Perraud A and Christou N: 5-Fluorouracil resistance mechanisms in
colorectal cancer: From classical pathways to promising processes.
Cancer Sci. 111:3142–3154. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Uppada SB, Gowrikumar S, Ahmad R, Kumar B,
Szeglin B, Chen X, Smith JJ, Batra SK, Singh AB and Dhawan P: MASTL
induces colon cancer progression and chemoresistance by promoting
Wnt/β-catenin signaling. Mol Cancer. 17:1112018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun Y, Jiang Y, Huang J, Chen H, Liao Y
and Yang Z: CISD2 enhances the chemosensitivity of gastric cancer
through the enhancement of 5-FU-induced apoptosis and the
inhibition of autophagy by AKT/mTOR pathway. Cancer Med.
6:2331–2346. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang Y, Hou L, Li L, Li L, Zhu L, Wang Y,
Huang X, Hou Y, Zhu D, Zou H, et al: Dichloroacetate restores
colorectal cancer chemosensitivity through the
p53/miR-149-3p/PDK2-mediated glucose metabolic pathway. Oncogene.
39:469–485. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qu X, Zhai Y, Wei H, Zhang C, Xing G, Yu Y
and He F: Characterization and expression of three novel
differentiation-related genes belong to the human NDRG gene family.
Mol Cell Biochem. 229:35–44. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vaes N, Lentjes MHFM, Gijbels MJ,
Rademakers G, Daenen KL, Boesmans W, Wouters KAD, Geuzens A, Qu X,
Steinbusch HPJ, et al: NDRG4, an early detection marker for
colorectal cancer, is specifically expressed in enteric neurons.
Neurogastroenterol Motil. 29:2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kisiel JB, Klepp P, Allawi HT, Taylor WR,
Giakoumopoulos M, Sander T, Yab TC, Moum BA, Lidgard GP, Brackmann
S, et al: Analysis of DNA methylation at specific loci in stool
samples detects colorectal cancer and high-grade dysplasia in
patients with inflammatory bowel disease. Clin Gastroenterol
Hepatol. 17:914–921 e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chu D, Zhang Z, Zhou Y, Li Y, Zhu S, Zhang
J, Zhao Q, Ji G, Wang W and Zheng J: NDRG4, a novel candidate tumor
suppressor, is a predictor of overall survival of colorectal cancer
patients. Oncotarget. 6:7584–7596. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng J, Li Y, Zhu S, Li J, Zhao Q, Ji G,
Wang W and Chu D: NDRG4 stratifies the prognostic value of body
mass index in colorectal cancer. Oncotarget. 7:1311–1322. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Milczarek M, Rossowska J, Klopotowska D,
Stachowicz M, Kutner A and Wietrzyk J: Tacalcitol increases the
sensitivity of colorectal cancer cells to 5-fluorouracil by
downregulating the thymidylate synthase. J Steroid Biochem Mol
Biol. 190:139–151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lu P, Xu M, Xiong Z, Zhou F and Wang L:
Fusobacterium nucleatum prevents apoptosis in colorectal cancer
cells via the ANO1 pathway. Cancer Manag Res. 11:9057–9066. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang CM, Kang MK, Jung WJ, Joo JS, Kim YJ,
Choi Y and Kim HP: p53 expression confers sensitivity to
5-fluorouracil via distinct chromatin accessibility dynamics in
human colorectal cancer. Oncol Lett. 21:2262021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Palma S, Raffa CI, Garcia-Fabiani MB,
Ferretti VA, Zwenger A, Perez Verdera PV, Llontop A, Rojas Bilbao
E, Cuartero V, Abba MC and Lacunza E: RHBDD2 overexpression
promotes a chemoresistant and invasive phenotype to rectal cancer
tumors via modulating UPR and focal adhesion genes. Biochim Biophys
Acta Mol Basis Dis. 1866:1658102020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yao RQ, Ren C, Xia ZF and Yao YM:
Organelle-specific autophagy in inflammatory diseases: A potential
therapeutic target underlying the quality control of multiple
organelles. Autophagy. 17:385–401. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang X, Zhou T, Li W, Zhang T, Che N and
Zu G: Clinicopathological and prognostic significance of C/EBP
homologous protein (CHOP) in advanced gastric cancer. Pathol Res
Pract. 214:1105–1109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ji DB and Wu AW: Organoid in colorectal
cancer: Progress and challenges. Chin Med J (Engl). 133:1971–1977.
2020. View Article : Google Scholar : PubMed/NCBI
|