Introduction
As a major form of lung cancer, non-small cell lung cancer (NSCLC) is the leading cause of tumor mortality in humans, with >500,000 casualties per year worldwide (1,2). Although early screening and treatment of NSCLC has evolved in recent years, its high fatality rate has not yet notably declined. The negligence in the diagnosis of NSCLC can mainly be attributed to the lack of a standard, effective, economical, diagnostic method to screen early tumors (3). It has been reported that timely diagnosis, as well as treatment in the early stage, increases the 5-year survival rate to 60-80% for patients with NSCLC (4). Therefore, it is a requirement to discover effective molecular markers specific to early-stage lung cancer.
DNA methylation is an important regulatory mechanism in oncogenesis. In mammals, DNA methyltransferases (DNMTs) promotes DNA methylation, while implementing s-adenosyl methionine as a methyl donor to methylate the carbon atom at position 5 of the pyrimidine ring of cytosine nucleotides that form CpG islands (5). The DNMT family, especially DNMT1, DNMT3A and DNMT3B, serve essential roles in the initiation and maintenance of DNA methylation (6,7). Lin et al (8) discovered that hypermethylation of tumor suppressor gene such as FHIT and P16 core regions in tumor tissues is closely associated with DNMT proteins, which are upregulated in a variety of tumors. Especially in patients with NSCLC and a history of smoking, the expression of DNMT1, DNMT3A and DNMT3B proteins are notably upregulated, and patients with high expression of DNMT1 protein have been indicated to exhibit a poor prognosis (9). Kim et al (9) investigated 102 cases of NSCLC, and revealed that DNMT1 and DNMT3B mRNA was increased, with the concomitant hypermethylation of related tumor suppressor genes.
MicroRNAs (miRNAs/miRs) are a cluster of small non-coding RNAs with 17-25 nucleotides in length, and are capable of regulating gene expression in the post-transcriptional phase by directly binding to the 3′-untranslated region (3′-UTR) of the target mRNA (10). As important regulatory factors, miRNAs participate in various physiological and pathological processes, including cell proliferation, differentiation, apoptosis and the occurrence of metastasis and regression of tumors (11,12). Given their flexibility in tumor physiology and pathology, miRNAs may serve as markers for tumor prognosis. The function of certain miRNAs and their regulatory mechanisms have been elucidated. For example, through recruiting DNMT3A and DNMT3B to regulate target gene demethylation, miR-29 restrains the proliferation of hepatocellular carcinoma cells (13), and experimental results have confirmed that miR-29 is a tumor suppressor gene in hepatocellular carcinoma (14). miR-152 along with miR-185 regulate the cisplatin sensitivity of ovarian tumor cells by targeting DNMT1 in vivo and in vitro (15). miR-152 belongs to the miR-148/152 family, and it has been indicated to be downregulated in the cytoplasm in NSCLC (16). Our previous study used bioinformatics analysis and revealed that miR-152-3p was a cisplatin resistance-associated miRNA (17). The target genes of miR-152-3p were initially predicted using TargetScan and PicTar databases, which uncovered a binding site for miR-152-3p on the mRNA of neural cell adhesion molecule 1 (NCAM1) (17). However, the function of miR-152-3p in NSCLC and whether it is associated with DNMT-mediated methylation, as well as its regulatory mechanisms remain elusive.
The present study conducted in-depth molecular biological studies on DNMT3B and miR-152-3p in NSCLC cell lines, aiming to determine whether DNMT3B can regulate miR-152-3p and whether there is a regulatory pathway for the inhibition of proliferation in NSCLC cells. The current study discovered a regulatory network of the pathogenesis of NSCLC based on miR-152-3p methylation, with the aim to provide a novel therapeutic approach for NSCLC gene therapy.
Materials and methods
Cell culture and transfections
The lung adenocarcinoma cell line A549, as well as the cisplatin-resistant cell lines A549/DDP and A549/ADM (ATCC), which have been subjected to mycoplasma testing and short tandem repeat profiling, were conserved in the laboratory. All cells were cultivated in DMEM (Hyclone; Cytiva) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and penicillin (100 U/ml)/streptomycin (0.1 mg/ml) in a cell incubator at a constant temperature of 37°C with 5% CO2. Cells were inoculated into 24-well plates at a density of 5×104 cells/well, and cultured for 18-24 h before transfection. Overexpression empty vector pcDNA3.1, short hairpin (sh)-DNMT3B, sh-NC(pcRNAU6.1-SHH-shRNA-1, GENE ID 6469; Shanghai GeneChem, http://www.genechem.com.cn), mimics negative control (NC) and inhibitor NC, miR-152-3p mimics and inhibitor [General Biosystems (Anhui) Corporation Ltd.] along with a pcDNA-NCAM1 overexpression plasmid (Shanghai GeneChem) were transfected into cells that were 60-80% confluent with Lipofectamine 3000® reagent (Thermo Fisher Scientific, Inc.). The transfection volume ratio of mimics/inhibitor or shRNA/OE-NCAM1 to Lipofectamine 3000 or empty vectors was 1:1 or 1:2 according to the cell status and resistance to transfection. The transfection was performed at room temperature for 15 min and then the cells were placed in cell incubator at a constant temperature of 37°C with 5% CO2 for 4-6 h. The final concentration of mimics and inhibitor was 30 nM. The transfected cells were then cultured for 48 h before further experiments and control group was untransfected cells. Cell transfection efficiency is presented in Figs. S1 and S2. The sequences of miR-152-3p mimics and inhibitor and corresponding NCs are listed in Table I. The three target sequence for DNMT3B were GCTGTCCGAACTCGAAATAACGGCAGTAGGAAATTAGAATCAGCCCATTTGACTTGGTGATTG.
 |
Table I.
Primers for RT-quantitative PCR of miR-152-3p and NCAM1 and sequences of miR-152-3p mimics and inhibitor.
|
Table I.
Primers for RT-quantitative PCR of miR-152-3p and NCAM1 and sequences of miR-152-3p mimics and inhibitor.
| Primer |
Sequence (5′-3′) |
| hsa-miR-152-3p-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCAAGT |
| hsa-miR-152-3p-F |
GCGCTCAGTGCATGACAGA |
| hsa-miR-152-3p-R |
GTGCAGGGTCCGAGGT |
| hsa-miR-152-3p mimics-sense |
UCAGUGCAUGACAGAACUUGG |
| hsa-miR-152-3p mimics-antisense |
CCAAGUUCUGUCAUGCACUGA |
| mimics NC-sense |
UCACAACCUCCUAGAAGAGUAGA |
| mimics NC-antisense |
UCUACUCUUUCUAGGAGGUUGUGA |
| hsa-miR-152-3p inhibitor |
CCAAGUUCUGUCAUGCACUGA |
| inhibitor NC |
UCUACUCUUUCUAGGAGGUUGUGA |
| hsa-NCAM1-F |
CTCACCCCAAACCAGCAG |
| hsa-NCAM1-R |
GGTGGCCTCTGACTCACT |
| DNMT3B-F |
TACAATAGGATAGCCAAGTTAAAG |
| DNMT3B-R |
GCAGCTTCTGGCG |
| U6-F |
CTCGCTTCGGCAGCACA |
| U6-R |
AACGCTTCACGAATTTGCGT |
| GAPDH-F |
CCAGGTGGTCTCCTCTGA |
| GAPDH-R |
GCTGTAGCCAAATCGTTGT |
Cell proliferation assay
MTT assay was used to examine the proliferation of the cells. Non-transfected A549 cells or cells transfected with mimics/inhibitor, shDNMT3B or OE-NCAM1 cells in the logarithmic growth phase were re-seeded into 96-well plates, cultured for 48 h and cell proliferation was assessed. A total of 20 µl of an aqueous solution of MTT (5 mg/ml) was added to each well and the plates were incubated for 4 h at 37°C in a humidified atmosphere containing 5% CO2. After, the medium was removed from the wells, 100 µl DMSO was added to each well and the absorbance was measured at a wavelength of 570 nm at 24, 48 and 72 h. Three experiments in quadruplicate were conducted independently.
