Introduction
Colorectal cancer (CRC) is the third-most common
cancer type in males and the second-most common cancer type in
females, and has the second-highest overall mortality rate
worldwide (1). Major risk factors
for CRC include a Western dietary pattern, sedentary lifestyle and
increasing obesity due to lifestyle factors related to economic
growth and industrialization. CRC has a relatively higher incidence
in economically developed countries, which may be associated with a
higher life expectancy, education and human developmental index due
to better diagnosis of CRC (2).
Furthermore, preventable lifestyle habits such as smoking, alcohol
consumption and obesity contribute to an increased risk of tumour
formation (3). Abdominal fat is
sub-divided into two compartments: Visceral adipose tissue (VAT)
and subcutaneous adipose tissue. VAT is responsible for the
secretion of pro-inflammatory adipokines (such as TNF); in
addition, it is infiltrated with macrophages, leading to chronic
inflammation and promoting tumour growth (4). Furthermore, insulin resistance
related to obesity has an important role in the promotion of
carcinogenesis. Chronic inflammation is also a condition
responsible for the elevated risk of CRC in patients with
ulcerative colitis and Crohn's disease (3). In general, cancer is a disease
associated with ageing; the risk of CRC markedly increases after
the age of years 50. However, a higher incidence in individuals
aged <40 years has also been observed (4).
Approximately 50% of patients in stage I–III develop
metastases mostly localized to the liver (5); furthermore, ~20% of patients already
have liver metastases at the time-point of diagnosis (6). Patients with stage I disease have a
5-year survival rate of 90%; however, a marked decline is observed
in patients with stage IV disease (6). Understanding molecular pathway
abnormalities is crucial for improving the diagnosis, prognosis and
treatment of CRC, as it is a heterogeneous multifactorial disease
with a range of different prognoses and responses to therapy
(7). During the last decade, the
development of novel targeted therapies and chemotherapeutics has
led to improved clinical outcomes for patients with advanced
metastatic disease. Despite the current screening methods and
prognostic factors, there are still numerous patients that are not
profiting from novel treatment strategies (4), which makes the early diagnosis of CRC
recurrence key for improving prognosis and reducing mortality.
Therefore, there is a significant requirement for a representative,
sensitive and easily detectable biomarker (8).
MicroRNAs (miRNAs/miRs) are a class of small,
non-coding, single-stranded RNAs consisting of approximately 18–25
nucleotides (9). They regulate the
translation of protein-coding genes at the post-transcriptional
level by binding to complementary sequences in the 3-untranslated
regions of their target mRNAs (10), leading to inhibition of translation
or degradation of the mRNA (11).
In addition to tumour tissue, miRNAs have been identified in
numerous biological sample types, including plasma, serum, saliva,
faeces, urine, cerebrospinal and amniotic fluid (12). Circulating miRNAs are actively
released into the bloodstream from tumour cells and may be detected
in the form of exosomes, free or bound to proteins (11). miRNAs may either function as tumour
suppressors through inhibition of oncogene expression or as
oncogenic miRNAs by inhibiting the expression of tumour suppressor
genes. Due to their localization within fragile sites in the
genome, the expression of miRNAs may be dysregulated by different
genetic alterations, including deletions, amplifications,
translocations and point mutations, and also DNA hyper-methylation
and hypo-methylation (10).
Altered miRNA expression has been identified in various types of
cancer, e.g., pancreatic cancer, hepatocellular cancer, breast
cancer, CRC and lung cancer (13).
miRNAs have important roles in cellular processes and are also
involved in mechanisms of cancer development, such as cell
proliferation, differentiation, apoptosis, angiogenesis and
epithelial-mesenchymal transition. Several miRNAs have been
suggested as candidates for CRC diagnosis; however, it is still
difficult to draw clear conclusions (12).
Carcinoembryonic antigen (CEA) is a widely used
marker for CRC recurrence detection recommended by the European
Group on Tumour Markers and the American Society of Clinical
Oncology (14,15). Furthermore, CEA may be detected
from peripheral venous blood, which makes CEA a suitable marker;
however, its sensitivity is not sufficient in certain cases
(16). The plasma level of CEA
should be established pre-operatively to monitor the dynamics of
its concentration after surgery, which is not always accomplished.
In addition, CEA may only be present at low concentrations in
patients with poorly differentiated tumours. miRNAs are easily
detectable and stable biomarkers; however, further research is
required to establish them as reliable biomarkers for local and
distant recurrence of CRC. Implementation of miRNAs in clinical
practice remains a challenge due to their diversity in affecting
numerous molecular and cellular processes (17).
Apart from cancer, miRNAs have important roles in
numerous molecular networks associated with inflammatory and
autoimmune disorders. There is evidence that a connection exists
between cancer and chronic inflammation (18), in which miR-155, miR-210 and miR-21
are involved. It has been indicated that upregulation of miR-155
results from oncogenic and inflammatory stimuli and triggers
malignant transformation. Furthermore, miR-155 overexpression is
frequently associated with high cytokine production (19). Studies have indicated that miR-21
is involved in both negative and positive feedback loops
controlling inflammation. This has been demonstrated in different
cell types under different conditions; therefore, this may be an
explanation for the opposing effects of miR-21 on inflammation
(20). Inflammation itself is
linked with wound healing, which, along with systemic inflammatory
response syndrome, is considered to be a normal post-operative
condition. The pro-inflammatory microenvironment is also present
during chemotherapy because of cellular senescence due to the
acquired pro-inflammatory senescence-associated secretory phenotype
of these cells. The same miRNAs have been indicated to be involved
in wound healing, as similar mechanisms and pathways are
responsible for inflammation. These miRNAs include the mentioned
miR-21 and miR-155 (19), as well
as miRNAs involved in angiogenesis, such as miR-16, miR-103,
miR-191 and miR-21, the expression of which has been investigated
in a study (20).
The aim of the present study was to evaluate a panel
of seven circulating miRNAs in pre-operative, post-operative and
3-month follow-up samples to investigate changes in the level of
selected miRNAs immediately after surgery and a longer time
interval after the operation.
Materials and methods
Patients
Patients with diagnosed CRC or colorectal adenoma
who underwent surgical intervention at the Clinic of Surgery and
Transplant Centre, Jessenius Faculty of Medicine in Martin,
Comenius University in Bratislava, (Martin, Slovakia) were enrolled
in the present study. Patients were informed about the study and
signed the informed consent form approved by the Ethical Committee
at Jessenius Faculty in Martin, Comenius University in Bratislava
(Martin, Slovakia) in accordance with the Declaration of Helsinki.
The cohort included 110 patients with CRC or colorectal adenoma
selected according to the following criteria: i) All patients were
diagnosed with a defined clinical stage; ii) disease was confirmed
by routine histopathological examination; and iii) none of the
patients had any other disease or known malignancies that may have
affected miRNA plasma levels.
Blood sample collection
Blood samples were collected between January 2018
and August 2020. For each patient, peripheral blood samples were
collected two or three times and processed as soon as possible, no
later than 1 h after collection. Pre-operative and post-operative
blood samples were taken one day prior to surgery and 3–7 days
after surgery, respectively. Another set of post-operative blood
samples was collected during the follow-up period (3 months after
surgical intervention). However, the follow-up group did not
include blood samples from all patients (n=20). Patients in the
follow-up group did not receive any chemotherapy at the time of
sampling.
Blood samples were collected into K3EDTA
tubes and transported 1 h after collection. The blood plasma was
immediately separated from 10 ml of whole blood by two-step
centrifugation at 850 × g for 10 min at 4°C and 18,620 × g for 10
min at 4°C to completely remove components including cell debris,
residual platelets and microvesicles. The plasma samples were
stored at −80°C until use.
Quality control of plasma samples,
total RNA extraction and reverse transcription (RT)
The thawed plasma samples were visually inspected
and the presence of free haemoglobin was measured by determining
the absorbance at 414 nm by a Nanodrop® 1000
(ThermoFisher Scientific, Inc.). Samples with an absorbance between
0.039 and 0.25 were included in the study. Only patients with
pre-operative/post-operative (n=60) and/or follow-up blood samples
(n=14) were included in the study. Total RNA including the miRNA
fraction was extracted from 200 µl of plasma using an miRNeasy
Serum/Plasma Advanced kit (Qiagen GmbH) following the
manufacturer's protocol. RNA spike-in controls (RNA Spike-in kit;
Qiagen GmbH) were added into the lysis buffer to provide control of
RNA extraction. Total RNA was resuspended in 20 µl of RNase-free
water and stored at −80°C until RT. A total of 10 µl of RT reaction
mixture contained, in addition to common ingredients (miRCURY LNA
RT kit; Qiagen GmbH), 1 µl of total RNA and 0.5 µl of RNA synthetic
spike-ins. RT was performed in a thermal cycler for 1 h at 42°C,
followed by inactivation for 5 min at 95°C. The obtained cDNA was
stored at −20°C until analysis, no longer than 5 weeks, and diluted
at a 1 : 30 ratio prior to analysis.
Measurement of selected miRNAs by
RT-quantitative (q)PCR
Based on a review of the literature (4–26), various
circulating miRNAs (miR-106a-5p, miR-210-5p, miR-155-5p, miR-21-5p,
miR-103a-3p, miR-191-5p, miR-16-5p) were selected and their levels
were analysed. Quantification of each miRNA was performed in
duplicate using an miRCURY LNA miRNA PCR System (Qiagen GmbH). All
pipetting steps were performed on a Bravo liquid handling station
(Agilent Technologies, Inc.) and run in duplicate/triplicate on a
LightCycler (LC)480 instrument (Roche Diagnostics GmbH). Rapid
quantification analysis was performed using LC480 instrument
software and quantification cycle (Cq) values were calculated by
the second derivative method (27). Secondary analysis and quality
control of RNA isolation and RT through synthetic spike-ins were
performed using the GeneGlobe data analysis tool (Qiagen GmbH). The
web tool (https://geneglobe.qiagen.com/sk/analyze) was also used
for rough data analysis, although no suitable reference genes were
found with the geNorm or Normfinder algorithms, as all tested
groups had different average arithmetic means.
Statistical analysis
The data were visualized and analysed using R
version 3.5.2 (28). Expression
and fold changes (FC) were computed using the standard formula
(29) for a paired design.
Furthermore, normalization of Cq values gene by gene (ΔCq) and
between groups is important to minimize the technical variability
and the FC is the expression ratio of the miRNA (−ΔΔCq) between two
conditions. The null hypothesis that the population median FC is
equal to and/or is equal for two levels of a categorical clinical
parameter (e.g., sex) was tested by the Wilcoxon rank-sum test. The
analogous null hypothesis for a categorical clinical parameter with
>2 levels (e.g., Cq) was tested by the Kruskal-Wallis test. The
association between two clinical parameters (e.g., sex and Cq) was
examined by the χ2 test. P<0.05 was considered to
indicate a statistically significant difference.
