Introduction
Oral squamous cell carcinoma (OSCC) accounts for
>90% of the total oral cancer cases (1). The prognosis of patients with OSCC is
affected by tumor recurrence and metastasis; metastasis is the main
causative factor for mortality in 90% of all cancer-related deaths
(1). Cancer stem cells (CSCs) are
implicated in tumor progression, metastasis, recurrence and the
high mortality rate (2).
CSCs are a minority subpopulation of cells within a
tumor that can sustain long-term tumor maintenance and growth
through their ability of self-renewal and their high
differentiation potential (3).
CSCs have been investigated as prognostic markers in OSCC (4), and are considered the main factor
responsible for the failure of traditional anticancer therapies
(such as surgery, radiotherapy, chemotherapy or combined
therapies). Tumor recurrence is a major issue in OSCC therapy and
the elimination of CSCs is a target of therapies (5). Thus, various antitumor strategies
have been developed to eliminate CSCs, including focusing on the
identification of specific CSC markers, such as aldehyde
dehydrogenase (ALDH), CD44, CD133, octamer-binding transcription
factor 4 (Oct4), sex-determining region Y-box 2 (Sox2), homeobox
protein NANOG, keratins, E-cadherin and integrins, phosphorylated
STAT3 and spalt-like transcription factor 4 (6). Other studies have focused on
molecular signaling pathways involved in the CSC niche, such as
autophagy, apoptosis or the differentiation of CSCs (7–9). The
search for novel cancer therapeutic strategies is still
ongoing.
Zinc-finger protein 750 (ZNF750) is a potential
antitumor gene that may be used to eliminate CSCs. ZNF750 has been
validated as a lineage-specific tumor suppressor gene in squamous
cell carcinoma (10). Previous
studies have revealed that the decreased or deleted expression of
ZNF750 is responsible for cell proliferation, invasion and
migration in esophageal squamous cell carcinoma and OSCC (11,12).
Furthermore, the overexpression of ZNF750 has been demonstrated to
decrease the cancer stem cell marker CD44+ cell
population in OSCC (11). In
addition, RNA sequence analysis (GSE134835) has revealed that
ZNF750 functions as a tumor suppressor gene, suppressing the
expression of histone lysine methyltransferase enhancer of zeste
homolog 2 (Ezh2) in the CAL-27 OSCC cell line (13). Ezh2 has been implicated in
promoting cancer cell proliferation, migration, invasion and
metastasis, as well as CSC self-renewal (14).
However, to the best of our knowledge, the effects
of ZNF750 on Ezh2-mediated CSC renewal have not yet been
investigated. The identification of novel antitumor strategies with
which to reduce or eliminate CSCs would be invaluable for cancer
treatment. CSC-like cells comprise only a fraction of the total
tumor cell population, which display stem cell-like characteristics
with an efficient self-renewal ability and can be defined by
functional analysis (15).
Therefore, the present study aimed to examine the effects of ZNF750
on the renewal and invasive abilities of CSC-like cells derived
from OSCC cells.
Materials and methods
Cell lines and plasmids
The present study used 293T cells and the OSCC cell
line CAL-27 purchased from Shanghai Zhong Qiao Xin Zhou
Biotechnology Co., Ltd. and Procell Life Science & Technology
Co., Ltd., respectively. OSCC cell line TCA-83 was provided from Dr
Zhen Meng (Peking University School and Hospital of Stomatology,
Beijing, China). All experiments were performed with
mycoplasma-free cells. The cells were cultured in DMEM complemented
medium with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin, followed by incubation at 37°C in a
humidified incubator containing 5% CO2. The lentiviral
packaging plasmids, psPax2 and pRSV-Rev, and the VSV-G
envelope-encoding plasmid, pMD2.G, were provided from Dr Padraig
Strappe (Central Queensland University, Queensland, Australia). The
pLVX-PGK-Puro lentiviral vector backbone (oe-control) and
lentiviral vector pLVX-ZNF750-PGK-Puro (oe-ZNF750) were purchased
from Biowit Technologies, Ltd. The Hu6-CMV-puro lentiviral vector
backbone (sh-control) and Hu6-shZNF750-CMV-puro (sh-ZNF750), which
were used to knockdown the ZNF750 gene, were purchased from
Shanghai GeneChem Co., Ltd. The lentiviral vector backbones
(oe-control and sh-control) were used as a control for
investigating the changes in the oe-ZNF750 and sh-ZNF750 groups,
respectively.