Colony formation assay
A total of ~500 A549 cells transiently transfected with sh-DNMT3B, miR-152-3p mimics/inhibitor, OE-NCAM1 or their respective NCs were re-seeded in six-well plates without soft agar and cultured in freshly prepared complete DMEM medium at a constant temperature of 37°C with 5% CO2 for 10-14 days. Then the medium was discarded and cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were stained with crystal violet for 15 min at room temperature and colonies (consisting cells about 60 and diameter 1-2 mm) in each well were counted under a light microscope (CKX41; Olympus Corporation) manually at ×10 magnification.
Transwell invasion assay
Transwell chambers (Millipore Sigma) were filled with 100 µl Matrigel (BD Biosciences) diluted in serum-free DMEM and precoated at 37°C for 30 min and 5×104 A549 cells transfected with mimics/inhibitor, OE-NCAM1 or respective NCs in serum-free DMEM were seeded in the upper chamber. Complete DMEM was placed in the lower Transwell chamber, and the cells were incubated for 48 h in a cell incubator at 37°C. The cells that did not invade the basal membrane in the invasion chamber were removed with a cotton swab. The cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Subsequently, the outside of the Transwell chamber was stained with 0.1% crystal violet for 10 min at room temperature. Invaded cells were observed under an inverted light microscope (Ts2-FL; Nikon Corporation). The invaded cells were manually counted from 10 random fields at ×100 magnification.
Flow cytometry
A549 cells transfected with miR-152-3p mimics/inhibitor or respective NCs were harvested and the evaluation and assessment of apoptosis was based on staining 100 µl cells in 5 µl Annexin V-phycoerythrin as well as 5 µl 7-aminoactinomycin D (BioLegend, Inc.) prior to flow cytometry for 15 min at room temperature. Cellular samples were examined using the CytoFLEX flow cytometer (Beckman Coulter) and the data was analyzed by CytExpert 2.0 (Beckman Coulter, Inc.).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA in A549 cells was extracted with TRIzol® reagent (Thermo Fisher Scientific). cDNA (Super Script IV; cat. no. 18091050, Thermo Fisher Scientific, Inc.) was synthesized using OligodT primers according to manufacturer's instruction. Target gene mRNA levels were examined using a fluorescent quantitative PCR kit (PerfectStart Green qPCR SuperMix; Beijing Transgen Biotech Co., Ltd.). The thermocycling conditions were as follows: 94°C for 30 sec, followed by 40 cycles of 94°C for 5 sec, 60°C for 15 sec and 72°C for 10 sec. The relative expression levels of NCAM1, DNMT3B mRNA and miR-152-3p were calculated using the 2−ΔΔCq (18) with GAPDH or U6 as the internal reference, respectively. The primers used are listed in Table I.
Western blotting
Whole A549 cells extracts were prepared using RIPA buffer (Beyotime Institute of Biotechnology). The protein concentration was determined by the bicinchoninic acid (BCA) method. Protein (50 µg) from each group was loaded and separated by SDS-PAGE electrophoresis (12% running and 5% stacking gel) before being transferred to a PVDF membrane (EMD Millipore). The membrane was blocked in 5% BSA-PBST solution at room temperature for 60 min and incubated with NCAM1 (Abcam; cat. no; cat. no. ab75813; 1:500), cleaved-caspase-3 (Abcam; cat. no. ab32351; 1:1,000), cleaved-poly (ADP-ribose) polymerase (PARP) (CST; cat. no. 9532S; 1:500) and β-actin antibodies (PROTEINTECH; cat. no. 60009-1-Ig, 1:5,000) at 4°C overnight. The secondary antibodies Anti-Rabbit IgG (H+L;KPL, Inc.; cat. no. 074-1506, 1:5,000) and Anti-Mouse IgA + IgG + IgM (H+L; KPL, Inc.; cat. no. 074-1807, 1:5,000) were incubated at room temperature for 60 min. ECL kit (GENVIEW; cat. no. GE2301) was used to visualization according to the manufacturer's instruction. A Clinx ChemiScope 6000 (Clinx Science Instruments) imaging system was used to observe the bands and ImageJ (v.1.8.0; National Institutes of Health) was used for densitometry.
Luciferase assay
The wild type (WT) or mutant (MUT) 3′-UTRs of NCAM1 were subcloned into the pGL3 vector. The 0.16 mg/ml plasmid containing NCAM1-3′-UTR-WT/NCAM1-3′-UTR-MUT (Promega Corporation; cat. no. E1751), as well as 5 pmol/mlmiR-152-3p mimics/mimics NC were co-transfected into 293T cells (ATCC) using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.; cat. no. L3000015). 293 T cells were cultured in DMEM (Hyclone; Cytiva) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin in a cell incubator at a constant temperature of 37°C with 5% CO2. The luciferase activity was assessed 48 h after transfection using Dual-Luciferase Reporter Assay System (Promega Corporation; cat. no. E1910) according to manufacturer's instruction. Method of normalization for Firefly luciferase activity was by comparison with Renilla luciferase activity.
Methylation specific polymerase (MSP) chain reaction
Genomic DNA extracted from A549, A549/DDP and A549/ADM cells was bisulfite-treated using Bisulfite Conversion kit (Abcam; cat. no. ab117126) according to the manufacturer's instructions. The methylation level of miR-152-3p was measured via an MSP chain reaction. The reaction conditions were as follows: Pre-denaturation at 95°C for 5 min; 35 cycles at 94°C for 20 sec, 60°C for 30 sec, 72°C for 20 sec; and final extension at 72°C for 5 min. The primers used for MSP chain reaction are listed in Table II.
 |
Table II.
Primers for MSP chain reaction of the miR-152-3p promoter.
|
Table II.
Primers for MSP chain reaction of the miR-152-3p promoter.
| Primer type |
Primer name |
Sequence (5′-3′) |
| Methylated |
MSP-M-miR-152-1-Forward |
TTTGAGATTTAAAGGGTGATTTGAC |
| |
MSP-M-miR-152-1-Reverse |
TCCTACTTCCTACCTACTCTCCGA |
| Unmethylated |
MSP-U-miR-152-1-Forward |
TTTGAGATTTAAAGGGTGATTTGAT |
| |
MSP-U-miR-152-1-Reverse |
CTACTTCCTACCTACTCTCCAAC |
Bisulfite sequencing PCR
Genomic DNA from A549, A549/DDP and A549/ADM cells was prepared as aforementioned for the MSP reaction. A549 cells were treated with or without 40 µM thioguanine (Sigma-Aldrich; Merck KGaA) and 10 µM nanaomycin A (Sigma-Aldrich; Merck KGaA) for 24 h before obtaining genomic DNA. The core region of miR-152-3p amplified by TaKaRa EpiTaq HS in Fly Fast Pfu Mix (TransGen Biotech Co., Ltd.) was cloned into pUCm-T vector (TsingKe Biological Technology), and at least five respective clones were sent to company (Shanghai GeneChem) and sequenced for the amplified fragment using Sanger sequencing. The PCR reaction conditions were as follows: Pre-denaturation at 98°C for 2 min; 30 cycles at 98°C for 10 sec, 55°C for 30 sec, 72°C for 30 sec; and final extension at 72°C for 2 min. The whole bisulfate sequencing process was DNA extraction, DNA bisulfate conversion, PCR amplification for the target gene region, amplified fragment cloned to T vector and colonies send for Sanger Sequencing. Bisulfite sequencing PCR primers were synthesized by Invitrogen; Thermo Fisher Scientific, Inc. (Table III).
 |
Table III.