Analysis of involvement of
differentially expressed miRNAs in signalling pathways associated
with CRC relapse
The involvement of differentially expressed miRNAs
(miR-106a-5p, miR-21-5p and miR-16-5p) in signalling pathways
associated with CRC relapse was analysed through the online
software DIANA mirPath v.3 (http://snf-515788.vm.okeanos.grnet.gr/) from the Kyoto
Encyclopedia of Genes and Genomes (KEGG) based on analysis of
functional pathway enrichment, as well as numerous segments of Gene
Ontology (GO) analysis in the Homo sapiens species (30). The online analysis tool was used
for combining the available in silico-predicted targets
(DIANA-microT-CDS and/or TargetScan v6.2 algorithm) and
high-quality experimentally supported interactions (DIANA-TarBase
v7.0 algorithm), and P<0.05 was considered to indicate
statistical significance. In the KEGG analysis, gene intersections
with all three miRNAs included were preferred and in the GO
analysis, category intersections for merging the results with false
discovery rate correction were chosen.
Results
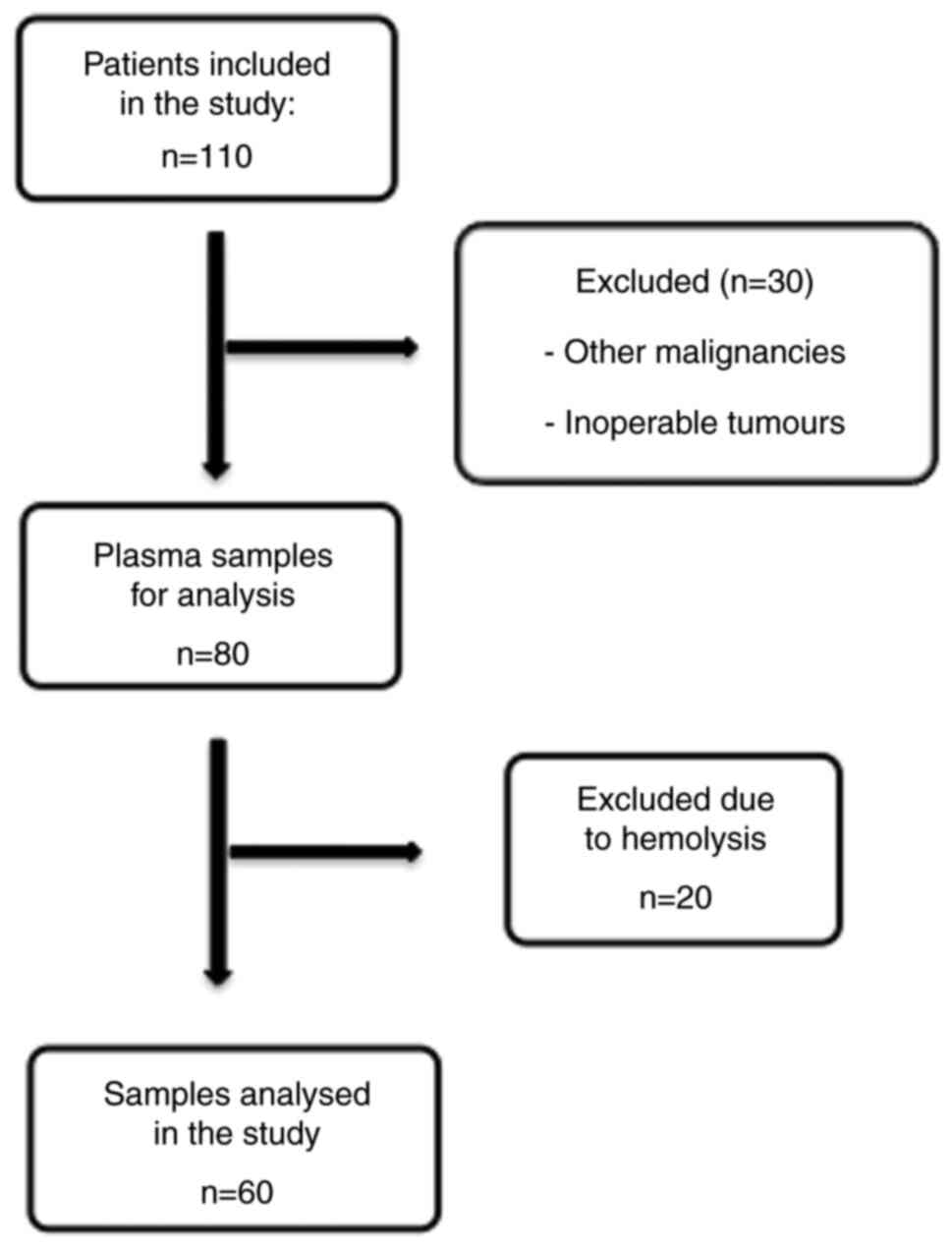
Patient characteristics
Overall, 110 patients of Caucasian ethnicity with
diagnosed CRC or adenoma were recruited for the present study. As
indicated in Fig. 1, 30 patients
were excluded due to other malignancies or inoperable tumours. The
plasma samples of the remaining 80 patients were analysed.
Extraction of RNA including the miRNA fraction was performed on 60
samples without haemolysis. If haemolysis was present in the first
or second sample taken at different time points, the patient was
excluded from analysis. It is known that the circulating miRNA
profile may be significantly influenced by haemolysis of red blood
cells, which does not mirror the physiological release from cells,
including cancer cells (31).
The patient characteristics are presented in
Table I. The patients' mean age at
the time-point of diagnosis was 69 years [interquartile range
(IQR), 63–73 years] for males (n=41) and 68 years (IQR, 58–72
years) for females (n=19). Carcinoma was present in almost 83% of
males and 78% of females. Most patients were at stage II or III and
in 54% of males and 74% of females, tumours were localized in the
proximal colon. Table II contains
additional information on neoadjuvant treatment and follow-up.
Disease recurrence was present in 40% of males and in 31% of
females. Regrettably, it was not possible to acquire follow-up
blood samples from 78.1% of males and 73.7% of females due to low
compliance of the patients. Furthermore, the situation was
complicated by the coronavirus pandemic. The information regarding
patient relapse is incomplete, as patients were also managed at
other hospitals and there is no national central register in
Slovakia.
 | Table I.Characteristics of patients included
in the study. |
Table I.
Characteristics of patients included
in the study.
| Item | Males (n=41) | Females (n=19) |
|---|
| Age, years | 69 (63; 73) | 68 (58; 72) |
| Tumor type |
|
|
|
Carcinoma | 34 (82.9) | 15 (78.0) |
|
Adenoma | 2 (4.9) | 2 (11.0) |
|
WNL | 5 (12.2) | 2 (11.0) |
| Tumor
localization |
|
|
| Rectum,
distal colon | 19 (46.0) | 5 (26.0) |
|
Proximal colon | 22 (54.0) | 14 (74.0) |
| Stage |
|
|
| I | 7 (17.1) | 2 (10.5) |
| II | 10 (24.4) | 3 (15.8) |
|
III | 10 (24.4) | 9 (47.4) |
| IV | 4 (9.7) | 2 (10.5) |
|
Adenomas/polyps | 10 (24.4) | 3(15.8) |
| pT |
|
|
| 0 | 1 (2.4) | 0 (0.0) |
| 1 | 6 (14.6) | 0 (0.0) |
| 2 | 6 (14.6) | 2 (10.5) |
| 3 | 15 (36.6) | 8 (42.1) |
| 4 | 7 (17.2) | 4 (21.1) |
|
Adenomas/polyps | 6 (14.6) | 5 (26.3) |
| pN |
|
|
| 0 | 22 (53.7) | 7 (36.8) |
| 1 | 8 (19.5) | 3 (15.8) |
| 2 | 9 (21.9) | 4 (21.1) |
|
Unknown | 2 (4.9) | 5 (26.3) |
| pM |
|
|
| 0 | 2 (4.9) | 2 (10.5) |
| 1 | 4 (9.8) | 3 (15.8) |
| x | 35 (85.3) | 11 (57.9) |
|
Adenomas/polyps | 0 (0.0) | 3 (15.8) |
| Grade |
|
|
| 1 | 14 (34.1) | 2 (10.5) |
| 2 | 12 (29.3) | 7 (36.8) |
| 3 | 4 (9.8) | 3 (15.8) |
|
Adenomas/polyps | 11 (26.8) | 7 (36.8) |
| Microsatellite
stability |
|
|
|
MSS | 32 (78.1) | 8 (42.1) |
|
MSI | 1 (2.4) | 5 (26.3) |
|
Adenomas/polyps | 8 (19.5) | 6 (31.6) |
| Relapse |
|
|
|
Absent | 15 (36.6) | 9 (47.4) |
|
Present | 10 (24.4) | 4 (21.1) |
|
Unknown | 16 (39.0) | 6 (31.5) |
 | Table II.Distribution of male and female
patients according to neoadjuvant treatment and 3 months of
follow-up. |
Table II.
Distribution of male and female
patients according to neoadjuvant treatment and 3 months of
follow-up.
| Item | Males, n (%) | Females, n (%) |
|---|
| Neoadjuvant
therapy |
|
|
|
Yes | 5 (12.2) | 1 (5.3) |
| No | 36 (87.8) | 18 (94.7) |
| Follow-up after 3
m |
|
|
|
Carcinoma | 9 (21.9) | 4 (21.0) |
|
Adenoma | 0 (0.0) | 1 (5.3) |
| No 3 m
follow-up | 32 (78.1) | 14 (73.7) |
| Relapse |
|
|
|
Absent | 15 (36.6) | 9 (47.4) |
|
Present | 10 (24.4) | 4 (21.1) |
| No
information available | 16 (39.0) | 6 (31.5) |
Evaluation of Cq values determined by
miRNA expression analysis of plasma
Most of the selected miRNAs (miR-106a, −21, −103a,
−191 and −16) were detected in plasma samples with calculated Cq
values between 22 and 35 using the second derivative method. A
small portion of analysed miRNAs (miR-210-5p and miR-155-5p) had
Cq>35, which means weak plasma expression and these miRNAs were
not included in the general conclusions of the study. Secondary
analysis using the GeneGlobe data analysis tool revealed that RNA
isolation and RT were performed correctly. During the data
normalization process, differences in average arithmetic mean
between tested groups from 0.5 to 1 Cq were indicated. No
combinations of selected miRNAs were appropriate for data
normalization without calculation of the stability factor due to
missing Cq values for certain samples. Normalization to the
synthetic spike-in Caenorhabditis elegans miR-39
(cel-miR-39) was also not appropriate, as it is not affected by the
biological condition of the patient and input concentration of
miRNA. Therefore, in FC calculations, the median of all measured
miRNAs except spike-ins was used, as recommended in the literature
(32). In the statistical analysis
of FC and the P-value calculation, post-operative and follow-up
samples were compared to pre-operative samples.
