Introduction
Gastric cancer is a common tumor of the digestive system, which can occur in any part of the stomach; its incidence rate ranks second among all malignant tumors, while its mortality rate ranks third, with both rates increasing every year in China in the last 10 years (1,2). Epidemiological studies showed that ~400,000 new cases of gastric cancer occur in China every year, resulting in high socioeconomic pressure (3,4). Most patients with early stages of gastric cancer show no obvious symptoms; however, a few patients experience nausea, vomiting or upper gastrointestinal symptoms similar to those caused by ulcers, making it difficult to diagnose (5). Pain and weight loss are the most common clinical symptoms associated with advanced stages of gastric cancer (5). At the time of diagnosis, most patients with gastric cancer have middle- or late-stage disease. Surgical treatment and chemotherapy are the main treatment methods; however, since 70% of advanced gastric cancer cells can be transferred through the lymphatic system (6,7), the current treatment methods are not sufficient (7). Therefore, in-depth studies on treatment modalities are of great significance for the clinical management of gastric cancer.
Tumor stem cells are the main cause of tumor proliferation and if they are not eradicated properly it will lead to multiple tumor recurrence (7). Thus, treatment schemes centered on the stemness of tumor cells could be the core of an effective anticancer strategy (8,9). Tumorigenesis is a dynamic and complex process, wherein tumor stem cells, with genetic and functional characteristics, play an important role. The metastatic potential of clustered tumor stem cells is greater compared with that in single stem cells and is the key target of anticancer treatment (10,11). Cancer stem cells can drive tumor growth and metastasis. A study found that when tissue cells, formed by tumors, were transplanted into the colons of mice, metastasis and tumor formation were observed in the liver. After removal of the targeted stem cell populations, it was found that metastatic liver tumor number was significantly decreased and the tumors shrank, suggesting that stemness is a prerequisite for cancer metastasis (12). As cells become malignant, changes in metabolic pathways occur accordingly. Metabolism, the ability to use energy, is a characteristic of all living things. The metabolism of cancer cells is extremely vigorous and the mutated cells continue to invade surrounding tissues for unrestricted growth. The Warburg effect is the metabolic basis of tumor (cancer gene) growth, tumor progression and metastasis, and tumor treatment resistance, while the Warburg effect and fat synthesis are the main pathways of tumor metabolism (7,13). Thus, targeting cancer cell metabolism has also become a new strategy for cancer treatment (14).
Kallikrein 6 (KLK6) is a member of the KLK family of serine proteases, which is involved in extracellular matrix remodeling, tumor invasion and nervous system plasticity (15). The KLK6 gene was found to be significantly overexpressed in gastric cancer tissues, suggesting that its expression status may be a strong indicator of prognosis in patients with gastric cancer (12). Inhibition via short hairpin (sh)RNA of KLK6 expression suppressed the proliferation and invasion of gastric cancer cells (16). KLK6 could also be used as a biomarker in the early stages of tumor metastasis (17–19). Our previous study reported that KLK6 suppressed HGC-27 gastric cancer cell growth by inhibiting epithelial-mesenchymal transition (EMT) (Zhou et al, unpublished data); however, the mechanism of action underlying the effect of KLK6 is still unclear. Tumor stemness indicates that the tumor cells have the characteristics of self-renewal and infinite proliferation, which play an important role in the survival, proliferation, metastasis and recurrence of tumor cells. The stemness genes of tumor cells can affect cancer metastasis by regulating EMT-related pathways. The stronger the stemness of the tumor cells, the stronger their drug resistance (20,21). Therefore, the maintenance of tumor cell stemness is of great significance to tumor cells (22,23). The PI3K/AKT/mTOR signal transduction pathway has been proved to play an important role in the proliferation, metastasis and apoptosis of a variety of cancer cells by regulating gene expression. Blocking the PI3K/AKT/mTOR signaling pathway has become a new target for the treatment of a variety of cancer cells (24). Porta et al (25) found that inhibition of PI3K/AKT/mTOR pathway could treat prostate cancer. Fattahi et al (26) observed that inhibition of PI3K/AKT/mTOR pathway can inhibit the proliferation and invasion of gastric cancer cells.
In the present study, the effect of KLK6 on the stemness and metabolism of gastric cancer cells was investigated. This could determine the mechanism of action of KLK6 and provide a theoretical basis for the treatment of gastric cancer.
Materials and methods
Cell culture and transfection
The human gastric carcinoma cell line, HGC-27, was purchased from the Type Culture Collection of the Chinese Academy of Sciences. The cells were cultured in RPMI-1640 medium (HyClone; Cytiva), supplemented with 10% fetal bovine serum (FBS; Bioswamp Life Science Lab) at 37°C in a humidified incubator with 5% CO2.
The full-length cDNA of human KLK6 (gene ID, 5653) was obtained from the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/gene/5653). KLK6 mRNA was amplified using PCR (forward primer, 5′-GCTCTAGATGAAGAAGCTGATGGTG-3′ and reverse, 5′-CGGGATCCTCACTTGGCCTGAATGGT-3′) and ligated into the pCDH vector (Addgene, Inc.) to produce pCDH-KLK6 [overexpression (OV)-KLK6] plasmids. The interference fragments (sh-RNA-1, 5′-AGAATAAGTTGGTGCATGG-3′; sh-RNA-2, 5′-CAGATGGTGATTTCCCTGAC-3′; sh-RNA-3, 5′-GATCAAAGGAGAAGCCAGGA-3′ and sh-RNA-4, 5′-CAGATACACGAACTGGATCC-3′) were ligated into the pSICOR vector (Addgene, Inc.) to produce pSICOR-shKLK6 plasmids. The HGC-27 cells were transfected with sh-RNA (1–4), OV-KLK6 and their corresponding negative controls (sh-NC and OV-NC) using Lipofectamine® 2000 (cat. no. 11668-027; Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions at 37°C. After 24 h [relevant pre-experiments were performed and the results showed that although a 24-h transfection time was relatively short, the overexpression and knockdown of KLK6 was stably expressed after transfection (data not shown)], transfection efficiency was confirmed using western blot analysis and reverse transcription-quantitative PCR (RT-qPCR). To further clarify that KLK6 regulated cell stemness and metabolism via the PI3K/AKT/mTOR axis, a PI3K/AKT inhibitor, PI3K-in-1 (cat. no. HY-12068, MedChemExpress) was added to each group.
HGC-27 cells were respectively divided into six groups: Control (HGC-27 cells), sh-NC, sh-KLK6, ov-NC, OV-KLK6, sh-KLK6 + PI3K-in-1 (25 µM), OV-KLK6 + PI3K-in-1 groups.