Primers for bisulfite sequencing PCR of the miR-152-3p promoter.
|
Table III.
Primers for bisulfite sequencing PCR of the miR-152-3p promoter.
| Primer |
Sequence (5′-3′) |
| BSP-Forward |
TAGAGTAGGGTTAGGGGGAGTAGTTA |
| BSP-Reverse |
ATAAACTCCAAAAACATACCCATCA |
Chromatin immunoprecipitation (ChIP) assay
Briefly, 1×107 A549 and A549/DDP cells were collected in 20 ml PBS, subsequently crosslinked with 1% formaldehyde for 10 min at room temperature, and immunoprecipitation was performed with DNMT1 (Abcam; cat. no; cat. no. ab19905, 1:100), DNMT3A (Abcam; cat. no; cat. no. ab226261, 1:100), DNMT3B antibody (Abcam; cat. no; cat. no. ab227883, 1:100), positive ChIP antibody Histone H3 (cat. no. D2B12) XP Rabbit mAb (Cell Signaling Technology; cat. no. 4620S; 1:500) and negative ChIP antibody/Normal Rabbit IgG Antibody (Cell Signaling Technology; cat. no. 2729; 1:500) following the manufacturer's instructions of the SimpleChIP Enzymatic Chromatin IP kit according to the manufacturer's instruction (Cell Signaling Technology; cat. no. 9003). DNA fragments obtained by ChIP were detected by PCR Mix (Thermo Fisher Scientific; cat. no. 10572014) according to the manufacturer's instructions. PCR reaction conditions were: Pre-denaturation at 95°C for 5 sec; 30 cycles at 95°C for 30 sec, 60°C for 2 min, 72°C for 1 min; and final extension at 72°C for 5 min. The primers used for BSP are listed in Table IV. PCR product (10 µl) was used for 2% agarose gel electrophoresis and visualized with 0.5 µg/ml ethidium bromide for 15 min at room temperature. Positive control represents the tested sample + positive ChIP antibody; negative control represents the tested sample with negative ChIP antibody.
 |
Table IV.
Primers for Chip-PCR.
|
Table IV.
Primers for Chip-PCR.
| Chip-PCR amplification primer |
Product size:154 bp; Tm:56°C |
| ChIP-miR-152-F1 |
CCCCAGTCAACTAGCTGT |
| ChIP-miR-152-R1 |
AACACTGTGCGTGTATTTTAC |
Bioinformatics Analysis
Patient cohort GSE74190 with 30 LUSC patients and 44 adjacent tissues was download from dbDEMC database (Experiment ID: EXP00310; http://www.biosino.org/dbDEMC/experiment/detail/EXP00310C). Gene expression profiles of 483 LUAD patients/59 normal samples and 486 LUSC patients/338 normal samples were downloaded from the TCGA database (https://cancergenome.nih.gov/). The expression of miR-152-3p and NCAM1 RNA sequencing data between lung cancer and normal tissues was analyzed using the edgeR package in R (Version 4.0, R: http://www.r-project.org/). P<0.05 was considered to indicate a statistically significant difference.
Statistical analysis
All data with three experimental repeats are presented as the mean ± SD. GraphPad Prism 8.0 (GraphPad Software, Inc.) was used for statistical analysis using one-way ANOVA with Tukey's as post hoc test for multiple group comparisons or two tailed unpaired Student's t-test for comparisons between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
DNMT3B binds to and methylates the miR-152-3p core promoter region
To examine the methylation level of the miR-152-3p promoter, methylated and non-methylated primers were used to amplify the bisulfite-treated samples of the three cell lines simultaneously. A549 cells demonstrated a relatively high level of methylation in the miR-152-3p region compared with A549/DDP and A549/ADM cells (Fig. 1A). In addition, in the bisulfite sequencing experiment the methylation levels of miR-152-3p in A549/ADM (25%) and A549/DDP (29%) groups decreased when compared with the A549 group (35%), and the overall trend was consistent with the MSP results (Fig. 1B). A ChIP assay was carried out to assess whether DNMT3B and DNMT1 can bind to the core region of miR-152-3p. There were prominent bands in the experimental group, indicating that DNMT3B bound to the core region of the miR-152-3p gene in A549 and A549/DDP cells (Fig. 1C). As the ChIP assay results for DNMT1 and DNMT3A were negative (Fig. 1C) and the methylation level of miR-152-3p was not significantly altered in A549 cells treated with the DNMT1 inhibitor thioguanine, it was concluded that DNMT1 did not bind to miR-152-3p. On the other hand, treatment with the DNMT3B inhibitor nanaomycin A resulted in a decrease in the methylation level of the miR-152-3p core region (Fig. 1D). When A549 cells were treated with nanaomycin A, miR-152-3p was upregulated and NCAM1 was downregulated (Fig. 1E). Based on these results, it was concluded that DNMT3B could bind to and methylate the core region of miR-152-3p.
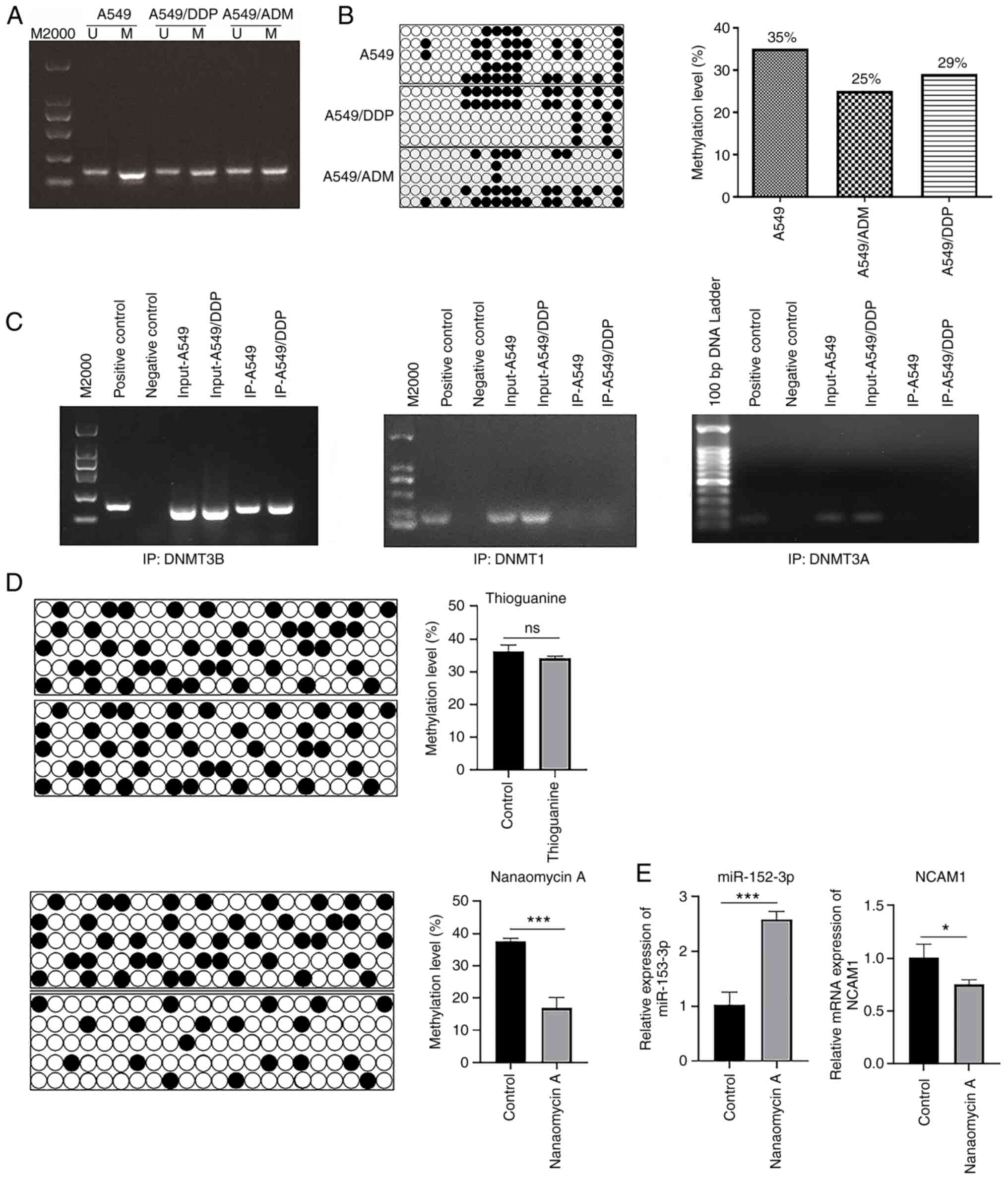 |
Figure 1.
