Introduction
Ovarian cancer is a diverse set of diseases that includes epithelial ovarian cancer, endometrioid carcinoma, adenocarcinoma and other types of cancer. Ovarian adenocarcinoma (OA) is a common malignancy accounting for ~4% of female cancer cases in the United States (1). According to the latest GLOBOCAN statistics, in 2018, OA accounted for 3.2% of all new cancer cases and caused 3.3% of cancer-related deaths (2). The overall 5-year survival rate of patients with OA is ~46% (3). However, for patients diagnosed with tumor metastasis to distant organs, such as the liver and lungs, the 5-year survival rate drops to <20% (4). Although aging, obesity, hormone therapy and a family history of OA are known to be major risk factors for OA, the exact molecular mechanism of OA remains elusive, limiting the development of novel therapeutic approaches (5,6).
Studies examining the molecular pathogenesis of OA have characterized a considerable number of molecular pathways involved in the onset of this disease (7–9). In addition, some critical molecular factors, such as angiogenesis-associated factors, have proven to be promising targets for the development of novel therapies, such as targeted therapy that can be applied to treat cancer by regulating gene expression (10). With their ability to regulate gene expression, non-coding RNA (ncRNA) molecules, such as microRNAs (miRNAs/miRs) and long ncRNAs (lncRNAs), are implicated in a variety of human cancer types, including OA (11,12). Thus, ncRNA molecules may represent potential targets for cancer therapy, and functional characterization of such ncRNA targets is required. A recent study has reported that MYCN opposite strand (MYCNOS) is an oncogenic lncRNA in liver cancer (13). Bioinformatics analysis revealed a potential interaction between MYCNOS and miR-152, which can play a tumor-suppressive role by targeting mitogen-activated protein kinase kinase 7 (MKK7) (14). MKK7 modulates JNK signaling by interacting with scaffold proteins, such as JNK-interacting protein 1, 2 and 3 or filamin A (15). MKK7 plays a key role in hepatic development. For example, MKK7 total knockout mice were reported to exhibit severely disorganized livers and reduced hepatoblast numbers, and died at E12.5-E13.5 (16).
The aim of the present study was to examine the interaction between MYCNOS and miR-152 and its effect on MKK7 expression in OA.
Materials and methods
Patients with OA and follow-up
A total of 62 female patients with OA (age range, 39–68 years; mean age, 54.2±6.7 years) who were admitted to The Fourth Affiliated Hospital of Nanchang University (Jiangxi, China) between April 2013 and April 2015 were enrolled in the present study. According to the American Joint Committee on Cancer classification system (17), the patients were stratified into stage I or II (n=28) and stage III or IV (n=34) cases. All patients were diagnosed using a histopathological biopsy. Patients who had recurrent OA, other severe clinical disorders or were initiated on therapy were excluded. From the day of admission, the 62 patients were followed up monthly to record their survival. Patients who died of causes other than OA were excluded. The present study was approved by The Ethics Committee of The Fourth Affiliated Hospital of Nanchang University (approval no. NC1607AC98), and a written informed consent form was signed by all patients.
Tissue collection
Prior to therapy, OA and paired non-tumor tissue samples were collected from all patients using fine needle aspiration. All tissue samples were confirmed via histopathological analysis and stored in a liquid nitrogen tank before subsequent analysis.
OA cells and cell transfection
Caov-3 and SKOV3 cell lines were purchased from The Cell Bank of Type Culture Collection Committee of Chinese Academy of Sciences (Shanghai, China) and used as OA cell models. In a 5% CO2 incubator, Caov-3 cells were cultured at 37°C in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). Moreover, SKOV3 cells were cultured at 37°C in low-glucose DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.).
The MYCNOS and MKK7 expression vectors were constructed using the plasmid pcDNA3.1 backbone (Invitrogen; Thermo Fisher Scientific, Inc.). The miR-152 mimic and negative control (NC) were synthesized by Invitrogen (Thermo Fisher Scientific, Inc.). Cells were transfected with 1 µg expression vector or 40 nM miRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The empty pcDNA3.1 vector or NC miRNA were used as NCs. Untransfected cells were also used as a control group. Following 48 h of transfection at 37°C, cells were subjected to experimentation.
The MYCNOS luciferase vector was constructed using the pGL3 plasmid vector (Promega Corporation). Short hairpin RNA (shRNA) molecules specifically targeting MYCNOS were synthesized by Invitrogen (Thermo Fisher Scientific, Inc.) and cloned into the MYCNOS luciferase vector to perform dual luciferase reporter assays. The cells were co-transfected as aforementioned with the MYCNOS luciferase vector and NC miRNA or miR-152 mimic. Luciferase activity was measured 48 h later using the Dual-Luciferase Reporter assay system (Promega Corporation) according to manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity. The sequences of the shRNAs and miRNAs used in the present study are as follows: shMYCNOS: 5′-CCGGTGGCAATTGCTTGTCATTAAACTCGAGTTTAATGACAAGCAATTGCCATTTTTG-3′; sh-NC, 5′-CACCGCTATGATATCGTCTGTTTCAAGAGAACAGACGATATCATAGCTTTTTTG-3′; miR-152: 5′-UCAGUGCAUGACAGAACUUGGAAGUUCUGUCAUGCACUGAUU-3′; NC miRNA: 5′-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3′.
RNA preparation
RNA was isolated from tissue samples and cells (Caov-3 and SKOV3) using RiboZol™ (Invitrogen; Thermo Fisher Scientific, Inc.) and treated with DNase I for 100 min at 37°C to completely remove genomic DNA. miRNA was isolated and purified using the miRNeasy Serum/Plasma Advanced kit (Qiagen GmbH) according to the manufacturer's instructions. A volume of 1 µl of isolation spike-in mix containing synthetic cel-miR-54 (1×107 copies/µl), spike-A (2×105 copies/µl), spike-B (4×103 copies copies/µl) and, when appropriate, 1 µl GlycoBlue Co-precipitant (15 mg/ml) (Invitrogen; Thermo Fisher Scientific, Inc.) were added into each sample at the lysis step. RNA was eluted into 20 µl nuclease-free water and stored at −80°C. lncRNA was extracted and quantified via chromatin isolation using RNA purification, as previously described (18).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted and reverse transcribed using the SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen; Thermo Fisher Scientific, Inc.). For RT, RNA was mixed with oligo-(dT) and specific primers and preincubated for 2 min at 42°C. After the addition of 250 U SuperScript II reverse transcriptase, the samples were incubated at 42°C for 100 min. For random-hexamer-primed RT reactions, RNA was mixed with 500 ng random hexamer primers, and the mixture was preincubated at 25°C for 2 min. Following the addition of reverse transcriptase, the reaction was maintained at 25°C for 10 min, then incubated at 42°C for 100 min. To determine the expression levels of MYCNOS and MKK7 mRNA, qPCR was performed using the FastStart Universal SYBR Green Master mix (Bio-Rad Laboratories, Inc.). The following thermocycling conditions were used in the experiments: PCR initial activation at 95°C for 15 min, followed by 40 cycles of denaturation at 94°C for 15 sec, annealing at 55°C for 30 sec and an extension at 70°C for 30 sec. The mRNA expression levels of MYCNOS and MKK7 were normalized to those of GAPDH and calculated using the 2−ΔΔCq method (19). Each reaction was set up in duplicate. The primers used for RT-qPCR were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast) and were synthesized by AUGCT Biotechnology. The sequences used were as follows: MYCNOS forward, 5′-TCCGACAGCTCAAACACAGAC-3′ and reverse, 5′-CCAGCTTTGCAGCCTTCTC-3′; MKK7 forward, 5′-TCGAGCTCTAGGTGGCGTCATCCTT-3′ and reverse, 5′-GGGCTGATATCCAGGTTGAGGTCGA-3′; and GAPDH forward, 5′-CAGCCTCAAGATCATCAGCA-3′ and reverse, 5′-TGTGGTCATGAGTCCTTCCA-3′.
