Introduction
Malignant pleural mesothelioma (MPM) is an aggressive neoplasm that arises from mesothelial cells that line the serosal membrane of the pleura (1). MPM requires accurate and early diagnosis as early treatment improves patient prognosis (2). However, it is difficult to distinguish MPM from benign mesothelial proliferative disorders in routine practice due to overlaps in morphology (3). Thus, genomic-based ancillary assays, such as fluorescence in situ hybridization (FISH) to detect homozygous deletion (homo-d) of the 9p21 locus and immunohistochemistry (IHC) analysis to detect loss of BRCA1-associated protein 1 (BAP1) expression and methylthioadenosine phosphorylase [MTAP; the protein product of the MTAP gene located in the telomere side of cyclin-dependent kinase inhibitor 2A (CDKN2A) in the 9p21 locus (3)] proteins, are effective tools for detecting characteristic genetic abnormalities that are essential in the diagnosis of MPM (3,4).
The CDKN2A gene, located at the 9p21 locus (5), is a tumor suppressor gene which encodes the p16 protein (5). Homo-d of CDKN2A is one of the most frequent genetic abnormalities in malignant mesothelioma and is found in 50–65.8% of MPM cases (6,7). CDKN2A FISH is a common assay used to detect this deletion, and a 222 kilobase (kb) CDKN2A probe (long probe, L-probe) is a commercially available and the most frequently used FISH probe, which covers an area of 222 kb, including the CDKN2A, CDKN2B and MTAP genes. Another commercially available probe is the 57 kb CDKN2A probe (short probe, S-probe) that covers an area of 57 kb, confined to the CDKN2A and CDKN2B genes (6,7). FISH reveals heterozygous deletion (hetero-d) signals in addition to homo-d and normal signals (8). Recently, CDKN2A FISH with L-probe revealed high proportions of hetero-d, while S-probe yielded high proportions of homo-d, with low hetero-d (8).
Microdeletion, defined as a deletion <200 kb, can induce false negative results of signal deletion in FISH assays, owing to redundant probe reactivity (9–13). Some of the high hetero-d signals may result from microdeletions in MPM (8). It was hypothesized that an S-probe can effectively detect microdeletion status of the CDKN2A gene, and thus increase homo-d rate in MPM, which has a high hetero-d and low homo-d status with L-probe (8).
The present study aimed to evaluate the effectiveness of S-probe in diagnosing MPM. In routine practice, it is important to confirm homo-d of the CDKN2A gene for an accurate diagnosis of MPM, to start treatment and predict prognosis. Borderline cases are infrequently found in which FISH with L-probe reveals high hetero-d and low homo-d; however, it is worth performing FISH with S-probe if homo-d of CDKN2A is detected (2). Thus, the aforementioned hypothesis was investigated in four cases of MPM, in which CDKN2A FISH with L-probe yielded high hetero-d and low homo-d. The results of FISH with S-probe were assessed in association with the deletion range evaluated via quantitative (q)PCR analysis.
Materials and methods
Case selection
CDKN2A FISH results were reviewed in 106 MPM cases that were diagnosed at the Department of Pathology of Fukuoka University Hospital (Fukuoka, Japan) between January 2017 and December 2019, including consultation cases. A total of five cases of MPM exhibited high hetero-d and low homo-d pattern in CDKN2A FISH. A total of four cases were used in the present study, owing to the presence of redundant tissue, for further investigation. All four samples were obtained from male patients, the mean age at diagnosis was 68.5 years (age range, 63–80 years). Pathological diagnoses were performed according to the 2021 World Health Organization classification of thoracic tumors (1). The mesothelial origin was confirmed by positive reactivity for Wilms tumor 1, calretinin or podoplanin (D2-40) and negative reactivity for epithelial cell adhesion molecule (BerEP4), thyroid transcription factor-1, claudin-4 or carcinoembryonic antigen. The present study was approved by the Institutional Review Board of Fukuoka University (approval no. 11-7-11). The requirement for informed consent was waived as the anonymous use of redundant tissues is part of the standard treatment agreement with patients, when no objection is expressed.
IHC staining
For immunostaining, 4-µm-thick sections of formalin-fixed paraffin embedded (FFPE) tissues were used. Sections were deparaffinized and rehydrated with xylene, alcohol and tap water. IHC analysis was performed on an automated DAKO Omnis platform using the DAKO EnVision FLEX, High pH (Omnis https://www.agilent.com/ja-jp/product/dako-omnis-solution-for-ihc-ish/dako-omnis/dako-omnis-75902, code GV800), according to the manufacturer's instructions. Heat-epitope retrieval was performed using a high pH for 30 min at 97°C. Protein blocking was performed using Protein block Serum-Free (Agilent Technologies, Inc.) for 20 min at 25°C. The slides were subsequently incubated using one of the following antibodies: Mouse monoclonal anti-calretinin antibody (clone Calret1, cat. no. M7245, Dako; Agilent Technologies, Inc., ready to use; room temperature for 40 min), mouse monoclonal anti-BAP1 antibody (clone C-4, cat. no. sc-28383, Santa Cruz Biotechnology, Inc.; 1:100 dilution; room temperature for 1 h) or mouse monoclonal anti-MTAP antibody (clone 2G4, cat. no. H00004507-M01J, Abnova; 1:100 dilution; room temperature for 45 min). Endogenous peroxidase was inactivated using peroxidase blocking reagent (Dako; Agilent Technologies, Inc.) for 3 min at room temperature. Subsequently, slides were incubated with a peroxidase-labeled polymer (EnVision Flex/HRP, cat. no. DM842; Agilent Technologies, Inc.), at room temperature for 20 min). The slides were visualized with 3,3′-diaminobenzidine at room temperature for 5 min and counterstained with hematoxylin at room temperature for 5 min. Analysis was performed using an Olympus BX 53 light microscope (magnification, ×400). Each specimen was evaluated by two authors (YO and KN).