DNMT3B binds to and methylates the miR-152-3p core region. (A) Bisulfite treated DNA samples from A549, A549/DDP and A549/ADM cells were amplified with methylated and non-methylated primers at the same time, and the amplification level was detected by PCR. (B) Bisulfite sequencing was used to detect the methylation level of miR-152-3p in A549, A549/DDP and A549/ADM cells. Five respective clones from each group were sequenced. (C) ChIP assay detected the binding of DNMT3B, DNMT3A and DNMT1 proteins to the core region of miR-152-3p. (D) Bisulfite sequencing results of the methylation levels in the core region of miR-152-3p in A549 cells were detected after treatment with DNMT3B/DNMT1 methylase inhibitors in three independent experiments. (E) Relative mRNA expression levels of miR-152-3p and NCAM1 in A549 cells treated with DNMT3B methylase inhibitor. Two-tailed Student's t-test was used for statistical analysis. *P<0.05, ***P<0.001. DNMT, DNA methyltransferase; miR, microRNA; IP, immunoprecipitation, ChIP, chromatin IP; NCAM1, neural cell adhesion molecule 1; U, unmethylated; M, methylated.
|
miR-152-3p targets and inhibits NCAM1
In the dual luciferase reporter assay, the inhibitory effects of miR-152-3p mimics on the luciferase activity of NCAM1-3′-UTR-WT decreased the relative luciferase activity compared with mimics NC. However, no changes were observed in the luciferase activity of NCAM1-3′-UTR-MUT (Fig. 2C). RT-qPCR and western blot results illustrated that miR-152-3p mimics could inhibit NCAM1 expression while miR-152-3p inhibitor promoted the expression of NCAM1 compared with respective NC in A549 cells (Fig. 2A and B). The expression level of miR-152-3p was higher in A549/DDP cells than in A549 cells, and the expression level of NCAM1 was lower in A549/DDP cells than in A549 cells (Fig. 2D). These results indicated that NCAM1 expression was associated with miR-152-3p and that miR-152/3p may be involved in the cisplatin resistance by targeting NCAM1. Searching in dbDEMC, a database of differently expressed miRNA of human cancers, it was found that the expression level of miR-152-3p in 30 lung squamous cell carcinoma (LUSC) cancer samples increased compared with that in 44 adjacent normal tissues (Fig. 2E). In addition, NCAM1 expression decreased in patients with NSCLC obtained from The Cancer Genome Atlas (TCGA; Fig. 2F). Overall, the aforementioned results indicated that miR-152-3p could target and inhibit NCAM1 in lung cancer.
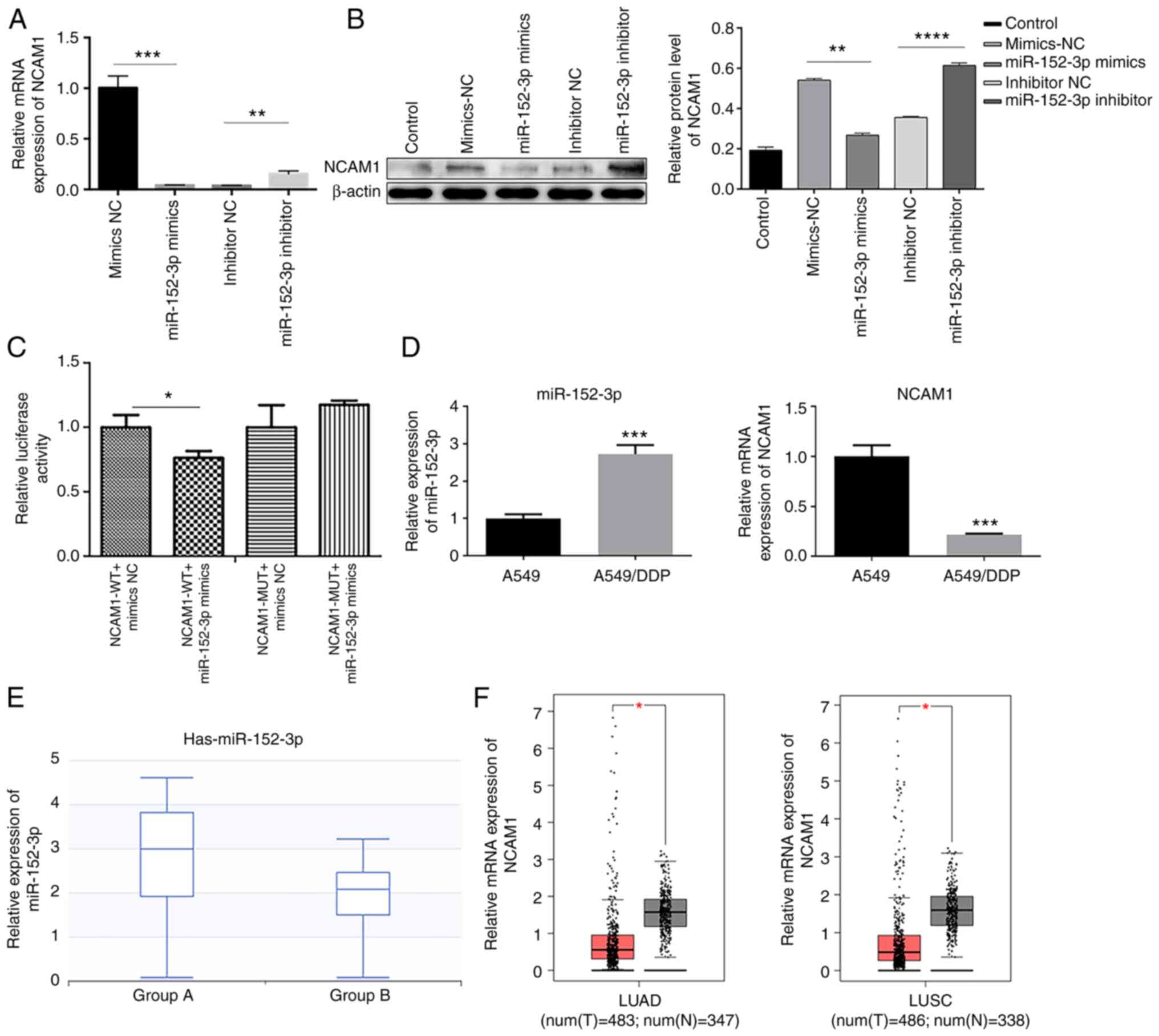 |
Figure 2.