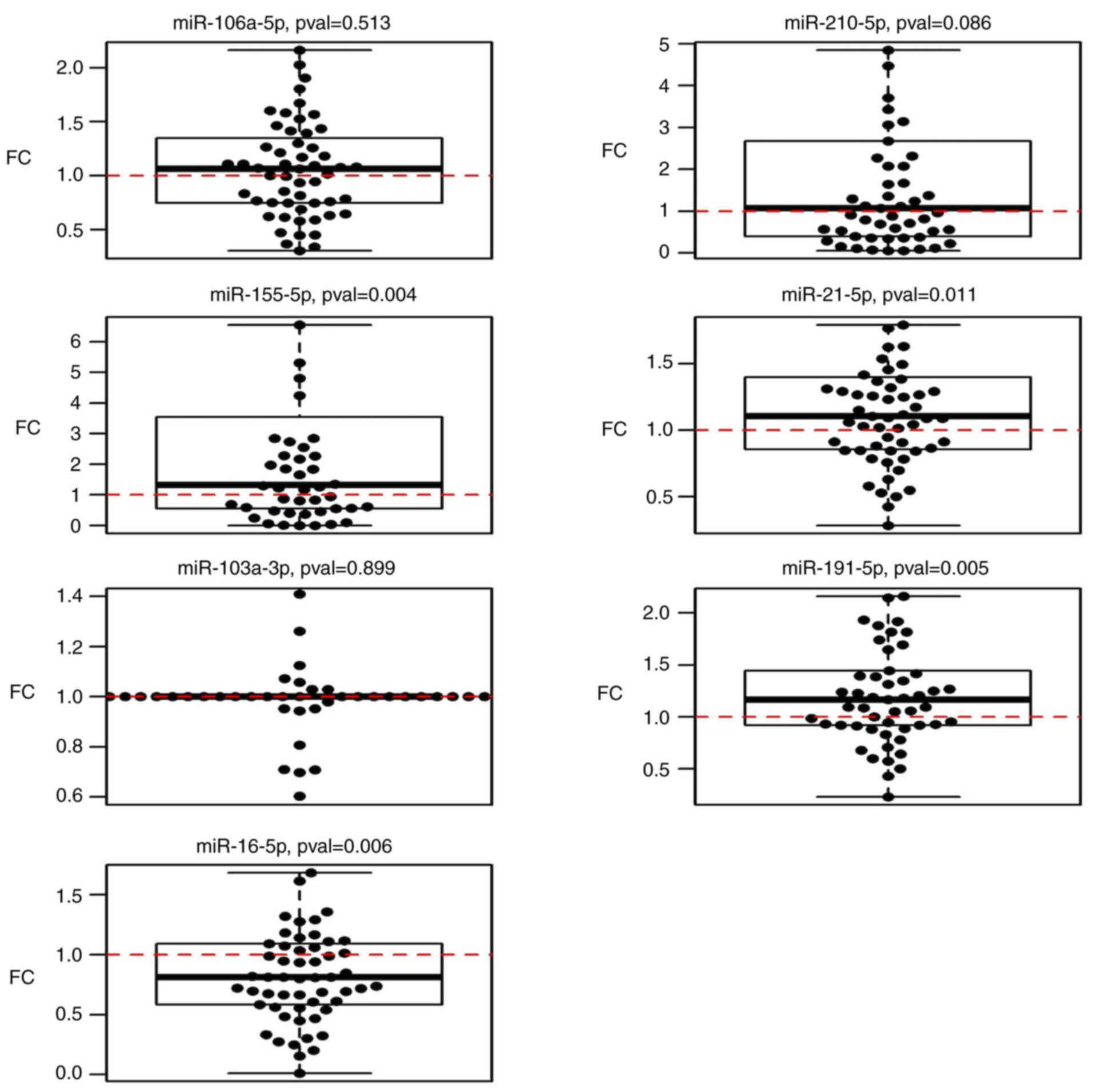
Circulating miRNA expression in post-
vs. pre-operative samples from patients with CRC
The results of the miRNA analysis of samples
acquired pre- and post-operatively indicated significant
upregulation of miR-21-5p (P=0.010), miR-155-5p (P=0.004) and
miR-191-5p (P=0.005), and downregulation of miR-16-5p (P=0.006)
(Table III; Fig. 2).
 | Table III.Fold-changes in the levels of
selected miRNAs in the post-operative and follow-up samples
collected 3 months after surgery compared with pre-operative
stages. |
Table III.
Fold-changes in the levels of
selected miRNAs in the post-operative and follow-up samples
collected 3 months after surgery compared with pre-operative
stages.
| miRNA | Post-operative
samples (n=60) | P-value | Follow-up samples
(n=14) | P-value |
|---|
| miR-106a-5p | 1.04 (0.91,
1.18) | 0.513 | 3.48 (1.54,
5.88) | 0.005 |
| miR-210-5p | 1.47 (0.94,
2.29) | 0.086 | 3.15 (0.8,
12.03) | 0.130 |
| miR-155-5p | 1.9 (1.26,
3.67) | 0.004 | 3.46 (0.88,
8.24) | 0.110 |
| miR-21-5p | 1.15 (1.04,
1.3) | 0.010 | 0.65 (0.46,
1.43) | 0.140 |
| miR-103a-3p | 1.01 (0.83,
1.26) | 0.900 | 0.69 (0.54,
1.04) | 0.052 |
| miR-191-5p | 1.2 (1.06,
1.37) | 0.005 | 1.48 (1.01,
2.17) | 0.048 |
| miR-16-5p | 0.83 (0.71,
0.94) | 0.006 | 1.73 (1.16,
2.65) | 0.003 |
Comparison of miRNA expression with
histopathological parameters indicated significant upregulation of
miR-210-5p in the circulation of patients who underwent neoadjuvant
therapy (P=0.012). When excluding patients with neoadjuvant therapy
from the data, miR-106a-5p expression was significantly increased
with rising severity of the stage (P=0.033). No association with
tumour type, stage, size, nodal status, metastases, grade or
relapse was obtained. No significant associations were observed
between miR-21-5p, miR-103a-3p, miR-155-5p, miR-191-5p and
miR-16-5p expression and histopathological parameters. All P-values
are listed in Table SI.
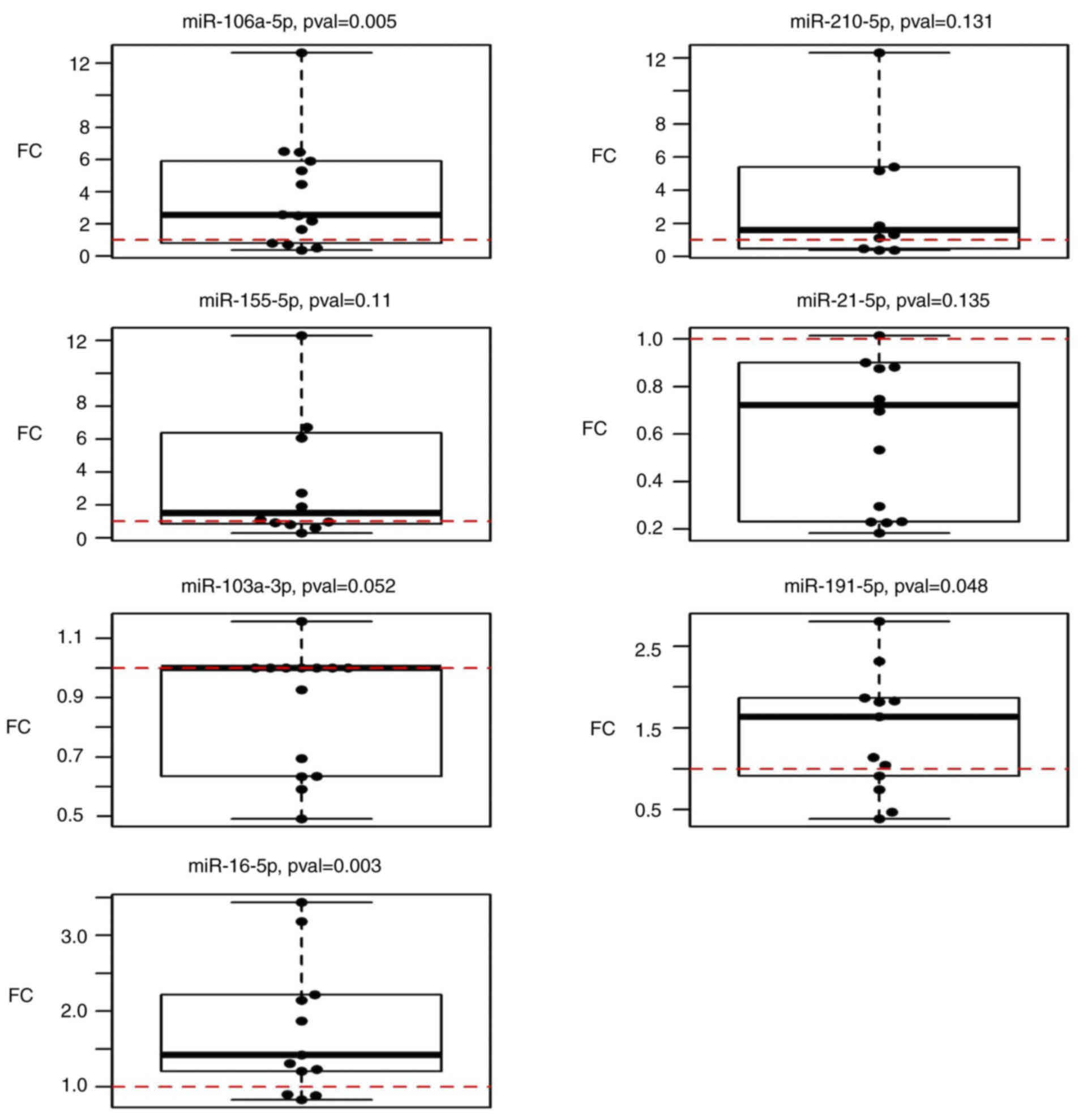
Circulating miRNA expression in plasma
of patients with CRC in follow-up samples compared to pre-operative
levels
The expression of selected miRNAs in follow-up
samples (n=14) was compared to that in pre-operative plasma samples
(n=60). The results suggested that miR-106a-5p, miR-191-5p and
miR-16-5p were significantly upregulated (P=0.005, 0.048 and 0.003,
respectively) in follow-up samples taken 3 months after surgery
when compared to the condition before surgery (Fig. 3). However, miR-210-5p (P=0.1309),
miR-155-5p (P=0.1099), miR-21-5p (P=0.1353) and miR-103a-3p
(P=0.052) did not exhibit any statistically significant changes
(Table III).
