Introduction
Nasopharyngeal carcinoma (NPC) is prevalent in East
and Southeast Asia, including south China (1,2). Due
to the early onset of lymphatic metastasis, as well as a high
recurrence rate, more efficient therapeutic methods are required
(1,3). Radiotherapy is currently the primary
treatment type for non-metastatic NPC, which differs from advanced
NPC (3,4), which at present, is primarily treated
with chemoradiotherapy using a platinum-based reagent. Previous
studies have shown that combination therapy of platinum with other
drugs may significantly increase the efficacy of chemotherapy in
advanced NPC. For example, a clinical trial has shown that the
addition of gemcitabine and cisplatin-based induction chemotherapy
to basic chemoradiotherapy significantly improved survival rates in
locoregionally-advanced NPC (5).
Other drugs have also been reported to enhance the efficacy of
platinum-based chemotherapy in NPC (6–8).
However, the incidence of adverse effects following combination
treatment is still a concern.
Numerous studies have investigated natural products
and compounds as novel treatments for different cancer types, or to
enhance the efficacy of classic chemotherapeutic drugs such as
cisplatin (9,10). Cordycepin (3-deoxyadenosine) is a
compound extracted from the Cordyceps genus of ascomycete
fungi, which is used in traditional Chinese medicine (11). Previous studies have demonstrated
the anticancer characteristics of cordycepin in different cancer
cell types, including hepatocellular carcinoma, oral cancer and
lung cancer (12–15); however, the underlying mechanisms
remain unclear (16,17).
The aim of the present study was to investigate the
effects of cisplatin on NPC cells, (namely, whether it has the same
effects as in other cancer types), using transcriptome sequencing
to elucidate the underlying molecular mechanisms of cordycepin
treatment in NPC.
Materials and methods
Reagents and cell culture
Cordycepin and cisplatin were purchased from
MedChemExpress. Immediately before use, cordycepin was dissolved in
media to generate a 10 mM stock solution, and cisplatin was
dissolved in N,N-dimethylformamide to generate a 10 mM stock
solution. Both stock solutions were stored at −20°C. The Human
C666-1 NPC cell line, which is an EBV-positive NPC cell line taken
from undifferentiated NPC tissue, was obtained from the cell line
database (Shanghai FuHeng Biological Technology Co., Ltd.), and
were cultured in RPMI-1640 media (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; Invitrogen;
Thermo Fisher Scientific, Inc.), penicillin (100 U/ml) and
streptomycin (100 U/ml) (both Gibco; Thermo Fisher Scientific,
Inc.) in a humidified incubator at 37°C (5% CO2).
Cell viability assay
C666-1 cells were seeded into a 96-well plate
(5×103 cells per well) and treated with increasing
concentrations of cordycepin (0, 250, 500, 750 and 1,000 µM for
24–72 h, or cisplatin for 24 h at 37°C. Following treatment, cell
viability was assessed using the Cell Counting Kit 8 (CCK-8) assay
(APExBIO Technology LLC); 10 µl CCK-8 solution was added, and the
cells were incubated for 4 h at 37°C. Optical density was detected
at 450 nm using a microplate reader (Thermo Fisher Scientific,
Inc.).
Colony formation assay
C666-1 cells were counted, seeded into 12-well
plates in triplicate (800 cells per well), cultured in RPMI
(supplemented with 10% fetal bovine serum), and treated with
cordycepin for up to 14 days as aforementioned. Then, the cells
were washed twice with PBS and fixed using methanol for 10 min at
4°C. After two additional washes with PBS, the cells were stained
with crystal violet for 30 min at room temperature. The cells were
then washed with double-distilled water (ddH2O) to
remove the crystal violet, and the colony numbers were counted
using ImageJ software (version 1.52; National Institutes of
Health), set to the area of colonies above 5 pixel^2 as
1 colony.
Wound-healing assay
C666-1 cells were seeded into 12-well plates and
incubated in serum-free medium at 37°C for 18 h. The cell
monolayers were scratched with a 10-µl pipette tip and washed with
serum-free RPMI-1640 media (Gibco; Thermo Fisher Scientific, Inc.)
to remove cells detached from the plates. The cells were incubated
in the presence or absence of cordycepin for 48–72 h (as
aforementioned) in medium containing 10% FBS. Then, the medium was
replaced with PBS and images of the cells were captured using a
fluorescence inverted microscope (Leica Microsystems, Inc.;
magnification, ×20) in brightfield mode. The results were
quantified using ImageJ software (version 1.52).
Transwell migration assay
To assess cellular migration, 5×104 cells
were seeded into Transwell inserts in a 24-well plate, with
serum-free medium in the upper chambers, and RPMI containing 10%
FBS added to the lower chambers. The cells were incubated for 24–48
h at 37°C, washed once with PBS, and then fixed with 4%
paraformaldehyde for 10 min at room temperature. The cells were
stained with 0.1% crystal violet for 30 min at room temperature,
and then washed with ddH2O. The non-migrated cells were
removed with a cotton swab, and the stained cells were observed by
a fluorescence inverted microscope (Leica Microsystems, Inc.;
magnification, ×20).
RNA extraction, library construction,
and sequencing
Total RNA of each cordycepin-treated and control
sample was extracted using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), following the manufacturer's
instructions. The quality of RNA was assessed on an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc.) and checked using
RNase-free agarose gel electrophoresis. Then mRNA was enriched and
purified using oligo (dT) beads. The purified mRNA was cut into
short fragments using fragmentation buffer (Shanghai Yeasen
Biotechnology Co., Ltd.) and reverse transcribed into cDNA using
random primers. Second-strand cDNA were synthesized using DNA
polymerase I, RNase H, dNTP and buffer. Then the cDNA fragments
were purified using the QIAquick PCR extracting kit (Qiagen). After
performing end repair and adding poly (A), the cDNA fragments were
ligated using Illumina sequencing adapters. The ligation products
were enriched via PCR amplification to construct the cDNA library
template (NovaSeq 6000 S4 Reagent Kit v1.5; 300 cycles; cat. no.
20028312). Finally, the library (10 pM per sample) was sequenced by
150 bp paired end sequencing using the Illumina Novaseq 6000
(Illumina Inc.) by Guangzhou Gene Denovo Biotechnology Co. Ltd.
Transcriptome mapping, annotation and
differential expression analyses
Sequencing reads were edited for quality and cleaned
using fastp (version 0.18.0, http://github.com/OpenGene/fastp). Clean data were
mapped to the Homo sapiens (human) genome (GRCh38.p13) using
HISAT2 (version 2.4, http://daehwankimlab.github.io/hisat2/). The mapped
reads of each sample were assembled using StringTie (version 1.3.1,
http://ccb.jhu.edu/software/stringtie) using a
reference-based approach. For each transcription region, a FPKM
(fragment per kilobase of transcript per million mapped reads)
value was calculated to quantify its expression abundance and
variation using RSEM software (http://deweylab.biostat.wisc.edu/rsem). Differentially
expressed genes were identified using DESeq2 (http://www.bioconductor.org/) and edgeR (http://www.rproject.org/) package, with a threshold
false discovery rate <0.05, and absolute value of the log2 fold
change ≥1. All expressed genes were functionally annotated against
the NCBI non-redundant protein database using the BLAST algorithm
with a cut-off E-value ≤10−5. The genes were also
subjected to classification and enrichment analyses of Gene
Ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathways. GO and KEGG classification were performed using
Gene Ontology database (http://geneontology.org/) and the KEGG automatic
annotation server (https://www.genome.jp/kegg/), respectively.
