Introduction
Hepatocellular carcinoma (HCC) is one of the most
common hepatic malignancies and remains the leading cause of
cancer-associated mortality worldwide (1,2).
Surgical resection has been considered as the most effective
therapeutic option for patients with HCC; however, it may produce
new metastases and promote the growth of existing micro-metastasis
(3). It is therefore crucial to
determine the molecular mechanisms underlying HCC and develop novel
efficient therapeutic strategies for HCC.
A large part of human genomes cannot encode proteins
and most of the transcripts are non-coding RNAs. Long non-coding
RNAs (lncRNAs) are defined as highly conserved transcripts of
>200 nucleotides in length that are not translated into proteins
(4). lncRNAs, as novel epigenetic
regulatory molecules, are implicated in various cellular and
biological processes, and the abnormal expression of lncRNAs are
closely associated with tumorigenesis, metastasis and apoptosis by
competing for corresponding miRNAs (5). Research on the functional role and
molecular mechanisms of lncRNAs have provided a new insight for
cancer therapies. Recent studies reported that some lncRNAs are key
regulators in HCC (6,7). For example, the expression of the
lncRNA SOX9 antisense RNA 1 (SOX9-AS1) is increased in HCC and
might have a prognostic significance in patients with HCC.
Furthermore, SOX9-AS1 can facilitate HCC progression and metastasis
through Wnt/β-catenin pathway (8).
In addition, the lncRNA DSCAM antisense RNA 1 is upregulated in HCC
and can accelerate the progression of HCC via sponging micro-RNA
(miR)-338-3p (9). A previous study
identified the functional lncRNA cytoskeleton regulator RNA
(CYTOR), also known as Linc00152, which is situated on human
chromosome 2p11.2 and which abnormal expression is related to the
inflammatory response and apoptosis (10). Previous studies have reported that
CYTOR plays a pivotal role in the development of various cancers,
including colorectal cancer (11),
breast cancer (12) and lung cancer
(13), in which it functions as an
oncogenic lncRNA. However, the biological role and underlying
mechanism of CYTOR in HCC cell proliferation and apoptosis remain
unknown.
The present study determined CYTOR expression in HCC
tissues and its correlation with the clinicopathological
characteristics of patients with HCC. The effects and molecular
mechanism of CYTOR on HCC cell proliferation, apoptosis and tumor
growth were also investigated. The findings from the present study
may accelerate the understanding of HCC pathogenesis and the
development of effective therapies.
Materials and methods
Clinical samples
HCC tumor tissues (n=78) and adjacent non-tumor
tissues (n=78) were collected from patients with HCC who underwent
hepatectomy at Tianjin First Central Hospital between February 2019
and February 2020. The histopathological diagnosis was performed
according to the Tumor-Node-Metastasis Staging Guide (14) (based on the extent of the tumor, the
extent of spreading to lymph nodes and the presence of metastasis).
The samples were immediately frozen and stored at −80°C. None of
the patients enrolled in this study had received any preoperative
chemotherapy or radiotherapy prior to surgery. The
clinicopathological characteristics of the patients are described
in Table I. All patients have signed
written informed consents for the use of their tissues in this
research. The protocol was approved by the Clinical Research Ethics
Committee of Tianjin First Central Hospital (approval no.
2019N013YY).
 | Table I.Association between CYTOR expression
and the clinicopathological characteristic of patients with
hepatocellular carcinoma. |
Table I.
Association between CYTOR expression
and the clinicopathological characteristic of patients with
hepatocellular carcinoma.
|
|
| CYTOR
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | Number | Low (n=37) | High (n=41) | P-value |
|---|
| Age, years |
|
|
| 0.802 |
|
<55 | 41 | 20 | 21 |
|
|
≥55 | 37 | 17 | 20 |
|
| Sex |
|
|
| 0.934 |
|
Female | 32 | 15 | 17 |
|
|
Male | 46 | 22 | 24 |
|
| Liver
cirrhosis |
|
|
| 0.431 |
| No | 60 | 27 | 33 |
|
|
Yes | 18 | 10 | 8 |
|
| Tumor size, cm |
|
|
| 0.017a |
| <5
cm | 46 | 28 | 18 |
|
| ≥5
cm | 32 | 9 | 23 |
|
| Lymphovascular
invasion |
|
|
| 0.026a |
| No | 34 | 21 | 13 |
|
|
Yes | 44 | 16 | 28 |
|
| AFP, ng/ml |
|
|
| 0.838 |
|
<200 | 41 | 19 | 22 |
|
|
≥200 | 37 | 18 | 19 |
|
| TNM stage |
|
|
| 0.006b |
|
I–II | 56 | 32 | 24 |
|
|
III–IV | 22 | 5 | 17 |
|
The Cancer Genome Atlas (TCGA)
program
The gene expression profiles of 180 patients with
HCC, including 180 pairs of tumor and adjacent non-tumor tissues,
were download from the TCGA dataset (http://www.oncolnc.org/). The survival analysis of
CYTOR was visualized by GraphPad Prism version 6.0 software
(GraphPad Software Inc.)
Cell lines and cell culture
The normal adult liver epithelial THLE-2 cell line
and the human HCC cell lines Huh7, MHCC97-L, MHCC97-H and SK-Hep-1
were obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. The cells were cultured in Dulbecco's
Modified Eagle's Medium (DMEM; Gibco, CA, USA) supplemented with
10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 mg/ml
streptomycin and 100 U/ml penicillin (Invitrogen; Merck KGaA) and
placed at 37°C in a humidified incubator containing 5%
CO2.
Transient transfection
To suppress CYTOR expression, the short hairpin
(sh)RNAs targeting CYTOR (sh-CYTOR) were inserted into pLKO.1
plasmid (GenePharma Co., Ltd.) and 4 µg sh-CYTOR were transfected
into Huh7 cells. Scramble shRNA was used as control (sh-NC). The
target sequences of CYTOR shRNAs were as follow: sh#1:
CTGGAAACCTCTTGACTCT and sh#2: CAGGAAGCTCTATGACACA. To overexpress
CYTOR expression, full-length CYTOR cDNA was cloned into pcDNA3.1
plasmid (pcDNA-CYTOR) (GenePharma Co., Ltd.) and then 4 µg
pcDNA-CYTOR were transfected into Huh7 cells, and the pcDNA3.1
empty vector was used as the control (pcDNA-NC). To regulate
miR-125b expression in cells, 50 nM miR-125b mimic
(5′-UCCCUGAGACCCUUUAACCUGUGA-3′), 50 nM normal control (NC) mimic
(5′-UGACAACCUGGUAGAAAGAGACUUC-3′), 50 nM miR-125b inhibitor
(5′-UCCCUGAGACCCUUAACCUGUG-3′) and 50 nM NC inhibitor
(5′-UCGGCCUUUUGCUCACAGACCA-3′), designed and synthesized by
Shanghai R&S Biotechnology Co., Ltd., were also transfected
into Huh7 cells. Cells were transiently transfected with the above
vectors using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturers' protocol
at 37°C. After 48 h transfection, cells were harvested for further
experiments.
RNA extraction and reverse
transcription quantitative (RT-q)PCR
Total RNA from HCC tissues or Huh7 cells was
extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). For miRNA analysis, RNA reverse transcription
into cDNA was performed using TaqMan MicroRNA Reverse Transcription
Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
RT-qPCR reactions were performed using TaqMan miRNA assay kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). U6 was used
as the internal control. For mRNA analysis, RNA reverse
transcription into cDNA was performed using M-MLV reverse
transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) and
RT-qPCR reactions were performed with SYBR-Green Real-Time PCR
Master Mixes (Thermo Fisher Scientific, Inc.). GAPDH was used as
the internal control. The relative expression levels were
normalized to endogenous controls and were expressed as
2−ΔΔCq (15). The
sequences of the primers used were as follows: PCNA, forward,
5′-AACCGGTTACTGAGGGCGAG-3′, reverse, 5′-AAAGTCTAGCTGGTTTCGGCT-3′;
CYTOR, forward, 5′-AGAATGAAGGCTGAGGTGTG-3′, reverse,
5′-CAGCGACCATCCAGTCATTTA-3′; miR-125b, forward,
5′-TCCCTGAGACCCTAACTTGTGA-3′, reverse,
5′-AGTCTCAGGGTCCGAGGTATTC-3′; GAPDH, forward,
5′-GAAGGTGAAGGTCGGAGTC-3′, reverse, 5′-GAA GATGGTGATGGGATTTC-3′;
and U6, forward, 5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′.