Western blot analysis
Protein isolation was carried out using RIPA buffer, and protein concentrations were measured using a BCA assay kit (both from Invitrogen; Thermo Fisher Scientific, Inc.). Protein samples were denatured at 95°C for 10 min, separated via SDS-PAGE on 10% gels, then transferred to PVDF membranes. The membranes were blocked with PBS containing 5% non-fat milk at 25°C for 2 h and incubated with primary antibodies against GAPDH (cat. no. ab9485; 1:2,500; Abcam) or MKK7 (cat. no. ab4762; 1:500; Abcam) for 12 h at 4°C. The membranes were then incubated with HRP-conjugated IgG secondary antibody (cat. no. ab6721; 1:1,000; Abcam) at 25°C for another 2 h. Protein bands were visualized using ECL reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and analyzed using ImageJ software (version 1.48; National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
Caov-3 and SKOV3 cells were seeded into 96-well plates at a density of 5×103 cells/well. After being cultured for 24 h in an incubator with 5% CO2 for complete adherence, the cells were incubated with the CCK-8 solution (Sigma-Aldrich; Merck KGaA) at 37°C for 2 h. The optical density values were determined at 450 nm and at 24, 48, 72 and 96 h after incubation, and growth curves were plotted. The experiment was repeated three times.
Statistical analysis
Gene expression levels in paired tissue samples are presented as the average of three technical replicates and were compared using paired Student's t-tests. The data from the transfection experiments are presented as the mean ± SD of three biological replicates and were compared using one-way ANOVA followed by Tukey's post hoc test. All statistical analyses were performed using GraphPad Prism software (version 7.0; GraphPad Software, Inc.). The 62 patients with OA were divided equally into high- and low-expression groups using the median MYCNOS levels as the cutoff value. The survival curves were plotted for both groups based on the 5-year follow-up data and compared using the log-rank test. P<0.05 was considered to indicate a statistically significant difference.
Results
MYCNOS upregulation is associated with poor survival in patients with OA
OA and paired non-tumor tissues were collected from patients with OA (n=62), followed by the determination of MYCNOS mRNA levels using RT-qPCR. MYCNOS expression was found to be significantly upregulated in OA tissues compared with that in non-tumor tissue samples (Fig. 1A; P<0.05). The 62 patients were grouped into high- and low-MYCNOS expression groups (n=31 per group). Analysis of survival curves according to MYCNOS expression indicated that overall survival was significantly reduced in the high-expression group compared with the low-expression group (Fig. 1B; hazard ratio = 2.246; 95% CI: 0.28–4.12; P=0.0239). Therefore, MYCNOS expression is upregulated in OA tissue and associated with poor survival in patients with OA.
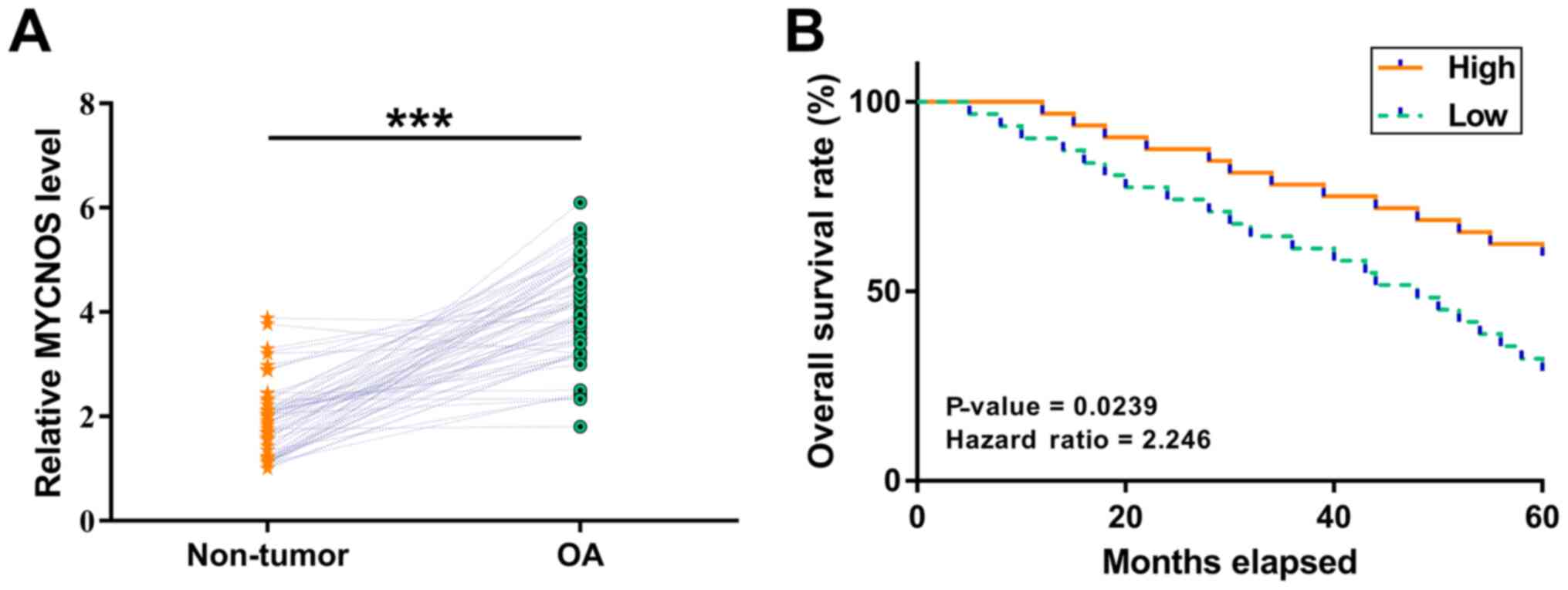 |
Figure 1.