Western blotting
Cells were harvested and lysed in RIPA buffer
(Sangon Biotech Co., Ltd.) supplemented with protease inhibitor (1%
phenylmethylsulfonyl fluoride) at 4°C, for 30 min, and then
centrifuged at 10,309 × g for 15 min 4°C. The protein
concentrations of the lysates were measured on a spectrophotometer
using a BCA Protein Assay Kit (Sangon Biotech Co., Ltd.). The
remainder were added to a 5X loading buffer at 1:4 (Sangon Biotech
Co., Ltd.) and heated at 95°C for 5 min. Next, 50 µg protein per
lane was electrophoretically separated by 10% SDS-PAGE, and
transferred to PVDF membranes. The membranes were blocked using 5%
non-fat milk for 1 h at room temperature, and the incubated with
primary antibodies against GAPDH (1:2,000; cat. no. AP0063;
Bioworld Technology, Inc.) ERK1/2 (1:1,000; cat. no.137F5; Cell
Signaling Technologies, Inc.), p-ERK1/2 (1:1,000; cat. no.9101;
Cell Signaling Technologies, Inc.) and β-catenin (1:1,000; cat. no.
ab16051; Abcam) in 1X TBS with 0.05% Tween (TBS-T), at 4°C
overnight. The membranes were then incubated with HRP-conjugated
secondary antibodies (anti-rabbit; 1:5,000; cat. no. 7074; Cell
Signaling Technologies, Inc.) in 1X TBS-T at room temperature for 1
h. The proteins were visualized using a chemiluminescence (ECL)
reagent (Pierce ECL Western Blotting Substrate; Thermo Fisher
Scientific, Inc.).
Reverse transcription-quantitative
(RT-q) PCR
RNA sequencing (RNA-seq) results were validated
using RT-qPCR. Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) per the
manufacturer's instructions. Reverse transcription and qPCR were
performed using an RT Kit (Hunan Accurate Bio-Medical Co., Ltd.)
and TB Green PCR Master Mix (Takara Bio, Inc.), respectively,
according to the manufacturers' protocols. The qPCR reaction was
carried out using a three-step method on a LightCycler 480
Instrument II (Roche Diagnostics): Initial denaturation at 95°C for
5 min, then amplification at 95°C for 5 sec, 58°C for 30 sec and
72°C for 20 sec (a total of 40 cycles). The oligonucleotide primers
are displayed in Table I; GAPDH
was used as the housekeeping gene to normalize the expression
levels of mRNA, which were quantified using the 2−ΔΔCq
method (18).
 | Table I.Primers for qPCR. |
Table I.
Primers for qPCR.
| Name | Sequence
(5′-3′) |
|---|
| SELE |
|
|
Forward |
TCAAGGGCAGTGGACACAGCAA |
|
Reverse |
GGAAACTGCCAGAAGCACTAGG |
| MUC20 |
|
|
Forward |
AGAGTGGCAGAAAGGCTGATGC |
|
Reverse |
CTGATGTCCGTTAGCCTCTCCT |
| ACTL10 |
|
|
Forward |
GCCAGTTTCAGCGTGGGTAACG |
|
Reverse |
CAGCGTTTTGGGCATCTTCTGC |
| EFHD1 |
|
|
Forward |
GAGGGTGTCAAAGGTGCCAAGA |
|
Reverse |
TGAGTTTCTGGAAGGCTGCCTG |
| RGS9 |
|
|
Forward |
CAACGATGCCATCATGTCAGGC |
|
Reverse |
CCATCGTTCCACTCGCATCTTG |
| GAPDH |
|
|
Forward |
AACATCATCCCTGCCTCTACTG |
|
Reverse |
CCTCCGACGCCTGCTTCAC |
Statistical analysis
Each experiment was repeated three times
independently, and the data are expressed as the mean ± standard
deviation. The data were analyzed using GraphPad Prism 6.02
(GraphPad Software, Inc.); differences between two groups were
analyzed using the unpaired Student's t-test, and differences among
≥3 groups were compared using one-way ANOVA followed by Dunnett's
multiple comparisons test. P<0.05 was considered to indicate a
statistically significant difference.
Results
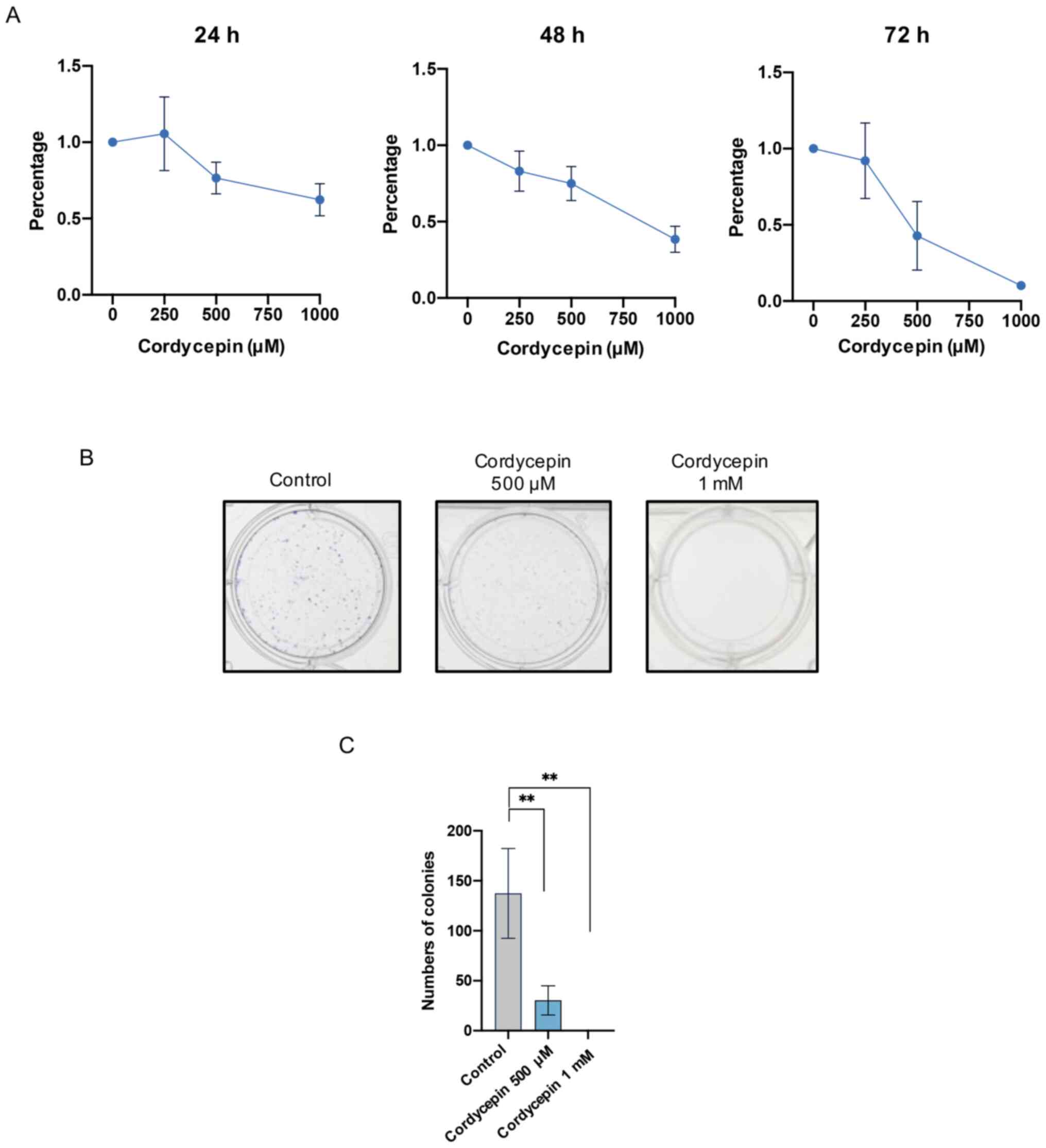
Cordycepin inhibits the proliferation
of NPC cells
C666-1 cells, EBV-positive NPC cells taken from
undifferentiated NPC tissue (19),
were used to investigate the effects of cordycepin on NPC cell
proliferation. Increasing concentrations of cordycepin (250, 500
and 1,000 µM) were added to the culture media for 24, 48 and 72 h,
and C666-1 cell viability was determined using a CCK-8 assay. The
results demonstrated that cordycepin decreased the viability of
C666-1 cells in a dose-dependent manner, with an IC50 of 1.37 mM at
24 h, 842.5 µM at 48 h and 546.9 µM at 72 h (Fig. 1A). Furthermore, a colony formation
assay revealed that 500 µM cordycepin significantly inhibited
colony formation, and that 1 mM cordycepin completely inhibited
clone formation (Fig. 1B and C).