BAP1 and MTAP staining results were scored as loss or retained. BAP1 loss in tumor cells was defined as complete loss of nuclear staining. MTAP loss in tumor cells was defined as complete loss of cytoplasmic staining in the area where internal positive control cells (lymphocytes, histiocytes, fibroblasts, pneumocytes or endothelial cells) exhibited cytoplasmic and nuclear MTAP staining (14).
FISH assay
CDKN2A FISH was performed using 4-µm-thick FFPE tissue sections. Sections were deparaffinized, rehydrated in 100, 90 and 70% ethanol, washed with 2× saline sodium citrate (SSC) (UltraPure™ 20X SSC, Invitrogen; Thermo Fisher Scientific, Inc.), incubated in pretreatment solution (PathVysion HER2 DNA probe kit; 30 min at 80°C), and digested with pepsin solution (Sigma-Aldrich; Merck KGaA, for 3 h at 37°C). Refixation was performed using 10% formalin (10 min at room temperature); sections were subsequently washed with 2× SSC, dehydrated in ethanol, dried and exposed to either of two DNA probe: 222 kb Vysis LSI CDKN2A/CEP9 probe alone (Abbott Japan LLC) or 57 kb SureFISH 9p21.3 CDKN2A (Agilent Technologies, Inc.) probe and 367 kb SureFISH Chr9 CEP FISH probe (Agilent Technologies, Inc,). Denaturation (10 min at 80°C) and hybridization (24 h at 37°C) were performed in ThermoBrite (Abbott Japan LLC.). After 24 h, the specimens were treated with 2× SSC containing 0.3% NP 40 (Nonidet P-40, nacalai tesque) (2 min at 72°C), 2× SSC containing 0.1% NP 40 (5 min at room temperature) and 2× SSC (5 min at room temperature). Nuclei were counterstained with DAPI using antifade reagent for 24 h at 4°C (VECTASHIELD Mounting Medium, Vector Laboratories, Inc.). Analysis was performed using a fluorescence microscope (magnification, ×1,000) (Axio imager Z1; Carl Zeiss AG) and Isis FISH imaging system (Metasystems, http://metasystems-international.com/en/products/isis). FISH signal was classified under three signal patterns: Normal (two red and two green signals), hetero-d (one red and two green signals) and homo-d (no red and two green signals), and >100 tumor cells were assessed for each case.
DNA extraction
A total of 6/7 sections of 10-µm FFPE-cut specimens were used for each case. Before extracting DNA, macrodissection was performed to purify the tumor samples, with the exception of case 4 where the sample was too small. Genomic DNA was extracted using a RecoverAllTM Total Nucleic Acid Isolation kit (Ambion; Thermo Fisher Scientific, Inc.; cat. no. AM1975), according to the manufacturer's instructions, with one modification: Protease digestion was performed for 20–24 h. To confirm the quality of extracted DNA samples, the positive control (β-globin) primer (forward, 5′-ACACAACTGTGTTCACTAGC-3′ and reverse, 5′-CAACTTCATCCACGTTCACC-3′) region was amplified via PCR using KOD FX Neo (Toyobo, Ltd., cat. no. KFX-201). The PCR reaction protocol was as follows: 94°C for 15 min, followed by 35 cycles of denaturation at 98°C for 10 sec, annealing at 55°C for 30 sec, and extension at 68°C for 30 sec. The PCR products were detected by electrophoresis of 3% agarose gel with GelRed™ Nucleic Acid Gel Stain (10,000×) in DMSO (Biotium, Inc.), visualized by ultraviolet light.
qPCR
The extracted DNA from tumor tissue samples was used for qPCR. The chromosome map of the 9p21 region and the targeting regions of the two commercially available CDKN2A FISH probes are presented in Fig. 1. The seven primer sets for analyzing the deleted region and the reference gene region were designed by custom oligo primers (Sigma-Aldrich; Merck KGaA) (Table I). Primer A was designed for the MTAP gene, primers B-D were designed for the non-coding region between the CDKN2A and MTAP genes and primers E and F were designed for the CDKN2A gene. The copy number of the regions was quantified via qPCR analysis of genomic DNA using KOD SYBR® qPCR Mix (Toyobo Life Science). The PCR reaction protocol was as follows: 98°C for 15 min, followed by 40 cycles of denaturation at 98°C for 10 sec, annealing at 60°C (primer A, primer F, RPS6) or 62°C (primers B, C, D and E) for 10 sec, and extension at 68°C for 30 sec. Melting curve analysis was also performed for non-specific products. The copy number was determined by the relative standard curve method using the RPS6 gene (9p22) (15) as the reference gene according to the previous reports (9,15), and the calculated numbers were evaluated as follows: ≥0.8 as no deletion, >0.5 and <0.8 as a one-allele deletion, and ≤0.5 as a two-allele deletion.
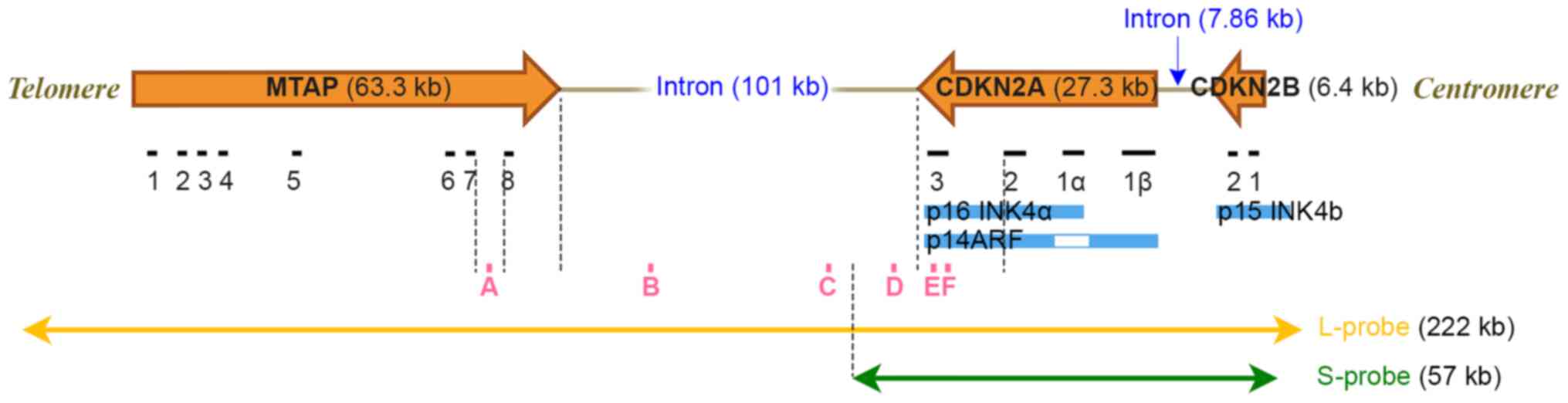 |
Figure 1.