MYCNOS overexpression is associated with poor survival in patients with OA. OA and paired non-tumor tissue samples were collected from patients with OA (n=62). (A) MYCNOS mRNA expression levels were measured using reverse transcription-quantitative PCR. The data are presented as the mean values of three technical replicates. ***P<0.001. (B) The patients with OA were grouped into MYCNOS high- and low-expression groups using the median MYCNOS expression level as the cutoff value. The survival curves were plotted for both groups based on the 5-year follow-up data and compared using the log-rank test. n=31 in each group. MYCNOS, MYCN opposite strand; OA, ovarian adenorcarcinoma.
|
MYCNOS directly interacts with miR-152
The interaction between MYCNOS and miR-152 was predicted using the IntaRNA2.0 online RNA-RNA prediction program (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp). It was observed that MYCNOS and miR-152 may form strong base-pairing (Fig. 2A). Dual luciferase reporter assays demonstrated that the miR-152 group exhibited significantly lower luciferase activity compared with the NC group (Fig. 2B; P<0.05), demonstrating the direct interaction between MYCNOS and miR-152.
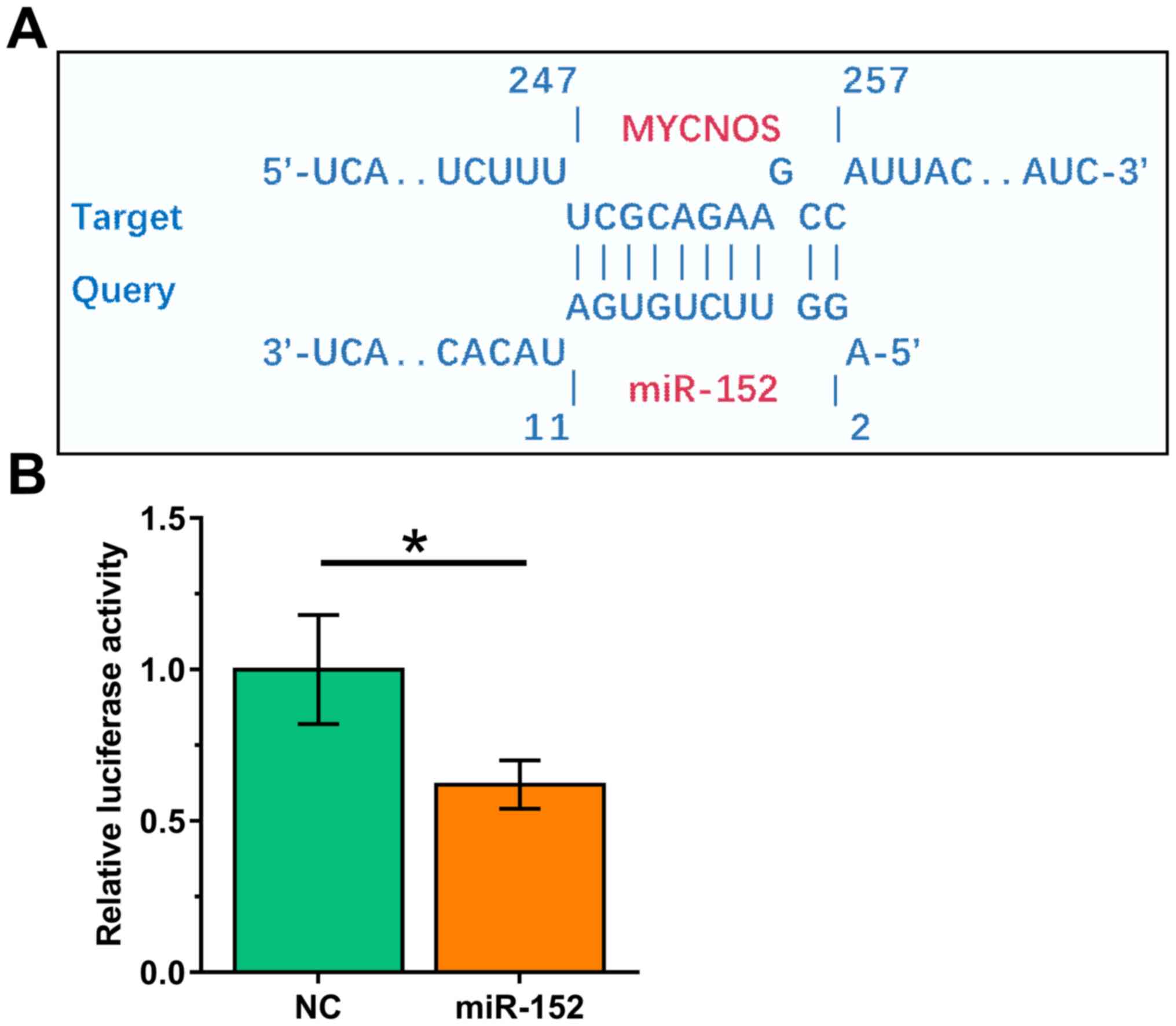 |
Figure 2.
MYCNOS directly interacts with miR-152. (A) The interaction between MYCNOS and miR-152 was predicted using the IntaRNA2.0 online RNA-RNA prediction program. (B) Caov-3 cells were co-transfected with MYCNOS luciferase vector + NC miRNA (NC group) or MYCNOS luciferase vector + mimic of miR-152 (miR-152 group). Luciferase activity was measured at 48 h post-transfection. *P<0.05. MYCNOS, MYCN opposite strand; miR/miRNA, microRNA; NC, negative control.
|
MYCNOS and miR-152 overexpression does not affect their mutual expression levels
To examine the interaction between MYCNOS and miR-152, Caov-3 and SKOV3 cells were transfected with MYCNOS expression vector or miR-152 mimic. Overexpression of MYCNOS and miR-152 was confirmed using RT-qPCR (Figs. 3A and 4A; P<0.05). The expression levels of miR-152 remained unchanged in MYCNOS-overexpressing cells (Figs. 3B and 4B). Similarly, miR-152 mimic transfection did not significantly alter MYCNOS expression levels (Figs. 3C and 4C).