These results indicate that cordycepin inhibited the proliferation
of NPC cells.
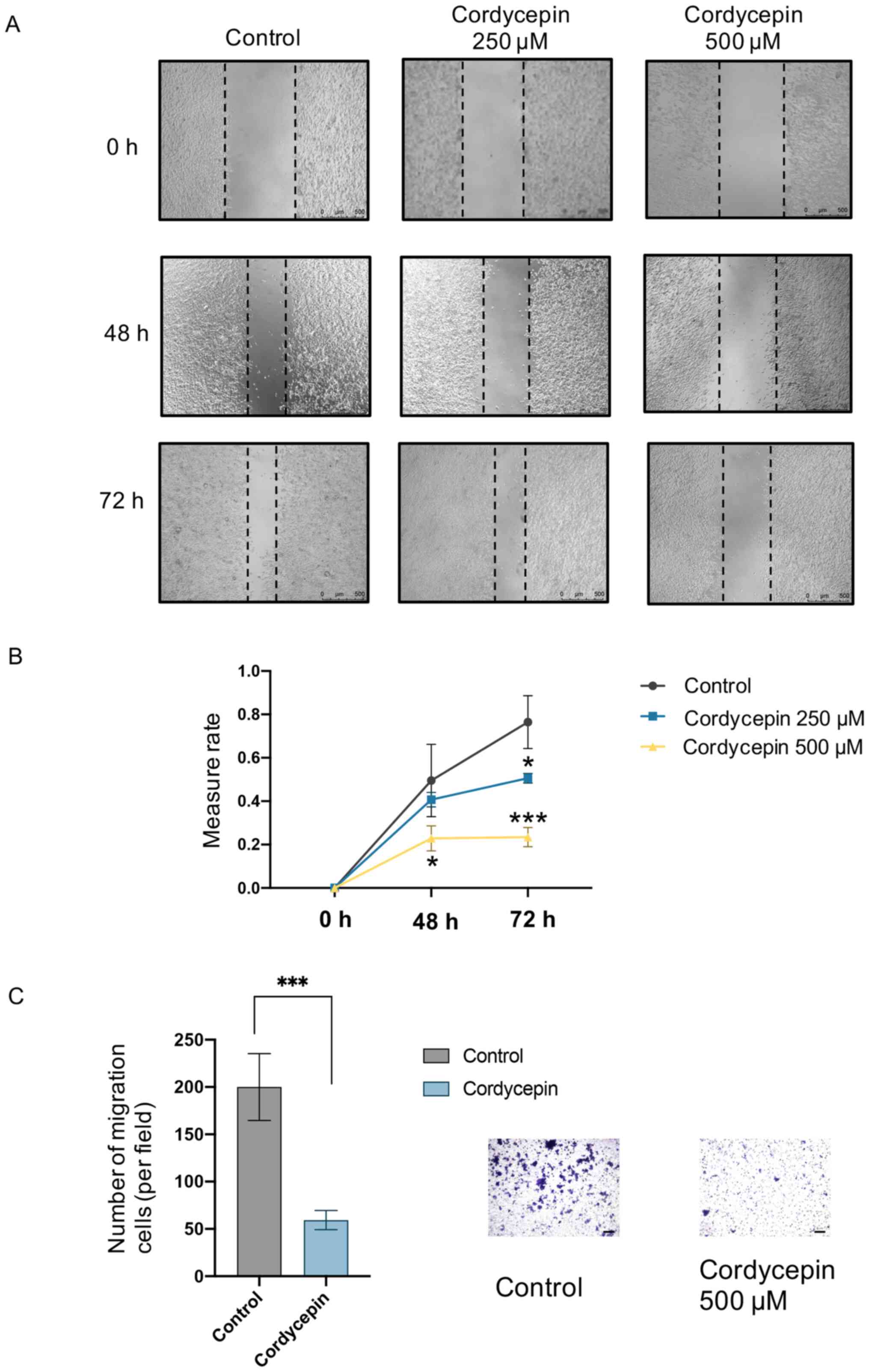
Cordycepin inhibits the migration of
NPC cells
To investigate the effects of cordycepin on the
migration of C666-1 cells, wound-healing and Transwell assays were
performed. Cells treated with 250 and 500 µM cordycepin showed
inhibited wound-healing ability (Fig.
2A and B). Furthermore, 500 µM cordycepin significantly
inhibited C666-1 cell migration through the Transwell insert
membrane (Fig. 2C). These results
indicate that cordycepin inhibited the migration of NPC cells.