Cell counting Kit-8 (CCK-8)
In total, 3×103 cells were seeded in a 96-well plate using RPMI 1640 medium containing 10% FBS and treated for 24 h. To evaluate cell viability, 10 µl of CCK-8 solution was added to each well and the cells were cultured at 37°C for 4 h. The optical density (OD) was measured using a microplate reader (Multiskan FC; Thermo Fisher Scientific, Inc.) at 450 nm.
Microsphere formation
The transfected HGC-27 cells were suspended in serum-free culture medium at a concentration of 1×105 cells/ml. Microsphere culture medium (DMEM/FBS:B27 at 1:50, 20 ng/ml epidermal growth factor, 20 ng/ml basic fibroblast growth factor, 5 µg/ml insulin and 10 µg/ml transferrin) was added to each well of a 6-well ultra-low-adsorption culture plate (Corning, Inc.). Images were captured using a light microscope on day 5 (DM IL LED; Leica Microsystems GmbH).
Flow cytometry
The HGC-27 cells (1×106) were resuspended in 100 µl flow cytometry buffer in an Eppendorf tube and 2 µl CD133-fluorescein isothiocyanate (FITC) (cat. no. 11-1331-80) or CD44-FITC (cat. no. 11-0441-82) (both from eBioscience; Thermo Fisher Scientific, Inc.) was added (27). The cells were incubated in the dark for 35 min at 4°C and 400 µl flow cytometry dye buffer (cat. no. PAB180076; Bioswamp Life Science Lab) was added to each tube. The cells were then subjected to flow cytometry (NovoCyte; ACEA Bioscience, Inc.) and the results were analyzed using NovoCyte software (version 2.0, ACEA Bioscience, Inc.).
Biochemical analysis
The levels of glucose (cat. no. F006), ATP (cat. no. A095-1) and lactic acid (cat. no. A019-2) (all from Nanjing Jiancheng Bioengineering Institute) were evaluated using the respective biochemical detection kits according to the manufacturer's instructions.
RT-qPCR
Total RNA was extracted from 1×105 HGC-27 cells and tumors using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's procedures and cDNA was synthesized using a reverse transcriptase kit (cat. no. 639505; Takara Bio, Inc.) according to the manufacturer's instructions. RT-qPCR was performed with a real-time system (CFX-Connect 96; Bio-Rad Laboratories, Inc.) using the SYBR Green PCR kit (cat. no. KM4101; Roche Diagnostics), and the GAPDH gene was used as the control. Each reaction was performed in duplicate. The following thermocycling conditions were used: Initial denaturation at 95°C for 3 min followed by denaturation at 95°C for 5 sec, annealing at 56°C for 10 sec and extension at 72°C for 25 sec for 39 cycles, followed by a final step at 65°C for 5 sec and 95°C for 50 sec. The relative gene expression levels were determined using the 2−ΔΔCq method (28). The primers were designed and purchased from Nanjing Kingsy Biotechnology Co., Ltd. and are listed in Table I.
 |
Table I.
Primer sequences.
|
Table I.
Primer sequences.
| Primer name |
Sequence (5′-3′) |
| KLK6 |
|
| Forward |
GAACTCATCCAGCCCCTT |
| Reverse |
CATCCCCAGCACACAACA |
| GAPDH |
|
| Forward |
CCACTCCTCCACCTTTG |
| Reverse |
CACCACCCTGTTGCTGT |
Animals
A total of 30 nude Balb/c mice (male; 6 weeks of age) weighing 18–20 g were purchased from Changzhou Cavens Biological Co., Ltd. (animal certification no. 1107301911000025). The animals were housed in a pathogen-free environment in opaque polypropylene cages, with a standard 12 h light/dark cycle, at 22±3°C and 50–60% humidity. Food and water were available ad libitum. The mice were allowed to adapt to the aforementioned conditions for 7 days prior to experimentation.
Xenograft mouse model
The mice were randomized into five groups (n=6 per group): Control, sh-NC, sh-KLK6, OV-NC and OV-KLK6. The HGC-27 cells (1×106 cells) were inoculated subcutaneously into the right axillary region of each mouse, according to a previous study (29). Either 10 µg sh-KLK6 or 10 µg OV-KLK6 was injected into the same subcutaneous site, where the tumor cells were implanted, 15, 18, 21 and 24 days after subcutaneous inoculation. The mice in the control, OV-NC (empty vector) and sh-NC (empty vector) groups were injected with an equal volume of PBS on the same days. Tumor formation was monitored every 2 days, beginning at day 8, by measuring the tumor length (L) and width (W) using calipers. The tumor volume (V) was calculated as follows: V=(L × W2)/2. After 27 days, all the mice were sacrificed by cervical dislocation, and the tumors were dissected and collected for follow-up experiments. Animal death was confirmed as the absence of a heartbeat and breath.
Immunohistochemistry
Sections were fixed with 10% formalin for 48 h at 4°C, paraffin-embedded tissue blocks were placed at −20°C for at least 30 min to increase the hardness before slicing. The tissue blocks were sliced at a thickness of 5-µm. Before primary antibody incubation, the sections were heated at 60°C in an incubator 1 h, dewaxed with xylene, hydrated with ethyl alcohol (100, 95, 85 and 75%) and subjected to antigen retrieval (0.01 M sodium citrate buffer) at 125°C for 15 min. PBS solution was then used for washing three times for 3 min each and endogenous peroxidase activity was eliminated using 3% H2O2 for 10 min. Then, 10% goat serum (cat. no. I1726; Bioswamp Life Science Lab) was added to each group and incubated for 30 min at 4°C. The sections were then incubated overnight at 4°C with primary antibodies against Nanog (rabbit; 1:50; cat. no. PAB43881; Bioswamp Life Science Lab) and Oct-4 (rabbit; 1:50; cat. no. PAB35586; Bioswamp Life Science Lab). Next, the sections were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (1:1,000; ab6721; Abcam). The sections were counterstained with Harris' hematoxylin (4°C for 3 min) and evaluated by visual assessment of the staining intensity using a light microscope at ×200 magnification.