CSC-like cell enrichment and
identification
CSCs in OSCC are possibly isolated via cell-surface
markers or specific practical features. Yet, no specific marker or
CSC feature can specifically isolate oral CSC populations from OSCC
(5). Spheroid assays are used to
determine the self-renewal ability of CSCs (16). It has been reported that tumor
spheres extracted from OSCC cells exhibit higher stem-like features
(5), for example they show greater
volume of expression of pluripotent transcription agents, such as
Oct2, Sox2, Nanog and Kruppel-like factor 4 (17,18).
Therefore, in the present study, CSC-like cells were enriched from
tumor spheres following several cycles of passage.
For spheroid formation assay, CAL-27 cells (5,000
cells/well) and TCA-83 cells (5,000 cells/well) was cultured in an
ultra-low adherent six-well plate (Corning, Inc.) with serum-free
DMEM (Boster Biological Technology) supplemented with B27
serum-free supplements (1:50; Invitrogen; Thermo Fisher Scientific,
Inc.), 20 ng/ml basic fibroblast growth factor and 20 ng/ml human
recombinant epidermal growth factor (all from PeproTech, Inc.). The
first-generation spheroids were dissociated to single cells, and
reseeded for a second and third round of spheroid formation to
obtain CSC-like cells (the five cycles of passage cells were used
in the following study). The spheroid formation was imaged and
counted under a light microscope (CKX71; Olympus Corporation)
following 7 days of culture. The rate of tumor sphere
formation=number of tumor spheres/input cells ×100%. Following
tertiary generation sphere formation, tumor spheres were
dissociated using trypsin digestion and the cells were re-plated to
verify whether the enriched CSC-like cells from the tumor spheres
exhibited stem cell properties. In addition, the effects of ZNF750
on ALDH activity, cell viability and CSC markers were further
investigated. For investigating the effects of ZNF750 on tumor
malignant biological behavior, CSC-like cells enriched from CAL-27
cells were used in the corresponding experiments.
Lentiviral gene transfer
The lentiviral particles were produced by using the
3rd generation system as previously described (11). Briefly, the three plasmids 7 µg
psPax2, 3 µg pRSV-Rev and 3 µg VSV-G were co-transfected with a
lentiviral vector (7 µg) into 293T cells using Lipofectamine
2000® (Thermo Fisher Scientific, Inc.). Lipofectamine
2000®/DNA complexes (2.5:1) were added to the 293T cells
with the addition of caffeine (final concentration, 4 mM) to
achieve a higher titer lentivirus (19). The lentivirus-containing
supernatant was harvested at 48 and 72 h post-transfection,
centrifuged at 4°C, 1,751 × g for 10 min, filtered through the
Steriflip-HV0.45 µm PVDF Filter Unit (MilliporeSigma) and
concentrated using the one-step virus concentration solution,
PEG-it™ (System Biosciences, LLC). The pellet was resuspended in
cold PBS. The CSC-like cells were randomly divided into four groups
(oe-control, oe-ZNF750, sh-control and sh-ZNF750 groups),
transduced with viral stock (at MOI of 10) that expressed ZNF750 or
silenced the ZNF750 gene, and supplemented with polybrene (3 µg/ml,
Sigma-Aldrich; Merck KGaA) for 6 h. Cells were collected for
subsequent experimentation after transduction 5 days later.