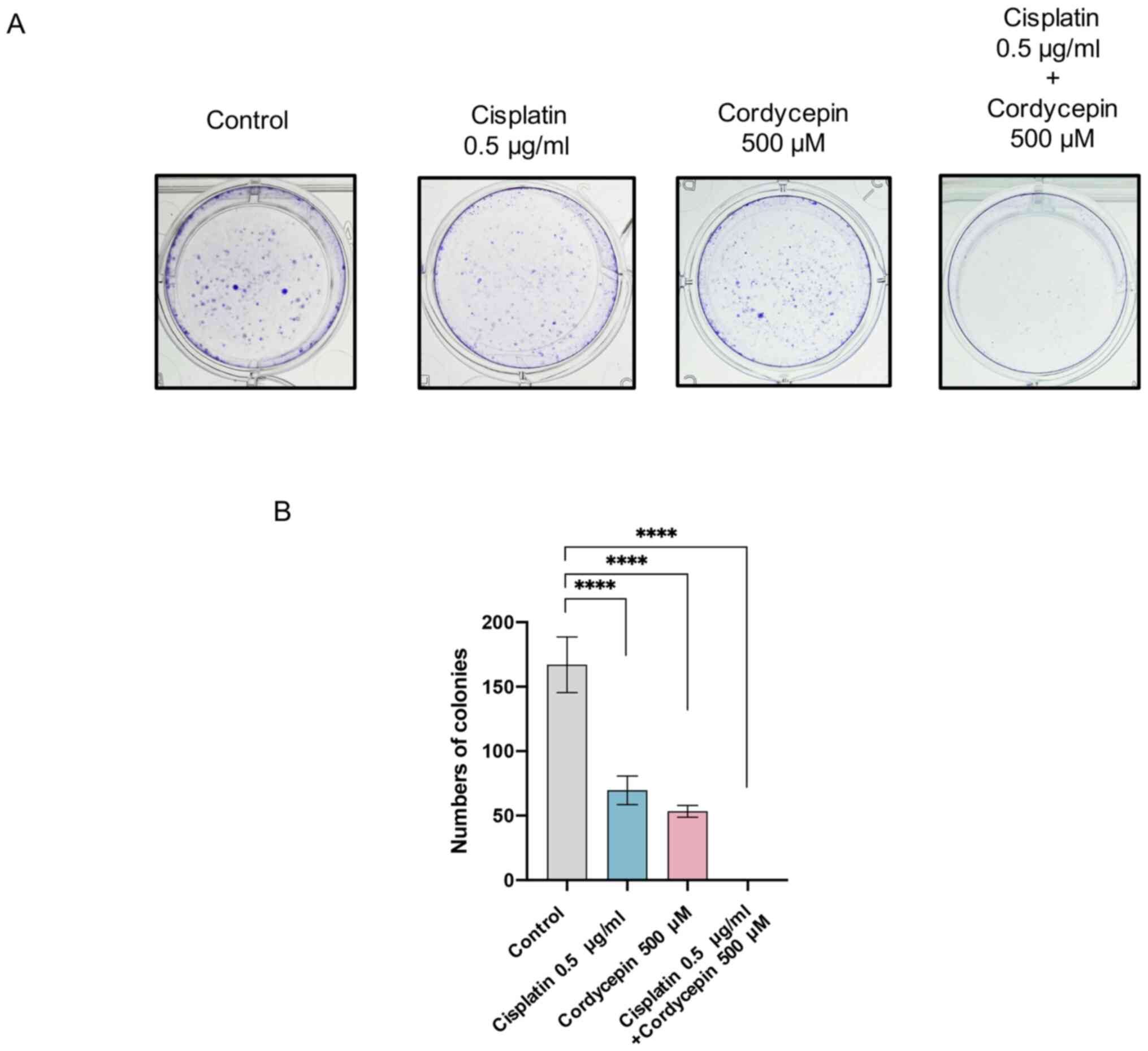
Cordycepin enhances the effects of
cisplatin on NPC cells
Next, the potential for cordycepin to enhance the
chemotherapeutic effects of cisplatin was investigated in NPC
cells. An additional colony formation assay was performed, and a
relatively low concentration of cisplatin that did not affect cell
viability (0.5 µg/ml) was used. The assay revealed that the colony
formation ability of NPC cells was completely inhibited following
combined treatment with cordycepin and cisplatin (Fig. 3). This result indicates that
cordycepin enhanced the inhibitory effects of cisplatin on NPC
cells.
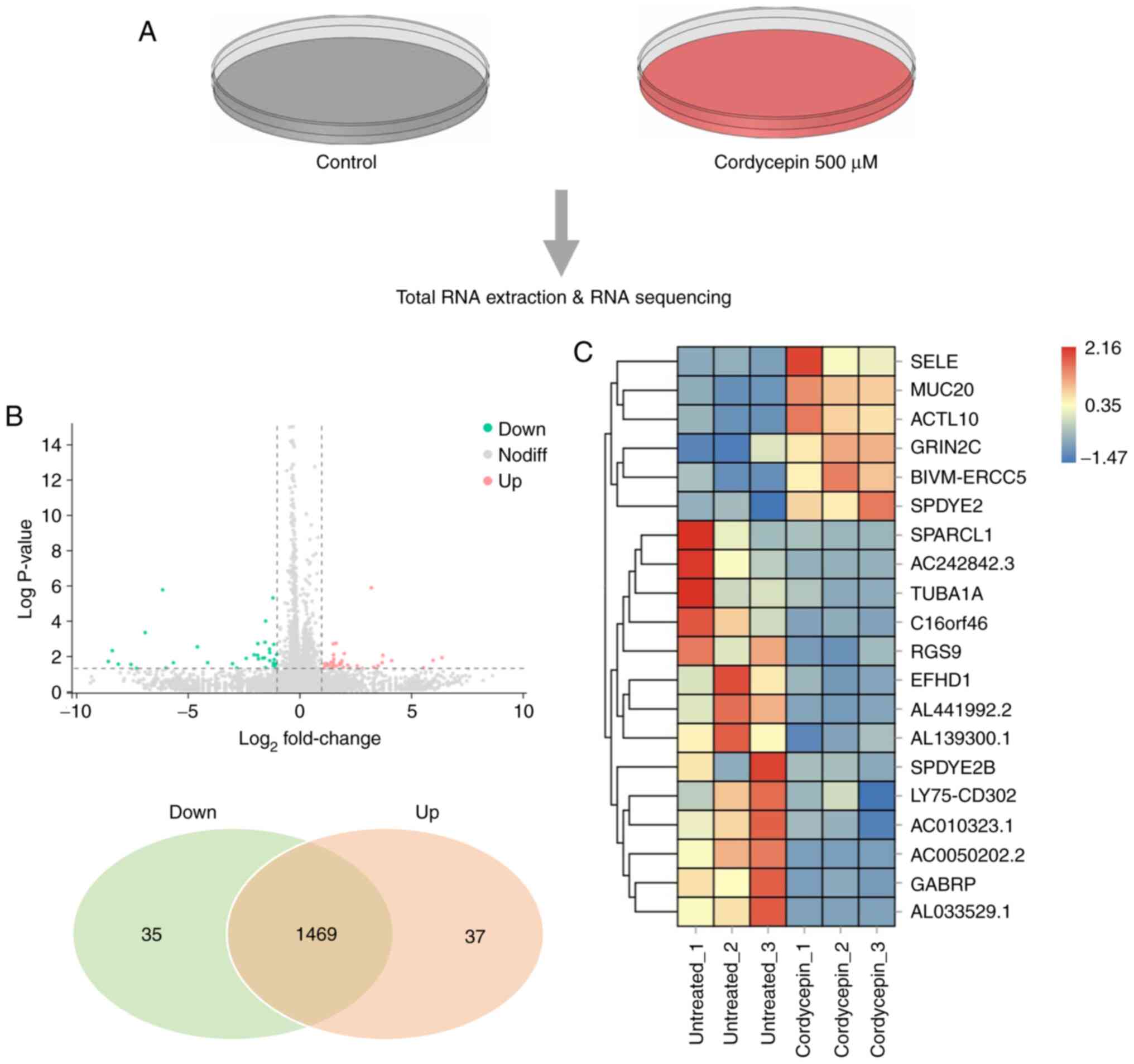
Changes in the global gene expression
profile of C666-1 cells after cordycepin treatment
To investigate how cordycepin regulates downstream
signaling pathways, transcriptome RNA-seq analyses were performed
to compare cells treated with 500 µM cordycepin for 48 h with
untreated control cells (Fig. 4A).