Western blot analysis
Total protein was extracted from the HGC-27 cells and gastric tumor tissues by using radioimmunoprecipitation assay lysis buffer (cat. no. W1689; Bioswamp Life Science Lab), and their concentration was measured using a bicinchoninic acid protein assay kit (cat. no. PAB180007; Bioswamp Life Science Lab). Total protein (20 µg/lane) was separated on a 12% gel using SDS-PAGE and transferred to polyvinylidene fluoride membranes. The membranes were blocked with a buffer containing 5% non-fat milk in PBS with 0.05% Tween-20 for 2 h, and the following primary antibodies were used: Nanog (1:2,000; cat. no. PAB43881), Oct-4 (1:1,000; cat. no. PAB35586), SOX2 (1:1,000; cat. no. PAB30154), Notch1 (1:1,000; cat. no. PAB35376), hexokinase (HK)1 (1:1,000; cat. no. PAB30519), HK2 (1:1,000; cat. no. PAB30271), GLUT1 (1:1,000; cat. no. PAB40969), phosphorylated (p)-PI3K (1:1,000; cat. no. PAB43641-P), PI3K (1:1,000; cat. no. PAB30084), p-AKT (1:1,000; cat. no. PAB43181-P), AKT (1:1,000; cat. no. PAB30596), p-mTOR (1:1,000; cat. no. PAB36313-P), mTOR (1:1,000; cat. no. PAB30674) and GAPDH (1:1,000; cat. no. PAB36269) (all from Bioswamp Life Science Lab). After three washes with PBS/0.05%Tween-20, the membranes were incubated with horseradish peroxidase-conjugated secondary goat anti-rabbit IgG (1:20,000; cat. no. SAB43714; Bioswamp Life Science Lab) at 4°C, 1 h. The protein bands were visualized using an enhanced chemiluminescence color detection kit (cat. no. Tanon-5200; Tanon Science and Technology Co., Ltd.) and the images were obtained using a Tanon Gis digital image analysis system and analyzed using Tanon Gis software (version 4.2) (both Tanon Science and Technology Co., Ltd.).
Statistical analysis
The data are presented as the mean ± standard deviation. To analyze the differences between groups, one-way analysis of variance followed by Tukey's post hoc test was performed to compare differences between multiple groups using SPSS version 22 (IBM Corp.). P<0.05 was considered to indicate a statistically significant difference. All experiments are repeated three times.
Results
Evaluation of transfection efficiency
To detect the transfection efficiency of the interference and overexpression vectors, the mRNA and protein expression level of KLK6, in each group, was determined, as shown in Fig. 1A-D, respectively. Compared with that in the control group, the mRNA and protein expression level of KLK6 in the OV group was significantly increased (P<0.05), whereas that in the four sh-RNA groups was significantly decreased (P<0.05). As sh-RNA-3 reduced the mRNA and protein expression level of KLK6 to a more significant level, it was used to silence KLK6 in the following experiments. To eliminate the effect of empty vector or the transfection procedure on the experimental results, the cell viability of the control and the empty vector groups following transfection was investigated. The results showed that there was no significant difference between the control group and the empty group (P>0.05; Fig. 1E).
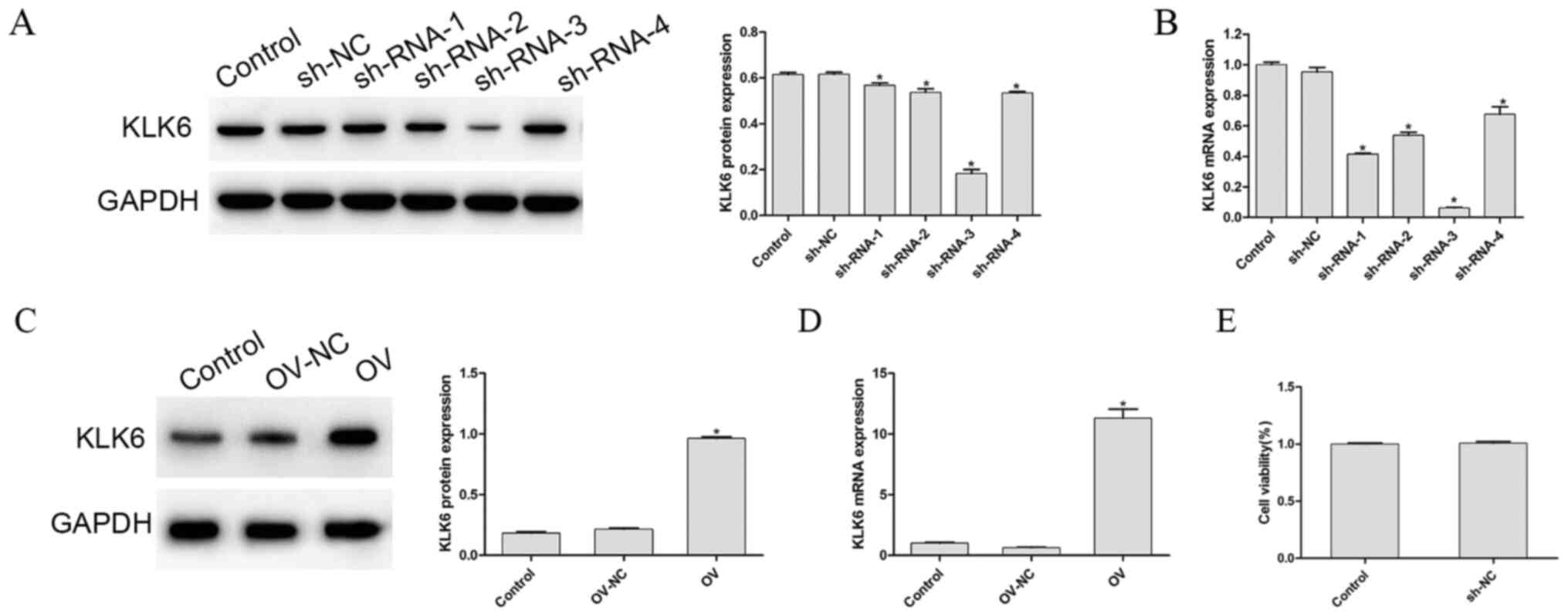 |
Figure 1.
Protein and mRNA expression level of KLK6 following transfection. (A) Protein and mRNA expression level of KLK6 was observed in cells transfected with (A and B) control, sh-NC and sh-RNA (1–4) groups, and (C and D) control, OV and OV-NC groups. (E) Cell Counting Kit-8 assay was used to determine the cell viability of the control and NC group. *P<0.05 vs. control. n=3. OV, overexpression; NC, negative control; sh, short hairpin; KLK6, kallikrein 6.
|
KLK6 silencing attenuates the stem cell-like properties of HGC-27 cells
As shown in Fig. 2A, compared with that in the control and the OV group, the ability of HGC-27 cells to form microspheres was enhanced by OV-KLK6. However, when the KLK6 gene was silenced, the microsphere-forming ability was weakened compared with that in the sh-NC group.