Western blotting
Following various transfections, Huh7 cells were
lysed in RIPA buffer (Beyotime Institute of Biotechnology) for 10
min on ice. The concentration of proteins was measured using the
BCA method (Beyotime Institute of Biotechnology). Proteins (50 µg)
were separated by 10% SDS-PAGE and transferred onto PVDF membranes.
Membranes were blocked with 5% skimmed milk for 2 h at room
temperature and incubated with primary antibodies against Bax
(Abcam; cat. no. ab182733; 1:2,000), Bcl-2 (Abcam; cat. no.
ab182858; 1:2,000), cleaved-caspase-3 (Abcam; cat. no. ab214430;
1:2,000), pro-caspase-3 (Abcam; cat. no. ab32150; 1:1,000),
cleaved-caspase-9 (Abcam; cat. no. ab2324; 1:1,000), pro-caspase-9
(Abcam; cat. no. ab138412; 1:1,000), poly ADP ribose polymerase
(PARP) (Cell Signaling Technology, Inc.; cat. no. 9532, 1:1,000),
SEMA4C (Abcam; cat. no. ab135856; 1:500) and GAPDH (Cell Signaling
Technology, Inc. cat. no. 97166; 1:1,000) overnight at 4°C.
Membranes were then incubated with horseradish peroxidase-labeled
secondary antibodies (Abcam; cat. nos. ab205718 and ab205719;
1:2,000) for 2 h at temperature. Enhanced chemiluminescence reagent
(Thermo Fisher Scientific, Inc.) was used to detect the signal on
the membrane. The data were analyzed via densitometry using ImageJ
software (version 1.8.0; National Institutes of Health) and
normalized to expression of the internal control (GAPDH).
Cell Counting Kit-8 (CCK-8) assay
CCK-8 kit assay (Beyotime Institute of
Biotechnology) was used to detect cell viability. Briefly,
2×103 Huh7 cells were seeded into a 96-well plate. Then,
10 µl CCK-8 solution was added in each well at four time points
(24, 48, 72 and 96 h) and incubated for another 2 h at 37°C. The
absorbance was read at 450 nm on a microplate reader.
EdU immunoflurescence assay
EdU immunoflurescence assay was carried out for
detection of cell proliferation. Briefly, Huh7 cells were cultured
with EdU solution and incubated for 2 h at 37°C. Then, the medium
was removed and cells were fixed with PBS containing 4%
paraformaldehyde for 10 min at room temperature. Subsequently,
cells were incubated with 70% ethanol and stained using the
Cell-Light™ EdU Apollo® 488 In Vitro Imaging Kit
(RiboBio Co., Ltd.). Cells were visualized using fluorescence
microscopy (magnification, ×100). EdU analysis was performed using
ImageJ software v.1.50 (National Institutes of Health).
Bioinformatics analysis
Putative CYTOR or miR-125b targets were predicted
using StarBase version 3.0 (http://starbase.sysu.edu.cn/) or TargetScan
(http://www.targetscan.org/vert_60/)
RNA immunoprecipitation (RIP)
assay
Following transfection with miR-125b mimic or
inhibitor for 48 h, Huh7 cells were lysed with RIPA lysis buffer
(Beyotime Institute of Biotechnology) for 10 min on ice following
the manufacturers' instructions and incubated with RIP
immunoprecipitation buffer mixed with magnetic beads conjugating
human anti-Argonaute2 (Ago2) antibody or negative control mouse IgG
(EMD Millipore) at 4°C overnight. A protein-RNA complex was
captured and digested with 0.5 mg/ml proteinase K containing 0.1%
SDS to extract RNA. The magnetic beads were repeatedly washed with
RIP washing buffer to remove non-specific adsorption as much as
possible. RT-qPCR analysis was conducted to estimate the enrichment
of CYTOR and miR-125b. Input and IgG served as controls in RIP
assay.
Luciferase reporter assay
The wild and mutant type of CYTOR or SEMA4C
containing the binding site for miR-125b were cloned into
pGL3-reporter vectors, designed as pGL3-CYTOR-WT and pGL3-CYTOR-MuT
or pGL3-SEMA4C-WT and pGL3-SEMA4C-MuT. Huh7 cells were
co-transfected with luciferase reporter vectors and miR-125b mimic,
miR-125b inhibitor or negative controls using Lipofectamine 2000.
The miR quantities and sequences were as follows: 50 nM, miR-125b
mimic, 5′-UCCCUGAGACCCUUUAACCUGUGA-3′; 50 nM, miR-125b inhibitor,
5′-UCCCUGAGACCCUUAACCUGUG-3′; 50 nM, NC mimic,
5′-UGACAACCUGGUAGAAAGAGACUUC-3′; and 50 nM, NC inhibitor
5′-UCGGCCUUUUGCUCACAGACCA-3′. After transfection for 48 h, the
luciferase activity was measured by a Dual-Luciferase Reporter
Assay System (Promega Corporation) and normalized to Renilla
luciferase activity.
Mice xenograft assay
A total of 36 BALB/c male nude mice (6–8 weeks old)
were purchased from the Animal Center of the Chinese Academy of
Science. All animal experiments were performed in the animal
laboratory of Tianjin First Central Hospital and the protocol was
approved by the Animal Care and Use Committee (approval no.
2019N046YY). Mice were randomly divided into 3 groups of 6 mice as
follows: Control group, sh-NC group and sh-CYTOR#1 group. Huh7
cells stably transfected with sh-CYTOR#1 or negative control were
amplified and a total of 200 µl (2.5×107 cells/ml) was
subcutaneously injected into the dorsal sides of the nude mice. The
xenograft tumor size was measured every 7 days for 28 days
according to the following formula: Volume = (length ×
width2)/2. Four weeks later, mice were euthanized with
2% pentobarbital sodium (120 mg/kg bodyweight) and the tumors were
collected for immunohistochemistry imaging.
Immunohistochemistry
Tumors were fixed in 10% formalin (48 h, room
temperature), embedded in paraffin and cut into 4 µm-thick slides.
The slides of paraffin-embedded xenograft tissues were probed with
specific rabbit anti-SEMA4C antibody (Abcam; cat. no. ab121334;
1:200) and mouse anti-Ki67 (Cell Signaling Technology, Inc.; cat.
no. 9449; 1:200) for 48 h at 4°C, followed by incubation with the
goat anti-rabbit or goat anti-mouse secondary antibodies (Abcam;
cat. nos. ab6721 and ab6728; 1:1,000) for 1 h at 37°C. Then, the
complexes were detected using HRP-streptavidin conjugates,
visualized using 3,3′-diaminobenzidine (DAB; Wuhan Boster
Biological Technology, Ltd.) and quantified with Image ProPlus
(IPP) v7.0 software (Media Cybernetics, Inc.).
Statistical analysis
The data were presented as the means ± standard
deviation of three independent experiments. The differences between
two or more groups were analyzed using two-tailed Student's t-tests
or one -way ANOVA followed by Turkey's post-hoc test, respectively.
Statistical analysis was performed using SPSS Statistics 20.0
software (IBM Corp.) and GraphPad Prism version 6.0 software
(GraphPad Software Inc.). The association between CYTOR expression
and the clinicopathological characteristics of patients with HCC
was analyzed by χ2 test. The correlation between CYTOR
expression and miR-125b or SEMA4C expression was evaluated using
Pearson's correlation coefficient. Survival analysis was analyzed
by the Kaplan-Meier method, followed by difference analysis between
the survival curves using log-rank test. P<0.05 was considered
to indicate a statistically significant difference.