In total, 20,295 genes were identified, and the expression levels
of 1,541 genes were altered to varying degrees. Among the
differentially expressed genes, 72 were significantly different
(Log2|FoldChange |> 2), P<0.05), including 35 downregulated
and 37 upregulated genes (Fig.
4B). The top upregulated and downregulated genes are presented
in Fig. 4C. This experiment showed
global gene expression alterations following cordycepin
treatment.
Functional annotation of
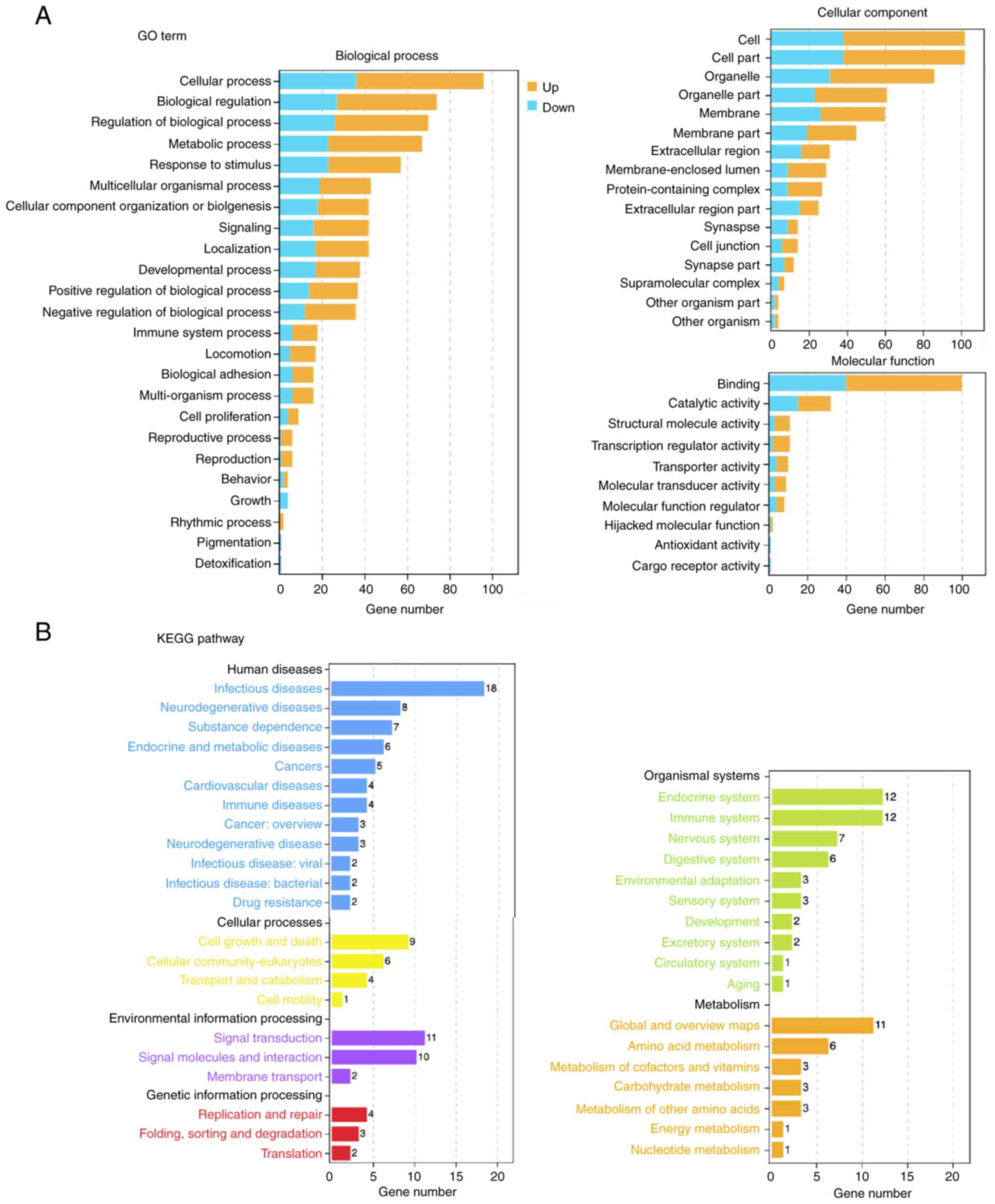
differentially expressed genes after cordycepin treatment
To map changes in the downstream signaling pathways,
GO and KEGG pathway enrichment analyses were performed on all
significantly differentially expressed genes (above a 1.5-fold
change). An overview of the top functions is provided in Fig. 5. Both up- and downregulated genes
were enriched, revealing a global map of signaling transduction
changes in the cells.
A detailed enrichment figure reveals KEGG pathways
that could affect the proliferation and migration of cancer cells,
including ‘cell adhesion molecules’, ‘PD-L1 expression and PD-1
checkpoint pathway in cancer’, ‘T cell receptor signaling pathway’,
‘TNF signaling pathway’, and ‘VEGF signaling pathway’. GO term
enrichment results highlighted biological processes related to cell
proliferation and migration, such as “regulation of the JNK
cascade’, ‘positive regulation of the stress-activated MAPK
cascade’ and ‘cell adhesion’ (Fig.
6A).