Chromosome map of the 9p21 region and CDKN2A FISH probe targeting areas. The L-probe covers areas up to 222 kb, which includes the CDKN2A, CDKN2B and MTAP genes. The S-probe only covers areas up to 57 kb, which includes the CDKN2A and CDKN2B genes. A, B, C, D, E and F are the locations of each PCR product used in quantitative PCR analysis. CDKN, cyclin-dependent kinase inhibitor; MTAP, methylthioadenosine phosphorylase; L-probe, long probe; S-probe, short probe.
|
 |
Table I.
Primer sets.
|
Table I.
Primer sets.
| Set |
Primer |
Sequence (5′-3′) |
Target region (gene) |
Product size (bp) |
| A |
F |
GGGCACTTTGTGACTCTTCTTAACC |
MTAP |
105 |
| |
R |
GGAGACTTTGGGGTATGGTCCTC |
|
|
| B |
F |
TACAGGTAGGGAAGCAGGCAATC |
Non-coding region between |
90 |
| |
R |
TATGGGTATGCTCTGGTCTTCTGC |
MTAP and CDKN2A |
|
| C |
F |
GCGATTGCTGCCAAGATACCC |
Non-coding region between |
89 |
| |
R |
TCCACAGGCCTTTGTCTCCAG |
MTAP and CDKN2A |
|
| D |
F |
TTCCGGACACACTGGGTCAC |
Non-coding region between |
99 |
| |
R |
GTTTACAGGCAAGTGAGGTTGTCC |
MTAP and CDKN2A |
|
| E |
F |
GCCTGTTTTCTTTCTGCCCTCTGC |
CDKN2A |
89 |
| |
R |
GACTGATGATCTAAGTTTCCCGAGGTTTCTC |
|
|
| F |
F |
CCACATCTTCACGCCTTCGC |
CDKN2A |
83 |
| |
R |
AGGGTTGCAAGAAGAAAACGAGTG |
|
|
| RPS6 |
F |
CTACTGAGTAAGGGGCATTCCTGT |
The reference gene |
102 |
| |
R |
GAGAACGCTCAGATTTGCATCCAC |
|
|
Results
Clinicopathological characteristics
The clinicopathological characteristics are summarized in Table II. All four samples were obtained from male patients, the mean age at diagnosis was 68.5 years (age range, 63–80 years). A total of two samples were obtained following pleural biopsy, one following extrapleural pneumonectomy and one following transbronchial lung biopsy (TBLB) from a metastatic lesion. All cases were epithelioid MPM in terms of histological subtype (Fig. 2A, D, G and J). Mesothelial phenotype was confirmed by positive immunostaining for calretinin (Fig. 2B, E, H and K). IHC analysis of BAP1 protein (Fig. 2C, F, I and L) was as follows: Three cases with BAP1 loss and one with BAP1 retained. The mean period from diagnosis to mortality was 2.6 years (3–90 months).
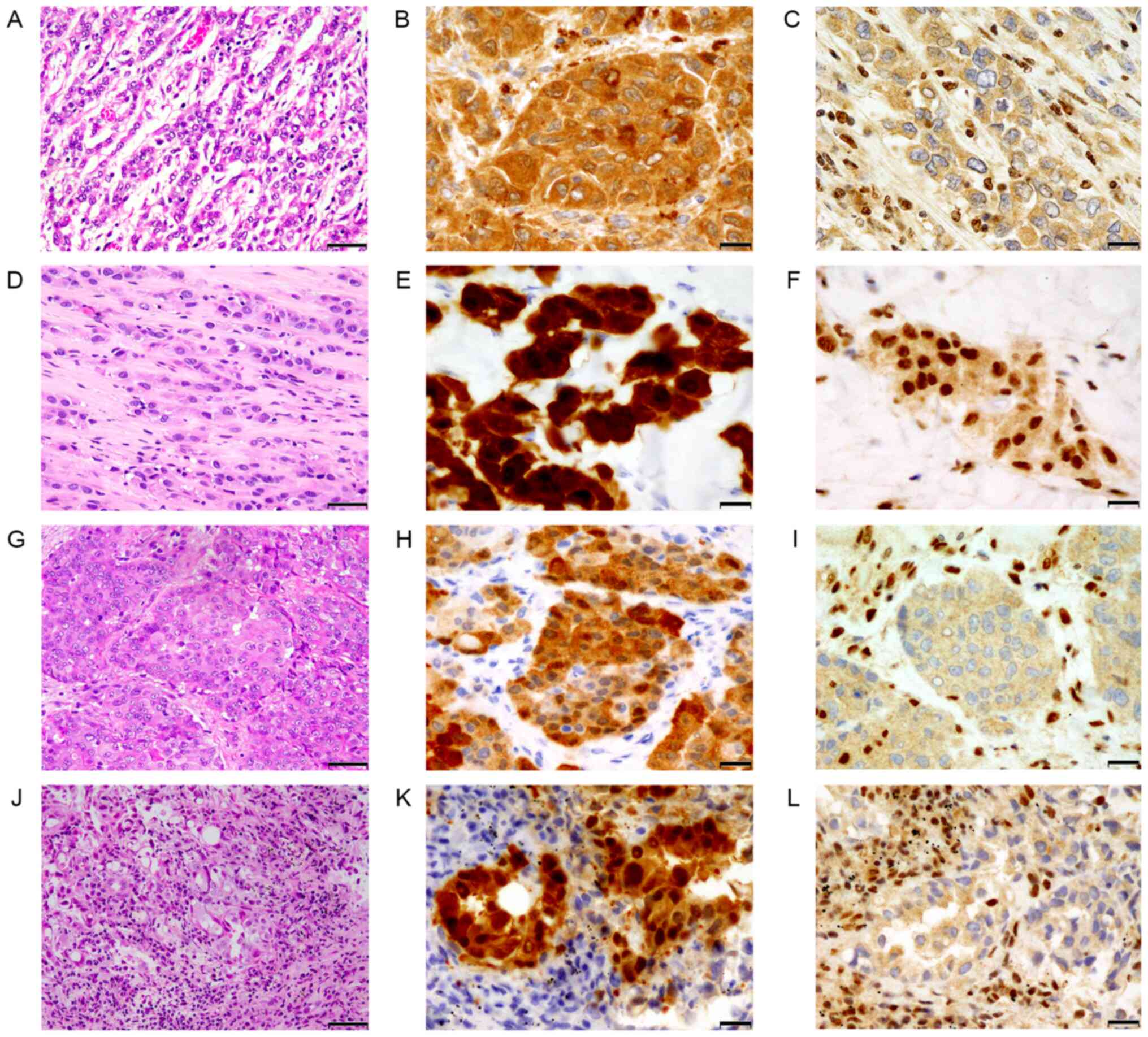 |
Figure 2.