Results
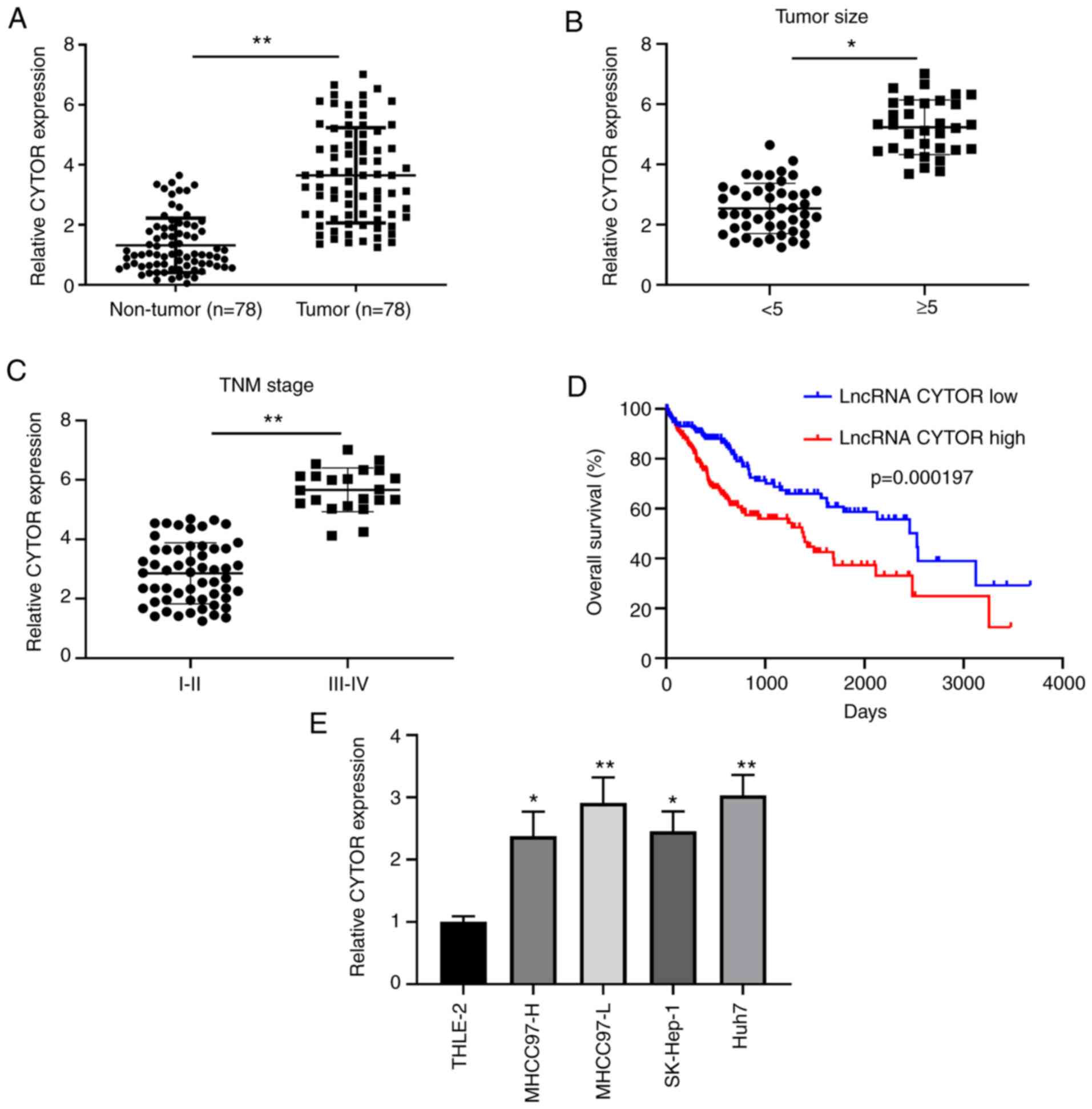
CYTOR is upregulated in HCC tissues
and associated with the poor survival rate of patients with
HCC
To investigate the role of CYTOR in HCC, CYTOR
expression was evaluated in 78 matched pairs of HCC and adjacent
non-tumor tissues by RT-qPCR. The results demonstrated that CYTOR
expression was significantly increased in HCC tumor tissues
compared with adjacent non-tumor tissues (Fig. 1A). Furthermore, CYTOR expression was
significantly higher in HCC tissues with tumor size ≥5 cm compared
with tumor tissues of <5 cm (Fig.
1B). In addition, the expression of CYTOR was significantly
higher in stage I–II tumors compared with III–IV stage tumors
(Fig. 1C). To further confirm the
association between CYTOR expression and the clinicopathological
characteristics of patients with HCC, the 78 patients were divided
into two groups based on the median level of CYTOR in HCC tissues.
The results demonstrated that CYTOR exhibited a significant
association with tumor size, lymphovascular invasion and
Tumor-Node-Metastasis stage (Table
I). Kaplan-Meier survival analysis from the TCGA database
demonstrated that patients with HCC and low CYTOR expression
exhibited a higher overall survival rate compared with patients
with high CYTOR expression (Fig.
1D). Consistent with CYTOR expression in HCC tissues, CYTOR
expression was significantly increased in HCC cell lines compared
with the normal adult liver epithelial THLE-2 cells (Fig. 1E). These findings indicated that
CYTOR upregulation was closely associated with the poor prognosis
of patients with HCC and that it may be involved in HCC
progression.
CYTOR enhances the proliferation and
inhibits the apoptosis of HCC cells
The functional significance of CYTOR in HCC cells
was explored. Huh7 cells were selected for subsequent experiments
as they displayed the highest CYTOR among all HCC cells. CYTOR was
upregulated or downregulated by transfection with pcDNA-CYTOR or
shRNA-CYTOR, respectively. The results from RT-qPCR confirmed that
CYTOR was effectively overexpressed in Huh7 cells after
transfection with pcDNA-CYTOR, while CYTOR was effectively
downregulated in Huh7 cells following transfection with
shRNA-CYTOR. The sh-CYTOR#1 was selected for further experiments
due to its stronger inhibitory effects compared with sh-CYTOR#2
(Fig. 2A). The results from CCK-8
assay demonstrated that HCC cell viability was significantly
increased in pcDNA-CYTOR transfected cells, whereas it was
inhibited in sh-CYTOR#1 transfected cells compared with control
cells (Fig. 2B). Furthermore, CYTOR
overexpression could significantly increase the expression of
proliferating cell nuclear antigen (PCNA), which is a good
indicator of cell proliferation. Conversely, decreasing CYTOR
attenuated the proliferation of Huh7 cells compared with control
groups (Fig. 2C). Similarly, results
from EdU assay indicated that the number of positive cells in
pcDNA-CYTOR group was significantly enhanced, whereas it was
significantly decreased in the sh-CYTOR#1 group compared with
control groups (Fig. 2D). In
addition, transfection with pcDNA-CYTOR significantly increased
Bcl-2 expression and decreased the expression of the
apoptotic-related proteins, including Bax, cleaved-caspase-3,
cleaved-caspase-9 and PARP in Huh7 cells. Transfection with
sh-CYTOR#1 had the opposite effects (Fig. 2E). Taken together, these findings
suggested that CYTOR could stimulate the proliferation and
attenuate the apoptosis of Huh7 cells.