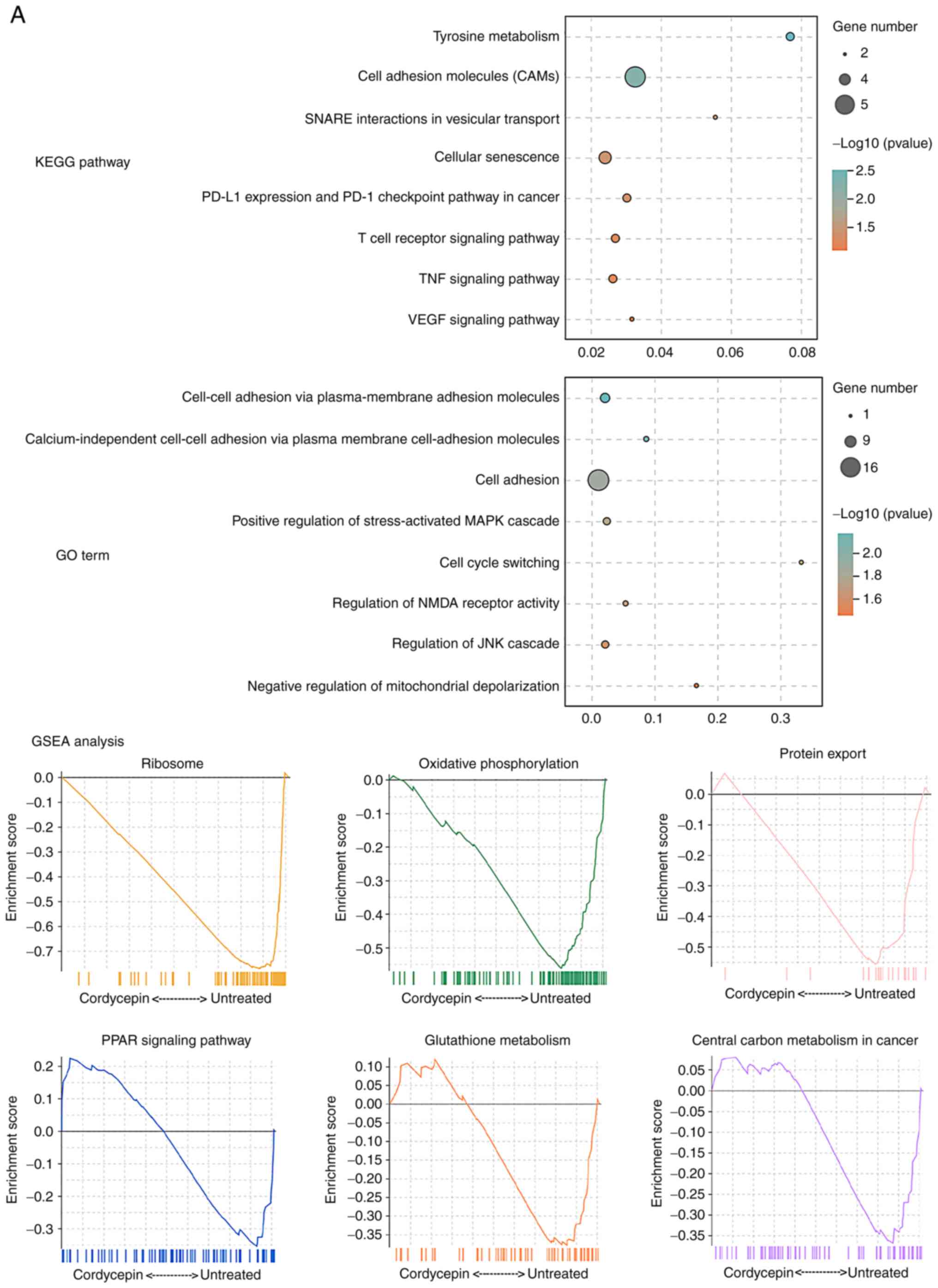
Next, Gene Set Enrichment Analysis of the KEGG
pathways was performed to identify those that were the most
inhibited by cordycepin treatment. These included ‘ribosome’,
‘oxidative phosphorylation’, ‘protein export’, ‘PPAR signaling
pathway’, ‘glutathione metabolism’ and ‘central carbon metabolism
in cancer’ (Fig. 6B).
These analyses indicate downstream signaling pathway
alterations that are regulated by cordycepin.
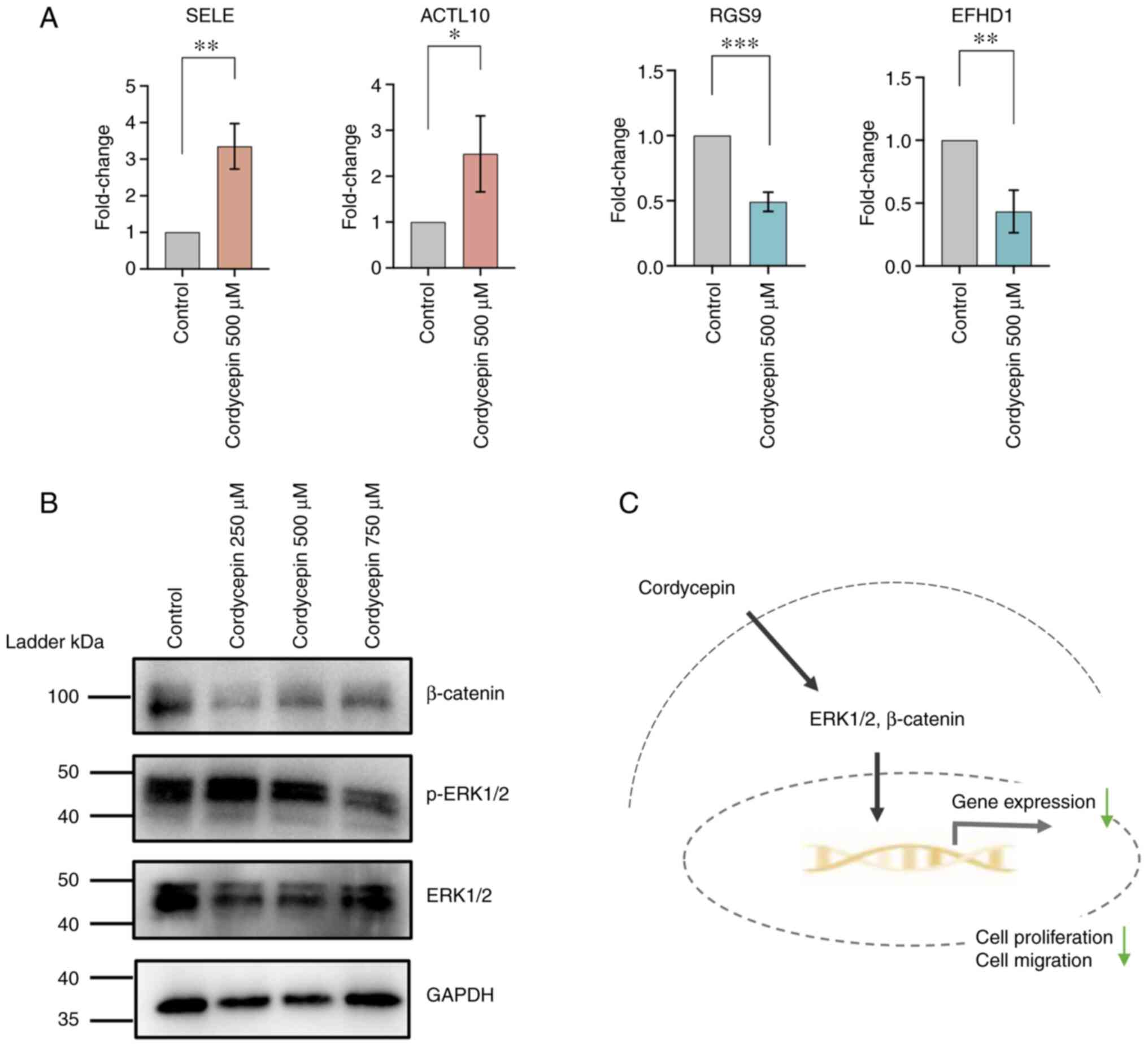
Cordycepin may inhibit NPC cells
through the ERK and β-catenin signaling pathways
To verify the RNA-seq data, four significantly
differentially expressed genes identified from RNA-seq were
selected and verified via RT-qPCR. Selectin E and actin like 10
were upregulated by 500 µM cordycepin treatment, while regulator of
G protein signaling 9 and EF-hand domain family member D1 were
downregulated, which was consistent with the sequencing results
(Fig. 7A). To investigate whether
cordycepin affects the MAPK/ERK and Wnt signaling pathways, western
blotting was performed to evaluate the protein expression levels of
ERK1/2 and phosphorylated-ERK1/2 (p-ERK) at different
concentrations of cordycepin for 48 h. The results showed a similar
expression level of p-ERK at 500 µM cordycepin compared with the
control group, while significant inhibition of ERK and p-ERK1/2 was
found after treatment with 750 µM cordycepin, which is close to the
IC50 value at this time point (Fig.