Histology of malignant pleural mesothelioma (A-C, case 1; D-F, case 2; G-I, case 3 and J-L, case 4). (A, D, G and J) The tumor exhibited proliferation of atypical epithelioid tumor cells with eosinophilic cytoplasm, arranged in cords or small nests. (B, E, H and K) Tumor cells were positive for calretinin. (C, I and L) Cases 1, 3 and 4 exhibited BAP1 loss, (F) whereas case 2 retained BAP1 expression. Olympus BX 53 light microscope. (A, D, G and J) Magnifications: ×200 scale bar, 50 µm). (B, C, E, F, H, I, K and L) Magnification: ×400 scale bar, 20 µm. BAP1, BRCA1-associated protein 1.
|
 |
Table II.
Clinicopathological characteristics of the four MPM cases assessed in the present study.
|
Table II.
Clinicopathological characteristics of the four MPM cases assessed in the present study.
| Characteristic |
Case 1 |
Case 2 |
Case 3 |
Case 4 |
| Age, years |
62 |
78 |
68 |
66 |
| Sex |
Male |
Male |
Male |
Male |
| Histology |
Epithelioid |
Epithelioid |
Epithelioid |
Epithelioid |
| MTAP IHC |
Retained |
Retained |
Retained |
Retained |
| BAP1 IHC |
Loss |
Retained |
Loss |
Loss |
| Source of tissue |
EPP |
Pleural biopsy |
Pleural biopsy |
TBLB |
| Treatment |
EPP + radiation; CDDP + PEM; GEM + VNR |
CBDCA + PEM |
Chemotherapy |
None |
| Time from diagnosis to mortality |
7 years and 6 months |
2 years and 2 months |
5 months |
3 months |
MTAP IHC and CDKN2A FISH results
A total of 3/4 cases exhibited strong nuclear and cytoplasmic or cytoplasmic only expression for MTAP protein (Fig. 3A, G and J), while case 2 exhibited relatively weak cytoplasmic reactivity for MTAP (Fig. 3D). With the L-probe, CDKN2A FISH revealed high hetero-d signals in all cases (cases 1–4; 73.3, 37.1, 59.2 and 64.8%, respectively) (Fig. 3B, E, H and K) compared with homo-d signals (cases 1–4; 12.1, 12.4, 25.4 and 22.2%, respectively). In the hetero-d signals with L-probe, one red signal was slightly smaller than two CEP9 green signals. However, the CDKN2A FISH with S-probe exhibited high homo-d in all cases (cases 1–4; 96.8, 90.0, 87.5 and 82.6%, respectively) (Fig. 3C, F, I and L) compared with low hetero-d (cases 1–4; 0.0%, 1.2, 1.2 and 4.3%, respectively). In the control tissues, CDKN2A FISH with S-probe exhibited normal signals (Fig. S1). The comparative line graph of homo-d (solid line) and hetero-d (dotted line) demonstrated the increased proportions of homo-d and decreased proportions of hetero-d in FISH with S-probe compared with L-probe (Fig. 4).