miR-152-3p targets and inhibits NCAM1. (A) Reverse transcription-quantitative PCR demonstrated the effects of miR-152-3p mimics and inhibitor on NCAM1 transcription level in A549 cells. (B) Western blotting revealed the effects of miR-152-3p mimics and inhibitor on NCAM1 protein levels in A549 cells. One-way ANOVA was used for statistical analysis. (C) Relative luciferase activity in the indicated groups from three independent tests. Two-tailed Student's t-test was used for statistical analysis. (D) Relative mRNA expression of miR-152-3p and NCAM1 in A549 and A549/DDP cells. (E) Relative expression of miR-152-3p in lung squamous cell carcinoma samples. Group A referred to the expression of miR-152-3p in 30 patients with LUSC and group B to that of 44 adjacent normal tissues. (F) Relative mRNA expression of NCAM1 in LUAD/LUSC and normal tissues from The Cancer Genome Atlas dataset. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 as indicated or vs. A549. miR, microRNA; NC, negative control; NCAM1, neural cell adhesion molecule 1; WT, wild type; MUT, mutant; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; T, tumor; N, normal.
|
miR-152-3p inhibits the proliferation and invasion of human NSCLC A549 cells
MTT assay revealed that miR-152-3p mimics decreased the proliferation of A549 cells (Fig. 3A), and the colony formation assay demonstrated that miR-152-3p mimics reduced the colony numbers of A549 cells compared with mimics NC (Fig. 3B). On the other hand, miR-152-3p inhibitor exhibited the opposite effect. The two assays verified that miR-152-3p impeded lung adenocarcinoma cell proliferation. Transwell assay demonstrated that cells were less invasive in the mimics group, but the invasion capacity increased in the inhibitor group compared with the NC group (Fig. 3C). With respect to cell apoptosis, flow cytometry demonstrated that miR-152-3p mimics promoted A549 cell apoptosis while inhibitor inhibited apoptosis (Fig. 3D). Western blotting demonstrated that the protein level of NCAM1 was decreased and apoptosis-related proteins (cleaved-caspase-3 and cleaved-PARP) increased in cells transfected with miR-152-3p mimics. Consistently, with miR-152-3p inhibitor, NCAM1 expression was increased and cleaved-caspase-3 and cleaved-PARP expression was decreased (Fig. 3E). The aforementioned findings indicated that miR-152-3p reduces cell proliferation and invasion in lung cancer and may target NCAM1.
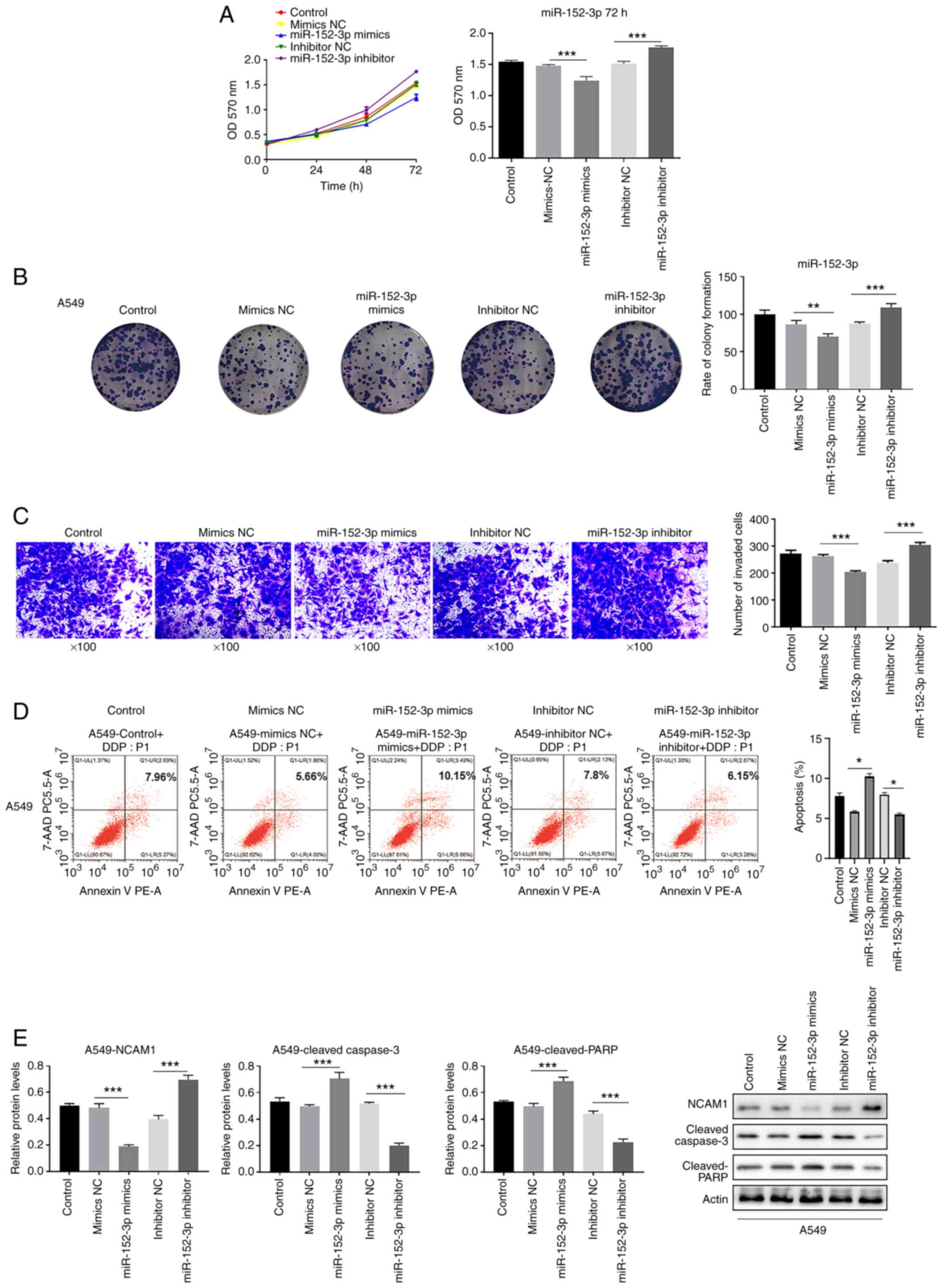 |
Figure 3.
miR-152-3p inhibits proliferation and invasion in A549 cells. (A) Proliferation of A549 cell lines transfected with miR-152-3p mimics/inhibitor at 24, 48 and 72 h; the y-axis indicates the OD value at 570 nm by MTT assay. Histogram indicates the OD value at 72 h. (B) Colony formation rate of A549 cells transfected with miR-152-3p mimics or inhibitor was detected using colony formation assay. (C) Number of invaded cells was detected via Transwell invasion assay. (D) Apoptotic rate was detected via flow cytometry from three independent experiments in triplicate. (E) Western blotting revealed the expression of proteins NCAM1, cleaved-caspase-3 and cleaved-PARP in A549 cells in three independent repeats. One-way ANOVA was used for statistical analysis. *P<0.05, **P<0.01, ***P<0.001. miR, microRNA; NC, negative control; NCAM1, neural cell adhesion molecule 1; OD, optical density; PE, phycoerythrin; PARP, poly(ADP-ribose) polymerase; 7-AAD, 7-aminoactinomycin D.
|
miR-152-3p inhibits the proliferation of A549 cells through NCAM1
In order to explore whether miR-152-3p exerts its effect on the inhibition of the proliferation of A549 cells through NCAM1, cell proliferation was examined. In A549 cells, overexpression of NCAM1 attenuated the inhibitory effect of miR-152-3p and accelerated cell proliferation in A549 cells (Fig. 4A and B). Furthermore, overexpression of NCAM1 significantly accelerated cell invasion in A549 cells compared with OE-NC (Fig. 4C). After transfection with NCAM1 overexpression plasmid, NCAM1 expression was increased and cleaved-caspase-3 and cleaved-PARP expression was decreased (Fig. 4D) compared with OE-NC. In conclusion, overexpression of NCAM1 was demonstrated to attenuate the effect of miR-152-3p in A549 cells, suggesting that miR-152-3p inhibited cell growth by downregulating NCAM1.