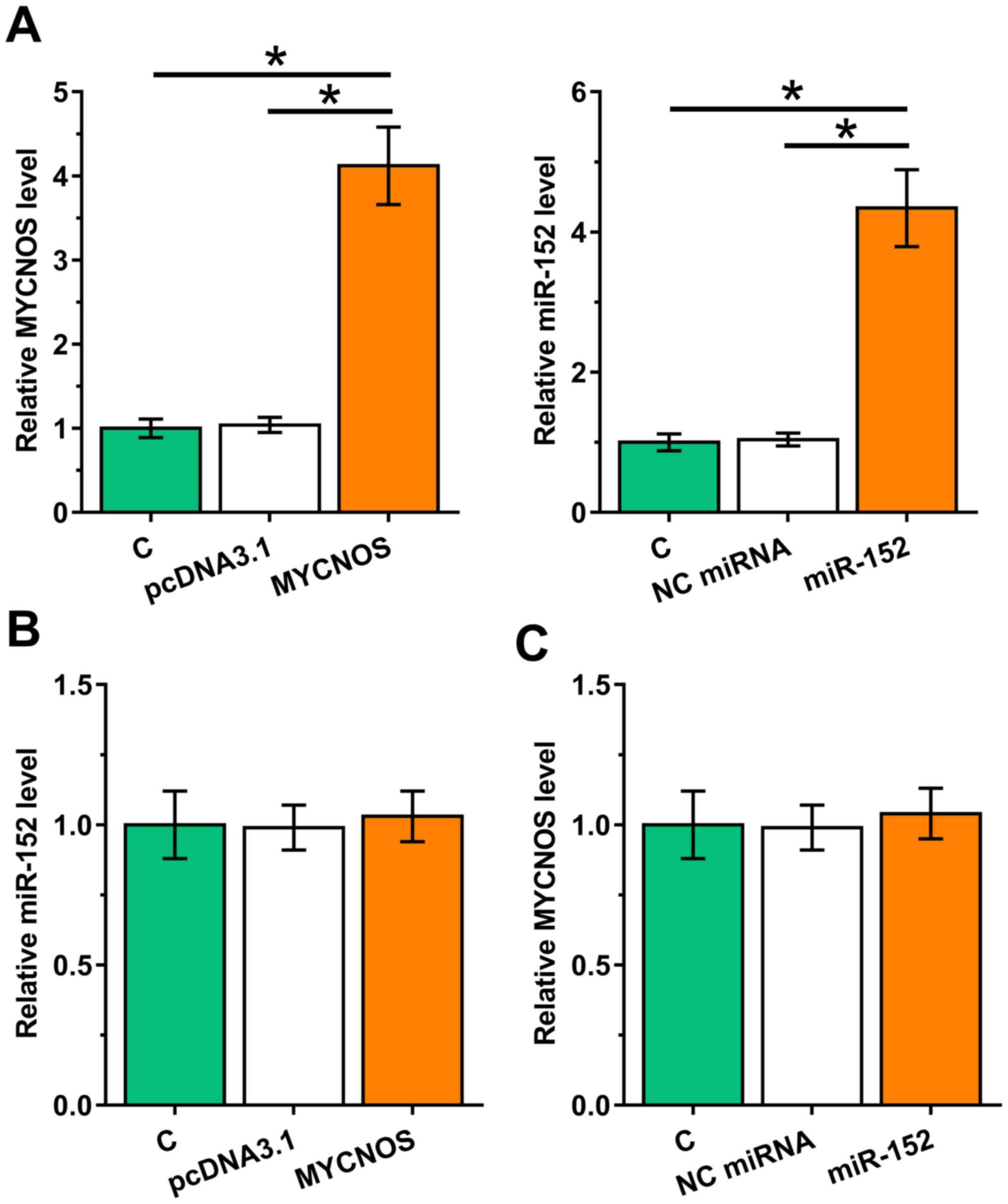 |
Figure 3.
Overexpression of MYCNOS and miR-152 does not mutually affect their expression levels in Caov-3 cells. To explore the interaction between MYCNOS and miR-152, Caov-3 cells were transfected with MYCNOS expression vector or miR-152 mimic. (A) Overexpression of MYCNOS and miR-152 was confirmed using RT-qPCR. (B and C) The effect of (B) MYCNOS overexpression on miR-152 expression and (C) miR-152 mimic transfection on MYCNOS expression were evaluated using RT-qPCR. The data are presented as the mean ± SD. *P<0.05. MYCNOS, MYCN opposite strand; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; C, untransfected control cells.
|
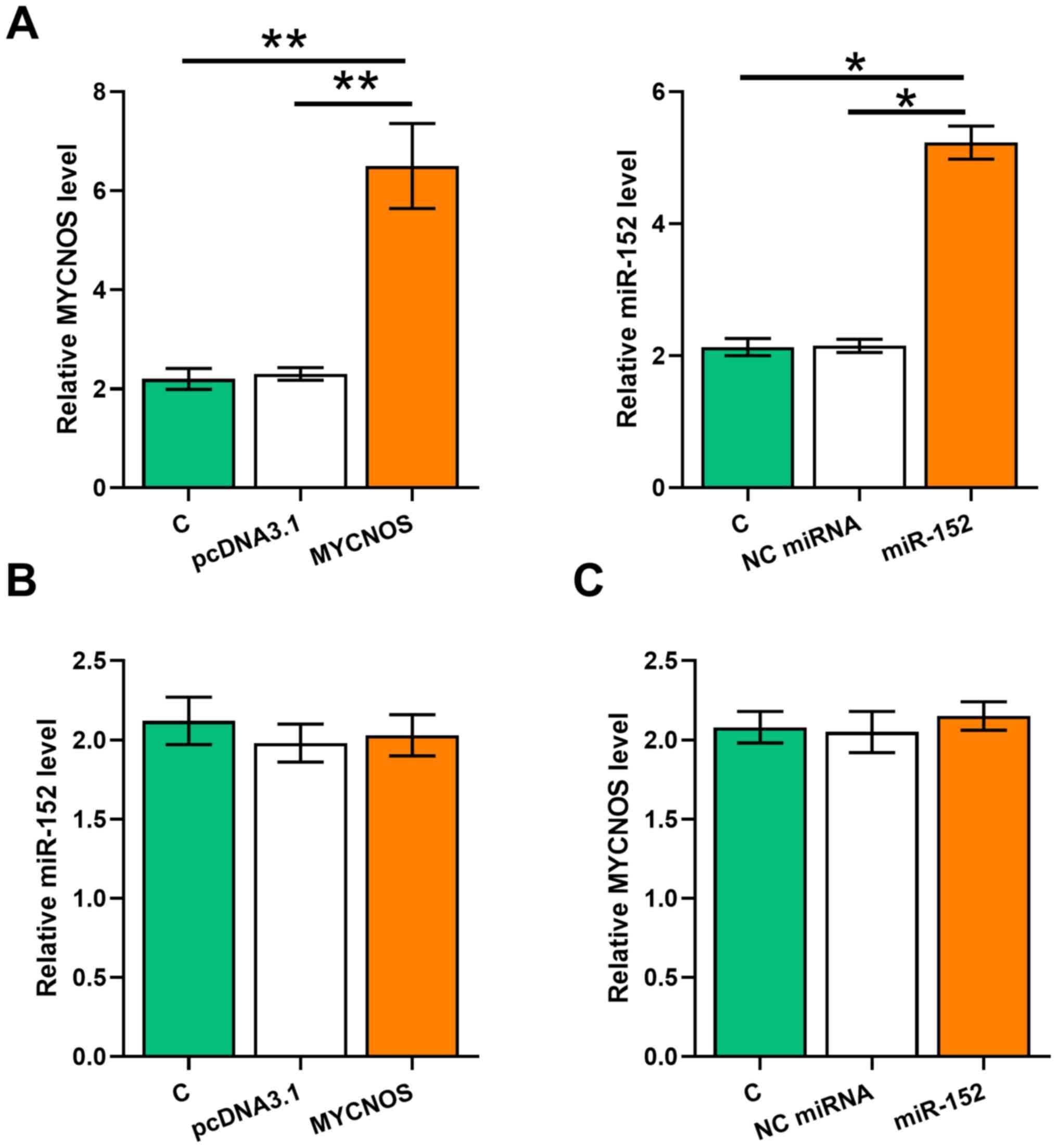 |
Figure 4.