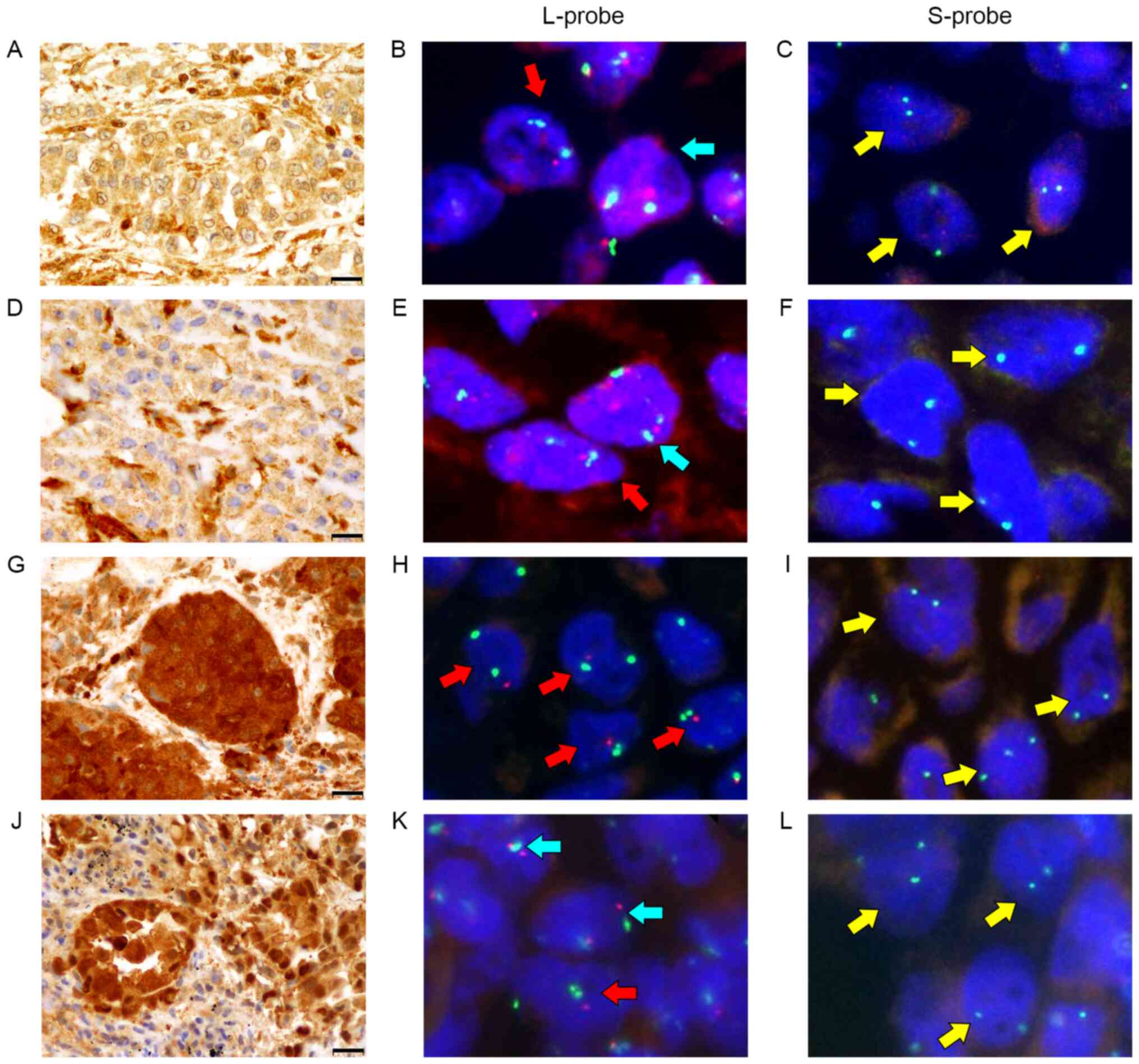 |
Figure 3.
MTAP immunostain and two cyclin-dependent kinase inhibitor 2A fluorescence in situ hybridization probes (L-probe and S-probe). Summary of the four cases (A-C, case 1; D-F, case 2; G-I, case 3 and J-L, case 4) are presented. (A, G and J) Cases 1, 3, 4 exhibited strong MTAP expression in both the nuclei and cytoplasm, (D) while case 2 exhibited weak cytoplasmic positivity for MTAP protein. (B, E, H and K) All cases exhibited a high hetero-d signal (red arrow) compared with normal signal (blue arrow) and homo-d signal (yellow arrow) in L-probe. (C, F, I and L) All cases exhibited a high rate of homo-d signal in S-probe. (A, D, G and J) Olympus BX 53 light microscope (magnification, ×400; scale bar, 20 µm). (B, C, E, F, H, I, K and L) Axioplan2 imaging fluorescence microscope. MTAP, methylthioadenosine phosphorylase; L-probe, long probe; S-probe, short probe.
|
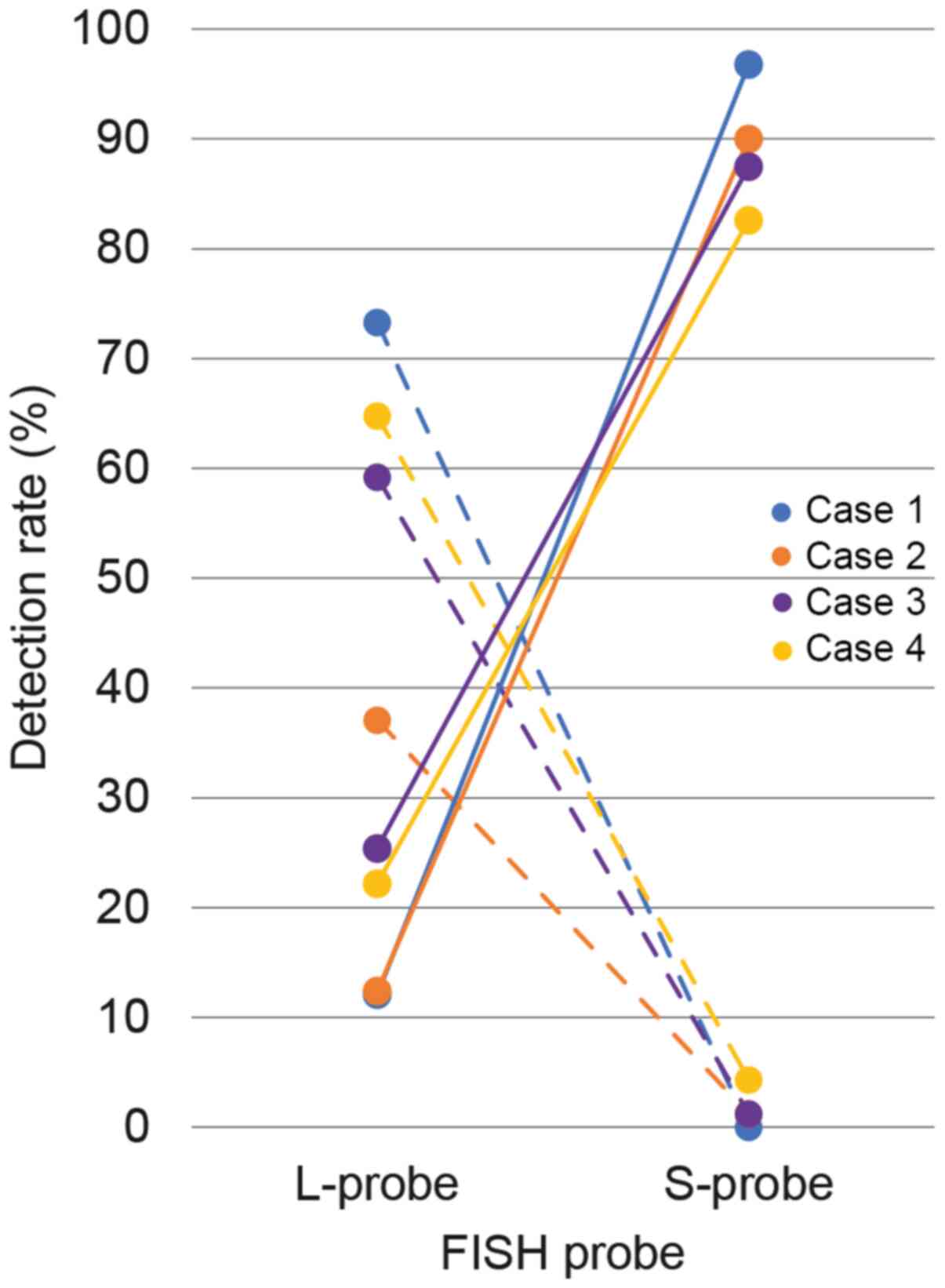 |
Figure 4.