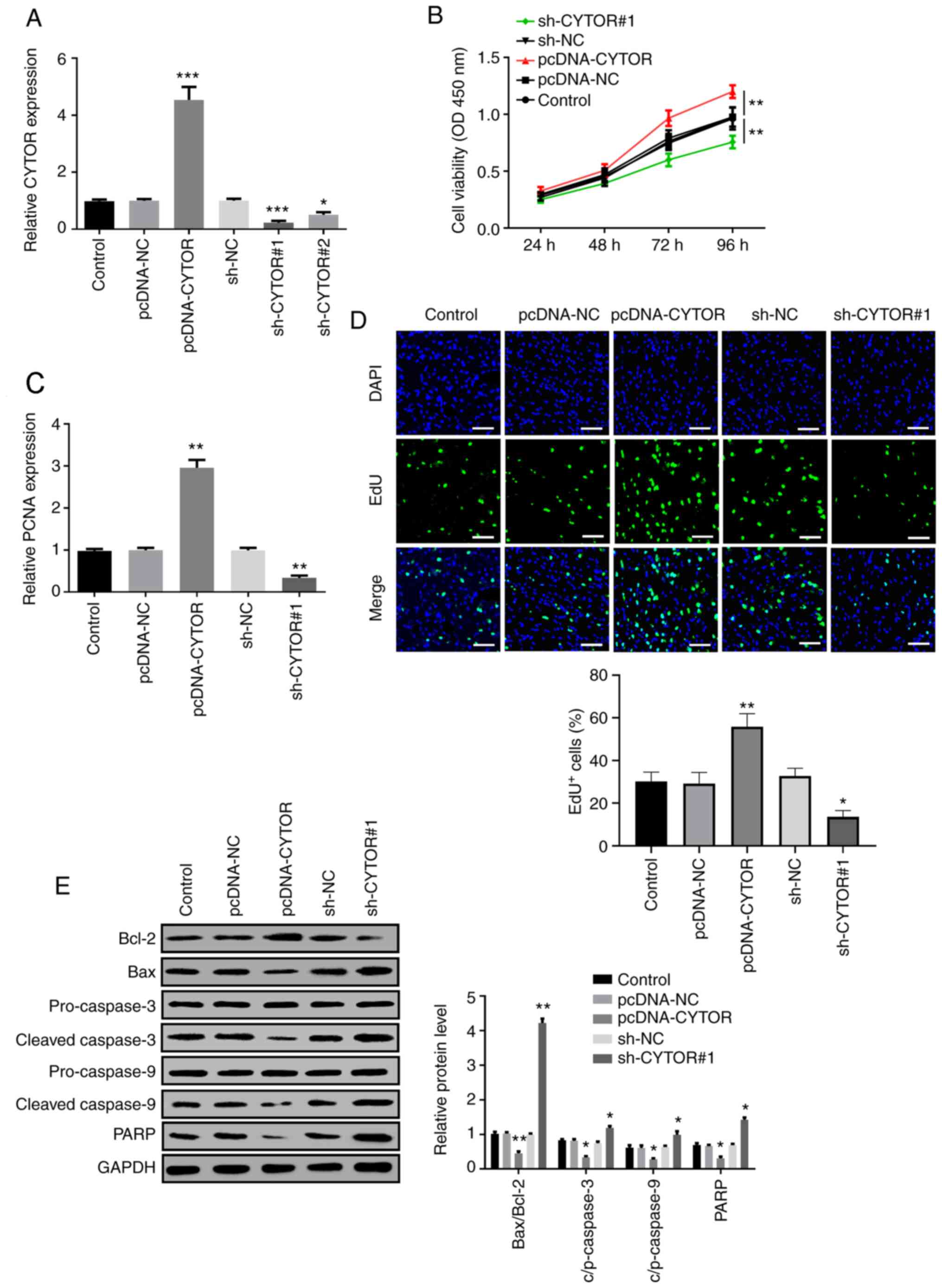 | Figure 2.CYTOR promotes the proliferation and
inhibit the apoptosis of HCC cells. (A) CYTOR overexpression or
knockdown was achieved by pcDNA-CYTOR or CYTOR shRNAs and the
transfection efficiency was verified by RT-qPCR. (B) Cell viability
was measured by CCK-8 assay. (C) PCNA expression was detected by
RT-qPCR and (D) cell proliferative cells measured by EdU assay
(scale bar, 20 µm) in Huh7 cells following transfection with
pcDNA-CYTOR or sh-CYTOR#1. (E) Expression of the apoptosis-related
proteins Bcl-2, Bax, cleaved/pro-caspase-3, cleaved/pro-caspase-9
and PARP was determined using western blotting. *P<0.05,
**P<0.01 and ***P<0.001. CYTOR, cytoskeleton regulator RNA;
RT-qPCR, reverse transcription quantitative PCR; NC, negative
control; sh, short hairpin; PARP, poly ADP ribose polymerase; OD
optical density. |
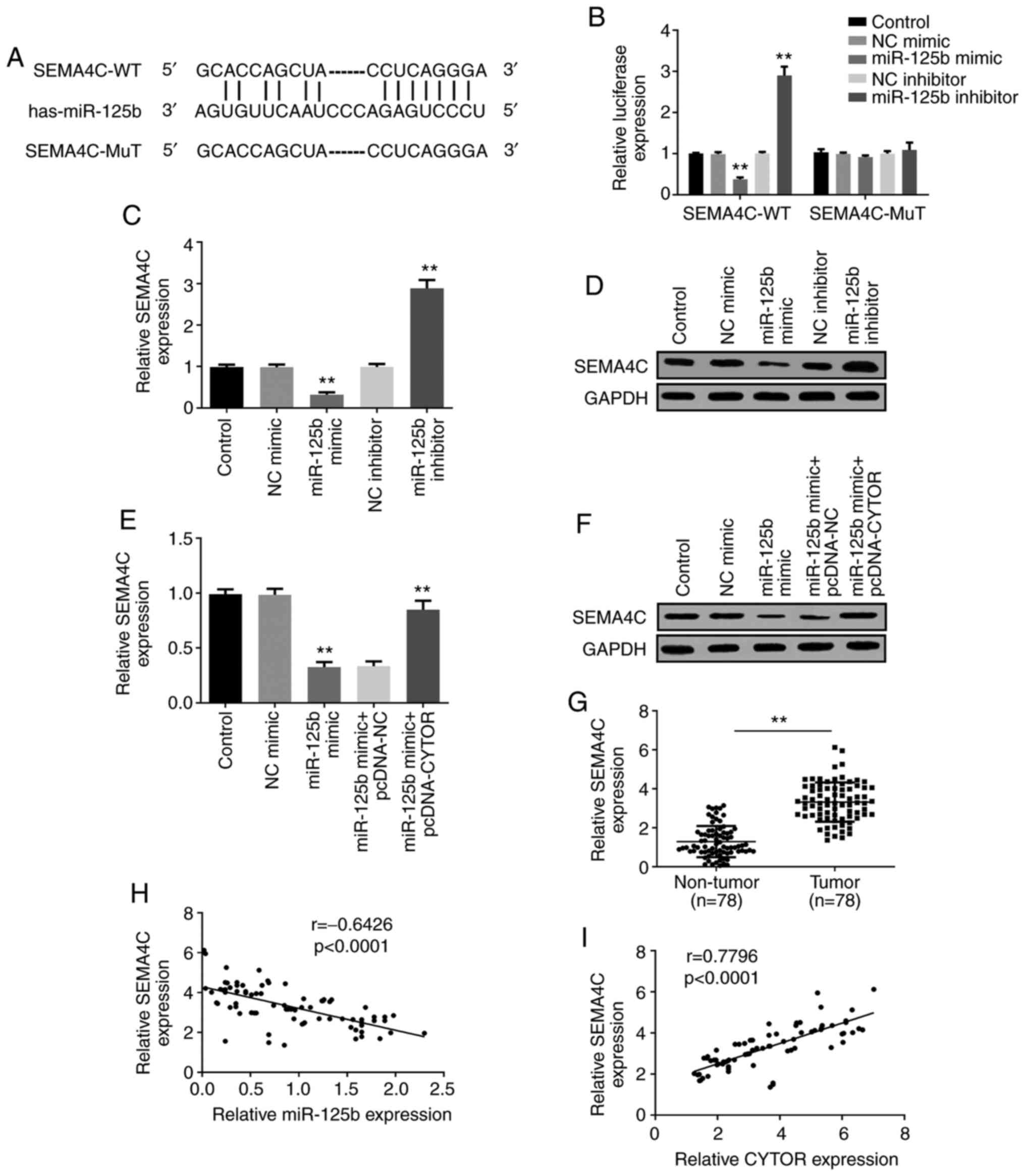
CYTOR directly interacts with
miR-125b
The underlying molecular mechanism of CYTOR on HCC
cells was further studied. Starbase prediction algorithm confirmed
that miR-125b was a potential binding target of CYTOR (Fig. 3A). Subsequently, miR-125b expression
was overexpressed or knocked-down following transfection with
miR-125b mimic or inhibitor in Huh7 cells, respectively, and the
successful transfection efficiencies were confirmed by RT-qPCR
(Fig. 3B). Luciferase reporter assay
and RIP assay were subsequently carried out to verify the
interaction between CYTOR and miR-125b. The results from luciferase
reporter assay demonstrated that miR-125b mimic significantly
decreased, while miR-125b inhibitor significantly increased the
luciferase activity of CYTOR-WT. However, neither miR-125b
overexpression nor suppression had any impact on the luciferase
activity of CYTOR-MuT (Fig. 3C). RIP
assay revealed that CYTOR and miR-125b expression was elevated in
anti-Ago2 group compared with that in control group (Fig. 3D). In addition, the enrichment of
CYTOR in anti-Ago2 group was facilitated by miR-125b mimic and
inhibited by miR-125b inhibitor (Fig.