7B). The protein expression of β-catenin was also arrested
(relative to the untreated control group) following cordycepin
treatment. Different expression levels of β-catenin were detected
at low (250 µM) and high (500 and 750 µM) concentrations, which
suggests different regulation mechanisms of signaling transduction.
These results indicate that cordycepin may inhibit proliferation
and migration of NPC cells through the ERK and β-catenin signaling
pathways.
Discussion
Natural products such as gingerol, curcumin and
gambogic have been widely studied, and are considered candidates
for treating different types of cancer (9,10).
Cordycepin is the primary compound extracted from Cordyceps
fungi, which has been used as a dietary supplement in some Asian
countries for hundreds of years (11). Furthermore, the anticancer
properties of cordycepin have been identified in various types of
malignancy, including breast, liver and lung cancer (13,15,17,20,21).
Cordycepin is an adenosine derivative that can
regulate cell functions through adenosine receptors, death
receptors or epidermal growth factor receptor (EGFR) (21–23).
The extract inhibits the proliferation, migration, invasiveness and
cell cycle of non-small-cell lung cancer cells (15,24).
In addition, drug-resistant lung cancer cell lines with EGFR
mutations are more sensitive to cordycepin treatment than those
without EGFR mutations, and this effect may be produced through an
interaction with AMPK, whereby cordycepin activates the signaling
pathway downstream of AMPK to induce apoptosis (15). Cordycepin also inhibits pancreatic
cancer cell proliferation by targeting fibroblast growth factor
receptor 2 to block the MAPK signaling pathway (25).
In the present study, the effects of cordycepin on
NPC cells were investigated, where it was found to inhibit the
proliferation of EBV-positive NPC cells, and to augment the killing
effects of low concentrations of cisplatin. Previous studies have
indicated that cordycepin enhances the apoptotic effects of
cisplatin in head and neck tumor cell lines, and that co-treatment
with the two drugs significantly increases the cleavage of caspase
3 8 and 9, and PARP, and activates the MAPK pathway (26). Cordycepin also enhances the
chemotherapeutic effects of cisplatin against esophageal cancer.
Co-treatment inhibited cellular proliferation, migration and
metastatic capacity, and induced the apoptosis of esophageal cancer
cells by repressing the expression of p-PI3K, p-AKT, caspase 3 and
Bcl2, while activating p-AMPK, cleaved caspase 3 and Bax (27). In the current study, the underlying
mechanisms of cordycepin treatment in NPC cells were investigated
via RNA-seq analysis. GO and KEGG enrichment analyses highlighted
proliferation- and migration-related pathways, including ‘cell
adhesion molecules’, ‘VEGF signaling pathway’, and ‘regulation of
the JNK cascade’, which also suggests that cordycepin may affect
NPC cell function through these downstream pathways. qPCR analysis
validated four of the most significantly differentially expressed
genes identified from RNA-seq, which supports the reliability of
the results.
Western blot analyses revealed that at 500 µM
cordycepin, the levels of p-ERK1/2 increased, while at 750µM
cordycepin, the levels of total ERK1/2 and p-ERK1/2 were reduced.
This suggests that 750 µM cordycepin inhibits proliferation by
arresting the ERK pathway. A previous study suggested that
cordycepin induced the apoptosis of head and neck squamous cell
carcinoma cells through the phosphorylation of ERK proteins
(26), which could explain the
early increase in p-ERK1/2; the present study observations for NPC
cells at 500 µM cordycepin are consistent with these findings.
Cordycepin may retard migration by inhibiting the expression of
β-catenin, a key constituent of the Wnt signaling pathway and a
promising drug target for various cancer types (28,29).
Finally, the results of the current study revealed that when
sufficient cordycepin is applied, it inhibits the expression of
ERK1/2 and β-catenin, which represses the downstream signaling
pathway to reduce the proliferation and migration of NPC cells
(Fig. 7C).
Although natural products, such as cordycepin, show
promising anticancer potential, the majority of studies have only
considered in vitro systems. A few studies have investigated
the effects of cordycepin in cancer-bearing mice. In a human oral
squamous cell carcinoma xenograft model, cordycepin (via
intraperitoneally injection) inhibited tumor growth without
affecting weight, or the function of the liver or kidney (30). Another study that used a xenograft
model of cholangiocarcinoma demonstrated the anticancer ability of
cordycepin in vivo (31).
However, further studies investigating the efficacy of cordycepin
for treating NPC in vivo are required.
The present study only explored the effects of
cordycepin in an NPC cell line, but not in normal control cells.
Future studies will investigate how cordycepin regulates downstream
molecules in EBV-positive and -negative NPC cells, and will compare
them to normal nasopharynx epithelial cell lines via transcriptome
and proteome experiments.
In conclusion, the present study demonstrated the
anticancer effects of cordycepin in EBV-positive NPC cells. The
combination of cordycepin and cisplatin may allow NPC treatment
that goes beyond single cisplatin chemotherapy. Moreover, the
inhibitory effects of cordycepin in NPC cells resulted from the
activation of the MAPK/ERK and β-catenin signaling pathways.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 82002885
and 52007001), the China Postdoctoral Science Foundation (grant no.