Comparative line graph of homo- and hetero-d rates between the L- and S-probes. The vertical line represents the detection rate of homo-d (solid line) and hetero-d (dashed line) signals, while the horizontal line represents the corresponding results of L- and S-probes. For the L-probe, the hetero-d rate was high compared with the homo-d signal for each case. However, the homo-d rate increased by the S-probe. L-probe, long probe; S-probe, short probe; homo-d, homozygous deletion; hetero-d; heterozygous deletion; FISH, fluorescence in situ hybridization.
|
qPCR
qPCR analysis revealed no allele deletions in the MTAP gene in all cases, whereas two-allele deletions were detected in the CDKN2A gene in 3/4 cases (Fig. 5). Case 4 exhibited one-allele deletion in the CDKN2A gene. Deletion status of the non-coding region between the CDKN2A and MTAP genes varied, two-allele deletions for case 2 and one-allele deletion for cases 1, 3 and 4, respectively.
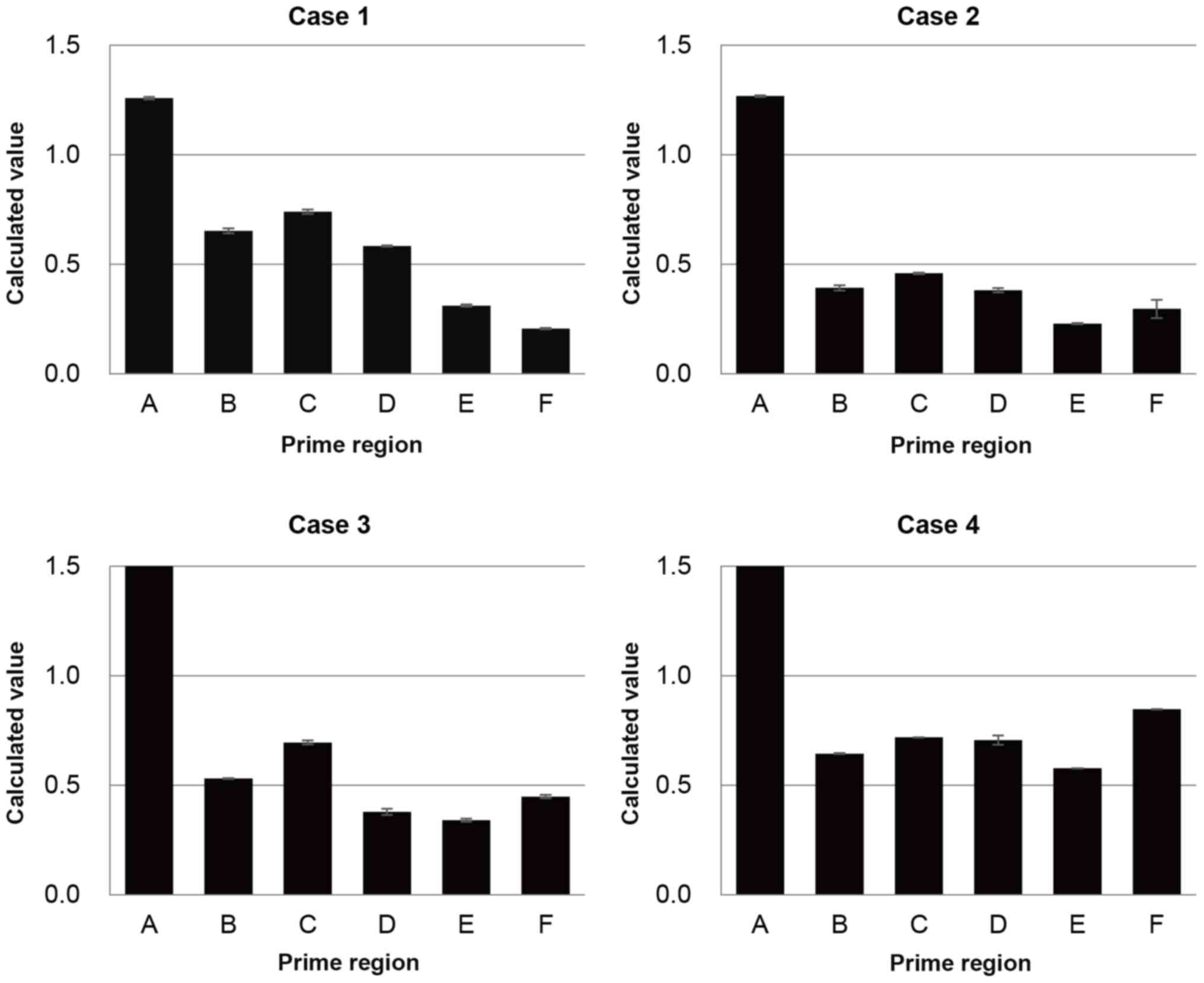 |
Figure 5.