Flow cytometry analysis
For the investigation of ALDH activity in the
CSC-like cells and the effects of ZNF750, the cells were stained
with Aldefluor™ reagent (Stemcell Technologies, Inc.) according to
the manufacturer's recommendations. The Aldefluor™ reagent system
was used to identify stem/progenitor cells on the basis of their
ALDH activity; cells expressing high levels of ALDH become brightly
fluorescent (ALDHbr) and can be identified and enumerated using a
flow cytometer. Briefly, 1×106 cells were suspended in 1
ml Aldefluor assay buffer containing the ALDH substrate, with or
without the ALDH specific inhibitor, diethylaminobenzaldehyde
(DEAB). DEAB is used to control for background fluorescence. The
cells were then incubated at 37°C for 40 min. ALDH+
cells were analyzed using a flow cytometer (FACSCalibur; BD
Biosciences) to compare the fluorescence intensity with the ALDH
inhibitor DEAB-treated negative control. BD FACSDiva software
version 8.0.1 (BD Biosciences) was used for analysis. A high
fluorescence was associated with a high ALDH activity
(ALDH+ cells).
Colony formation assay
To evaluate the effects of ZNF750 on the
self-renewal and tumor propagation potency of CSC-like cells
(enriched from CAL-27 cells), colony formation assays were
performed. The cells were seeded in six-well plates at a low
density (500 cells/well) in triplicate, and cultured at 37°C for 10
days. The plates were then washed with PBS and fixed with 4%
paraformaldehyde at room temperature for 30 min, followed by
staining with 0.5% crystal violet at room temperature for 1 min.
The plates were then washed with PBS, and images of each well were
captured (Nikon Corporation) and the colonies consisting of >50
cells were counted using AlphaView version 3.4.0 (ProteinSimple).
The experiment was repeated three times.
Cell viability assay
The effects of ZNF750 on the viability of the oral
CSC-like cells were detected using a Cell Counting Kit-8 (CCK-8;
Beyotime Institute of Biotechnology) assay according to the
manufacturer's instructions. Briefly, CCK-8 solution (50 µl) was
added to each well (1×105/ml) during the 2 h of culture
at 37°C, and the absorbance in each well at 48 and 72 h was
measured at 450 nm using a 96-well Multiskan MK3 microplate reader
(Thermo Fisher Scientific, Inc.). The experiment was repeated three
times.
cDNA preparation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the parental cells
(CAL-27 and TCA-83 cells) and CSC-like cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.). cDNA
was synthesized from 1 µg total RNA using the PrimeScript™ RT
reagent kit with gDNA Eraser, according to the manufacturer's
instructions, and amplified using the SYBR® Premix Ex
Taq™ II kit (all from Takara Biotechnology Co., Ltd.) on an ABI
7500 Sequence Detection System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). qPCR was performed to examine the expression of
stemness factors (Oct4 and Sox2), Ezh2, embryonic ectoderm
development (EED), SUZ12 polycomb repressive complex 2 subunit
(SUZ12), metastasis-related genes (MMP1, 3 and 13) and tissue
inhibitors of matrix metalloproteinases (TIMP1 and 2). All PCR
primers were designed for SYBR Green-based qPCR detection and
purchased from Shanghai Shenggong Biology Engineering Technology
Service, Ltd. The sequences of the PCR primers are presented in
Table I. The thermocycling
conditions were as follows: Initial incubation at 95°C for 30 sec,
then 40 cycles alternating in turn at 95°C for 5 sec and 60°C for
34 sec. Comparative gene expression analysis was performed using
the 2−ΔΔCq method (20)
with normalization to the level of the internal control gene,
β-actin (21).
 | Table I.Sequences of primers. |
Table I.