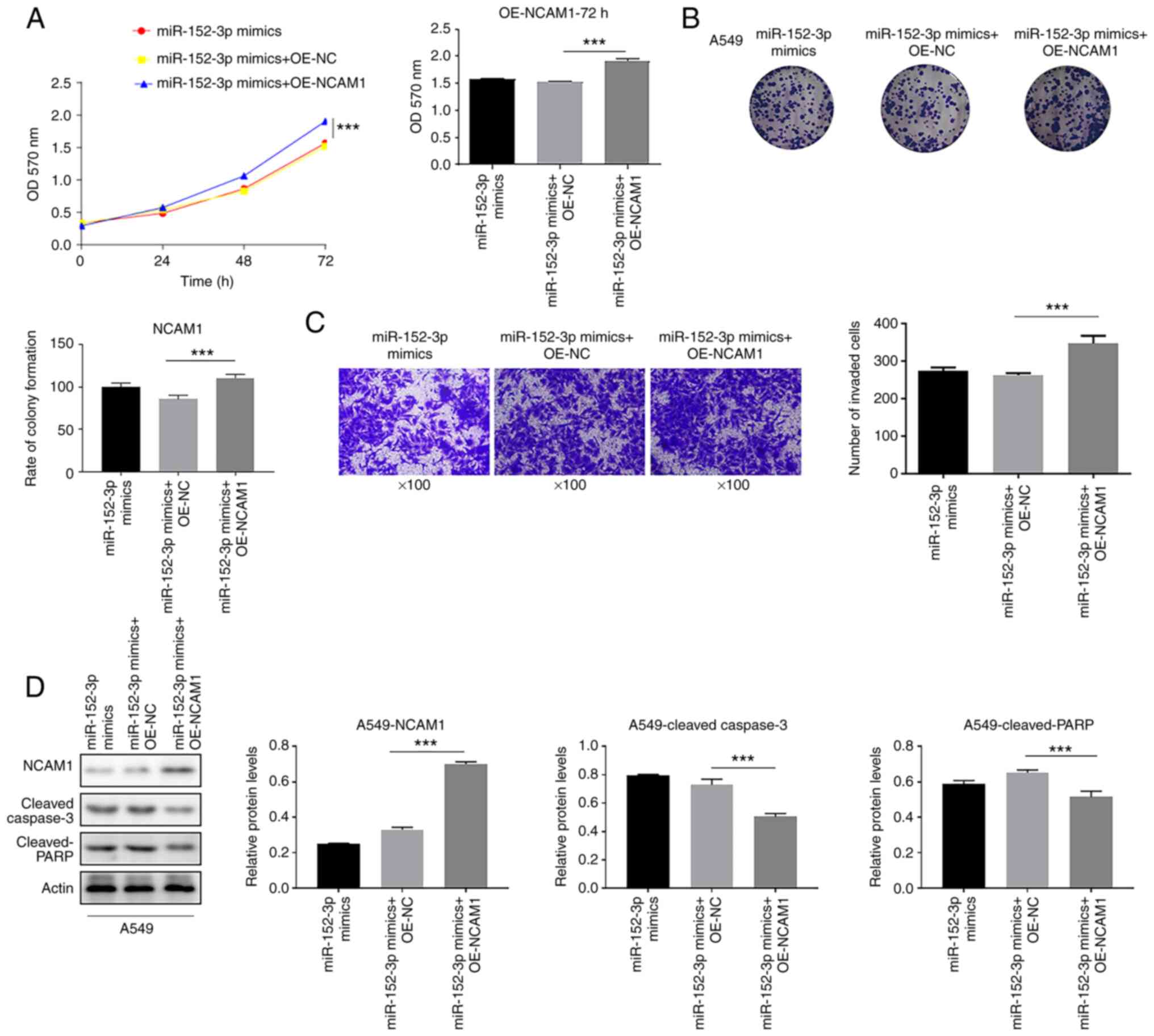 |
Figure 4.
miR-152-3p inhibits the proliferation of A549 cells through NCAM1. (A) The ability of miR-152-3p to inhibit cell proliferation in A549 cell lines was attenuated by NCAM1 overexpression, as indicated by the MTT assay. (B) Colony formation assay was performed to detect the cell proliferative ability of the indicated groups. (C) Number of invaded cells was detected via Transwell invasion assay in the indicated groups. (D) Expression levels of NCAM1, cleaved-caspase-3 and cleaved-PARP proteins in A549 cells detected via western blotting. One-way ANOVA was used for statistical analysis. ***P<0.001. miR, microRNA; NC, negative control; NCAM1, neural cell adhesion molecule 1; OD, optical density; PARP, poly (ADP-ribose) polymerase; OE, overexpression.
|
Knockdown of DNMT3B increases the level of miR-152-3p and delays cell proliferation
It was subsequently examined whether DNMT3B could affect the function on miR-152-3p, and it was revealed that in A549 cells, transfection with sh-DNMT3B reduced cell proliferation and colony formation, while miR-152-3p inhibitor attenuated this effect (Fig. 5A and B).
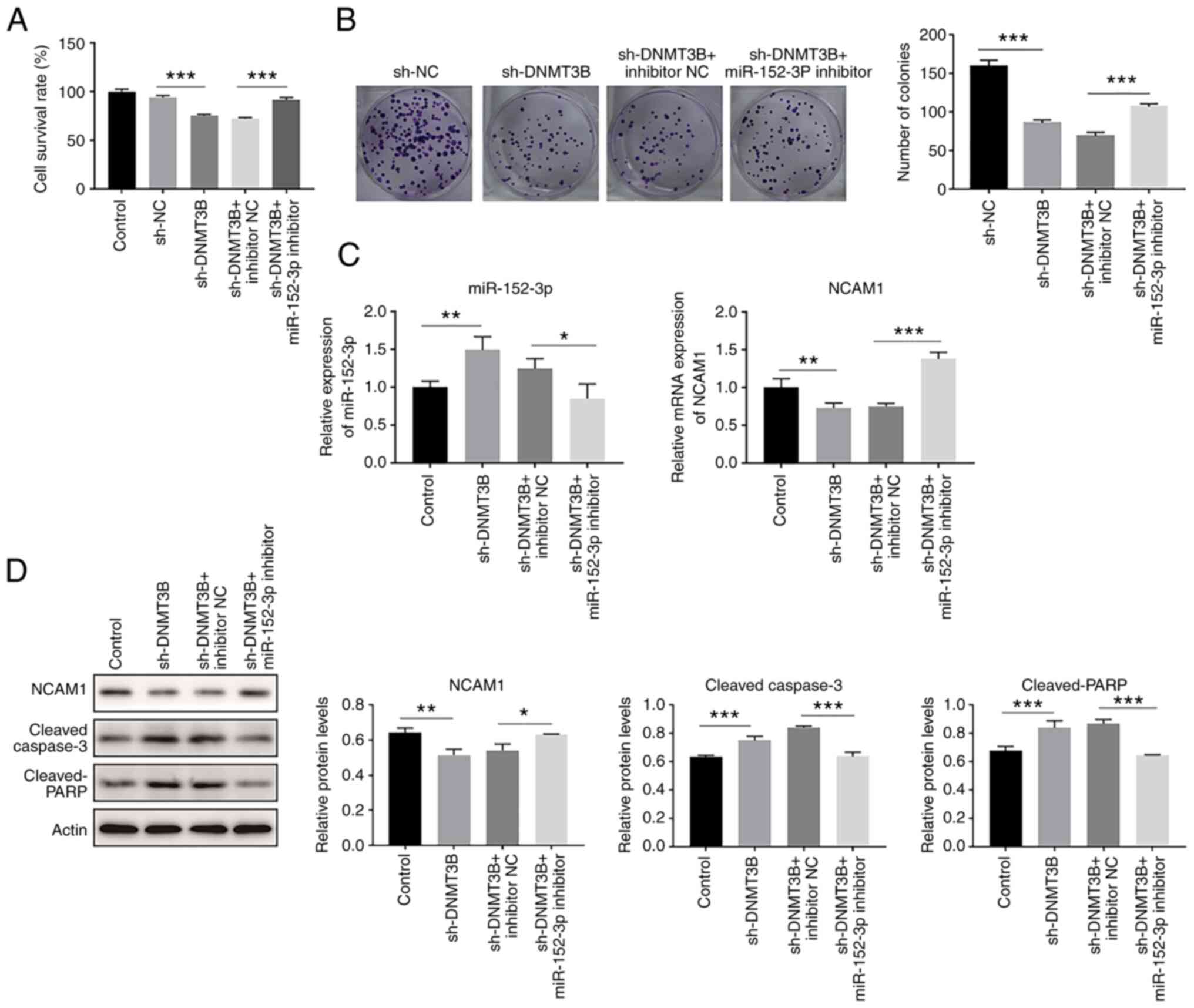 |
Figure 5.