Statistical analyses were performed to assess any
possible associations between miRNA expression and pathological
parameters. However, the expression of these seven miRNAs was not
statistically significantly associated with the TNM stage, stage,
grade or tumour type of the patients (data not shown).
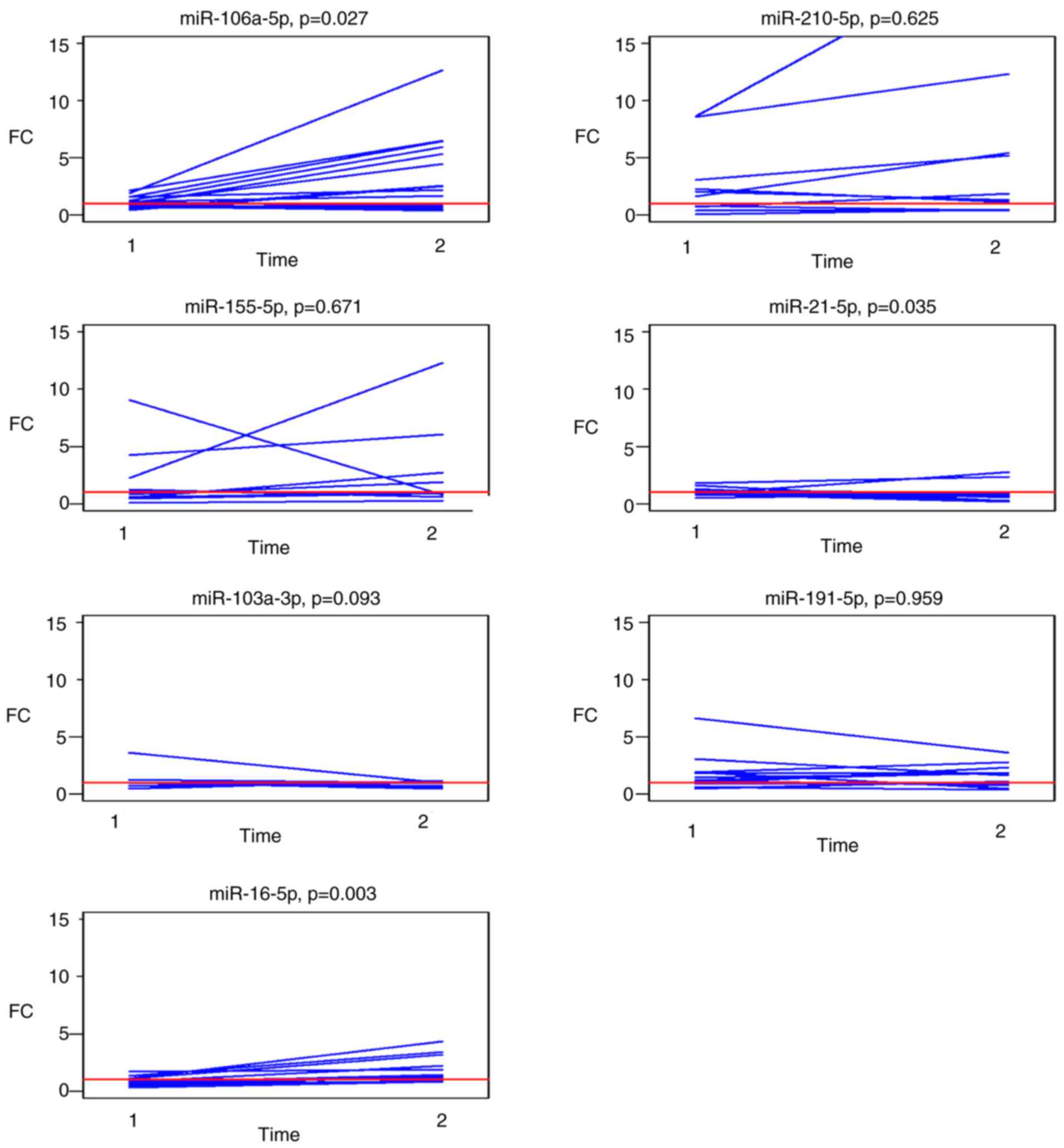
Circulating miRNA expression in paired
post-operative and follow-up samples
Changes in the expression levels of the selected
panel of miRNAs in paired post-operative and follow-up samples
compared to the pre-operative levels are graphically presented in
Fig. 4. Analysis of outliers for
patients with known histopathological data for each of the selected
miRNAs identified follow-up samples from two patients with
metastasis in the liver at the time of follow-up sampling with a
different profile of miR-210 and miR-21 expression
A summary of the median FCs in paired post-operative
and follow-up samples for each investigated miRNA is presented in
Table IV. Circulating miR-106a-5p
and miR-16-5p were significantly upregulated (P=0.027 and 0.003,
respectively) in the follow-up samples and miR-21-5p was
significantly downregulated (P=0.035).
 | Table IV.Differences in the post- and
follow-up vs. pre-operative levels of miRNAs from the selected
panel (fold changes). |
Table IV.
Differences in the post- and
follow-up vs. pre-operative levels of miRNAs from the selected
panel (fold changes).
| miRNA | Post-operative
samples (n=14) | Follow-up samples
(n=14) | P-value |
|---|
| miR-106a-5p | 1.09 (0.78,
1.50) | 2.54 (1.01,
5.77) | 0.027 |
| miR-210-5p | 1.5 (0.7, 2.3) | 1.6 (0.6, 5.4) | 0.600 |
| Unknown | 0 | 4 |
|
| miR-155-5p | 1.2 (0.6, 3.1) | 1.5 (0.9, 6.2) | 0.700 |
| Unknown | 2 | 2 |
|
| miR-21-5p | 1.02 (0.85,
1.22) | 0.72 (0.25,
0.90) | 0.035 |
| miR-103a-3p | 1.00 (1.00,
1.00) | 1.00 (0.65,
1.00) | 0.093 |
| miR-191-5p | 1.21 (0.93,
1.92) | 1.64 (0.91,
1.87) | >0.900 |
| Unknown | 1 | 1 |
|
| miR-16-5p | 0.81 (0.63,
1.10) | 1.42 (1.21,
2.22) | 0.003 |
| Unknown | 2 | 1 |
|
Signal and functional pathway
analysis
The target genes and function of the most
differentially expressed miRNAs associated with tumour removal,
wound healing and possible recurrence were analysed by KEGG pathway
analysis using the DIANA-mirPath v3.0 online web analysis tool.
miR-106a-5p was experimentally proven to have 1,160 target genes,
miR-16-5p has 2,886 target genes and miR-21-5p has 1,372 target
genes listed in DIANA-TarBase algorithm (accessed, 30 November
2020). In a gene intersection setting for merging the results of
the KEGG analysis (Table V), it
was indicated that all three miRNAs are significantly enriched in
pathways involving proteoglycans in cancer (hsa05205; 9 target
genes), the Hippo signalling pathway (hsa04390; 5 target genes),
focal adhesion (hsa04510; 11 target genes), signalling pathways
regulating pluripotency of stem cells (hsa04550; 7 target genes),
the prolactin signalling pathway (hsa04917; 5 target genes),
endocrine and other factor-regulated calcium reabsorption
(hsa04961; 2 target genes), pathways in cancer (hsa05200: 8 target
genes), CRC (hsa05210; 4 target genes), the FoxO signalling pathway
(hsa04068; 8 target genes), endometrial cancer (hsa05213; 3 target
genes), thyroid cancer (hsa05216; 3 target genes), lysine
degradation (hsa00310; 2 target genes) and the p53 signalling
pathway (hsa04115; 5 target genes). Three putative target genes
(CCND1, CTNNB1 and MAPK1) were indicated to be associated with 6 of
the 11 signalling pathways (proteoglycans in cancer, focal adhesion
and pathways in cancer, as well as colorectal, endometrial and
thyroid cancer).
 | Table V.KEGG analysis of the significantly
deregulated microRNAs in patients' plasma three months after
surgery with related target genes. |
Table V.
KEGG analysis of the significantly
deregulated microRNAs in patients' plasma three months after
surgery with related target genes.