Sequences of primers.
| Gene | Ref seq accession
number | Direction | Sequence
(5′-3′) |
|---|
| ZNF750 | NM_024702.3 | Forward |
GCCACCATCTACTCGCCTTA |
|
|
| Reverse |
GCAGGAAGTGTCTCGGGTCT |
| Oct4 | NM_002701.6 | Forward |
GTGAGAGGCAACCTGGAGAA |
|
|
| Reverse |
AACCACACTCGGACCACATC |
| Sox2 | NM_003106.4 | Forward |
CACAACTCGGAGATCAGCAA |
|
|
| Reverse |
TATAATCCGGGTGCTCCTTC |
| MMP1 | NM_002421.4 | Forward |
TGGGCTGAAAGTGACTGGGA |
|
|
| Reverse |
ATGGCATGGTCCACATCTGC |
| MMP3 | NM_002422.5 | Forward |
GGCCAGGGATTAATGGAGAT |
|
|
| Reverse |
TGAAAGAGACCCAGGGAGTG |
| MMP13 | NM_002427.3 | Forward |
CACTTTATGCTTCCTGATGACG |
|
|
| Reverse |
TCTGGCGTTTTTGGATGTTTAG |
| TIMP1 | NM_003254.3 | Forward |
CATCACTACCTGCAGTTTTGTG |
|
|
| Reverse |
TGGATAAACAGGGAAACACTGT |
| TIMP2 | NM_003255.5 | Forward |
GTCACAGAGAAGAACATCAACG |
|
|
| Reverse |
GATGTCGAGAAACTCCTGCTT |
| Ezh2 | NM_004456.4 | Forward |
AAATCAGAGTACATGCGACTGA |
|
|
| Reverse |
GTATCCTTCGCTGTTTCCATTC |
| EED | NM_003797 | Forward |
GTCCTGTGGTATGGATCATTCT |
|
|
| Reverse |
GTATCAAATCGCCTAACCATCG |
| SUZ12 | NM_015355 | Forward |
CAAACTGAAGCAAGAGATGACC |
|
|
| Reverse |
GCTATGGCAGAGTTTAAGATGC |
| GAPDH | NM_002046.5 | Forward |
TGCACCACCAACTGCTTAGC |
|
|
| Reverse |
GGCATGGACTGTGGTCATGAG |
Western blotting
Total protein was extracted from each group via
homogenization in ice-cold RIPA lysis buffer containing
phenylmethanesulfonyl fluoride (PMSF) (all from Beyotime Institute
of Biotechnology). Protein quantitation was performed by using a
bicinchoninic acid assay. Equal amounts of protein (15 µg) were
subjected to 10% SDS-PAGE and were transferred to polyvinylidene
difluoride (PVDF) membranes (MilliporeSigma) after electrophoresis.
Non-specific bindings to the membranes were blocked with 5% skimmed
milk in Tris-buffered saline with 0.05% Tween-20 at room
temperature for 1 h, followed by incubation with the following
antibodies: Anti-ZNF750 (1:1,000; cat. no. ab121685; Abcam),
anti-Ezh2 (1:1,000; cat. no. ab191250; Abcam), anti-MMP9 (1:500;
cat. no. 27306-1-AP; ProteinTech Group, Inc.) and monoclonal
antibody anti-β-actin (1:10,000; cat. no. 66009-1; ProteinTech
Group, Inc.) overnight at 4°C. The appropriate secondary antibodies
used were goat anti-mouse HRP-conjugated (1:10,000; cat. no.
SA00001-1; ProteinTech Group, Inc.) or goat anti-rabbit
HRP-conjugated (1:10,000; cat. no. SA00001-2; ProteinTech Group,
Inc.) at 4°C for 1 h. All primary and secondary antibodies were
diluted in TBS containing 0.1% Tween-20 at 4°C. The blots were
developed using the ECL western blotting kit (MilliporeSigma). The
semi-quantification of the protein bands was performed using the
AlphaView analysis system, version 3.4.0 (ProteinSimple). The
values of protein expression were normalized against β-actin.