Knockdown of DNMT3B increases miR-152-3p expression and delays cell proliferation. (A) Cell survival rate of A549 cell lines transfected with sh-DNMT3B and or miR-152-3p inhibitor, as indicated by the MTT assay. (B) Colony formation assay was used to detect the proliferative ability of the indicated groups. (C) Reverse transcription-quantitative PCR demonstrated the different expression levels of miR-152-3p and NCAM1 in the indicated groups. (D) Expression levels of NCAM1, cleaved-caspase-3 and cleaved-PARP proteins in A549 cells detected via western blotting. One-way ANOVA was used for statistical analysis. *P<0.05, **P<0.01, ***P<0.001. DNMT, DNA methyltransferase; miR, microRNA; NC, negative control; NCAM1, neural cell adhesion molecule 1; sh, short hairpin.
|
Upon transfection with sh-DNMT3B, miR-152-3p was increased and the mRNA expression of NCAM1 was decreased, and this effect could be reversed by miR-152-3p inhibitor treatment (Fig. 5C). Furthermore, NCAM1 protein expression was decreased and cleaved-caspase-3 and cleaved-PARP expression were increased after sh-DNMT3B transfection, while these effects were reversed when the cells were co-treated with miR-152-3p inhibitor (Fig. 5D). In conclusion, knockdown of DNMT3B could increase miR-152-3p, and thus inhibit cell proliferation.
Discussion
At present, miRNAs are considered to be a novel type of markers and class of therapeutic molecules. NSCLC cancer mortality has been ranked in the top two cancer mortalities worldwide, without any reliable biomarker for early diagnosis (19). Existing literature has reported that miR-152-3p expression was altered in a variety of cancers. For example, Chen et al (20) used tumor tissue sections along with cell lines to propose that gastrointestinal miR-152-3p expression was associated with tumor size as well as tumor stage. Further investigation revealed that miR-152-3p inhibited the growth and invasion of NSCLC via downregulation of the expression of the target genes fibroblast growth factor 2 and disintegrin and metalloproteinase domain-containing protein 17 (21,22). Although several studies on miR-152-3p in NSCLC tumors have been performed (21,22), there are few studies on the DNMT3B epigenetic regulation and miR-152-3p. The present study demonstrated that knockdown of DNMT3B could increase miR-152-3p and, thereby inhibiting cell proliferation. Since DNMT3B can bind to and methylate the core region of miR-152-3p, bioinformatics analysis was performed to investigate the in-depth mechanism of miR-152-3p in the progression of NSCLC, and NCAM1 was identified as a new target gene of miR-152-3p in lung cancer cells (17). Via RT-qPCR and western blotting experiments it was revealed that miR-152-3p could suppress NCAM1. The luciferase assay also indicated that NCAM1 is likely to be the target gene of miRNA-152-3p.
NCAM1 is part of a class of transmembrane molecules that can be classified into the immunoglobulin superfamily (23). It has been demonstrated that NCAM1 has crucial implications in cell adhesion, neuronal axon growth, as well as regulation of neuronal plasticity (23). NCAM1 has been indicated to be highly expressed in a considerable number of tumor cells, for instance, neuroblastoma (24), rhabdomyosarcoma (25) and small-cell lung cancer (SCLC) (24) cells. At present, it has been indicated that the development of immunotherapy strategies based on NCAM1 has excellent clinical application prospects (26). Previous research results revealed that NCAM1 was also highly expressed in human SCLC cells, and RNA interference NCAM1 expression could reduce the proliferation and metastatic ability of human SCLC cells (27). A recent study in nude mice revealed that suppressing miR-324-3p inhibited the tumor cell growth in human lung cells, and delayed proliferation in subcutaneous tumors in nude mice, while overexpression of NCAM1 could reverse this effect (28). Therefore, as a transmembrane protein highly expressed in human lung tumor cells, NCAM1 is a potential cell surface antigen for therapies targeting lung cancer. However, the role of NCAM1 in NSCLC cells has rarely been reported. The study of inhibitors targeting NCAM1 anchoring key molecules that affect the proliferation and metastatic potential of NSCLC cells will theoretically provide more effective data for the treatment of NSCLC.
The present study demonstrated that the core region in miR-152-3p was hypermethylated. Therefore, it was hypothesized that miR-152-3p may affect NCAM1 gene expression, which may be regulated via the methylation level of its promoter. DNA methylation level is regulated by DNA methyltransferases (DNMT1 (Abcam; cat. no. ab19905), DNMT3A (Abcam; cat. no. ab226261) and DNMT3B (Abcam; cat. no. ab227883). ChIP experiment revealed that the core region of miR-152-3p could form a complex with DNMT3B but not DNMT1, indicating that DNMT3B protein acted directly on the miR-152-3p promoter. Nanaomycin A (DNMT3B inhibitor) treatment resulted in demethylation of the miR-152-3p promoter and upregulated/downregulated miR-152-3p/NCAM1 mRNA expression, respectively, while thioguanine (DNMT1 inhibitor) failed to exhibit these effects, which further confirmed the direct action of DNMT3B on the miR-152-3p promoter. In addition, knockdown of DNMT3B could increase miR-152-3p and decrease NCAM1, hampering tumor cell proliferation in human lung cancer cells. In a future study, the mechanism related to the proliferation potential of NCAM1 will be explored via co-immunoprecipitation/mass-spectrometry experiments.
The current study demonstrated that inhibition of DNMT3B could diminish the methylation level detected in the miR-152-3p core region, but also that miR-152-3p was able to decrease NCAM1 expression. miR-152-3p could inhibit proliferation of A549 cells. Consistently, overexpression of NCAM1 could attenuate the effect of miR-152-3p, confirming that miR-152-3p reduced cell proliferation by downregulating NCAM1. Functionally, knockdown of DNMT3B could increase miR-152-3p expression and inhibit cell proliferation. In summary, it was demonstrated that DNMT3B negatively regulated the expression of miR-152-3p by regulating the methylation level in the core region of miR-152-3p, thereby controlling cell growth in human NSCLC cells (Fig. 6). miR-152-3p may be used as a novel therapeutic target for NSCLC, potentially providing a novel application of clinical individualized treatment.