| KEGG pathway | Pathway ID | P-value | Targeted genes |
|---|
| Proteoglycans in
cancer | hsa05205 |
6.6041×10−7 | STAT3, PDCD4, FRS2,
IGF1R, CCND1, CTNNB1, TIMP3, VEGFA, MAPK1 |
| Hippo signaling
pathway | hsa04390 |
8.3834×10−5 | YAP1, CCND2, CCND1,
CTNNB1, LATS1 |
| Focal adhesion | hsa04510 | 0.0076 | ITGB8, PAK2, CCND2,
IGF1R, ARHGAP35, CCND1, CTNNB1, VEGFA, MAPK1, TLN1, COL4A1 |
| Signaling pathways
regulating pluripotency of stem cells | hsa04550 | 0.0096 | STAT3, REST, IGF1R,
ZFHX3, CTNNB1, SKIL, MAPK1 |
| Prolactin signaling
pathway | hsa04917 | 0.0096 | STAT3, CCND2,
SOCS6, CCND1, MAPK1 |
| Endocrine and other
factor-regulated calcium reabsorption | hsa04961 | 0.0096 | CLTC, RAB11A |
| Pathways in
cancer | hsa05200 | 0.0096 | STAT3, IGF1R,
APPL1, CCND1, CTNNB1, VEGFA, MAPK1, COL4A1 |
| Colorectal
cancer | hsa05210 | 0.0096 | APPL1, CCND1,
CTNNB1, MAPK1 |
| FoxO signaling
pathway | hsa04068 | 0.0167 | STAT3, CCND2,
IGF1R, CCND1, PRKAB2, SOD2, MAPK1, CCNG2 |
| Endometrial
cancer | hsa05213 | 0.0167 | CCND1, CTNNB1,
MAPK1 |
| Thyroid cancer | hsa05216 | 0.027 | CCND1, CTNNB1,
MAPK1 |
| Lysine
degradation | hsa00310 | 0.0424 | WHSC1, KMT2C |
| p53 signaling
pathway | hsa04115 | 0.0424 | CCND2, CCND1,
SESN1, TNFRSF10B, CCNG2 |
Investigation of the biological function of target
genes regulated by differentially expressed miRNAs in the
subcategories biological process, cellular component or molecular
function, was performed through GO analysis. GO annotation results
for category intersection enrichment of target genes influenced by
up- or downregulated miRNAs in patients' follow-up samples are
provided in Table VI. The most
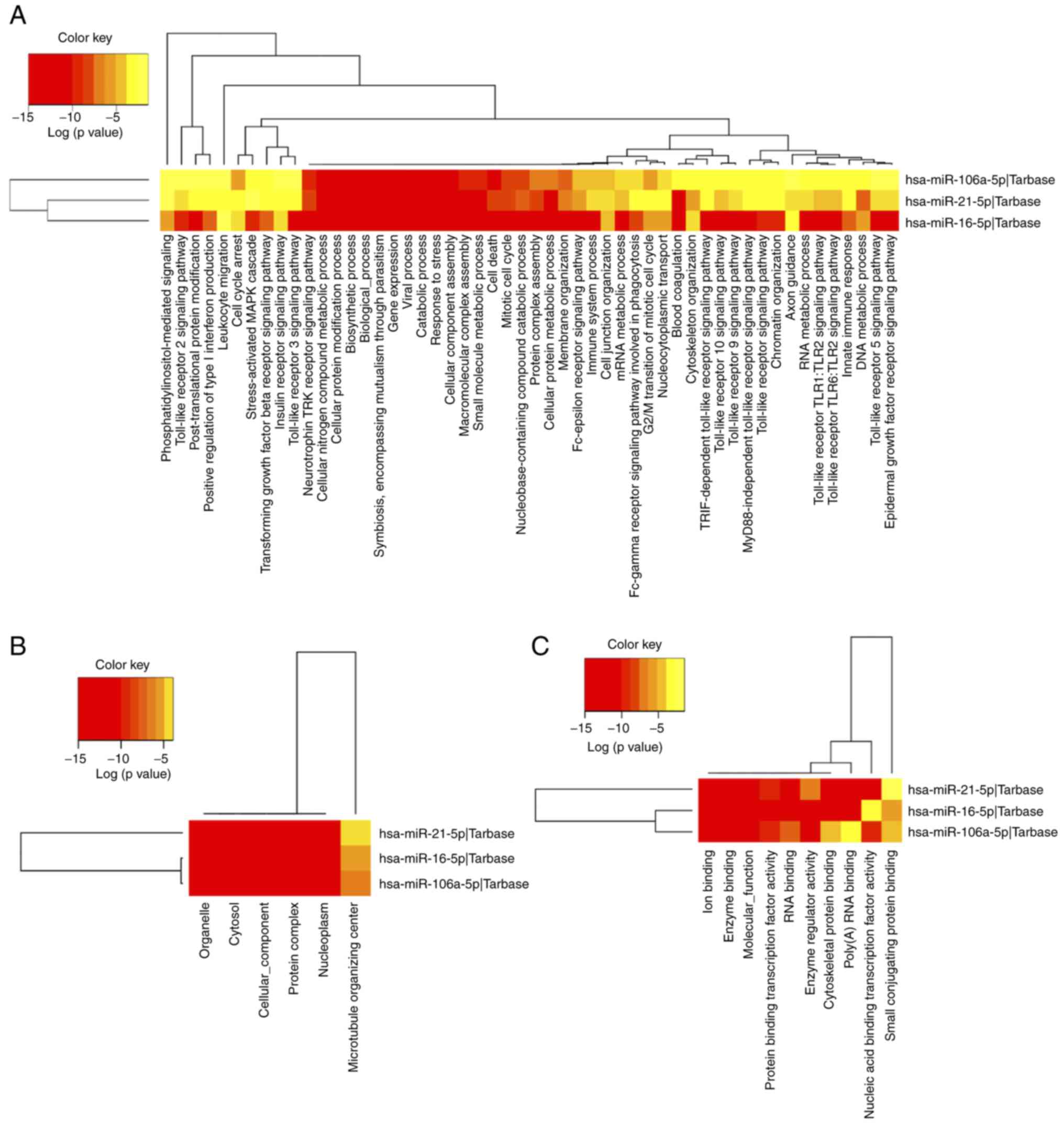
enriched GO annotations were ‘mitotic cell cycle’ (GO:0000278),
‘otein complex assembly’ (GO:0006461) and ‘cellular protein
modification process’ (GO:0006464). The top 10 GO terms in the
biological process category were presented and a full list of GO
terms is provided in Table SII. A
visualization of GO terms in each category is displayed in Fig. 5.
 | Table VI.GO annotation results of the target
genes of deregulated microRNAs in subcategories biological process
(top 10 GO terms were included), cellular component and molecular
function. |
Table VI.
GO annotation results of the target
genes of deregulated microRNAs in subcategories biological process
(top 10 GO terms were included), cellular component and molecular
function.
| A, Biological
process |
|---|
|
|---|
| GO ID | GO term name | P-value | Target gene
count |
|---|
| GO:0000278 | Mitotic cell
cycle |
<1×10−325 | 146 |
| GO:0006461 | Protein complex
assembly |
<1×10−325 | 239 |
| GO:0006464 | Cellular protein
modification process |
<1×10−325 | 744 |
| GO:0006950 | Response to
stress |
<1×10−325 | 625 |
| GO:0007596 | Blood
coagulation |
<1×10−325 | 150 |
| GO:0008150 |
Biological_process |
<1×10−325 | 3807 |
| GO:0008219 | Cell death |
<1×10−325 | 305 |
| GO:0009056 | Catabolic
process |
<1×10−325 | 605 |
| GO:0009058 | Biosynthetic
process |
<1×10−325 | 1079 |
| GO:0010467 | Gene
expression |
<1×10−325 | 286 |
|
| B, Cellular
component |
|
| GO ID | GO term
name | P-value | Target gene
count |
|
| GO:0005575 |
Cellular_component |
<1×10−325 | 3927 |
| GO:0005654 | Nucleoplasm |
<1×10−325 | 422 |
| GO:0005829 | Cytosol |
<1×10−325 | 880 |
| GO:0043226 | Organelle |
<1×10−325 | 2831 |
| GO:0043234 | Protein
complex |
<1×10−325 | 1092 |
| GO:0005815 | Microtubule
organizing center |
3.363976×10−14 | 140 |
|
| C, Molecular
function |
|
| GO ID | GO term
name | P-value | Target gene
count |
|
| GO:0000988 | Protein binding
transcription factor activity |
<1×10−325 | 165 |
| GO:0001071 | Nucleic acid
binding transcription factor activity |
<1×10−325 | 240 |
| GO:0003674 |
Molecular_function |
<1×10−325 | 3916 |
| GO:0003723 | RNA binding |
<1×10−325 | 641 |
| GO:0008092 | Cytoskeletal
protein binding |
<1×10−325 | 244 |
| GO:0019899 | Enzyme binding |
<1×10−325 | 475 |
| GO:0030234 | Enzyme regulator
activity |
<1×10−325 | 242 |
| GO:0043167 | Ion binding |
<1×10−325 | 1563 |
| GO:0044822 | Poly(A) RNA
binding |
<1×10−325 | 532 |
| GO:0032182 | Small conjugating
protein binding |
1.622062×10−9 | 40 |
Discussion
Numerous studies have focused on miRNAs and their
prognostic and predictive roles in CRC. Various studies had
conflicting results regarding the up- and downregulation of
selected miRNAs in assessing the prognosis and therapy response. In
the present study, a group of seven miRNAs were selected from the
literature that were significantly up- or downregulated
post-operatively and in follow-up samples when compared to the
pre-operative state.
Within the selected panel of circulating miRNAs, the
plasma expression of miR-106a-5p was not significantly changed in
post-operative samples. However, miR-106a-5p expression was
significantly increased in follow-up samples (P=0.005). By
contrast, certain studies have demonstrated higher plasma (6,33)
and/or serum (34) miR-106a
expression in patients with CRC than in healthy individuals and
lower miR-106 levels in post-operative samples (35). Li et al (34) also indicated that high miR-106a
expression in patients with CRC treated with adjuvant chemotherapy
is associated with poor therapeutic outcome and shorter
disease-free survival (DFS). In addition, increased levels of
miR-106a have been detected in CRC tissue compared to normal
adjacent tissue, and downregulation of miR-106a in CRC tissue has
been reported as a negative prognostic marker in patients with CRC
(36). On the other hand, in one
study, miR-106a levels in plasma of patients with CRC exhibited no
significant difference in the early post-operative period and
increased 1 month after surgery (37). The results of the present study are
in line with these results, although the pre-operative expression
was not compared with a healthy cohort.
Studies have suggested that miR-106a is involved in
the MAPK signalling pathway, focal adhesion, the FoxO signalling
pathway, CRC and other malignancies (38). miR-106a has been demonstrated to be
upregulated in metastatic CRC tissue and to be associated with
advanced TNM stage and lymph node metastasis. Furthermore, miR-106a
directly targets DLC1 and thus promotes CRC cell migration and
invasion through deregulation of the Wnt/β-catenin signalling
pathway (39). Upregulation of
miR-106a-5p in plasma of follow-up samples may be associated with
cell migration and proliferation during intestinal healing that is
accompanied by similar signalling pathways as cancer.
As mentioned in the Results, the expression level of
miR-155 and miR-210 in the present study was difficult to evaluate
due to higher Cq values (>35 Cq) and a high standard deviation
from triplicate experiments. A slightly higher expression level was
detected in post-operative samples and no change in follow-up
samples. High expression of miR-155 and miR-210 has been reported
in CRC tissue compared to normal adjacent tissue (40–42). The serum
levels of miR-155 and −210 decreased significantly 3 months after
surgery and chemotherapy and were elevated again at 12–18 months
after treatment and prior to the diagnosis of distant metastasis
and recurrence (5,26).
Sabry et al (43) found that upregulation of miR-210 is
associated with large tumour size, positive lymph node metastasis
and local invasion. Qu et al (42) also suggested that upregulation of
miR-210 is induced by hypoxia, which was confirmed by another study
(44). Concerning the expression
of miR-210, the present results did not reveal any statistically
significant changes; however, Sabry et al (43) determined higher expression of
miR-210 in patients with CRC and adenomas. Furthermore, the present
study indicated higher miR-210 expression in patients who underwent
neoadjuvant treatment. Jung et al (45) reported that patients with breast
cancer treated with neoadjuvant chemotherapy had elevated
circulating miR-210-5p and suggested that miR-210 may be used to
predict the outcome and monitor therapeutic response.
miR-155 has an important role in the immune system
due to targeting ~140 genes encoding immunomodulatory proteins,
inflammation-related proteins and tumour suppressor proteins
(46). miR-155 has been indicated
to be upregulated in numerous malignancies as well as CRC (47) where chronic inflammation has an
important role (48). Furthermore,
miR-155 downregulates core MMR proteins and induces microsatellite
instability (MSI) in CRC. Therefore, upregulation of miR-155 may
have a role in tumorigenesis by combining MSI and inflammatory
stimuli (49). Ulivi et al
(50) analysed a panel of
circulating miRNAs in relation to the outcome in patients with
metastatic CRC treated with bevacizumab. They determined that
miR-155 upregulation one month after bevacizumab treatment was
associated with significantly shorter progression-free survival and
overall survival (OS), and thus, miR-155 may be utilized in drug
response monitoring (50).