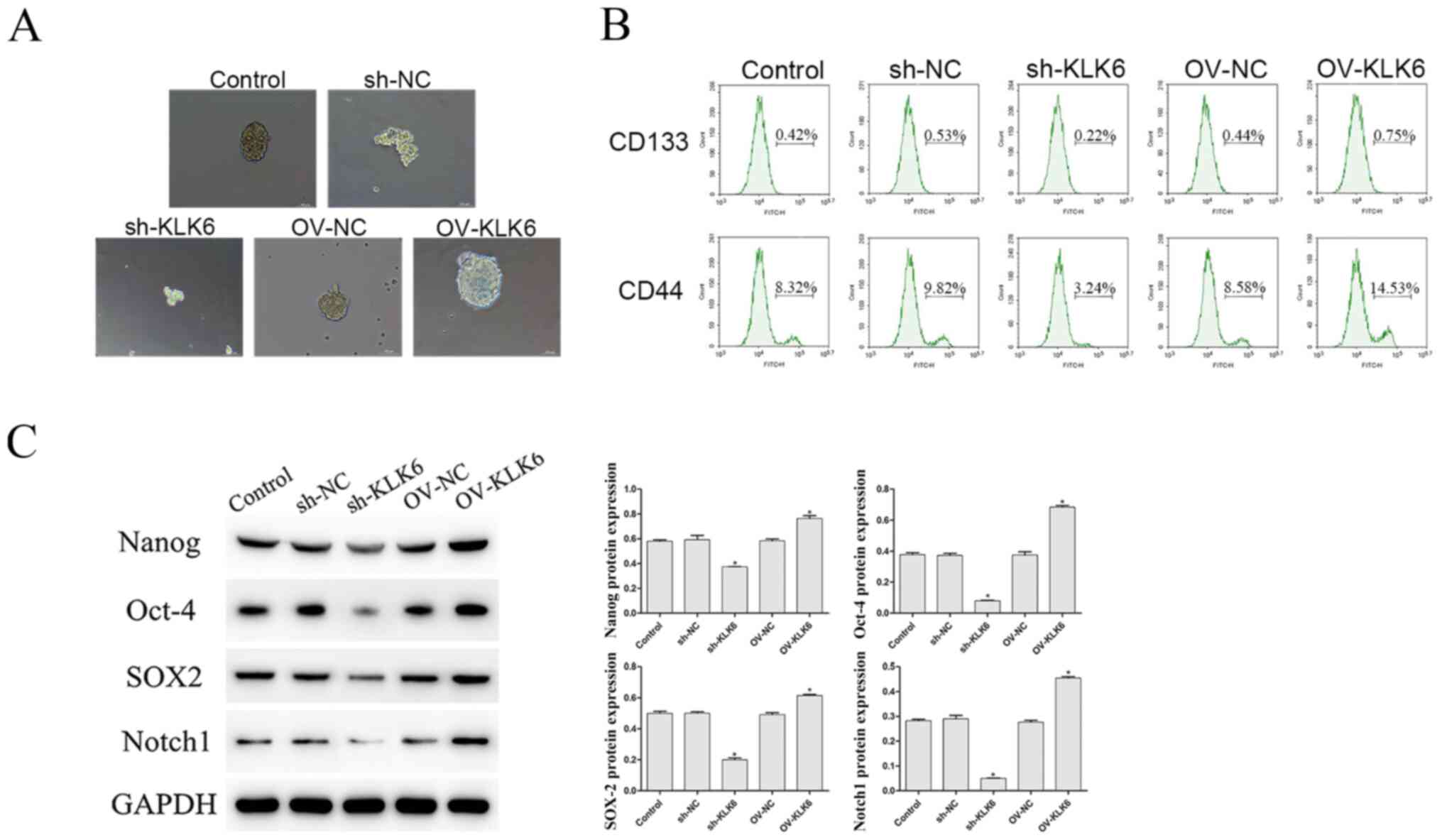 |
Figure 2.
Effect of KLK6 on the stem cell-like properties of the HGC-27 cells. (A) Microsphere formation ability was observed using a microscope. Scale bar, 50 µm. (B) Percentage of CD133+ and CD44+ cells was evaluated using flow cytometry. (C) Protein expression level of Nanog, Oct-4, SOX2 and Notch1 was observed using western blot analysis. *P<0.05 vs. control. n=3. OV, overexpression; NC, negative control; sh, short hairpin; KLK6, kallikrein 6.
|
In addition, the proportion of CD133+ and CD44+ cells was notably decreased by sh-KLK6 compared with that in the control and sh-NC groups (P<0.05) (Fig. 2B), while OV-KLK6 significantly increased the proportion of these cells (P<0.05).
The protein expression level of the stemness maintenance genes, Nanog, Oct-4, SOX2 and Notch1 was also investigated, and the results showed that the expression level of these genes were significantly increased and decreased by OV-KLK6 and sh-KLK6 compared with the control and NC (sh- and OV-) groups, respectively (both P<0.05) (Fig. 2C).
KLK6 silencing inhibits HGC-27 cell metabolism by regulating the PI3K/AKT/mTOR pathway
To evaluate the effect of KLK6 on the metabolism of the HGC-27 cells, cellular glucose uptake, and ATP and lactic acid content were measured. The results showed that, compared with the control and NC (sh- and OV-) groups, inhibition of KLK6 expression significantly decreased the uptake of glucose, and reduced the synthesis of ATP and lactic acid in the HGC-27 cells (P<0.05), while the levels were significantly increased by KLK6 overexpression (P<0.05; Fig. 3A). A PI3K inhibitor was added to the sh-KLK6 and OV-KLK6 groups, and the results showed that after the addition of PI3K-IN-1, the concentrations of ATP, lactic acid and glucose in sh-KLK6-PI3K-IN-1 group and OV-KLK6+PI3K-IN-1 group were decreased compared with the sh-KLK6 or OV-KLK6 group, respectively (Fig. 3A).