3E). Subsequently, RT-qPCR was used to investigate the
relationship between CYTOR and miR-125b expression. The results
demonstrated that CYTOR overexpression significantly decreased
miR-125b expression, whereas miR-125b was markedly increased in
sh-CYTOR#1 group (Fig. 3F),
suggesting a negative association between CYTOR and miR-125b
expression. Furthermore, miR-125b expression was detected in HCC
tissues and the results demonstrated that miR-125b was
significantly downregulated in HCC tissues compared with non-tumor
tissues (Fig. 3G). Pearson's
correlation analysis revealed that CYTOR expression was negatively
correlated with miR-125b in HCC tissues (Fig. 3H). These findings suggested that
CYTOR may directly interact with miR-125b in HCC.
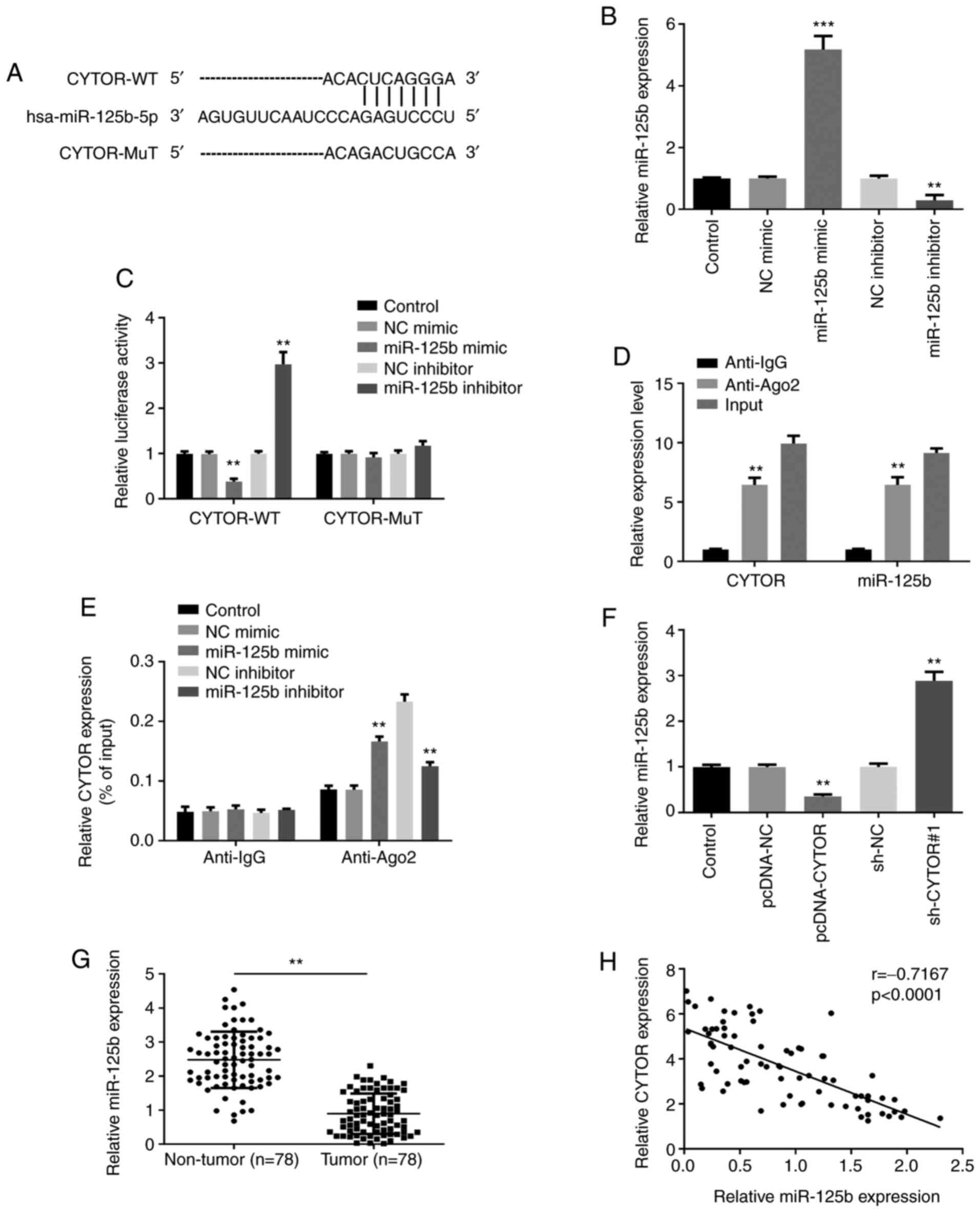 | Figure 3.CYTOR directly interacts with
miR-125b. (A) Bioinformatics analysis showed a predicted binding
site between CYTOR and miR-125b. (B) Transfection efficiency with
miR-125b mimic and inhibitor was confirmed by reverse transcription
quantitative PCR. (C) Luciferase reporter assay demonstrated that
miR-125b overexpression or knockdown could decrease or increase,
respectively, the luciferase activity of the CYTOR-WT. (D) RIP
assay showed that CYTOR and miR-125b expression were enriched in
anti-Ago2 group. (E) RIP assay demonstrated that the enrichment of
CYTOR in anti-Ago2 group can be enhanced by miR-125b mimic and
inhibited by miR-125b inhibitor. (F) CYTOR negatively regulated
miR-125b expression. (G) miR-125b was significantly downregulated
in HCC tumor tissues compared with adjacent non-tumor tissues
(n=78). (H) Correlation analysis revealed negative correlation
between CYTOR expression and miR-125b expression in HCC tissues
(n=78). **P<0.01 and ***P<0.001. CYTOR, cytoskeleton
regulator RNA; RT-qPCR, reverse transcription quantitative PCR; NC,
negative control; sh, short hairpin; miR, micro-RNA; WT, wild-type;
MUT, mutant; RIP, RNA immunoprecipitation. |
CYTOR sponges miR-125b to modulate HCC
cell proliferation and apoptosis
Whether miR-125b could be involved in the effect of
CYTOR on Huh7 cell proliferation and apoptosis was further
investigated. Huh7 cells were transfected with sh-CYTOR#1 alone or
with miR-125b inhibitor. The CCK-8 results demonstrated that Huh7
cell viability decrease by sh-CYTOR#1 could be partly rescued by
co-transfection with miR-125b inhibitor (Fig. 4A). The results from RT-qPCR
demonstrated that miR-125b inhibitor elevated the inhibitory effect
of sh-CYTOR#1 on HCC cell proliferation (Fig. 4B). Furthermore, the results from Edu
assay revealed that the number of positive cells in sh-CYTOR#1 +
miR-125b inhibitor group was significantly enhanced compared with
that in sh-CYTOR#1 group (Fig. 4C).