Overexpression of MYCNOS and miR-152 does not mutually affect their expression levels in SKOV3 cells. To explore the interaction between MYCNOS and miR-152, SKOV3 cells were transfected with MYCNOS expression vector or miR-152 mimic. (A) Overexpression of MYCNOS and miR-152 was confirmed using RT-qPCR. The effect of (B) MYCNOS overexpression on miR-152 expression and (C) miR-152 mimic transfection on MYCNOS expression were evaluated using RT-qPCR. The data are presented as the mean ± SD. *P<0.05, **P<0.01. MYCNOS, MYCN opposite strand; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; C, untransfected control cells.
|
MYCNOS sponges miR-152 to upregulate MKK7
In addition to being miRNA targets, lncRNAs may also serve as miRNA sponges. Thus, the effects of MYCNOS and miR-152 overexpression on MKK7, a confirmed target of miR-152, were explored using RT-qPCR and western blot analyses. It was observed that MKK7 expression significantly increased following MYCNOS overexpression, but was significantly reduced following miR-152 mimic transfection, both at the mRNA (Figs. 5A and 6A; P<0.05) and the protein levels (Figs. 5B and 6B; P<0.05). Moreover, MYCNOS overexpression suppressed the function of miR-152.
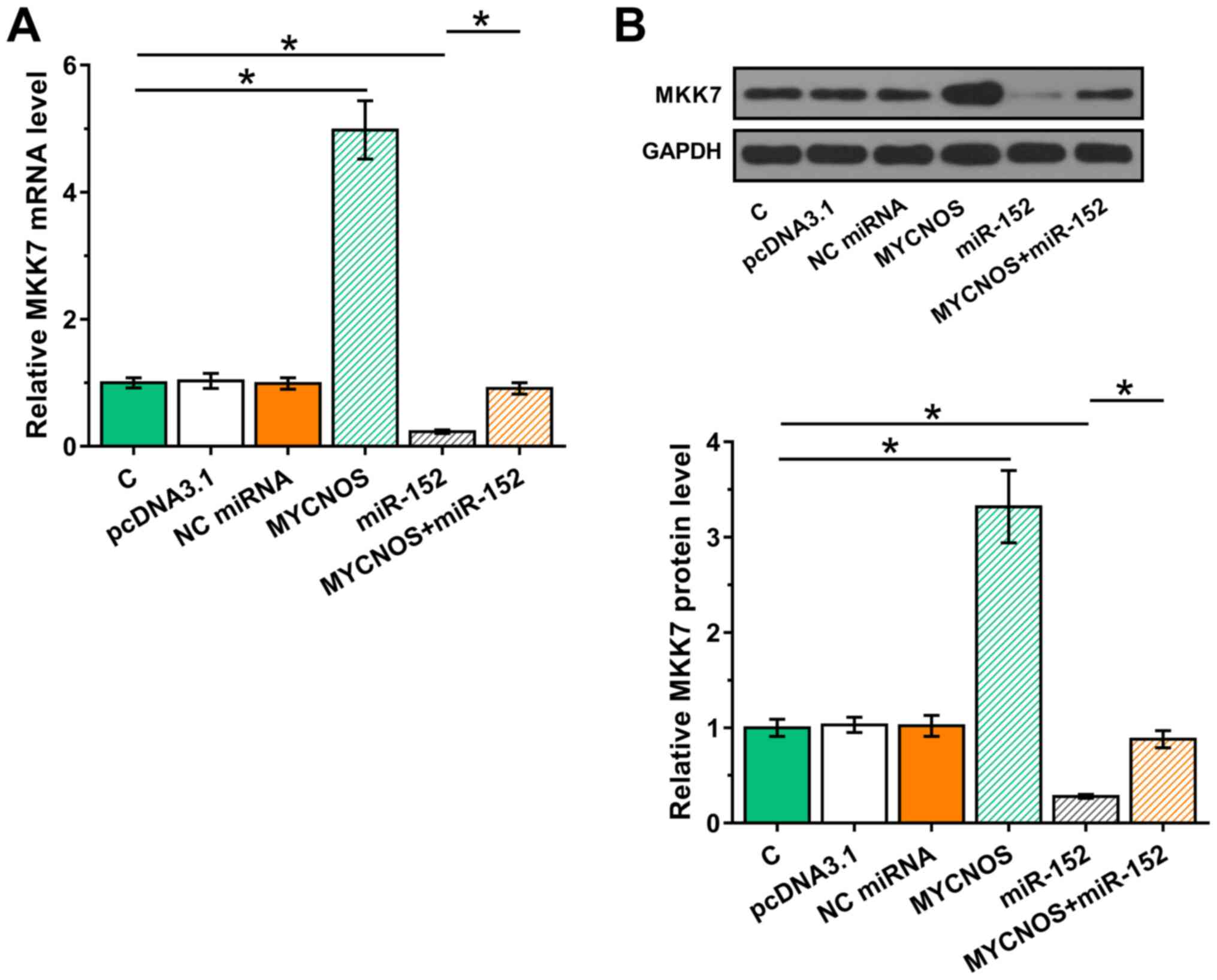 |
Figure 5.
MYCNOS sponges miR-152 to upregulate MKK7 expression in Caov-3 cells. The effect of MYCNOS and miR-152 overexpression on MKK7 expression levels were evaluated at the (A) mRNA and (B) protein levels using reverse transcription-quantitative PCR and western blotting, respectively. The data are presented as the mean ± SD. *P<0.05. MYCNOS, MYCN opposite strand; miR, microRNA; MKK7, mitogen-activated protein kinase kinase 7; NC, negative control; C, untransfected control cells.