qPCR analysis in the 9p21 locus. qPCR analysis was performed to detect the deletion status of the 9p21 regions. The MTAP gene (primer A) had no allele deletion, while two-allele deletions were observed for the CDKN2A gene (primer E and F) in all cases except for case 4. Deletion status of the non-coding region between the CDKN2A and MTAP genes (primer B-D) was two- or one-allele deletion for each case. qPCR, quantitative PCR; MTAP, methylthioadenosine phosphorylase; CDKN2A, cyclin-dependent kinase inhibitor 2A.
|
Discussion
To the best of our knowledge, the preset study was the first to demonstrate that CDKN2A FISH with S-probe is more effective in elucidating short homo-d in the 9p21 locus compared with L-probe in MPM. CDKN2A FISH with conventional L-probe revealed high hetero-d (range 37.1–73.3%) and low homo-d (range 12.1–25.4%) in all four cases with microdeletions, whereas that with S-probe yielded high proportions of homo-d (82.6–96.8%). Furthermore, qPCR analysis in these four cases demonstrated that 3/4 cases exhibited homo-d in the coding region of the CDKN2A gene, while case 4 exhibited hetero-d, according to the calculated value. In case 4, two-allele deletions were not confirmed via qPCR analysis despite the clear evidence of homo-d exhibited by FISH as the TBLB section was too small to allow macrodissection prior to qPCR. Thus, it was speculated that contamination of normal cells would affect the qPCR results. Based on these results, the size of deletions was ~27.3–142.5 kb at most. The results of the present study suggest it is worth performing CDKN2A FISH with S-probe when CDKN2A FISH with conventional L-probe results in high hetero-d and low homo-d.
The deletion size of the 9p21 locus found in MPM is usually 2–3 megabase in in vitro studies using cell lines; this range is large enough to accommodate the MTAP gene, as well as the CDKN2A and CDKN2B genes (16). However, in tumor specimens, the deleted region varies in size and range (9,10). Microdeletion in 9p21.3 is found in 32% of cases of acute lymphoblastic leukemia in adolescents and 10% of Ewing sarcoma (9,10). The smallest deletion is only 25 kb on the CDKN2A gene (10). Cases with CDKN2A microdeletion fail to exhibit high proportions of homo-d by FISH with L-probe, as the L-probe, which is designed to cover three genes in the 9p21 locus (CDKN2A, CDKN2B and MTAP) (10), is large enough to hybridize the non-deleted area next to the microdeleted CDKN2A gene, resulting in ‘pseudo’ hetero-d signals (9,13). In the present study, the false red signals were smaller compared with normal CEP9 green signals. Given the design of the L-probe and respective gene sizes, it is understandable that microdeletion cases exhibited high hetero-d and low homo-d with L-probe in CDKN2A FISH.
In the present study, MTAP expression was retained in all four cases. MTAP IHC is a useful surrogate assay for CDKN2A FISH as homo-d of MTAP always occurs in association with homo-d of CDKN2A (6,14). However, it would be favorable to perform CDKN2A FISH in cases with retained MTAP expression determined by IHC as retained MTAP protein expression does not necessarily rule out a microdeletion status of the CDKN2A gene (8,14).
Detection of CDKN2A deletion is important in routine practice as it effectively discriminates MPM from reactive mesothelial proliferations, and is associated with poor prognosis in MPM (3–6). There are several approaches used to circumvent the false negative results in detecting chromosomal microdeletion, including use of smaller probes (as used in the present study), enhancement of FISH signals (13), multiple ligation-dependent probe amplification (MLPA) assay (11) and array comparative genomic hybridization (CGH) (10). Practical use of S-probe can be somewhat challenging as the target red signal is small owing to the short probe length (9); thus, lower hybridization efficiency can result in false positive results for CDKN2A deletion (9). Therefore, careful establishment of an accurate FISH assay, which always reveals two red signals and two green signals in normal cells, is critical.
The present study is not without limitations. First, the sample size was too small. Prospective studies including S-probe performance in several MPM cases, that have high hetero-d and low homo-d status with L-probe, are warranted to verify the effectiveness of S-probe. Secondly, the S-probe can cause false negative results of signal deletion if the deletion range is too small (9–13). On such occasions, enhancement of FISH signals, MLPA assay and array CGH can be alternative approaches to detect CDKN2A gene deletion if MPM is highly suspected.
In the single institutional cohort assessed, 4.7% of MPM cases exhibited microdeletion of the CDKN2A gene, with high hetero-d and low homo-d detected by FISH with the conventional L-probe. The use of CDKN2A FISH with a short 57 kb probe can be an effective approach to detect a small homo-d in MPM. Furthermore, S-probe can increase the homo-d rate in MPM cases with high hetero-d with L-probe, and thus assist in solving diagnostic difficulties in cases involving high hetero-d with low homo-d.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms Hiroyo Fukagawa (Department of Pathology, Fukuoka University Hospital and School of Medicine, Fukuoka, Japan) for her assistance with qPCR analysis.