In addition, western blotting results showed that the effect of
sh-CYTOR#1 on the expression of the anti-apoptotic protein Bcl-2,
and the apoptotic-related proteins, Bax, cleaved-caspase-3,
cleaved-caspase-9 and PARP were partly reversed following
co-transfection with miR-125b inhibitor (Fig. 4D). These findings indicated that
CYTOR may modulate HCC cell proliferation and apoptosis by sponging
miR-125b.
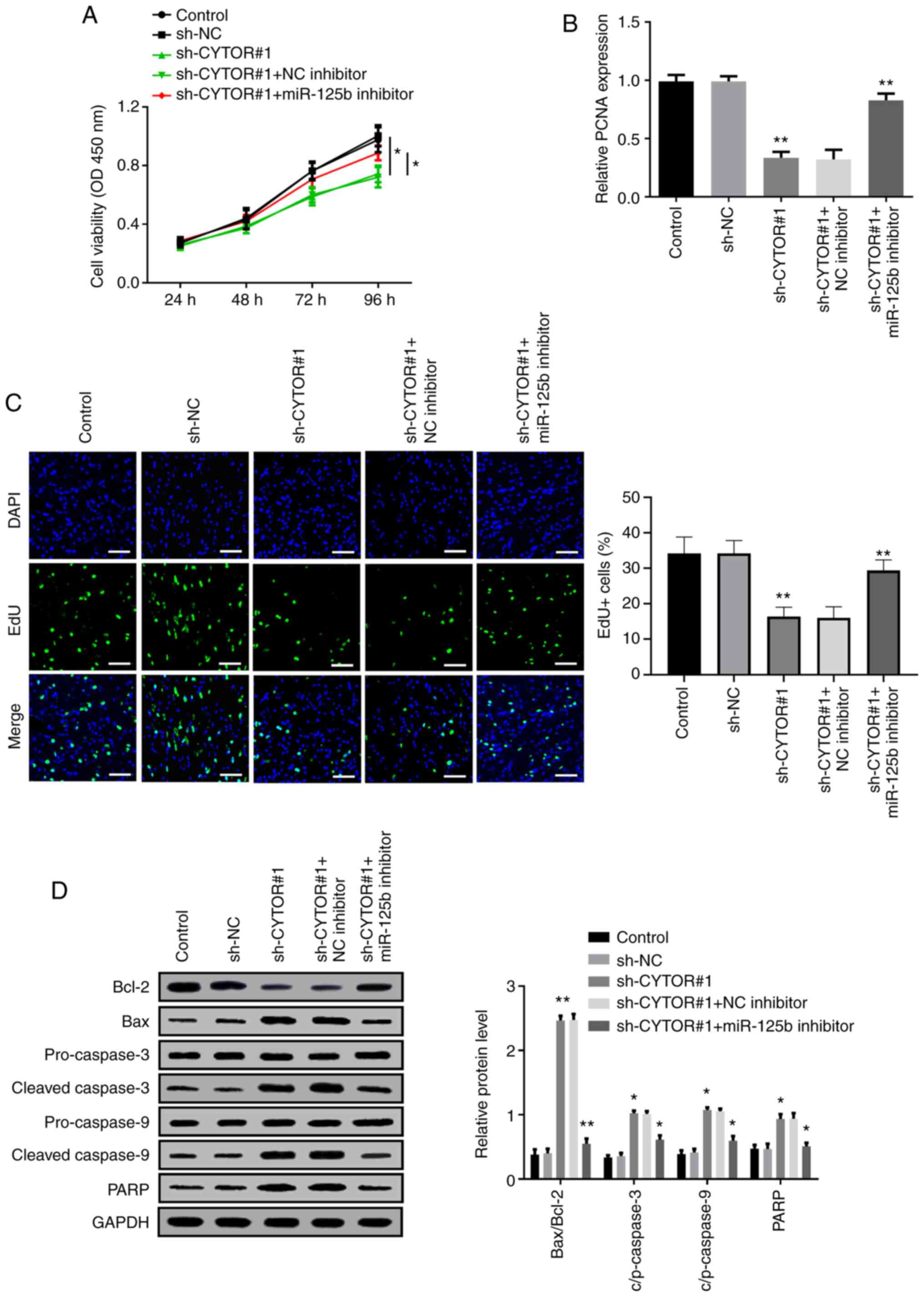 | Figure 4.CYTOR sponges miR-125b to modulate
HCC cell proliferation and apoptosis. (A) Decreased cell viability
induced following sh-CYTOR#1 transfection was restored by miR-125b
inhibitor. (B) Reverse transcription quantitative PCR analysis
demonstrated that PCNA expression was decreased by sh-CYTOR#1 and
rescued by miR-125b inhibitor. (C) Suppression of the proliferative
effect of sh-CYTOR#1 was enhanced by miR-125 inhibitor (scale bar,
20 µm). (D) Expression of the apoptosis-related proteins Bcl-2,
Bax, cleaved/pro-caspase-3, cleaved/pro-caspase-9 and PARP was
determined using western blotting. *P<0.05 and **P<0.01.
CYTOR, cytoskeleton regulator RNA; RT-qPCR, reverse transcription
quantitative PCR; NC, negative control; sh, short hairpin; miR,
micro-RNA; OD, optical density; PCNA, proliferating cell nuclear
antigen |
SEMA4C is the target of miR-125b and
CYTOR regulates SEMA4C through miR-125b in HCC
The potential target genes of miR-125b were
predicted by Targetscan, and SEMA4C was shown to share a binding
site with miR-125b (Fig. 5A). The
luciferase assay was performed to confirm this prediction and the
results demonstrated that the luciferase activity of SEMA4C-WT was
significantly decreased in miR-125b mimic-transfected Huh7 cells,
which was not the case in SEMA4C-MuT (Fig. 5B), indicating that SEMA4C may be a
downstream target gene of miR-125b. Furthermore, the results from
RT-qPCR and western blotting demonstrated that miR-125b mimic could
inhibit SEMA4C expression, whereas miR-125b inhibitor enhanced
SEMA4C expression (Fig. 5C and D).
Subsequently, the relationship between CYTOR, miR-125b and SEMA4C
was investigated, and the results demonstrated that SEMA4C
expression decrease by miR-125b mimic was rescued by transfection
with pcDNA-CYTOR (Fig. 5E and F),
suggesting that CYTOR could regulate SEMA4C expression through
targeting miR-125b. SEMA4C expression level in HCC tumor tissues
was significantly increased compared with non-tumor tissues
(Fig. 5G). The results from Pearson
correlation analysis demonstrated that SEMA4C expression was
negatively correlated with miR-125b expression; however, SEMA4C
expression was positively correlated with CYTOR expression in HCC
tissues (Fig. 5H and I). These
findings demonstrated that SEMA4C may be a target of miR-125b, and
that CYTOR may regulate SEMA4C expression by sponging miR-125b.