2021M692159), and the Sanming Project of Medicine in Shenzhen
(grant no. SZSM201612076).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ, YL, XS and HH were responsible for study design
and conceptualization. YZ performed most of the experiments. YZ and
HH confirm the authenticity of all the raw data. YZ, XM and WY were
responsible for data analysis. YZ wrote the manuscript. XS and HH
reviewed and edited the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NPC
|
nasopharyngeal carcinoma
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
EGFR
|
epidermal growth factor receptor
|
|
NSCLC
|
non-small-cell lung cancer
|
References
|
1
|
Chen YP, Chan ATC, Le QT, Blanchard P, Sun
Y and Ma J: Nasopharyngeal carcinoma. Lancet. 394:64–80. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee AW, Ng WT, Chan LL, Hung WM, Chan CC,
Sze HC, Chan OS, Chang AT and Yeung RM: Evolution of treatment for
nasopharyngeal cancer - success and setback in the
intensity-modulated radiotherapy era. Radiother Oncol. 110:377–384.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Colevas AD, Yom SS, Pfister DG, Spencer S,
Adelstein D, Adkins D, Brizel DM, Burtness B, Busse PM, Caudell JJ,
et al: NCCN Guidelines Insights: Head and Neck Cancers, Version
1.2018. J Natl Compr Canc Netw. 16:479–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Chen L, Hu GQ, Zhang N, Zhu XD,
Yang KY, Jin F, Shi M, Chen YP, Hu WH, et al: Gemcitabine and
Cisplatin Induction Chemotherapy in Nasopharyngeal Carcinoma. N
Engl J Med. 381:1124–1135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang YM, Qiao SQ, Lu L, Chen WP, Li SL
and Qi CH: Gemcitabine combined with cisplatin vs. taxane,
cisplatin, and fluorouracil in the treatment of locally advanced
nasopharyngeal carcinoma: A retrospective case-control study. Eur
Rev Med Pharmacol Sci. 24:7655–7663. 2020.PubMed/NCBI
|
|
7
|
Lu Y, Chen D, Liang J, Gao J, Luo Z, Wang
R, Liu W, Huang C, Ning X, Liu M, et al: Administration of
nimotuzumab combined with cisplatin plus 5-fluorouracil as
induction therapy improves treatment response and tolerance in
patients with locally advanced nasopharyngeal carcinoma receiving
concurrent radiochemotherapy: A multicenter randomized controlled
study. BMC Cancer. 19:12622019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin T, Qin WF, Jiang F, Jin QF, Wei QC,
Jia YS, Sun XN, Li WF and Chen XZ: Cisplatin and Fluorouracil
Induction Chemotherapy With or Without Docetaxel in Locoregionally
Advanced Nasopharyngeal Carcinoma. Transl Oncol. 12:633–639. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Y, Li S, Meng X, Gan RY, Zhang JJ and
Li HB: Dietary Natural Products for Prevention and Treatment of
Breast Cancer. Nutrients. 9:E7282017. View Article : Google Scholar
|
|
10
|
Luo H, Vong CT, Chen H, Gao Y, Lyu P, Qiu
L, Zhao M, Liu Q, Cheng Z, Zou J, et al: Naturally occurring
anti-cancer compounds: Shining from Chinese herbal medicine. Chin
Med. 14:482019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tuli HS, Sharma AK, Sandhu SS and Kashyap
D: Cordycepin: A bioactive metabolite with therapeutic potential.
Life Sci. 93:863–869. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu X, Shen H, Yin X, Long L, Xie C, Liu
Y, Hui L, Lin X, Fang Y, Cao Y, et al: miR-186 regulation of Twist1
and ovarian cancer sensitivity to cisplatin. Oncogene. 35:323–332.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo Z, Chen W, Dai G and Huang Y:
Cordycepin suppresses the migration and invasion of human liver
cancer cells by downregulating the expression of CXCR4. Int J Mol
Med. 45:141–150. 2020.PubMed/NCBI
|
|
14
|
Ho SY, Wu WS, Lin LC, Wu YH, Chiu HW, Yeh
YL, Huang BM and Wang YJ: Cordycepin Enhances Radiosensitivity in
Oral Squamous Carcinoma Cells by Inducing Autophagy and Apoptosis
Through Cell Cycle Arrest. Int J Mol Sci. 20:E53662019. View Article : Google Scholar
|
|
15
|
Wei C, Yao X, Jiang Z, Wang Y, Zhang D,
Chen X, Fan X, Xie C, Cheng J, Fu J, et al: Cordycepin Inhibits
Drug-resistance Non-small Cell Lung Cancer Progression by
Activating AMPK Signaling Pathway. Pharmacol Res. 144:79–89. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khan MA and Tania M: Cordycepin in
Anticancer Research: Molecular Mechanism of Therapeutic Effects.
Curr Med Chem. 27:983–996. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yoon SY, Park SJ and Park YJ: The
Anticancer Properties of Cordycepin and Their Underlying
Mechanisms. Int J Mol Sci. 19:E30272018. View Article : Google Scholar
|
|
18
|
Knapek KJ, Georges HM, Van Campen H,
Bishop JV, Bielefeldt-Ohmann H, Smirnova NP and Hansen TR: Fetal
Lymphoid Organ Immune Responses to Transient and Persistent
Infection with Bovine Viral Diarrhea Virus. Viruses. 12:E8162020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW,
Tsang YS, Wong N, Whitney BM and Lee JC: Nasopharyngeal carcinoma
cell line (C666-1) consistently harbouring Epstein-Barr virus. Int
J Cancer. 83:121–126. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang MM, Hong SY, Yang SH, Wu CC, Wang CY
and Huang BM: Anti-Cancer Effect of Cordycepin on FGF9-Induced
Testicular Tumorigenesis. Int J Mol Sci. 21:E83362020. View Article : Google Scholar
|
|
21
|
Wang Z, Wu X, Liang YN, Wang L, Song ZX,
Liu JL and Tang ZS: Cordycepin Induces Apoptosis and Inhibits
Proliferation of Human Lung Cancer Cell Line H1975 via Inhibiting
the Phosphorylation of EGFR. Molecules. 21:E12672016. View Article : Google Scholar
|
|
22
|
Chen Y, Yang SH, Hueng DY, Syu JP, Liao CC
and Wu YC: Cordycepin induces apoptosis of C6 glioma cells through
the adenosine 2A receptor-p53-caspase-7-PARP pathway. Chem Biol
Interact. 216:17–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee SY, Debnath T, Kim SK and Lim BO:
Anti-cancer effect and apoptosis induction of cordycepin through
DR3 pathway in the human colonic cancer cell HT-29. Food Chemical
Toxicol. 60:439–447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tao X, Ning Y, Zhao X and Pan T: The
effects of cordycepin on the cell proliferation, migration and
apoptosis in human lung cancer cell lines A549 and NCI-H460. J
Pharm Pharmacol. 68:901–911. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li XY, Tao H, Jin C, Du ZY, Liao WF, Tang
QJ and Ding K: Cordycepin inhibits pancreatic cancer cell growth in
vitro and in vivo via targeting FGFR2 and blocking ERK signaling.
Chin J Nat Med. 18:345–355. 2020.PubMed/NCBI
|
|
26
|
Chen YH, Wang JY, Pan BS, Mu YF, Lai MS,
So EC, Wong TS and Huang BM: Cordycepin enhances cisplatin
apoptotic effect through caspase/MAPK pathways in human head and
neck tumor cells. OncoTargets Ther. 6:983–998. 2013.PubMed/NCBI
|
|
27
|
Gao Y, Chen DL, Zhou M, Zheng ZS, He MF,
Huang S, Liao XZ and Zhang JX: Cordycepin enhances the
chemosensitivity of esophageal cancer cells to cisplatin by
inducing the activation of AMPK and suppressing the AKT signaling
pathway. Cell Death Dis. 11:8662020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krishnamurthy N and Kurzrock R: Targeting
the Wnt/beta-catenin pathway in cancer: Update on effectors and
inhibitors. Cancer Treat Rev. 62:50–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Su NW, Wu SH, Chi CW, Liu CJ, Tsai TH and
Chen YJ: Metronomic Cordycepin Therapy Prolongs Survival of Oral
Cancer-Bearing Mice and Inhibits Epithelial-Mesenchymal Transition.
Molecules. 22:E6292017. View Article : Google Scholar
|
|
31
|
Liu T, Zhu G, Yan W, Lv Y, Wang X, Jin G,
Cui M, Lin Z and Ren X: Cordycepin Inhibits Cancer Cell
Proliferation and Angiogenesis through a DEK Interaction via ERK
Signaling in Cholangiocarcinoma. J Pharmacol Exp Ther. 373:279–289.
2020. View Article : Google Scholar : PubMed/NCBI
|