In the present study, significant upregulation of
miR-21 in post-operative samples compared to the pre-operative
state and significant downregulation in follow-up samples compared
to paired post-operative samples was observed. Jin et al
(8) reported upregulation of
miR-21 in pre-operative samples, post-operative miR-21
downregulation and no difference in patients with recurrence.
miR-21 is an important molecule in epithelial-mesenchymal
transition (EMT) and its low expression is associated with
increased OS. Equivalently, higher serum miR-21 expression in
patients with CRC was reported to be associated with liver
metastasis and CRC recurrence (12), as well as poorer DFS and OS
(24). miR-21 expression in CRC
tumour tissue was indicated to be elevated compared with that in
normal adjacent tissue and miR-21 induces proliferation in CRC cell
lines (51). Upregulation of
miR-21 was also reported to be associated with a higher TNM stage
and BRAF mutation (52). miR-21
promotes carcinogenesis by inhibiting negative regulation of the
RAS/MEK/ERK pathway and also downregulates the expression of PTEN,
TPM1 and PDCD4, promoting tumour progression (53). The present study indicated a
slightly increased level of miR-21-5p expression immediately after
surgery but a significant reduction in paired samples taken 3
months after surgery. In a study with a similar in patients with
oesophageal squamous cell carcinoma, plasma expression of miR-21
was significantly reduced in post-operative samples taken 1 month
after oesophagectomy (54).
Furthermore, upregulation of miR-21 in post-operative samples may
reflect an increased inflammatory response and oxidative stress
with an increase in the oxidative stress response protein aldose
reductase (AR) and activation of PTEN-induced apoptosis and
controlled cell growth. Repression of AR led to inhibition of colon
cancer cell growth by downregulation of miR-21 expression and
upregulation of PTEN and FOXO3a expression (hsa04068). However, the
specific mechanism was not elucidated (55).
Hong et al (56) detected upregulation of miR-103 in
CRC cell lines. Zheng et al (57) determined upregulation of miR-103 in
CRC tissue compared to normal adjacent tissue. Furthermore, they
demonstrated that patients with high miR-103 expression had poorer
OS. Wang et al (58)
reported upregulation of miR-103a-3p in serum of patients with CRC
and in those with recurrence. There is currently a lack of studies
evaluating the role of miR-103a-3p in carcinogenesis due to its use
as a housekeeping gene as discussed below.
Another significantly upregulated miRNA in
post-operative samples of the present study was miR-191-5p. There
was also a weak significant association between the level of
miR-191 and tumour stage and microsatellite-stable cancers. The
function of miR-191 in CRC remains to be fully elucidated. However,
several studies have reported deregulation of miR-191 in various
cancer types and diseases. It has been indicated that miR-191
overexpression induces progression of hepatocellular carcinoma and
intrahepatic cholangiocarcinoma and promotes EMT in metastatic
bronchial epithelial cells (59).
Zhang et al (60) reported
that upregulation of miR-191 promoted proliferation in CRC and
reduced cell susceptibility to 5-fluorouracil. Qin et al
(61) demonstrated that high
expression of miR-191 is significantly associated with advanced TNM
stage, liver metastasis and unfavorable prognosis in patients with
CRC. On the other hand, Milanesi et al (62) and Chen et al (63) revealed that downregulation of
miR-191 in patients with KRAS-mutation CRC and low miR-191
expression is correlated with poor prognosis.
In the present study, the level of miR-16-5p
expression in patients' plasma after surgery was significantly
decreased (P=0.006) compared to the pre-operative state and
elevated in paired follow-up samples (P=0.003). Previous studies
have demonstrated a tumour-suppressive role of miR-16 in the
progression of several malignancies (64), including CRC (65). It has been indicated that
overexpression of miR-16 inhibited proliferation and induced
apoptosis by regulation of the p53/survivin signalling pathway
(66). Several studies have
revealed downregulation of miR-16 in malignant CRC tissue when
compared to adjacent tissue and identified an association between
low miR-16 expression and histological parameters such as advanced
TNM stage and poor histological grade, as well as a higher
incidence of lymph node metastases and tumour recurrence. Low
miR-16 expression is correlated with shorter DFS and OS (64,65,67).
Ostenfeld et al (68)
analysed miRNA profiles in epithelial-derived extracellular
vesicles (EVs) secreted by cancer cells and observed a
significantly reduced level of miR-16-5p post-operatively.
miR-16-5p, miR-103a-3p and miR-191-5p are commonly
used for qPCR data normalization, which is a critical step in each
gene expression experiment. Danese et al (25) aimed to identify the ideal reference
miRNAs in CRC from miRTarBase and came to the conclusion that
miR-16 and −103 target onco/tumour suppressor genes are not
suitable for normalization. The expression of miR-16-5p in blood
plasma is influenced by a varying level of haemolysis (29). Therefore, in the present study,
samples without haemolysis were carefully selected despite the
overall reduction in the number of samples in the test groups.
Proper data normalization is the most important part of each study
and may influence its overall results. In studies dealing with
circulating miRNA expression, the issue prevails that there are no
suitable housekeeping miRNAs expressed equally across the various
physiological conditions of patients. Other procedures advise
normalization to synthetic spike-ins added through the process of
sample preparation prior to analysis. However, the use of synthetic
spike-ins does not correspond to the physiological and biological
conditions under investigation and they do not have the same effect
as the housekeeping genes. Another obstacle was not having the same
input of RNA/miRNA concentration across the samples collected from
the patients at various time-points. Therefore, median
normalization was used, which is more frequently used in
next-generation sequencing (69,70)
and microarray (71) data
normalization.
Some oncomiRs were found to be downregulated 1 week
after surgical removal of the tumor (17,20).
It may be speculated that upregulation of oncomiRs in
post-operative samples may be associated with post-operative wound
healing. The present study demonstrated a decrease in these
oncomiRs later at a follow-up sampling. At present, intestinal
wound healing is only partially understood. It is predominantly
studied in inflammatory bowel disease as well as in Crohn's disease
and ulcerative colitis. During acute and chronic intestinal
inflammation, immune cells such as neutrophils and macrophages
induce local tissue damage by secreting tissue-degrading enzymes,
reactive oxygen radicals and pro-inflammatory cytokines. The
process promotes migration of myofibroblast cells to the site of
the defect, arranging contractility of the wound area and the
production of extracellular matrix (ECM) (72). ECM is composed of macromolecules
such as proteoglycans (pathway hsa05205), non-proteoglycan
polysaccharides, proteins such as collagen and elastin, and EVs.
ECM also has a major function in cell adhesion (hsa04510) (73) and intestinal healing is also
associated with intestinal stem cell differentiation and
proliferation (hsa04550) (74,75).
Notably, Hippo signalling (hsa04390) also has a role in regulating
the regeneration of organs such as the liver, heart, nervous system
and skin as well as intestine (76) and also prolactin (hsa04917) has
been suggested to have an important role in re-epithelialization
and promote wound healing (77).
According to a previous study, plasma and wound fluid levels of
proangiogenic proteins were elevated in patients after CRC
resection for 3 to 5 weeks, leading to the hypothesis that the
wound healing may stimulate tumour growth in residual tumour
deposits (78).
The tumour microenvironment (TME) is composed of
blood vessels, ECM, RNA, secreted proteins, small organelles and
various populations of stromal cells (fibroblasts, adipocytes,
pericytes and immune/inflammatory cells) (79). TME cells have a role in the
communication between them, which is performed in autocrine,
paracrine and/or endocrine ways. These cells produce growth factors
and cytokines which modify molecular and cellular processes and
thus alter the maturation of the TME (80).
In general, the TME participates in immune cell
recruitment and activation, ECM alterations and angiogenesis, which
contribute to tumour progression and wound healing (81). ECM proteins are an important
component of the TME and it is well-known that changes in the
composition of ECM proteins are responsible for the genesis of CRC
(82). Fibrous proteins,
proteoglycans, collagen and elastin as the major types of ECM
proteins provide structural support and elasticity in all tissues
(83). Various studies suggested
that expression of high levels of proinflammatory cytokines, such
as TNF produced by tumour-associated macrophages, induces neoplasm
growth and invasion (84).
Inflammation is involved in CRC development and progression and in
post-operative intestinal wound healing. Chronic inflammation is
linked to alterations in the TME through cytokines, chemokines and
growth factors (85). Li et
al (86) reported that elastin
recombinant protein increased the proliferation of CRC epithelial
cells, induced EMT, increased TNF secretion by bone marrow-derived
macrophages and reduced E-cadherin in CRC epithelial cells.
Inflammation is a trigger of ECM remodelling and furthermore, Li
et al (86) demonstrated
that proteins of ECM regulate inflammation.
In the present study, the panel of selected miRNAs
was investigated in plasma of patients with CRC at the
pre-operative stage, in the early post-operative period and 3
months after surgery. None of the patients received any
chemotherapy at the time of follow-up sampling. Significant
upregulation of miR-155-5p, miR-21-5p and miR-191-5p, and
downregulation of miR-16-5p was determined. In paired follow-up
samples, the most relevant upregulation was observed for
miR-106a-5p and miR-16-5p and downregulation for miR-21-5p. Pathway
analysis outlined their role in cancer development, but the same
pathways are also involved in controlled wound healing and
regeneration of intestinal epithelium (85,87).
Of note, the present study is limited by its sample size as well as
lack of samples in the follow-up group. Furthermore, functional
studies are necessary to confirm the present results in association
with the pathways identified by the in silico analysis. It
may be speculated that miRNA expression associated with intestinal
wound healing influences the expression levels in patients'
circulation (87) and the healing
process should be considered in studies focusing on disease
recurrence (45). For
post-operative sampling, a longer interval after surgery (4–6
weeks) (88) may help in the early
identification of patients with CRC recurrence in the case of
latent metastases. However, more studies are necessary to confirm
these results.