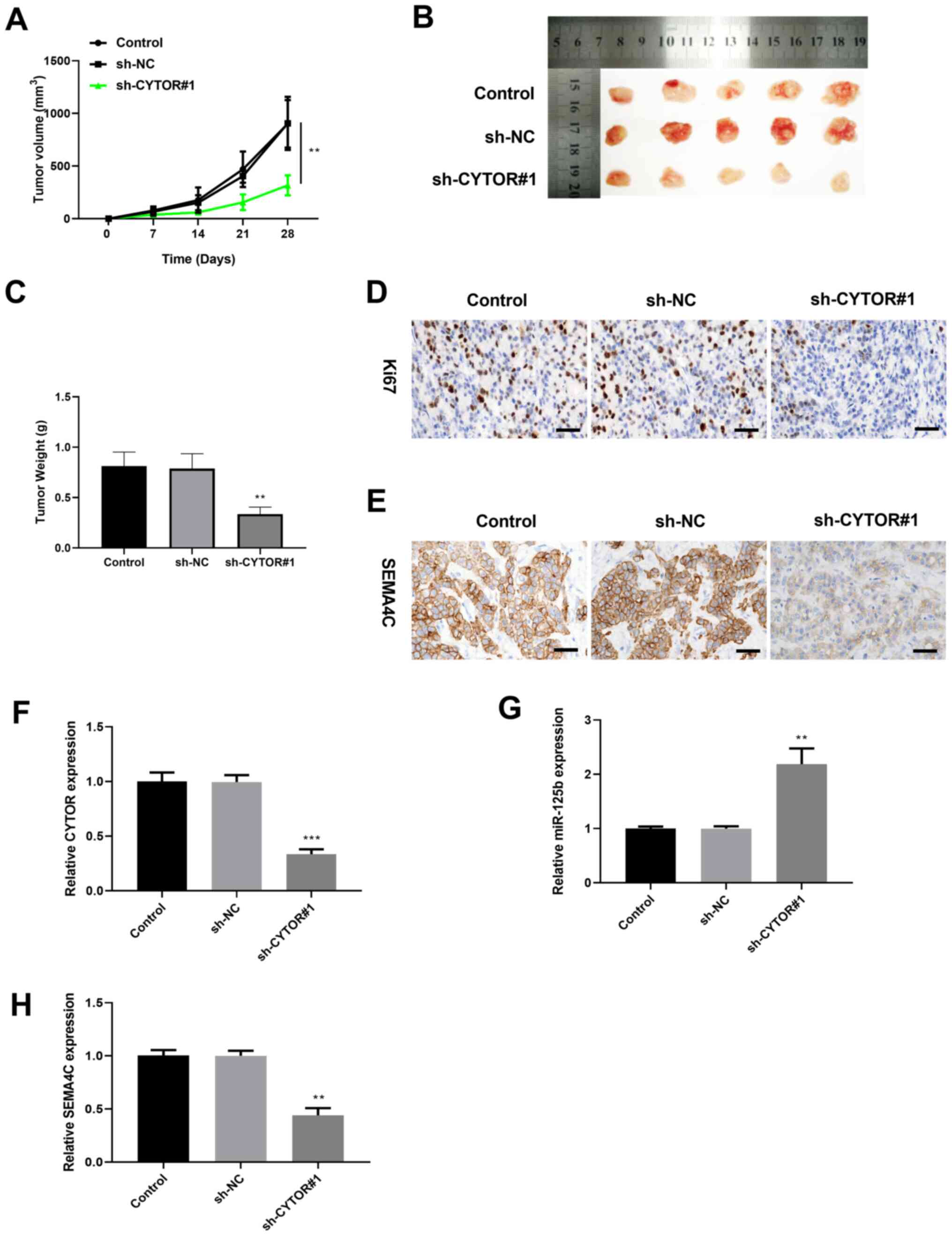
CYTOR knockdown inhibits tumor growth
in vivo
Huh7 cells transfected with sh-CYTOR#1 were injected
into nude mice to determine the effect of CYTOR on the HCC tumor
growth. As presented in Fig. 6A and
B, the tumor volume in the sh-CYTOR#1 group grew more slowly
during the 28 days observation in comparison with that in the sh-NC
group. Furthermore, the tumor weight in the sh-CYTOR#1 group was
significantly decreased compared with control group (Fig. 6C). The result from
immunohistochemistry demonstrated that the tumor cell proliferation
marker Ki67 and the expression of SEMA4C were decreased in the
sh-CYTOR#1 group compared with the control group (Fig. 6D and E). Furthermore, results from
RT-qPCR showed that CYTOR and SEMA4C expression levels were
significantly decreased, while miR-125b expression level was
significantly increased in tumor tissues of the sh-CYTOR#1 group
compared with tumor tissues of the sh-NC group (Fig. 6F-H). These findings indicated that
CYTOR knockdown could inhibit HCC tumor growth in vivo and
downregulate SEMA4C expression.
Discussion
HCC is the most common type of liver cancer and is
characterized by high morbidity and mortality rates worldwide
(16). Significant progress in
understanding the molecular mechanisms of HCC has been made,
especially when the competing endogenous RNA hypothesis was
presented (17). Currently, research
has focused on lncRNAs, which function primarily through
interactions with cellular macromolecules, such as proteins and
RNAs (18–20). Understanding the molecular mechanism
of some lncRNAs in HCC progression is therefore crucial.
The lncRNA CYTOR has been reported to modulate the
pathological process of various types of cancer. For example, CYTOR
serves an oncogene role in the development of colorectal cancer by
interacting with nucleolin and KH RNA binding domain containing,
signal transduction associated 1 (21). CYTOR knockdown was demonstrated to
attenuate the migration and invasion of colon cancer cells, while
CYTOR overexpression can enhance the metastatic properties of colon
cancer cells (22). Liang et
al (23) revealed that CYTOR is
upregulated in gastric cancer and HCC, and that elevated CYTOR
expression is closely associated with the poor prognosis of
patients with gastric cancer and HCC. The present study
demonstrated that CYTOR was upregulated in HCC tissues and that
CYTOR expression was associated with the poor prognosis of patients
with HCC. Furthermore, HCC cell proliferation was significantly
decreased following CYTOR knockdown whereas it was significantly
increased by CYTOR overexpression. The levels of apoptotic-related
proteins were significantly elevated after CYTOR knockdown, while
they were significantly decreased following CYTOR overexpression.
Taken together, these findings indicated that CYTOR may serve an
oncogenic role in HCC.
Numerous studies have confirmed that lncRNAs play
key roles in various types of cancer by sponging miRNAs and
consequently regulating gene expression (24,25). In
HCC, SOX9-AS1 was reported to stimulate tumor growth and metastasis
in HCC through SOX9 and the Wnt/β-catenin pathway (8). Furthermore, upregulation of cancer
susceptibility 15 (CASC15) can enhance the tumorigenicity and
epithelial to mesenchymal transition of HCC by increasing the
expression of twist family BHLH transcription factor 1 via sponging
to miR-33a-5p (26). Zhang et
al (27) reported that the
lncRNA X-inactive specific transcript sponges miR-497-5p to promote
cell proliferation and migration in HCC. To identify the underlying
mechanism of CYTOR in HCC, previous bioinformatics analysis
revealed that miR-125b, which was previously discovered as a tumor
suppressor in the modulation of HCC progression (28), might be considered as a potential
target of CYTOR. The present study reported a negative correlation
between CYTOR expression and miR-125b expression, and the results
from luciferase reporter assay confirmed that miR-125b may be a
direct target of CYTOR. Importantly, miR-125b inhibitor could
rescue the inhibitory effect of sh-CYTOR#1 on HCC cell
proliferation and attenuated the promoting effect of sh-CYTOR#1 on
cell apoptosis.
SEMA4C, a member of the semaphorin family, has been
reported to be highly expressed in several human cancers and to
stimulate the proliferation, migration and invasion of cancer cells
(29,30). SEMA4C was previously reported as a
functional target of miR-125b in lung cancer (31). The present study demonstrated that
miR-125b was bound to the 3′-UTR of SEMA4C and could negatively
modulate SEMA4C expression according to the starBase bioinformatics
database analysis and the luciferase assay. Previous studies have
reported that abnormal expression of SEMA4C exists in various
tumors. For instance, SEMA4C expression is significantly elevated
in breast cancer and cervical cancer (32,33). Liu
et al (34) demonstrated that
SEMA4C is upregulated in HCC tissues and promotes tumor growth and
cell metastasis in HCC, which is consistent with the results from
the present study showing that SEMA4C expression was significantly
increased in HCC tissues. The present study also revealed that
CYTOR promoted the expression of SEMA4C in HCC cells by decreasing
the expression of miR-125b.