|
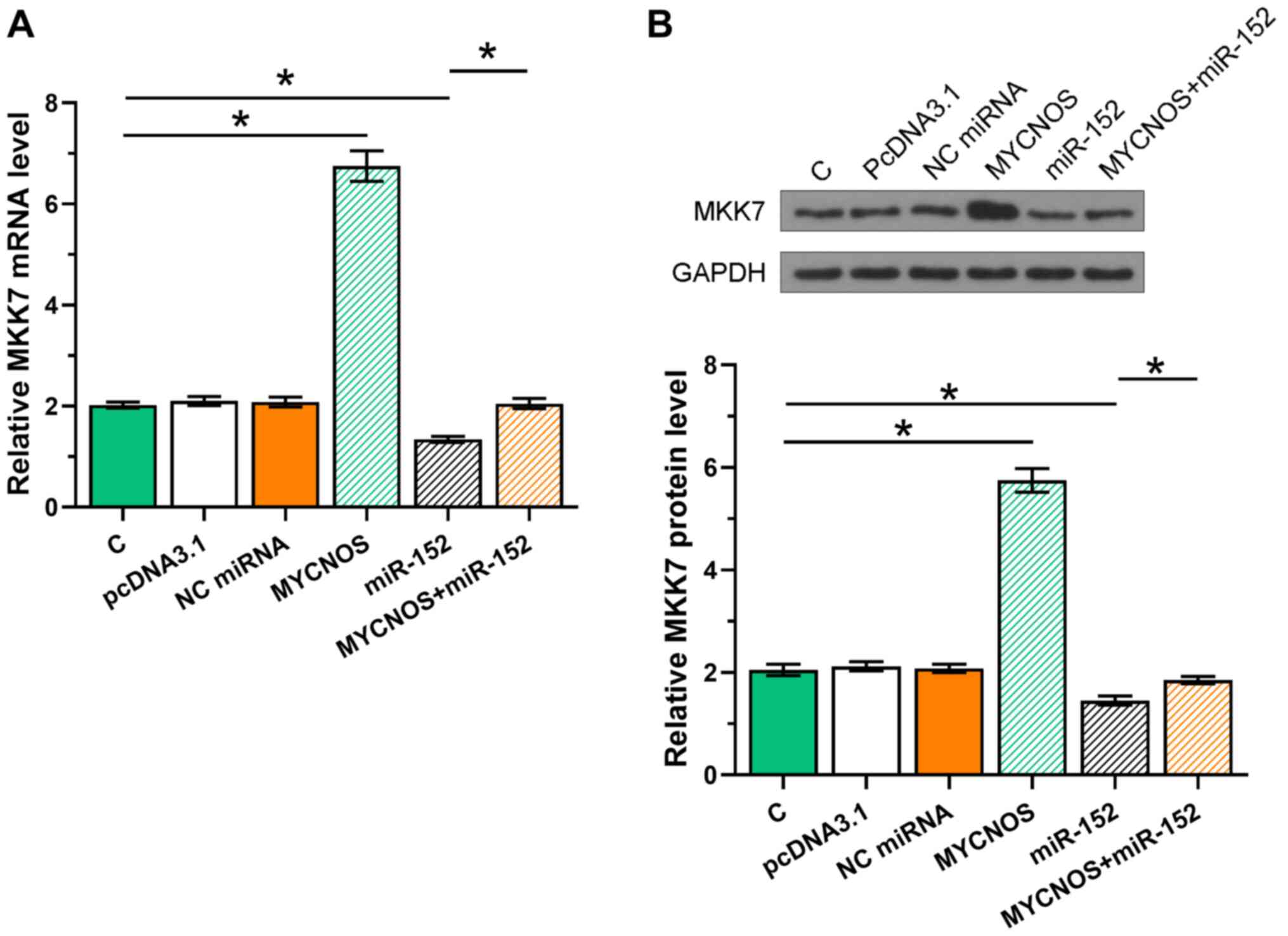 |
Figure 6.
MYCNOS sponges miR-152 to upregulate MKK7 expression in SKOV-3 cells. The effect of MYCNOS and miR-152 overexpression on MKK7 expression were evaluated at the (A) mRNA and (B) protein levels using reverse transcription-quantitative PCR and western blotting, respectively. The data are presented as the mean ± SD. *P<0.05. MYCNOS, MYCN opposite strand; miR, microRNA; MKK7, mitogen-activated protein kinase kinase 7; NC, negative control; C, untransfected control cells.
|
OA cell proliferation is regulated by the MYCNOS/ miR-152/MKK7 axis
The roles of MYCNOS, miR-152 and MKK7 in the regulation of cell proliferation were analyzed using CCK-8 assays. Cell proliferation increased following MYCNOS and MKK7 overexpression and was reduced following miR-152 mimic transfection. In addition, MYCNOS overexpression attenuated the effect of miR-152 overexpression (Figs. 7 and 8).
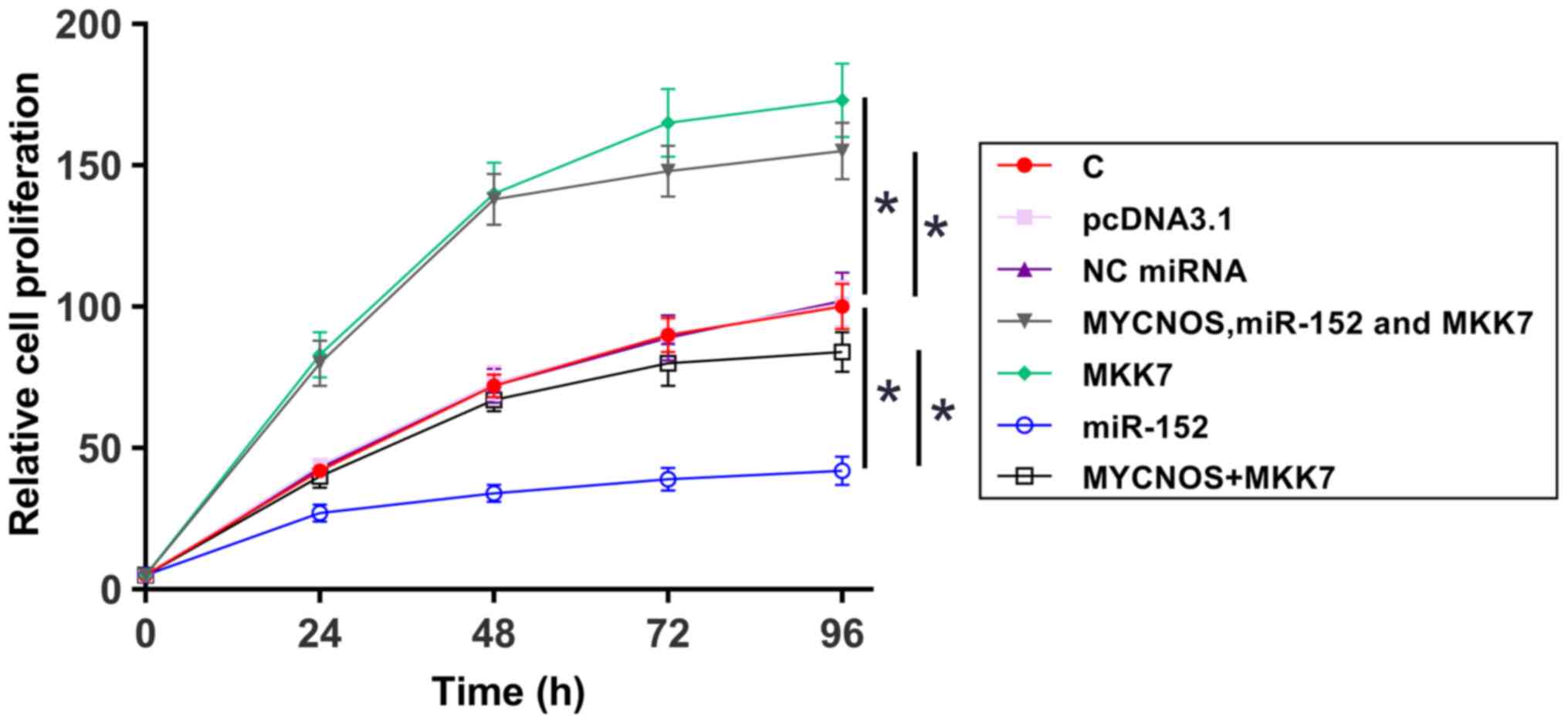 |
Figure 7.