The selected miRNAs seemed to be promising markers
for the monitoring of miRNA expression in the patients'
circulation. However, the present results require confirmation in a
larger cohort of patients. miRNA studies should also consider all
of the physiological conditions that are taking place in real time
in the organism and are not influenced by sample processing and
technical variations. Identifying appropriate biomarkers to
determine a higher risk of CRC recurrence and biomarkers to predict
patient response to adjuvant treatment is a novel way to monitor
and treat patients. Based on these results, it may be possible to
select patients in need of more intensive monitoring as well as
patients who are unlikely to benefit from adjuvant therapy.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank to Professor Zora
Lasabova (Department of Molecular Biology and Genomics, Jessenius
Faculty of Medicine in Martin, Comenius University, Bratislava,
Slovakia) for providing sample analysis and financial support.
Funding
This study was supported by the Project of Slovak Research and
Development Agency (grant no. APVV-16-0066) as well as by a grant
from The Ministry of Education, Science, Research and Sport of the
Slovak Republic (grant no. VEGA-1/0380/18) and by the Operational
Programme Integrated Infrastructure for the project ‘Integrative
strategy in development of personalized medicine of selected
malignant tumours and its impact on quality of life’ (grant no.
IMTS: 313011V446), co-financed by the European Regional Development
Fund.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Conceptualization, ZL and LL; methodology, ErK, VH
and ZK; software, VH, ZK, MG; validation, VH and ZK; formal
analysis, VH and MG; investigation, EvK, VH, MSa, BV, PM and MSm;
resources, ErK, VH, MSa, BV, PM and MSm; data curation, EvK, VH,
ZK, MG and PM; writing-original draft preparation, EvK and VH;
writing - review and editing, MSa, ErK, LL and ZL; supervision, ZL
and LL; project administration, ZL; funding acquisition, ZL. VH and
ErK confirm the authenticity of all the raw data. All authors have
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
All patients were informed about this study and
provided written informed consent. This research was approved by
the Ethical Committee at Jessenius Faculty of Medicine in Martin,
Comenius University in Bratislava (Martin, Slovakia).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization: International
Agency For Research on Cancer: Cancer Today. Data visualization
tools for exploring the global cancer burden in 2020. https://gco.iarc.fr/today/home
|
|
2
|
Song M and Chan AT: Environmental factors,
gut microbiota, and colorectal cancer prevention. Clin
Gastroenterol Hepatol. 17:275–289. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dong Y, Zhou J, Zhu Y, Luo L, He T, Hu H,
Liu H, Zhang Y, Luo D, Xu S, et al: Abdominal obesity and
colorectal cancer risk: Systematic review and meta-analysis of
prospective studies. Biosci Rep. Dec 12–2017.(Epub ahead of print).
doi: 10.1042/BSR20170945. View Article : Google Scholar
|
|
4
|
Keum N and Giovannucci E: Global burden of
colorectal cancer: Emerging trends, risk factors and prevention
strategies. Nat Rev Gastroenterol Hepatol. 16:713–732. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Balacescu O, Sur D, Cainap C, Visan S,
Cruceriu D, Manzat-Saplacan R, Muresan MS, Balacescu L, Lisencu C
and Irimie A: The impact of miRNA in colorectal cancer progression
and its liver metastases. Int J Mol Sci. 19:37112018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Riihimäki M, Hemminki A, Sundquist J and
Hemminki K: Patterns of metastasis in colon and rectal cancer. Sci
Rep. 6:297652016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin XH, Lu S and Wang AF: Expression and
clinical significance of miR-4516 and miR-21-5p in serum of
patients with colorectal cancer. BMC Cancer. 20:2412020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Strubberg AM and Madison BB: MicroRNAs in
the etiology of colorectal cancer: Pathways and clinical
implications. Dis Model Mech. 10:197–214. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jung G, Hernández-Illán E, Moreira L,
Balaguer F and Goel A: Epigenetics of colorectal cancer: Biomarker
and therapeutic potential. Nat Rev Gastroenterol Hepatol.
17:111–130. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cojocneanu R, Braicu C, Raduly L, Jurj A,
Zanoaga O, Magdo L, Irimie A, Muresan MS, Ionescu C, Grigorescu M
and Berindan-Neagoe I: Plasma and tissue specific miRNA expression
pattern and functional analysis associated to colorectal cancer
patients. Cancers (Basel). 12:8432020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ghareib AF, Mohamed RH, Abd El-Fatah AR
and Saadawy SF: Assessment of serum MicroRNA-21 gene expression for
diagnosis and prognosis of colorectal cancer. J Gastrointest
Cancer. 51:818–823. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wan TM, Iyer DN and Ng L: Roles of
microRNAs as non-invasive biomarker and therapeutic target in
colorectal cancer. Histol Histopathol. 35:225–237. 2020.PubMed/NCBI
|
|
14
|
Locker GY, Hamilton S, Harris J, Jessup
JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF and Bast RC Jr:
ASCO: ASCO 2006 update of recommendations for the use of tumor
markers in gastrointestinal cancer. J Clin Oncol. 24:5313–5327.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duffy MJ, van Dalen A, Haglund C, Hansson
L, Holinski-Feder E, Klapdor R, Lamerz R, Peltomaki P, Sturgeon C
and Topolcan O: Tumour markers in colorectal cancer: European Group
on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer.
43:1348–1360. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tan E, Gouvas N, Nicholls RJ, Ziprin P,
Xynos E and Tekkis PP: Diagnostic precision of carcinoembryonic
antigen in the detection of recurrence of colorectal cancer. Surg
Oncol. 18:15–24. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pesta M, Kucera R, Topolcan O, et al:
Plasma microRNA levels combined with CEA and CA19-9 in the
follow-up of colorectal cancer patients. Cancers (Basel).
11:E8642019. View Article : Google Scholar
|
|
18
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Piccinini AM and Midwood KS: Endogenous
control of immunity against infection: Tenascin-C regulates
TLR4-mediated inflammation via microRNA-155. Cell Rep. 2:914–926.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tili E, Michaille JJ and Croce CM:
MicroRNAs play a central role in molecular dysfunctions linking
inflammation with cancer. Immunol Rev. 253:167–184. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ristau J, Staffa J, Schrotz-King P, Gigic
B, Makar KW, Hoffmeister M, Brenner H, Ulrich A, Schneider M,
Ulrich CM and Habermann N: Suitability of circulating miRNAs as
potential prognostic markers in colorectal cancer. Cancer Epidemiol
Biomark Prev. 23:2632–2637. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen B, Xia Z, Deng YN, Yang Y, Zhang P,
Zhu H, Xu N and Liang S: Emerging microRNA biomarkers for
colorectal cancer diagnosis and prognosis. Open Biol. 9:180212
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rapado-González Ó, Álvarez-Castro A,
López-López R, Iglesias-Canle J, Suárez-Cunqueiro MM and
Muinelo-Romay L: Circulating microRNAs as promising biomarkers in
colorectal cancer. Cancers (Basel). 11:8982019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsukamoto M, Iinuma H, Yagi T, Matsuda K
and Hashiguchi Y: Circulating exosomal MicroRNA-21 as a biomarker
in each tumor stage of colorectal cancer. Oncology. 92:360–370.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Danese E, Minicozzi AM, Benati M, Paviati
E, Lima-Oliveira G, Gusella M, Pasini F, Salvagno GL, Montagnana M
and Lippi G: Reference miRNAs for colorectal cancer: Analysis and
verification of current data. Sci Rep. 7:84132017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen J, Wang W, Zhang Y, Chen Y and Hu T:
Predicting distant metastasis and chemoresistance using plasma
miRNAs. Med Oncol Northwood Lond Engl. 31:7992014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luu-The V, Paquet N, Calvo E and Cumps J:
Improved real-time RT-PCR method for high-throughput measurements
using second derivative calculation and double correction.
Biotechniques. 38:287–293. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Core R: Team: R: A Language and
Environment for Statistical Computing. R Foundation for Statistical
Computing. (Vienna, Austria). 2018.
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vlachos IS, Zagganas K, Paraskevopoulou
MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T and
Hatzigeorgiou AG: DIANA-miRPath v3.0: Deciphering microRNA function
with experimental support. Nucleic Acids Res. 43:W460–W466. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kirschner MB, Kao SC, Edelman JJ,
Armstrong NJ, Vallely MP, van Zandwijk N and Reid G: Haemolysis
during sample preparation alters microRNA content of plasma. PLoS
One. 6:e241452011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Goni R, García P and Foissac S: The qPCR
data statistical analysis. 9. 2009.
|
|
33
|
Chen WY, Zhao XJ, Yu ZF, Hu FL, Liu YP,
Cui BB, Dong XS and Zhao YS: The potential of plasma miRNAs for
diagnosis and risk estimation of colorectal cancer. Int J Clin Exp
Pathol. 8:7092–7101. 2015.PubMed/NCBI
|
|
34
|
Li J, Liu Y, Wang C, Deng T, Liang H, Wang
Y, Huang D, Fan Q, Wang X, Ning T, et al: Serum miRNA expression
profile as a prognostic biomarker of stage II/III colorectal
adenocarcinoma. Sci Rep. 5:129212015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
He Y, Wang G, Zhang L, Zhai C, Zhang J,
Zhao X, Jiang X and Zhao Z: Biological effects and clinical
characteristics of microRNA-106a in human colorectal cancer. Oncol
Lett. 14:830–836. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Díaz R, Silva J, García JM, Lorenzo Y,
García V, Peña C, Rodríguez R, Muñoz C, García F, Bonilla F and
Domínguez G: Deregulated expression of miR-106a predicts survival
in human colon cancer patients. Genes Chromosomes Cancer.
47:794–802. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F
and Yan D: Long non-coding RNA Fer-1-like protein 4 suppresses
oncogenesis and exhibits prognostic value by associating with
miR-106a-5p in colon cancer. Cancer Sci. 106:1323–1332. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Peng Q, Shen Y, Zhao P, Cheng M, Zhu Y and
Xu B: Biomarker roles identification of miR-106 family for
predicting the risk and poor survival of colorectal cancer. BMC
Cancer. 20:5062020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang C, Wang J, Liu H and Fu Z: Tumor
suppressor DLC-1 induces apoptosis and inhibits the growth and
invasion of colon cancer cells through the Wnt/β-catenin signaling
pathway. Oncol Rep. 31:2270–2278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cao H, Huang S, Liu A and Chen Z:
Up-regulated expression of miR-155 in human colonic cancer. J
Cancer Res Ther. 14:604–607. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu N, Jiang F, Han XY, Li M, Chen WJ, Liu
QC, Liao CX and Lv YF: MiRNA-155 promotes the invasion of
colorectal cancer SW-480 cells through regulating the
Wnt/β-catenin. Eur Rev Med Pharmacol Sci. 22:101–109.