This study presented some limitations. Firstly, the
relatively small sample size may lead to errors in the analysis of
clinicopathological characteristics and overall survival of
patients. Secondly, the small number of animals and human
observation errors may also lead to statistical errors. Thirdly,
the impact of the CYTOR/miR-125b/SEMA4C axis on cell invasion, cell
migration and cell cycle, in vitro and in vivo,
should be further investigate din the future
In summary, the findings form the present study
demonstrated that CYTOR was highly expressed in HCC tissues and
cells and could stimulate HCC cell proliferation and tumor growth
via modulating the miR-125b/SEMA4C axis. The CYTOR/miR-125b/SEMA4C
axis may therefore provide a new perspective for the development of
novel therapeutic strategy for HCC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Tianjin Science and
technology plan project (grant no. 19ZXDBSY00010).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QT performed the experiements, guaranteed the
integrity of the entire study and participated in writing the
manuscript. XY, LY, ZL and ZY collected the clinical samples,
performed the data analysis and statistical analysis. YZ designed
the study, supervised the research and participated in the writing
and reviewing of the manuscript. All authors read and approved the
final manuscript. QT and YZ confirmed the authenticity of all the
raw data.
Ethics approval and consent to
participate
The experiments using human samples were approved by
the Clinical Research Ethics Committee of Tianjin First Central
Hospital (approval no. 2019N013YY). Signed written informed
consents were obtained from the patients and/or guardians. The
animal protocol was approved by the Animal Care and Use Committee
(approval no. 2019N046YY).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Michelotti GA, Machado MV and Diehl AM:
NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol.
10:656–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu JJ, Xiao W, Dong SL, Liang HF, Zhang
ZW, Zhang BX, Huang ZY, Chen YF, Zhang WG, Luo HP, et al: Effect of
surgical liver resection on circulating tumor cells in patients
with hepatocellular carcinoma. BMC Cancer. 18:8352018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hu X, Jiang J, Xu Q, Ni C, Yang L and
Huang D: A systematic review of long noncoding RNAs in
hepatocellular carcinoma: molecular mechanism and clinical
implications. BioMed Res Int. 2018:81262082018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhan A, Soleimani M, Mandal SS and Long
Noncoding RNA: Long noncoding RNA and cancer: A new paradigm.
Cancer Res. 77:3965–3981. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang X, Yao B, Niu Y, Chen T, Mo H, Wang
L, Guo C and Yao D: Hypoxia-induced lncRNA EIF3J-AS1 accelerates
hepatocellular carcinoma progression via targeting
miR-122-5p/CTNND2 axis. Biochem Biophys Res Commun. 518:239–245.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng J, Yang G, Liu Y, Gao Y, Zhao M, Bu
Y, Yuan H, Yuan Y, Yun H, Sun M, et al: LncRNA PCNAP1 modulates
hepatitis B virus replication and enhances tumor growth of liver
cancer. Theranostics. 9:5227–5245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang W, Wu Y, Hou B, Wang Y, Deng D, Fu Z
and Xu Z: A SOX9-AS1/miR-5590-3p/SOX9 positive feedback loop drives
tumor growth and metastasis in hepatocellular carcinoma through the
Wnt/β-catenin pathway. Mol Oncol. 13:2194–2210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ji D, Hu G, Zhang X, Yu T and Yang J: Long
non-coding RNA DSCAM-AS1 accelerates the progression of
hepatocellular carcinoma via sponging miR-338-3p. Am J Transl Res.
11:4290–4302. 2019.PubMed/NCBI
|
|
10
|
Pang Q, Ge J, Shao Y, Sun W, Song H, Xia
T, Xiao B and Guo J: Increased expression of long intergenic
non-coding RNA LINC00152 in gastric cancer and its clinical
significance. Tumour Biol. 35:5441–5447. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li M, Wang Q, Xue F and Wu Y: lncRNA-CYTOR
Works as an oncogene through the CYTOR/miR-3679-5p/MACC1 axis in
colorectal cancer. DNA Cell Biol. 38:572–582. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu Y, Li M, Yu H and Piao H: lncRNA CYTOR
promotes tamoxifen resistance in breast cancer cells via sponging
miR 125a 5p. Int J Mol Med. 45:497–509. 2020.PubMed/NCBI
|
|
13
|
Zhang J and Li W: Long noncoding RNA CYTOR
sponges miR-195 to modulate proliferation, migration, invasion and
radiosensitivity in nonsmall cell lung cancer cells. Biosci Rep.
38:382018. View Article : Google Scholar
|
|
14
|
Thuluvath PJ, To C and Amjad W: Role of
locoregional therapies in patients with hepatocellular cancer
awaiting liver transplantation. Am J Gastroenterol. 116:57–67.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ruggieri A, Barbati C and Malorni W:
Cellular and molecular mechanisms involved in hepatocellular
carcinoma gender disparity. Int J Cancer. 127:499–504. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Böhmdorfer G and Wierzbicki AT: Control of
chromatin structure by long noncoding RNA. Trends Cell Biol.
25:623–632. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Yu H, Sun W, Kong J, Zhang L, Tang
J, Wang J, Xu E, Lai M and Zhang H: The long non-coding RNA CYTOR
drives colorectal cancer progression by interacting with NCL and
Sam68. Mol Cancer. 17:1102018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yue B, Liu C, Sun H, Liu M, Song C, Cui R,
Qiu S and Zhong M: A Positive Feed-Forward Loop between
LncRNA-CYTOR and Wnt/β-Catenin Signaling Promotes Metastasis of
Colon Cancer. Mol Ther. 26:1287–1298. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang J, Wei X, Liu Z, Cao D, Tang Y, Zou
Z, Zhou C and Lu Y: Long noncoding RNA CYTOR in cancer: A TCGA data
review. Clin Chim Acta. 483:227–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ,
Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al: LncRNA HULC enhances
epithelial-mesenchymal transition to promote tumorigenesis and
metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1
signaling pathway. Oncotarget. 7:42431–42446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Chen G, Yan Y and Fan Q: CASC15
promotes epithelial to mesenchymal transition and facilitates
malignancy of hepatocellular carcinoma cells by increasing TWIST1
gene expression via miR-33a-5p sponging. Eur J Pharmacol.
860:1725892019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Zhu Z, Huang S, Zhao Q, Huang C,
Tang Y, Sun C, Zhang Z, Wang L, Chen H, et al: lncRNA XIST
regulates proliferation and migration of hepatocellular carcinoma
cells by acting as miR-497-5p molecular sponge and targeting PDCD4.
Cancer Cell Int. 19:1982019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hua S, Quan Y, Zhan M, Liao H, Li Y and Lu
L: miR-125b-5p inhibits cell proliferation, migration, and invasion
in hepatocellular carcinoma via targeting TXNRD1. Cancer Cell Int.
19:2032019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li J, Wang Q, Wen R, Liang J, Zhong X,
Yang W, Su D and Tang J: MiR-138 inhibits cell proliferation and
reverses epithelial-mesenchymal transition in non-small cell lung
cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med.
19:2793–2805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Q, Wang Y, Lu X, Zhao Z, Zhu L, Chen
S, Wu Q, Chen C and Wang Z: MiR-125b regulates
epithelial-mesenchymal transition via targeting Sema4C in
paclitaxel-resistant breast cancer cells. Oncotarget. 6:3268–3279.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y and Huang S: Up-regulation of
miR-125b reverses epithelial-mesenchymal transition in
paclitaxel-resistant lung cancer cells. Biol Chem. Aug
20–2015.(Epub ahead of print). doi: 10.1515/hsz-2015-0153.
View Article : Google Scholar
|
|
32
|
Yang J, Zeng Z, Qiao L, Jiang X, Ma J,
Wang J, Ye S, Ma Q, Wei J, Wu M, et al: Semaphorin 4C Promotes
macrophage recruitment and angiogenesis in breast cancer. Mol
Cancer Res. 17:2015–2028. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song J and Li Y: miR-25-3p reverses
epithelial-mesenchymal transition via targeting Sema4C in
cisplatin-resistance cervical cancer cells. Cancer Sci. 108:23–31.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu J, Lin Y, Li F, Ye H, Zhou R, Jin Y, Li
B, Xiong X and Cheng N: MiR-205 suppresses tumor growth, invasion,
and epithelial-mesenchymal transition by targeting SEMA4C in
hepatocellular carcinoma. FASEB J. May 25–2018.doi:
10.1096/fj.201800113R.
|