Caov-3 cell proliferation is regulated by the MYCNOS/miR-152/MKK7 axis. The roles of MYCNOS, miR-152 and MKK7 in the regulation of Caov-3 cell proliferation were analyzed using a Cell Counting Kit-8 assay. The data are presented as the mean ± SD. *P<0.05. MYCNOS, MYCN opposite strand; miR, microRNA; MKK7, mitogen-activated protein kinase kinase 7; NC, negative control; C, untransfected control cells.
|
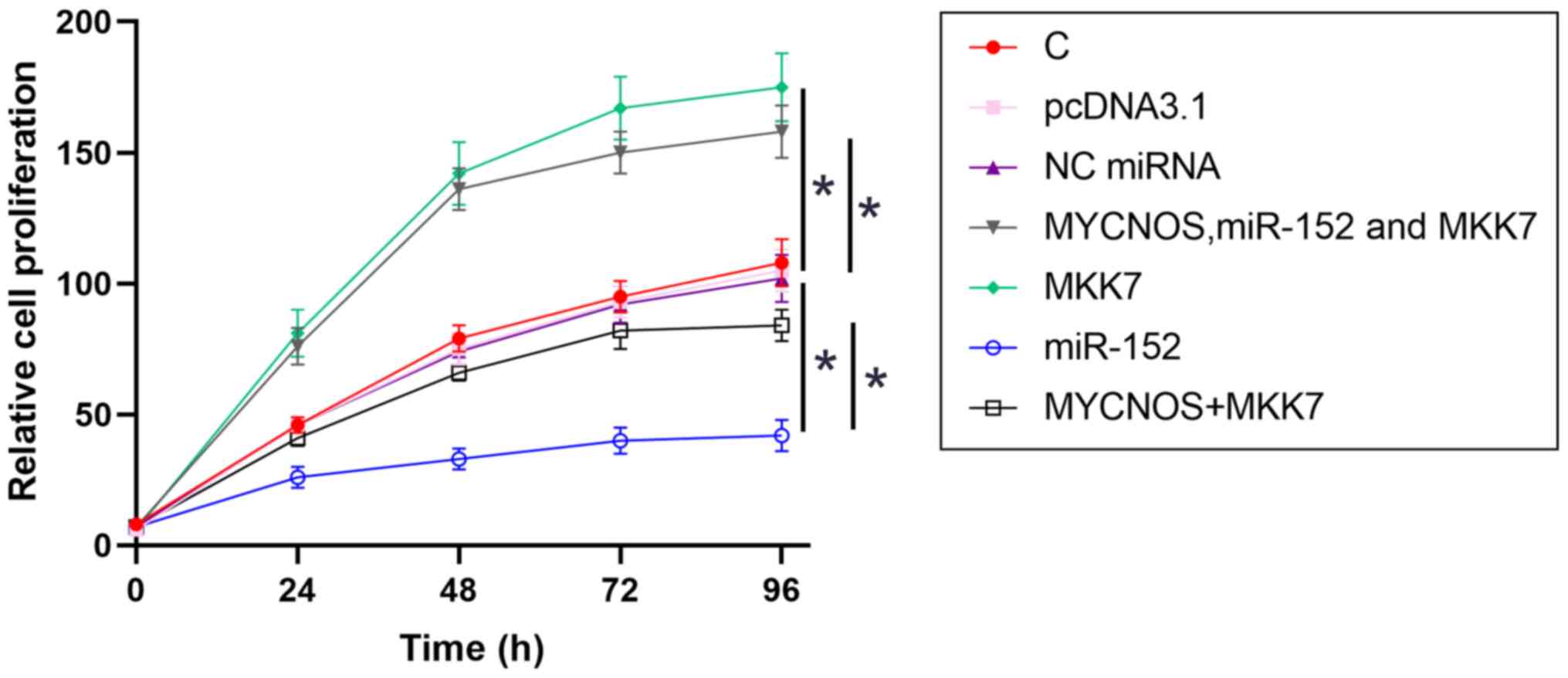 |
Figure 8.
SKOV3 cell proliferation is regulated by the MYCNOS/miR-152/MKK7 axis. The roles of MYCNOS, miR-152 and MKK7 in the regulation of SKOV3 cell proliferation were analyzed using a Cell Counting Kit-8 assay. The data are presented as the mean ± SD. *P<0.05. MYCNOS, MYCN opposite strand; miR, microRNA; MKK7, mitogen-activated protein kinase kinase 7; NC, negative control; C, untransfected control cells.
|
Discussion
OA treatment is hampered by the low rates of early diagnosis, owing to a lack of sensitive biomarkers (20). lncRNAs act as a crucial modulators of various physiological processes by affecting gene expression, regulating subcellular structures and altering specific protein localization (21). Several lncRNA molecules have been found to be aberrantly expressed in different types of cancer and they are associated with tumorigenesis, cancer development and clinical prognosis (22). It has been reported that the lncRNA MYCNOS regulates the expression of MYCN, a proto-oncogene, in human neuroblastoma cells (23). In a recent study, Yu et al (13) reported that MYCNOS was expressed at high levels in hepatocellular carcinoma and negatively regulated the expression of the tumor suppressor miR-340 to increase cancer cell proliferation and invasion. In another study, MYCNOS-01, an alternative transcript of MYCNOS, specifically upregulated MYCN at the protein level to promote the proliferation of rhabdomyosarcoma and neuroblastoma cells (24). However, the role of MYCNOS in OA remains unclear. To the best of our knowledge, the present study was the first to demonstrate that MYCNOS is upregulated in OA tissue and promotes OA cell proliferation. Therefore, MYCNOS may play an oncogenic role in OA by increasing cell proliferation. Moreover, MYCNOS upregulation in OA tissues was found to be associated with poor patient survival. This suggests that MYCNOS may serve as a prognostic factor for OA, and the measurement of MYCNOS expression prior to therapy may assist in the treatment of OA. However, more clinical trials are needed to confirm this hypothesis.
miRNAs are a class of short non-coding RNAs, 19–25 nucleotides in length, with multiple physiological and pathological functions, including regulation of cell proliferation, differentiation, proliferation and apoptosis (25). These small molecules regulate gene expression by degrading mRNA or inhibiting protein translation of target genes by binding to their 3′-untranslated region. Some miRNA molecules are also involved in drug resistance, acting as oncogenes or tumor suppressors (26). miR-152 is located at 17q21.32 in intron 1 of the COPI coat complex subunit ζ2 gene, serves as a tumor suppressor and is expressed at low levels in several cancer types (27). This miRNA has been found to play tumor-suppressive roles in different types of cancer, including OA (28,29). miR-152 is also expressed at low levels in OA (28). In addition, its overexpression increases the sensitivity of OA cells to chemotherapy (29). A recent study demonstrated that miR-152 targets MKK7 to suppress bladder cancer growth (30). Similarly, the present study indicated that miR-152 may also downregulate MKK7 to suppress OA cell proliferation. Previous studies have reported that the precursor of miR-152 was transported to the cytoplasm and further cleaved by Dicer to form a miR-152 duplex. In total, two different lengths and sequences of mature miRNA are produced from the opposite arms of the miR-152 duplex, namely miR-152-3p and miR-152-5p (31). The findings of the present study indicated that miR-152-3p and miR-152-5p may contribute to the upregulation of MKK7 in OA.