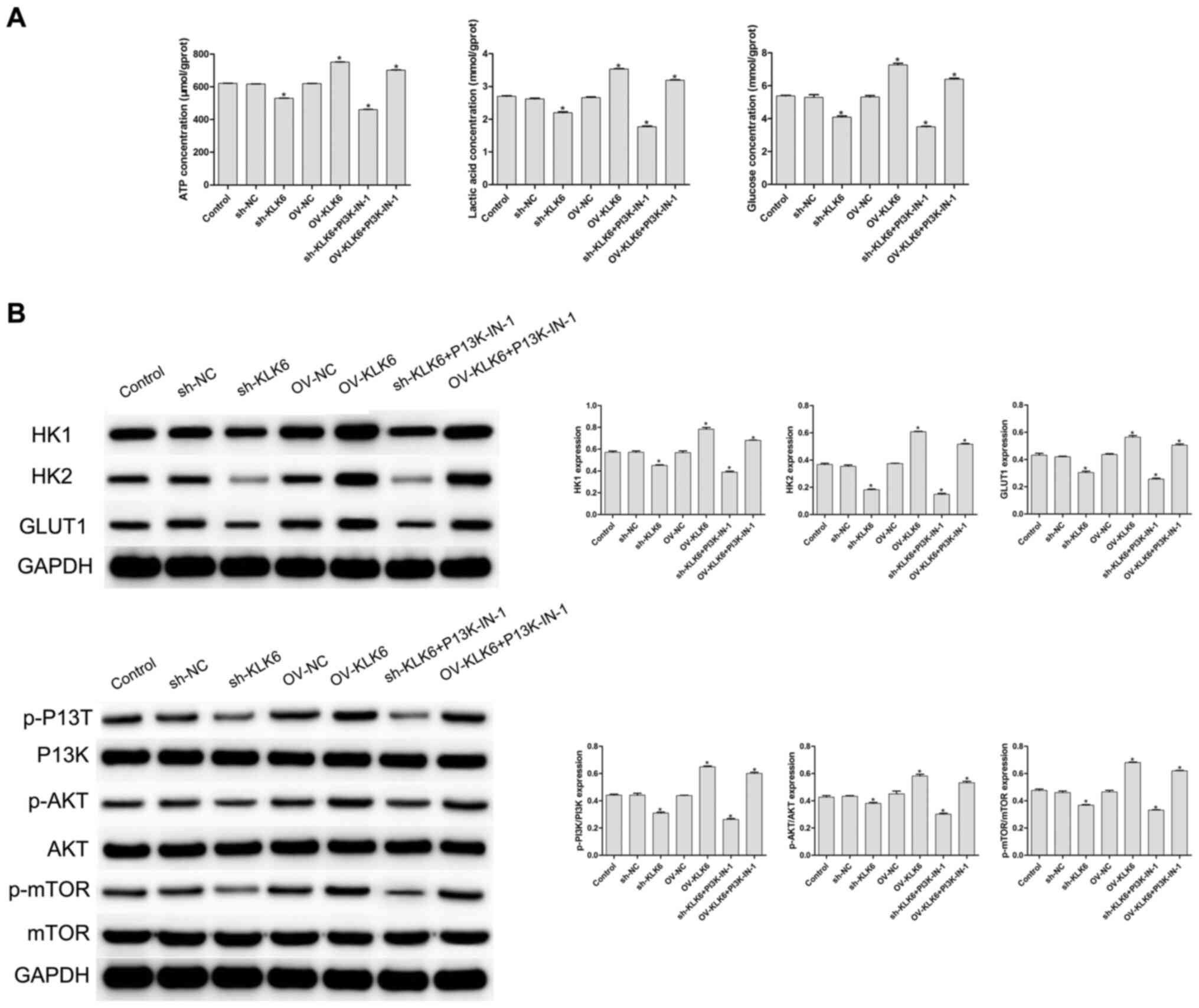 |
Figure 3.
Expression of KLK6 on the metabolism of the HGC-27 cells. (A) Levels of ATP, lactic acid and glucose were detected using biochemical kits. (B) Protein expression level of HK1, HK2, GLUT1, PI3K, p-PI3K, AKT, p-AKT, mTOR and p-mTOR was observed using western blot analysis. *P<0.05 vs. control. n=3. OV, overexpression; NC, negative control; sh, short hairpin; p, phosphorylated; KLK6, kallikrein 6; HK, hexokinase.
|
As shown in Fig. 3B, the protein expression levels of HK1, HK2 and GLUT1 were significantly increased by OV-KLK6, while they were significantly decreased by sh-KLK6 compared with the control and NC (sh- and OV-) groups (all P<0.05). To further investigate the potential mechanism of KLK6, the expression level of proteins associated with the PI3K/AKT/mTOR signaling pathway was detected using western blot analysis. Compared with that in the control group, the phosphorylation levels of PI3K, AKT and mTOR (p-PI3K, p-AKT and p-mTOR, respectively) were significantly decreased by sh-KLK6 (P<0.05), whereas they were significantly increased by OV-KLK6 (P<0.05).
When PIK3-IN-1 was administered, the expressions of HK1, HK2, GLUT1, p-PI3K, p-AKT and p-mTOR protein were decreased in the OV-KLK6+PIK3-IN-1 and sh-KLK6+PIK3-IN-1 groups compared with the OV-KLK6 and sh-KLK6 groups (P<0.05). The results suggested that KLK6 silencing inhibited the energy uptake of the HGC-27 cells and slowed down cell metabolism by regulating the PI3K/AKT/mTOR pathway.
KLK6 mediates the tumorigenicity and stemness of the HGC-27 cells by regulating the PI3K/AKT/mTOR pathway
In the aforementioned experiment, it was found that KLK6 could affect the stemness and energy metabolism of the HGC-27 cells. As tumor growth in vivo is a typical feature of gastric cancer (30), it was further investigated whether KLK6 mediated the growth of the HGC-27 cells in vivo (Fig. 4A). On the 27th day of the experiment, the tumor volume of the sh-KLK6 group was significantly lower compared with that in the control group (P<0.05), while that of the OV-KLK6 group was significantly increased (P<0.05). Compared with that in the control and NC groups, the mRNA expression level of KLK6 in the OV-KLK6 group was significantly increased (P<0.05), while it was significantly decreased in the sh-KLK6 group (P<0.05; Fig. 4A). Immunohistochemistry, western blot analysis and biochemical detection were also performed to further examine whether KLK6 affected the stemness and energy metabolism in vivo. Compared with that in the control group, the protein expression level of HK1, HK2 and GLUT1, and the levels of ATP, lactic acid and glucose in the sh-KLK6 group were significantly lower (P<0.05), and Oct-4 and Nanog expression decreased, while those in the OV-KLK6 group showed the opposite trend (P<0.05) (Fig. 4B). In addition, KLK6 silencing significantly inhibited the activity of the p-PI3K, p-AKT, and p-mTOR, while in OV-KLK6, those expression was significantly increased (P<0.05) (Fig. 4C), suggesting that KLK6 mediated the tumorigenicity and stemness of the HGC-27 cells by regulating the PI3K/AKT/mTOR signaling pathway.