Funding
The present study was supported by grants from the Research Center for Advanced Molecular Medicine, Fukuoka University (grant no. 171032), the Ministry of the Environment (grant nos. 190557 and 200598) and Grants-in Aid for Scientific Research (grant no. 24590495) from the Japanese Society for the Promotion of Science.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
YO performed the experiments, drafted and revised the manuscript and analyzed the data. MH conceptualized and supervised the present study, and drafted and revised the manuscript. SM curated the data, acquired the data, analyzed and interpreted the data, and drafted and revised the manuscript. AS and TT acquired the resources, curated the data, acquired the data, and analyzed and interpreted the data, and drafted and revised the manuscript. KN conceptualized and supervised the present study, was project administrator, acquired funding, and drafted and revised the manuscript. YO, MH, and KN confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Review Board of Fukuoka University (Fukuoka, Japan; approval no. 11-7-11). The requirement for informed consent was waived as the anonymous use of redundant tissues is part of the standard treatment agreement with patients, when no objection is expressed.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Glossary
Abbreviations
Abbreviations:
|
MPM
|
malignant pleural mesothelioma
|
|
FISH
|
fluorescence in situ hybridization
|
|
homo-d
|
homozygous deletion
|
|
IHC
|
immunohistochemistry
|
|
BAP1
|
BRCA1-associated protein 1
|
|
MTAP
|
methylthioadenosine phosphorylase
|
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A
|
|
kb
|
kilobase
|
|
hetero-d
|
heterozygous deletion
|
|
qPCR
|
quantitative PCR
|
|
TBLB
|
transbronchial lung biopsy
|
References
|
1
|
Sauter JL, Kadota K, Bueno R, Ladanyi M, Dacic S, Nowak AK, Gill RR, Schmitt F and Husain AN: Diffuse pleural mesothelioma. WHO Classification of Tumours: thoracic Tumours. IARC Press; Lyon: pp. 204–219. 2021
|
|
2
|
Rosen LE, Karrison T, Ananthanarayanan V, Gallan AJ, Adusumilli PS, Alchami FS, Attanoos R, Brcic L, Butnor KJ, Galateau-Sallé F, et al: Nuclear grade and necrosis predict prognosis in malignant epithelioid pleural mesothelioma: A multi-institutional study. Mod Pathol. 31:598–606. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chapel DB, Schulte JJ, Husain AN and Krausz T: Application of immunohistochemistry in diagnosis and management of malignant mesothelioma. Transl Lung Cancer Res. 9 (Suppl 1):S3–S27. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kinoshita Y, Hamasaki M, Matsumoto S, Yoshimura M, Sato A, Tsujimura T, Kamei T, Kawahara K and Nabeshima K: Genomic-based ancillary assays offer improved diagnostic yield of effusion cytology with potential challenges in malignant pleural mesothelioma. Pathol Int. 70:671–679. 2020.PubMed/NCBI
|
|
5
|
Illei PB, Rusch VW, Zakowski MF and Ladanyi M: Homozygous deletion of CDKN2A and codeletion of the methylthioadenosine phosphorylase gene in the majority of pleural mesotheliomas. Clin Cancer Res. 9:2108–2113. 2003.PubMed/NCBI
|
|
6
|
Sheffield BS, Hwang HC, Lee AF, Thompson K, Rodriguez S, Tse CH, Gown AM and Churg A: BAP1 immunohistochemistry and p16 FISH to separate benign from malignant mesothelial proliferations. Am J Surg Pathol. 39:977–982. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yoshimura M, Kinoshita Y, Hamasaki M, Matsumoto S, Hida T, Oda Y, Iwasaki A and Nabeshima K: Highly expressed EZH2 in combination with BAP1 and MTAP loss, as detected by immunohistochemistry, is useful for differentiating malignant pleural mesothelioma from reactive mesothelial hyperplasia. Lung Cancer. 130:187–193. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hamasaki M, Matsumoto S, Abe S, Hamatake D, Kamei T, Hiroshima K, Kawahara K, Sato A, Tsujimura T, Nakatani Y, et al: Low homozygous/high heterozygous deletion status by p16 FISH correlates with a better prognostic group than high homozygous deletion status in malignant pleural mesothelioma. Lung Cancer. 99:155–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Savola S, Nardi F, Scotlandi K, Picci P and Knuutila S: Microdeletions in 9p21.3 induce false negative results in CDKN2A FISH analysis of Ewing sarcoma. Cytogenet Genome Res. 119:21–26. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Usvasalo A, Savola S, Räty R, Vettenranta K, Harila-Saari A, Koistinen P, Savolainen ER, Elonen E, Saarinen-Pihkala UM and Knuutila S: CDKN2A deletions in acute lymphoblastic leukemia of adolescents and young adults: An array CGH study. Leuk Res. 32:1228–1235. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Martinez Ciarpaglini C, Gonzalez J, Sanchez B, Agusti J, Navarro L, Nieto G and Monteagudo C: The amount of melanin influences p16 loss in spitzoid melanocytic lesions: Correlation with CDKN2A status by FISH and MLPA. Appl Immunohistochem Mol Morphol. 27:423–429. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsutsumi Y, Chinen Y, Sakamoto N, Nagoshi H, Nishida K, Kobayashi S, Yokokawa Y, Taki T, Sasaki N, Yamamoto-Sugitani M, et al: Deletion or methylation of CDKN2A/2B and PVT1 rearrangement occur frequently in highly aggressive B-cell lymphomas harboring 8q24 abnormality. Leuk Lymphoma. 54:2760–2764. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mizuno Y, Chinen Y, Tsukamoto T, Takimoto-Shimomura T, Matsumura-Kimoto Y, Fujibayashi Y, Kuwahara-Ota S, Fujino T, Nishiyama D, Shimura Y, et al: A novel method of amplified fluorescent in situ hybridization for detection of chromosomal microdeletions in B cell lymphoma. Int J Hematol. 109:593–602. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hida T, Hamasaki M, Matsumoto S, Sato A, Tsujimura T, Kawahara K, Iwasaki A, Okamoto T, Oda Y, Honda H, et al: Immunohistochemical detection of MTAP and BAP1 protein loss for mesothelioma diagnosis: Comparison with 9p21 FISH and BAP1 immunohistochemistry. Lung Cancer. 104:98–105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sato A, Torii I, Tao LH, Song M, Kondo N, Yoshikawa Y, Hashimoto-Tamaoki T, Hasegawa S, Nakano T and Tsujimura T: Establishment of a cell line from a Japanese patient useful for generating an in vivo model of malignant pleural mesothelioma. Cancer Sci. 102:648–655. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng JQ, Jhanwar SC, Klein WM, Bell DW, Lee WC, Altomare DA, Nobori T, Olopade OI, Buckler AJ and Testa JR: p16 alterations and deletion mapping of 9p21-p22 in malignant mesothelioma. Cancer Res. 54:5547–5551. 1994.PubMed/NCBI
|



