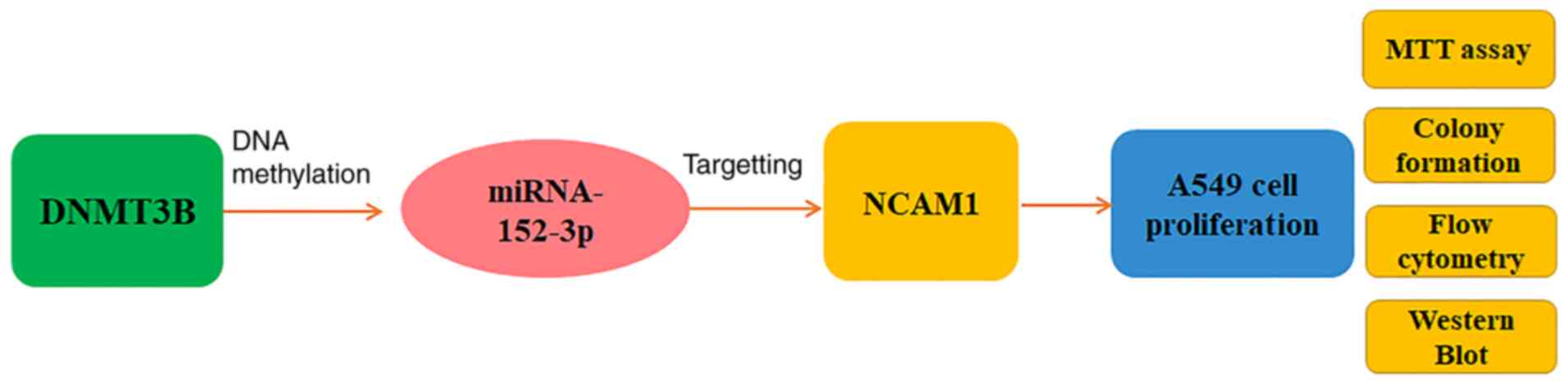 |
Figure 6.
Diagram of the DNMT3B/miR-152-3p/NCAM1 pathway. DNMT3B regulates the A549 cell proliferation through miR-152-3p/NCAM1 pathway tested by MTT assay, colony formation, apoptosis and western blotting.
|
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present work was supported by the Science and Technology Department of Guangxi Zhuang Autonomous Region (grant nos. 2019JJA140044 and 2019JJA140084) and Doctorate Awarding Unit Funding of the Affiliated Hospital of Youjiang Medical University for Nationalities [grant no. (2019) 48]. The funders had no role in the design of the study, the collection, analyses or interpretation of data, the writing of the manuscript or the decision to publish the results.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BY designed the experiment, monitored the progress of the experiment and analyzed the data. SQH acquired and interpreted the NSCLC patient data. HC and RL were involved in analyzing NSCLC patient data and revising the manuscript critically for important intellectual content. SHH and JZ performed the all the experiments in this study. YL made substantial contributions to the conception and design of the study, drafting the manuscript and giving final approval of the version to be published. BY, YL, SHH and JZ confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Mellema WW, Burgers SA and Smit EF: Tumor flare after start of RAF inhibition in KRAS mutated NSCLC: A case report. Lung Cancer. 87:201–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blanchon F, Grivaux M, Asselain B, Lebas FX, Orlando JP, Piquet J and Zureik M: 4-year mortality in patients with non-small-cell lung cancer: Development and validation of a prognostic index. Lancet Oncol. 7:829–836. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herbst RS, Morgensztern D and Boshoff C: The biology and management of non-small cell lung cancer. Nature. 553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
International Early Lung Cancer Action Program Investigators, . Henschke CI, Yankelevitz DF, Libby DM, Pasmantier MW, Smith JP and Miettinen OS: Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. 355:1763–1771. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Luczak MW and Jagodziński PP: The role of DNA methylation in cancer development. Folia Histochem Cytobiol. 44:143–154. 2006.PubMed/NCBI
|
|
6
|
Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A and MacLeod AR: DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 33:61–65. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Robertson KD: DNA methylation, methyltransferases, and cancer. Oncogene. 20:3139–3155. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT and Wang YC: Alteration of DNA methyltransferases contributes to 5CpG methylation and poor prognosis in lung cancer. Lung Cancer. 55:205–213. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J and Kim DH: Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer. 107:1042–1049. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kramann R and Moeller MJ: The next level of complexity: Post-transcriptional regulation by microRNAs. Kidney Int. 80:692–693. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 116:281–297. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The function of animal MicroRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang JY, Cheng H, Zhang HY, Ye YQ, Feng Q, Chen ZM, Zheng YL, Wu ZG, Wang B and Yao J: Suppressing microRNA-29c promotes biliary atresia-related fibrosis by targeting DNMT3A and DNMT3B. Cell Mol Biol Lett. 24:102019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, London CA, Kraybill W, Perrotti D, Croce CM, et al: MiR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci Signal. 6:ra632013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, Zhao R, Huang H, Wang X, Qiao Y, et al: MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: A novel epigenetic therapy independent of decitabine. Oncogene. 33:378–386. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Chen YY, Li SQ, Huang C and Qin YZ: Expression of miR-148/152 family as potential biomarkers in non-small-cell lung cancer. Med Sci Monit. 21:1155–1161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Huang S, Wei Z and Yang B: A putative competing endogenous RNA network in cisplatin-resistant lung adenocarcinoma cells identifying potentially rewarding research targets. Oncol Lett. 19:4040–4052. 2020.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Min L, Ren C, Xu X, Yang J, Sun X, Wang T, Wang F, Sun C and Zhang X: MiRNA-148a serves as a prognostic factor and suppresses migration and invasion through Wnt1 in non-small cell lung cancer. PLoS One. 12:e01717512017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C and Liu Z: Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastriontest Surg. 14:1170–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng Z, Ma R, Tan W and Zhang L: MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp Mol Med. 46:e1122014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Su Y, Wang Y, Zhou H, Lei L and Xu L: MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS Lett. 588:1983–1988. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rutishauser U, Acheson A, Hall AK, Mann DM and Sunshine J: The neural cell adhesion molecule (NCAM) as a regulator of cell-cell interactions. Science. 240:53–57. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Crossland DL, Denning WL, Ang S, Olivares S, Mi T, Switzer K, Singh H, Huls H, Gold KS, Glisson BS, et al: Antitumor activity of CD56-chimeric antigen receptor T cells in neuroblastoma and SCLC models. Oncogene. 37:3686–3697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Win KT, Lee MY, Tan TD, Tsai MP, Bahrami A, Raimondi SC and Chuang SS: Nasopharyngeal alveolar rhabdomyosarcoma expressing CD56: A mimicker of extranodal natural killer/T-cell lymphoma. Int J Clin Exp Pathol. 7:451–455. 2013.PubMed/NCBI
|
|
26
|
Amoureux MC, Coulibaly B, Chinot O, Loundou A, Metellus P, Rougon G and Figarella-Branger D: Polysialic acid neural cell adhesion molecule (PSA-NCAM) is an adverse prognosis factor in glioblastoma, and regulates olig2 expression in glioma cell lines. BMC Cancer. 10:912010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Aletsee-Ufrecht MC, Langley K, Rotsch M, Havemann K and Gratzl M: NCAM: A surface marker for human small cell lung cancer cells. FEBS Lett. 267:295–300. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song T, Zhou H, Wei X, Meng Y and Guo Q: Downregulation of microRNA-324-3p inhibits lung cancer by blocking NCAM1-MAPK axis through ALX4. Cancer Gene Ther. 28:455–470. 2021. View Article : Google Scholar : PubMed/NCBI
|




