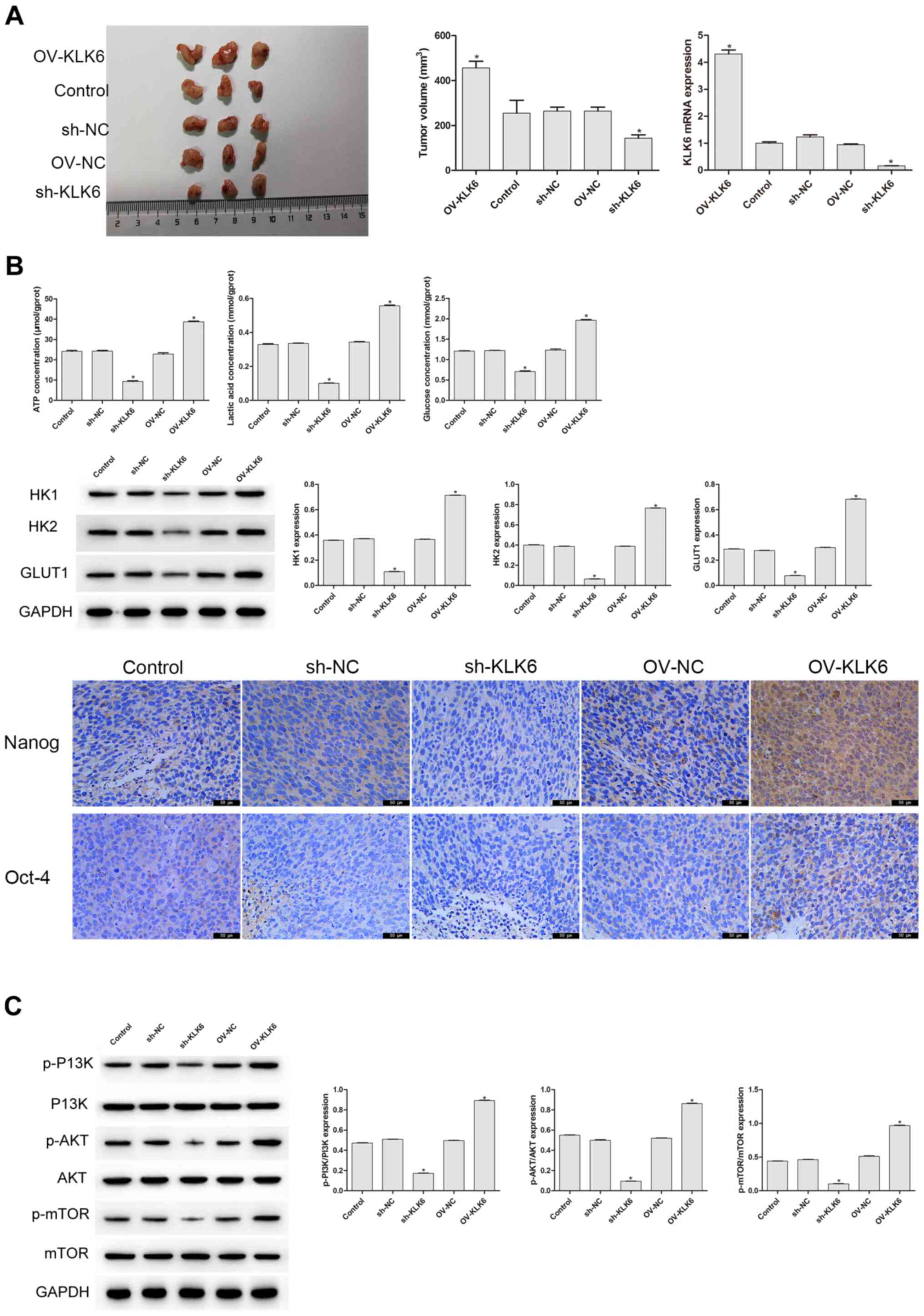 |
Figure 4.
KLK6 mediates the tumorigenicity and stemness of the HGC-27 cells in vivo. (A) sh-KLK6 attenuated the tumorigenicity and KLK6 mRNA expression level of the HGC-27 cells in vivo. (B) KLK6 silencing inhibited the levels of ATP, lactic acid, glucose and the protein expression levels of HK1, HK2, and GLUT1 in vivo, and the levels of Nanog and Oct-4 were detected using immunohistochemistry. (C) Effect of KLK6 on the proteins involved in the PI3K/AKT/mTOR signaling pathway was detected using western blot analysis. *P<0.05 vs. control. n=3. OV, overexpression; NC, negative control; sh, short hairpin; KLK6, kallikrein 6.
|
Discussion
Research has shown that cancer stem cells may play an important role in the invasion, metastasis, drug resistance, recurrence and other malignant behaviors of gastric cancer cells (31–35), and their tumorigenic activity has also been confirmed in a variety of malignant tumors (36), such as lung cancer (37), glioblastoma (38) and breast cancer (39). Giraud et al (40) confirmed that a relevant side population of the human gastric cancer cell line, HGC-27, could possess characteristics of tumor stem cells. An undifferentiated HGC-27 cell line was used in the present study and it was found that KLK6 silencing reduced the proportion of the cell population expressing CD44 and CD133, which are markers of tumor cell stemness (6). SOX2, Oct-4 and Nanog, which are involved in the process of tumorigenesis, invasion, metastasis and recurrence (41), are key regulators of tumor cell self-renewal, proliferation and differentiation, and also act as predictive biomarkers of poor prognosis for ovarian endometriosis (42), esophagus cancer (43) and etc. In the present study, it was found that KLK6 silencing, in turn, inhibited the protein expression level of the tumor cell stemness maintenance factors, SOX2, Oct-4 and Nanog, suggesting that KLK6 may suppress gastric cancer cell growth by inhibiting the stem-like properties and behaviors.
The glycometabolism pathway in tumor cells is different from that in normal cells. Normally, cells metabolize glucose using mitochondrial oxidative phosphorylation and glycolysis, both under anaerobic conditions. However, tumor cells prefer to produce energy and substances required by the cells using glycolysis, under either aerobic or anaerobic conditions (44,45). Glucose transporters (GLUTs) are carriers of glucose in mammalian cells, among which GLUT1 mainly transports glucose (46). An upregulation of GLUT1 was found to increase glucose uptake by tumor cells and promote cell proliferation (47). Hexokinases (HKs) are key enzymes that are involved in glycolysis and participate in the Warburg effect and in the immortalization of tumor cells (48). As key enzymes that affect tumor cell metabolism, HKs are essential in promoting tumorigenesis and tumor development, and their inhibition has become a new pharmacological strategy in cancer treatment (49). In the present study, KLK6 inhibition decreased glucose uptake, lactic acid production and ATP content in gastric cancer cells, and simultaneously suppressed the protein expression level of GLUT1, HK1 and HK2. It was also found that inhibition of KLK6 decreased the activity of proteins in the PI3K/AKT/mTOR signaling pathway, suggesting that KLK6 mediated energy metabolism in gastric cancer cells by regulating the PI3K/AKT/mTOR signaling pathway.