2018.PubMed/NCBI
|
|
42
|
Qu A, Du L, Yang Y, Liu H, Li J, Wang L,
Liu Y, Dong Z, Zhang X, Jiang X, et al: Hypoxia-inducible MiR-210
is an independent prognostic factor and contributes to metastasis
in colorectal cancer. PLoS One. 9:e909522014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sabry D, El-Deek SEM, Maher M, El-Baz MAH,
El-Bader HM, Amer E, Hassan EA, Fathy W and El-Deek HEM: Role of
miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in
colorectal carcinoma: Impact of HIF-1α-VEGF signaling pathway. Mol
Cell Biochem. 454:177–189. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ullmann P, Qureshi-Baig K, Rodriguez F,
Ginolhac A, Nonnenmacher Y, Ternes D, Weiler J, Gäbler K, Bahlawane
C, Hiller K, et al: Hypoxia-responsive miR-210 promotes
self-renewal capacity of colon tumor-initiating cells by repressing
ISCU and by inducing lactate production. Oncotarget. 7:65454–65470.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jung EJ, Santarpia L, Kim J, Esteva FJ,
Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC Jr, Park ST, et al:
Plasma microRNA 210 levels correlate with sensitivity to
trastuzumab and tumor presence in breast cancer patients. Cancer.
118:2603–2614. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wan J, Xia L, Xu W and Lu N: Expression
and function of miR-155 in diseases of the gastrointestinal Tract.
Int J Mol Sci. 17:7092016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rath T, Billmeier U, Waldner MJ, Atreya R
and Neurath MF: From physiology to disease and targeted therapy:
Interleukin-6 in inflammation and inflammation-associated
carcinogenesis. Arch Toxicol. 89:541–554. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Svrcek M, El-Murr N, Wanherdrick K, Dumont
S, Beaugerie L, Cosnes J, Colombel JF, Tiret E, Fléjou JF,
Lesuffleur T and Duval A: Overexpression of microRNAs-155 and 21
targeting mismatch repair proteins in inflammatory bowel diseases.
Carcinogenesis. 34:828–834. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ulivi P, Canale M, Passardi A, Marisi G,
Valgiusti M, Frassineti GL, Calistri D, Amadori D and Scarpi E:
Circulating plasma levels of miR-20b, miR-29b and miR-155 as
predictors of bevacizumab efficacy in patients with metastatic
colorectal cancer. Int J Mol Sci. 19:3072018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
You C, Jin L, Xu Q, Shen B, Jiao X and
Huang X: Expression of miR-21 and miR-138 in colon cancer and its
effect on cell proliferation and prognosis. Oncol Lett.
17:2271–2277. 2019.PubMed/NCBI
|
|
52
|
Mima K, Nishihara R, Yang J, Dou R, Masugi
Y, Shi Y, da Silva A, Cao Y, Song M, Nowak J, et al: MicroRNA MIR21
(miR-21) and PTGS2 expression in colorectal cancer and patient
survival. Clin Cancer Res. 22:3841–3848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Inamura K and Ishikawa Y: MicroRNA in lung
cancer: Novel biomarkers and potential tools for treatment. J Clin
Med. 5:362016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Komatsu S, Ichikawa D, Takeshita H,
Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H,
Shiozaki A, et al: Circulating microRNAs in plasma of patients with
oesophageal squamous cell carcinoma. Br J Cancer. 105:104–111.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Saxena A, Tammali R, Ramana KV and
Srivastava SK: Aldose reductase inhibition prevents colon cancer
growth by restoring phosphatase and tensin homolog through
modulation of miR-21 and FOXO3a. Antioxid Redox Signal.
18:1249–1262. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hong Z, Feng Z, Sai Z and Tao S: PER3, a
novel target of miR-103, plays a suppressive role in colorectal
cancer in vitro. BMB Rep. 47:500–505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zheng YB, Xiao K, Xiao GC, Tong SL, Ding
Y, Wang QS, Li SB and Hao ZN: MicroRNA-103 promotes tumor growth
and metastasis in colorectal cancer by directly targeting LATS2.
Oncol Lett. 12:2194–2200. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang DS, Zhong B, Zhang M-S and Gao Y:
Upregulation of serum miR-103 predicts unfavorable prognosis in
patients with colorectal cancer. Eur Rev Med Pharmacol Sci.
22:4518–4523. 2018.PubMed/NCBI
|
|
59
|
Xu W, Ji J, Xu Y, Liu Y, Shi L, Liu Y, Lu
X, Zhao Y, Luo F, Wang B, et al: MicroRNA-191, by promoting the EMT
and increasing CSC-like properties, is involved in neoplastic and
metastatic properties of transformed human bronchial epithelial
cells. Mol Carcinog. 54 (Suppl 1):E148–E161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang XF, Li KK, Gao L, Li SZ, Chen K,
Zhang JB, Wang D, Tu RF, Zhang JX, Tao KX, et al: miR-191 promotes
tumorigenesis of human colorectal cancer through targeting C/EBPβ.
Oncotarget. 6:4144–4158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qin S, Zhu Y, Ai F, Li Y, Bai B, Yao W and
Dong L: MicroRNA-191 correlates with poor prognosis of colorectal
carcinoma and plays multiple roles by targeting tissue inhibitor of
metalloprotease 3. Neoplasma. 61:27–34. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Milanesi E, Dobre M, Bucuroiu AI, Herlea
V, Manuc TE, Salvi A, De Petro G, Manuc M and Becheanu G:
miRNAs-based molecular signature for KRAS mutated and wild type
colorectal cancer: An explorative study. J Immunol Res.
2020:49271202020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen XY, Zhang J, Hou LD, Zhang R, Chen W,
Fan HN, Huang YX, Liu H and Zhu JS: Upregulation of PD-L1 predicts
poor prognosis and is associated with miR-191-5p dysregulation in
colon adenocarcinoma. Int J Immunopathol Pharmacol.
32:20587384187903182018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Qian J, Jiang B, Li M, Chen J and Fang M:
Prognostic significance of microRNA-16 expression in human
colorectal cancer. World J Surg. 37:2944–2949. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xiao G, Tang H, Wei W, Li J, Ji L and Ge
J: Aberrant expression of MicroRNA-15a and MicroRNA-16
synergistically associates with tumor progression and prognosis in
patients with colorectal cancer. Gastroenterol Res Pract.
2014:3645492014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ma Q, Wang X, Li Z, Li B, Ma F, Peng L,
Zhang Y, Xu A and Jiang B: microRNA-16 represses colorectal cancer
cell growth in vitro by regulating the p53/survivin signaling
pathway. Oncol Rep. 29:1652–1658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Diamantopoulos MA, Kontos CK, Kerimis D,
Papadopoulos IN and Scorilas A: Upregulated miR-16 expression is an
independent indicator of relapse and poor overall survival of
colorectal adenocarcinoma patients. Clin Chem Lab Med. 55:737–747.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ostenfeld MS, Jensen SG, Jeppesen DK,
Christensen LL, Thorsen SB, Stenvang J, Hvam ML, Thomsen A,
Mouritzen P, Rasmussen MH, et al: miRNA profiling of circulating
EpCAM(+) extracellular vesicles: Promising biomarkers of colorectal
cancer. J Extracell Vesicles. 5:314882016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Evans C, Hardin J and Stoebel D: Selecting
between-sample RNA-Seq normalization methods from the perspective
of their assumptions. Brief Bioinform. 19:776–792. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Dillies MA, Rau A, Aubert J,
Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G,
Castel D, Estelle J, et al: A comprehensive evaluation of
normalization methods for Illumina high-throughput RNA sequencing
data analysis. Brief Bioinform. 14:671–683. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Fundel K, Haag J, Gebhard PM, Zimmer R and
Aigner T: Normalization strategies for mRNA expression data in
cartilage research. Osteoarthritis Cartilage. 16:947–955. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Rieder F, Brenmoehl J, Leeb S, Schölmerich
J and Rogler G: Wound healing and fibrosis in intestinal disease.
Gut. 56:130–139. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Theocharis AD, Skandalis SS, Gialeli C and
Karamanos NK: Extracellular matrix structure. Adv Drug Deliv Rev.
97:4–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Howell JC and Wells JM: Generating
intestinal tissue from stem cells: Potential for research and
therapy. Regen Med. 6:743–755. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Xue X and Falcon DM: The role of immune
cells and cytokines in intestinal wound healing. Int J Mol Sci.
20:60972019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang Y, Yu A and Yu FX: The Hippo pathway
in tissue homeostasis and regeneration. Protein Cell. 8:349–359.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wang H, Li X, Lu J, Jones P and Xu W:
Prolactin may serve as a regulator to promote vocal fold wound
healing. Biosci Rep. 40:BSR202004672020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Shantha Kumara H, Yan X-H, Pettke E, Cekic
V, Gandhi ND, Bellini GA and Whelan RL: Plasma and wound fluid
levels of eight proangiogenic proteins are elevated after
colorectal resection. World J Gastrointest Oncol. 11:470–488. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Mizuno R, Kawada K, Itatani Y, Ogawa R,
Kiyasu Y and Sakai Y: The role of tumor-associated neutrophils in
colorectal cancer. Int J Mol Sci. 20:5292019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Roma-Rodrigues C, Mendes R, Baptista PV
and Fernandes AR: Targeting tumor microenvironment for cancer
therapy. Int J Mol Sci. 20:8402019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kasprzak A: The role of tumor
microenvironment cells in colorectal cancer (CRC) cachexia. Int J
Mol Sci. 22:15652021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Bonnans C, Chou J and Werb Z: Remodelling
the extracellular matrix in development and disease. Nat Rev Mol
Cell Biol. 15:786–801. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Salesse S, Odoul L, Chazée L, Garbar C,
Duca L, Martiny L, Mahmoudi R and Debelle L: Elastin molecular
aging promotes MDA-MB-231 breast cancer cell invasiveness. FEBS
Open Bio. 8:1395–1404. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Greten FR and Grivennikov SI: Inflammation
and Cancer: Triggers, Mechanisms, and Consequences. Immunity.
51:27–41. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Triner D and Shah YM: Hypoxia-inducible
factors: A central link between inflammation and cancer. J Clin
Invest. 126:3689–3698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Li J, Xu X, Jiang Y, Hansbro NG, Hansbro
PM, Xu J and Liu G: Elastin is a key factor of tumor development in
colorectal cancer. BMC Cancer. 20:2172020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Sommer K, Wiendl M, Müller TM, Heidbreder
K, Voskens C, Neurath MF and Zundler S: Intestinal mucosal wound
healing and barrier integrity in IBD-crosstalk and trafficking of
cellular players. Front Med (Lausanne). 8:6439732021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Monnet E and Smeak DD: Gastrointestinal
healing. In Gastrointestinal Surgical Techniques in Small Animals.
Monnet E and Smeak DD (eds). Wiley Blackwell. (Hoboken, NJ, USA, pp
1-8). 2020.
|