The aim of the present study was to explore the effect of the interaction between MYCNOS and miR-152 on MKK7 expression in OA. The results suggested that MYCNOS was upregulated in OA and may sponge miR-152 to upregulate MKK7 expression, thereby promoting OA cell proliferation. Interestingly, although MYCNOS and miR-152 could interact with each other, they did not mutually regulate their expression levels. Instead, MYCNOS overexpression inhibited the effect of miR-152 on MKK7 expression and OA cell proliferation, indicating that MYCNOS may sponge miR-152.
There were certain limitations to the present study. For example, the expression of MYCNOS, MKK7 and miR-152 in patients with OA and healthy controls, and the correlation of MYCNOS, MKK7 and miR-152 levels with clinical stage were not investigated. In addition, the sponging effect of MYCNOS on miR-152 by using the MYCNOS mutation plasmid was not studied. The lack of using a pGL3-MCYNOS plasmid with a mutated miR-152-binding site is a limitation of the present study that requires further investigation. Future studies should explore the specific effects of MYCNOS in OA in comparison between patients with OA and non-ovarian cancer subjects with healthy fallopian tubes.
In conclusion, MYCNOS was shown to be upregulated in OA and to predict poor patient survival. In addition, MYCNOS may sponge miR-152 to upregulate MKK7 expression, thereby promoting OA cell proliferation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GZ and JY conceived, designed and planned the study. GZ and DZ acquired the data. GZ and XC confirmed and analyzed the data. GZ and LL interpreted the results. DZ provided the study materials. GZ drafted the manuscript. XC provided administrative, logistical and technical support. GZ, DZ, XC, LL and JY reviewed and revised the manuscript for important intellectual content. GZ and LL confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Fourth Affiliated Hospital of Nanchang University (approval no. NC1607AC98).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Jayson GC, Kohn EC, Kitchener HC and Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vaughan S, Coward JI, Bast RC Jr, Berchuck A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R, Etemadmoghadam D, et al: Rethinking ovarian cancer: Recommendations for improving outcomes. Nat Rev Cancer. 11:719–725. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL: Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
La Vecchia C: Ovarian cancer: Epidemiology and risk factors. Eur J Cancer Prev. 26:55–62. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hunn J and Rodriguez GC: Ovarian cancer: Etiology, risk factors, and epidemiology. Clin Obstet Gynecol. 55:3–23. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sapiezynski J, Taratula O, Rodriguez-Rodriguez L and Minko T: Precision targeted therapy of ovarian cancer. J Control Release. 243:250–268. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duan P, Fan L, Gao Q, Silwal BM, Ren M, Shen Y and Qu W: Targeted Therapy of Ovarian Cancer with Angiogenesis Inhibitors. Curr Drug Targets. 18:1171–1178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim A, Ueda Y, Naka T and Enomoto T: Therapeutic strategies in epithelial ovarian cancer. J Exp Clin Cancer Res. 31:142012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee YT, Tan YJ and Oon CE: Molecular targeted therapy: Treating cancer with specificity. Eur J Pharmacol. 834:188–196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deb B, Uddin A and Chakraborty S: miRNAs and ovarian cancer: An overview. J Cell Physiol. 233:3846–3854. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tripathi MK, Doxtater K, Keramatnia F, Zacheaus C, Yallapu MM, Jaggi M and Chauhan SC: Role of lncRNAs in ovarian cancer: Defining new biomarkers for therapeutic purposes. Drug Discov Today. 23:1635–1643. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu J, Ou Z, Lei Y, Chen L, Su Q and Zhang K: lncRNA MYCNOS facilitates proliferation and invasion in hepatocellular carcinoma by regulating miR-340. Hum Cell. 33:148–158. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu H, Zhang M, Shi M, Zhang T, Zhang Z, Cui Q, Yang S and Li Z: A Survival-Related Competitive Endogenous RNA Network of Prognostic lncRNAs, miRNAs, and mRNAs in Wilms Tumor. Front Oncol. 11:6084332021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nakagawa K, Sugahara M, Yamasaki T, Kajiho H, Takahashi S, Hirayama J, Minami Y, Ohta Y, Watanabe T, Hata Y, et al: Filamin associates with stress signalling kinases MKK7 and MKK4 and regulates JNK activation. Biochem J. 427:237–245. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wada T, Joza N, Cheng HY, Sasaki T, Kozieradzki I, Bachmaier K, Katada T, Schreiber M, Wagner EF, Nishina H, et al: MKK7 couples stress signalling to G2/M cell-cycle progression and cellular senescence. Nat Cell Biol. 6:215–226. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edge SB and Compton CC: The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chu C, Quinn J and Chang HY: Chromatin isolation by RNA purification (ChIRP). J Vis Exp. 61:39122012.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lutz AM, Willmann JK, Drescher CW, Ray P, Cochran FV, Urban N and Gambhir SS: Early diagnosis of ovarian carcinoma: Is a solution in sight? Radiology. 259:329–345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi X, Sun M, Liu H, Yao Y and Song Y: Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qiu MT, Hu JW, Yin R and Xu L: Long noncoding RNA: An emerging paradigm of cancer research. Tumour Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vadie N, Saayman S, Lenox A, Ackley A, Clemson M, Burdach J, Hart J, Vogt PK and Morris KV: MYCNOS functions as an antisense RNA regulating MYCN. RNA Biol. 12:893–899. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
O'Brien EM, Selfe JL, Martins AS, Walters ZS and Shipley JM: The long non-coding RNA MYCNOS-01 regulates MYCN protein levels and affects growth of MYCN-amplified rhabdomyosarcoma and neuroblastoma cells. BMC Cancer. 18:2172018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malizia AP and Wang DZ: MicroRNAs in cardiomyocyte development. Wiley Interdiscip Rev Syst Biol Med. 3:183–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF and Pogribny IP: Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 7:2152–2159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang J, Wang Y, Guo Y and Sun S: Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 52:60–70. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou X, Zhao F, Wang ZN, Song YX, Chang H, Chiang Y and Xu HM: Altered expression of miR-152 and miR-148a in ovarian cancer is related to cell proliferation. Oncol Rep. 27:447–454. 2012.PubMed/NCBI
|
|
29
|
Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, Zhao R, Huang H, Wang X, Qiao Y, et al: miR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: A novel epigenetic therapy independent of decitabine. Oncogene. 33:378–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wallace MC, Preen DB, Short MW, Adams LA and Jeffrey GP: Hepatocellular carcinoma in Australia 1982–2014: Increasing incidence and improving survival. Liver Int. 39:522–530. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
You W, Zhang X, Ji M, Yu Y, Chen C, Xiong Y, Liu Y, Sun Y, Tan C, Zhang H, et al: miR-152-5p as a microRNA passenger strand special functions in human gastric cancer cells. Int J Biol Sci. 14:644–653. 2018. View Article : Google Scholar : PubMed/NCBI
|






