A limitation of the present study was that double staining was not performed in the flow cytometry assays; therefore, it was not confirmed whether the cells belonged to the same cell group. The effect of KLK6 on the stemness and metabolism of the HGC-27 gastric cancer cells and its potential mechanism was investigated; however, whether KLK6 affects other signaling pathways is still unclear and further research is therefore required.
In conclusion, KLK6 modulated stemness properties and the metabolic profile in gastric carcinoma cells, which may be achieved by regulating the PI3K/AKT/mTOR signaling pathway.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Authors' contributions
DZ designed the study and wrote the manuscript. YH and HL performed the experiments. WH performed the statistical analysis. All authors confirm the authenticity of all the raw data, and all authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
All the protocols and procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved (approval no. HLK-20181102-01) by the Animal Care and Use Committee at Wuhan Myhalic Biotechnology Co., Ltd. This is a third-party sharing public service platform for animal experiments, has established standard animal experiment barrier environment facilities, obtained an experimental animal use license in accordance with the law, and established Animal Ethics and Welfare Review Committee in accordance with the regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Yada T, Yokoi C and Uemura N: The current state of diagnosis and treatment for early gastric cancer. Diagn Ther Endosc. 2013:2413202013. View Article : Google Scholar
|
|
2
|
Li YX, Li XM, Zhang L, Zhang W, Chen C, Tao L, Zhao J, Li SG, Li F and Zhang WJ: Correlation between Helicobacter pylori infection and the development and prognosis of gastric cancer. Chin J Can Prev Trea. 2:19-22+36. 2015.(In Chinese).
|
|
3
|
Zuo TT, Zheng SR, Zeng HM, Zhang SW and Chen WQ: Epidemiological status of gastric cancer in China. Chin Cancer Clin l. 442017.
|
|
4
|
Cai M, Dai S, Chen W, Xia C, Lu L, Dai S, Qi J, Wang M, Wang M, Zhou L, et al: Environmental factors, seven GWAS-identified susceptibility loci, and risk of gastric cancer and its precursors in a Chinese population. Cancer Med. 6:708–720. 2017. View Article : Google Scholar
|
|
5
|
Karimi P, Islami F, Anandasabapathy S, Freedman ND and Kamangar F: Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar
|
|
6
|
Song Z, Wu Y, Yang J, Yang D and Fang X: Progress in the treatment of advanced gastric cancer. Tumour Biol. 39:10104283177146262017. View Article : Google Scholar
|
|
7
|
Smyth EC, Nilsson M, Grabsch HI, van Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648. 2020. View Article : Google Scholar
|
|
8
|
Wang ZD, Yang J and Guo QW: Tumor stem cells-a new target for tumor therapy. J Clin Exp Pathol. 29:427–430. 2013.
|
|
9
|
Vinogradov S and Wei X: Cancer stem cells and drug resistance: The potential of nanomedicine. Nanomedicine (Lond). 7:597–615. 2012. View Article : Google Scholar
|
|
10
|
Gopalan V, Islam F and Lam KY: Surface markers for the identification of cancer stem cells. Methods Mol Biol. 1692:17–29. 2018. View Article : Google Scholar
|
|
11
|
Kapeleris J, Zou H, Qi Y, Gu YS, Li J, Schoning J, Monteiro MJ and Gu W: Cancer stemness contributes to cluster formation of colon cancer cells and high metastatic potentials. Clin Exp Pharmacol Physiol. 47:838–847. 2020. View Article : Google Scholar
|
|
12
|
de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, et al: A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 543:676–680. 2017. View Article : Google Scholar
|
|
13
|
Liu H, Chen K, Wang L, Zeng X, Huang Z, Li M, Dong P and Chen X: miR-613 inhibits Warburg effect in gastric cancer by targeting PFKFB2. Biochem Biophys Res Commun. 515:37–43. 2019. View Article : Google Scholar
|
|
14
|
Qin XY, Xu HY and Lai LH: Targeting tumor metabolic pathways to develop anticancer agents. 12th national congress of Chinese society of biochemistry and molecular biology and 2018 national academic conference. 2018.
|
|
15
|
Nagahara H, Mimori K, Utsunomiya T, Barnard GF, Ohira M, Hirakawa K and Mori M: Clinicopathologic and biological significance of kallikrein 6 overexpression in human gastric cancer. Clin Can Res. 11:6800–6806. 2005. View Article : Google Scholar
|
|
16
|
Moreau ME, Garbacki N, Molinaro G, Brown NJ, Marceau F and Adam A: The kallikrein-kinin system: Current and future pharmacological targets. J Pharmacol Sci. 99:6–38. 2005. View Article : Google Scholar
|
|
17
|
Schrader CH, Kolb M, Zaoui K, Flechtenmacher C, Grabe N, Weber KJ, Hielscher T, Plinkert PK and Hess J: Kallikrein-related peptidase 6 regulates epithelial-to-mesenchymal transition and serves as prognostic biomarker for head and neck squamous cell carcinoma patients. Mol Cancer. 14:1072015. View Article : Google Scholar
|
|
18
|
Sells E, Pandey R, Chen H, Skovan BA, Cui HY and Ignatenko NA: Specific microRNA-mRNA regulatory network of colon cancer invasion mediated by tissue kallikrein-related peptidase 6. Neoplasia. 19:396–411. 2017. View Article : Google Scholar
|
|
19
|
Kaneko N, Kawano S, Yasuda K, Hashiguchi Y, Sakamoto T, Matsubara R, Goto Y, Jinno T, Maruse Y, Morioka M, et al: Differential roles of kallikrein-related peptidase 6 in malignant transformation and ΔNp63β-mediated epithelial-mesenchymal transition of oral squamous cell carcinoma. Oral Oncol. 75:148–157. 2017. View Article : Google Scholar
|
|
20
|
Plaks V, Kong N and Werb Z: The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 16:225–238. 2015. View Article : Google Scholar
|
|
21
|
Pastushenko I, Mauri F, Song Y, de Cock F, Meeusen B, Swedlund B, Impens F, Van Haver D, Opitz M, Thery M, et al: Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature. 589:448–455. 2021. View Article : Google Scholar
|
|
22
|
Pattabiraman DR and Weinberg RA: Tackling the cancer stem cells-what challenges do they pose? Nat Rev Dr Dis. 13:497–512. 2014. View Article : Google Scholar
|
|
23
|
Lytle NK, Barber AG and Reya T: Stem cell fate in cancer growth, progression and therapy resistance. Nat Rev Cancer. 18:669–680. 2018. View Article : Google Scholar
|
|
24
|
Shorning BY, Dass MS, Smalley MJ and Pearson HB: The PI3K-AKT-mTOR pathway and prostate cancer: At the crossroads of AR, MAPK, and WNT signaling. Int J Mol Sci. 21:45072020. View Article : Google Scholar
|
|
25
|
Porta C, Paglino C and Mosca A: Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 4:642014. View Article : Google Scholar
|
|
26
|
Fattahi S, Amjadi-Moheb F, Tabaripour R, Ashrafi GH and Akhavan-Niaki H: PI3K/AKT/mTOR signaling in gastric cancer: Epigenetics and beyond. Life Sci. 262:1185132020. View Article : Google Scholar
|
|
27
|
Wang C, Xie J, Guo J, Manning HC, Gore JC and Guo N: Evaluation of CD44 and CD133 as cancer stem cell markers for colorectal cancer. Oncol Rep. 28:1301–1308. 2012. View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
29
|
Jin JF, Ou XB, Ruan Y and Guo RZ: Establishment of a xenograft model in nude mice with human gastric cancer HGC-27 cells. J Zunyi Med Coll. 1:33–35. 2015.
|
|
30
|
Rao M, Zhu Y, Cong X and Li Q: Knockdown of CREB1 inhibits tumor growth of human gastric cancer in vitro and in vivo. Oncol Rep. 37:3361–3368. 2017. View Article : Google Scholar
|
|
31
|
Fagoonee S, Li H, Zhang H, Altruda F and Pellicano R: Gastric cancer as a stem-cell disease: Data and hypotheses. Panminerva Med. 56:289–300. 2014.
|
|
32
|
Hata M, Hayakawa Y and Koike K: Gastric stem cell and cellular origin of cancer. Biomedicines. 6:1002018. View Article : Google Scholar
|
|
33
|
Moharil RB, Dive A, Khandekar S and Bodhade A: Cancer stem cells: An insight. J Oral Maxillofac Pathol. 21:4632017. View Article : Google Scholar
|
|
34
|
Dawood S, Austin L and Cristofanilli M: Cancer stem cells: Implications for cancer therapy. Oncology (Williston Park). 28:1101–1107, 1110. 2014.
|
|
35
|
Yang LX, Qin SJ and Dong MQ: Advances in anti-tumor strategies based on targeted tumor stem cells. Shandong Med. 59:91–94. 2019.
|
|
36
|
Gao GL, Zhang XL, Sun ZL, Liu WY, Zhang P and Ye DX: Isolation and identification cancer stem cells in human gastric cancer cell line 7. Shanghai Med. 37:957-961+897. 2014.(In Chinese).
|
|
37
|
Maiuthed A, Chantarawong W and Chanvorachote P: Lung cancer stem cells and cancer stem cell-targeting natural compounds. Anticancer Res. 38:3797–3809. 2018. View Article : Google Scholar
|
|
38
|
Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL and Rich JN: Cancer stem cells in glioblastoma. Genes Dev. 29:1203–1217. 2015. View Article : Google Scholar
|
|
39
|
Troschel FM, Böhly N, Borrmann K, Braun T, Schwickert A, Kiesel L, Eich HT, Götte M and Greve B: miR-142-3p attenuates breast cancer stem cell characteristics and decreases radioresistance in vitro. Tumour Biol. 40:10104283187918872018. View Article : Google Scholar
|
|
40
|
Giraud J, Molina-Castro S, Seeneevassen L, Sifré E, Izotte J, Tiffon C, Staedel C, Boeuf H, Fernandez S, Barthelemy P, et al: Verteporfin targeting YAP1/TAZ-TEAD transcriptional activity inhibits the tumorigenic properties of gastric cancer stem cells. Int J Cancer. 146:2255–2267. 2020. View Article : Google Scholar
|
|
41
|
Ni T, Wang H, Zhan D, Tao L, Lv M, Wang W, Chu Z, Zhou Z, Sunagawa M and Liu Y: CD133+/CD166+ human gastric adenocarcinoma cells present the properties of neoplastic stem cells and emerge more malignant features. Life Sci. 269:1190212021. View Article : Google Scholar
|
|
42
|
Shariati F, Favaedi R, Ramazanali F, Ghoraeian P, Afsharian P, Aflatoonian B, Aflatoonian R and Shahhoseini M: Increased expression of stemness genes REX-1, OCT-4, NANOG, and SOX-2 in women with ovarian endometriosis versus normal endometrium: A case-control study. Int J Reprod Biomed. 16:ijrm.v16i12.3684. 2019.
|
|
43
|
Khosravi A, Jafari SM and Asadi J: Knockdown of TAZ decrease the cancer stem properties of ESCC cell line YM-1 by modulation of Nanog, OCT-4 and SOX2. Gene. 769:1452072021. View Article : Google Scholar
|
|
44
|
Yan JW, Zhong J, Wang GC and Feng F: Progress in research of targeting cancer glycolysis pathway for anticancer therapy. Chin J New Drugs. 23:550–556. 2014.
|
|
45
|
Zuo RJ: Warburg effect: Aerobic glycolysis during mammalian reproduction. Prog Biochemis Biophy. 42:132–139. 2015.
|
|
46
|
Wang YZ, Liu BL and Li XY: Progress in the study of glucose transporter 1 at the blood-brain barrier. Chin J Clin Pharm Ther. 19:1057–1063. 2014.
|
|
47
|
Chen R, Lin J, Yan W and Chen D: miR-522-3p promotes osteosarcoma cell growth by regulating glucose uptake and GLUT1 expression. Onco Targets Ther. 12:9053–9058. 2019. View Article : Google Scholar
|
|
48
|
Lis P, Dyląg M, Niedźwiecka K, Ko YH, Pedersen PL, Goffeau A and Ułaszewski S: The HK2 dependent ‘Warburg effect’ and mitochondrial oxidative phosphorylation in cancer: Targets for effective therapy with 3-bromopyruvate. Molecules. 21:17302016. View Article : Google Scholar
|
|
49
|
Xia HG, Najafov A, Geng J, Galan-Acosta L, Han X, Guo Y, Shan B, Zhang Y, Norberg E, Zhang T, et al: Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol. 210:705–716. 2015. View Article : Google Scholar
|


