Cell invasion, migration and wound
healing assays
To examine the effects of ZNF750 on the invasion of
oral CSC-like cells, the Transwell system with a filter
(polycarbonate membrane, 8-µm pore size) was used with Matrigel (BD
Biosciences) as the substrate for invasion. The cells
(1×105/ml) were dispersed onto a Matrigel-coated
polycarbonate membrane at a dilution of 1:6 in serum-free culture
DMEM (Boster Biological Technology) in the upper chamber. The lower
chamber comprised of DMEM containing 10% FBS. Cell invasion was
allowed to proceed for 24 h at 37°C in 5% CO2. After 24
h, the cells remaining on the upper surface of the filter
(non-invading cells) were removed using cotton swabs. Cells that
had invaded to the lower surfaces of the membrane were fixed with
4% formaldehyde at room temperature for 15 min and stained with
0.5% crystal violet (Sigma-Aldrich; Merck KGaA) for 1 min at room
temperature. Filters were visualized under a light microscope
(CKX71; Olympus Corporation). The assays were performed in
triplicate and the average numbers of invaded cells per field (five
randomly selected fields were counted) were assessed using
Image-Pro Plus 6 software (National Institutes of Health). Compared
with the cell invasion assay, the vertical cell migration assay
used a similar approach without Matrigel coating.
To evaluate the effects of ZNF750 on the lateral
migratory capacity of CSCs, a wound healing assay was performed.
The CSC-like cells (enriched from CAL-27 cells,
2×105/well) were cultured with DMEM medium containing 2%
FBS and seeded in six-well plates and cultured to almost 80%
confluency. The similar distance linear scratch wounds in each
group were created on the cell monolayer using a sterile 200 µl
pipette tip at 0 h, and the cells were then rinsed with PBS and
incubated at 37°C for 24 h. To visualize migrating cells and wound
healing, images were acquired using a light microscope at 0 and 24
h after scratching. Cell migration characteristics were measured
and analyzed using the manual tracking tool of Image-Pro Plus 6
software (National Institutes of Health) and calculated as a
percentage of wound healing according to the equation: Wound
healing (%)=[1-(wound area at T24 h/wound area at T0 h)] ×100%,
where T0 is the time immediately after wounding and T24 is the
detection time point. The cells in three wells of each group were
quantified. In total, three independent experiments were
performed.
Statistical analysis
Values are presented as the means ± SD of at least
three independent experiments. Data were analyzed using SPSS 23.0
software (SPSS, Inc.) with an unpaired Student's t-test for
differences between two groups. When comparing >2 groups, data
were analyzed using one-way ANOVA followed by a Tukey's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
CSC-like cell properties
The stem cell properties of CSC-like cells enriched
from CAL-27 and TCA-83 cells were investigated. Tumor spheres
exhibit higher levels of CSC-specific markers, including ALDH, and
higher expression levels of pluripotent transcription factors, such
as Oct4 and Sox2 (5). In the
present study, the spheroid assay indicated that the tumor sphere
formation abilities of the cells were enhanced after several cycles
of passage, demonstrated by the more rapid and larger sphere
formation at the same culture time points (Fig. 1A). With DEAB (ALDH inhibitor), the
ALDH activity was 0±0% and 0.1±0.1% in the parental cells (CAL-27
cells) and CSC-like cells, respectively. Without DEAB, the ALDH
activity was 0.1±0.1% in the CAL-27 cells, whereas it was increased
(17.8±1.0%) in the CSC-like cells (enriched from CAL-27 cells;
Fig. 1B; upper panel). Similar
results were also revealed in CSC-like cells enriched from TCA-83
cells without DEAB, as indicated by increased ALDH activity
(17.3±0.9%) compared with the TCA-83 cells (0.1±0.1%) (Fig. 1B; lower panel). Furthermore,
compared with their matched parental cells, the oral CSC-like cells
derived from tumor spheres exhibited a significantly increased cell
viability (Fig. 1C), as well as a
significantly increased Oct4 and Sox2 expression levels (Fig. 1D).
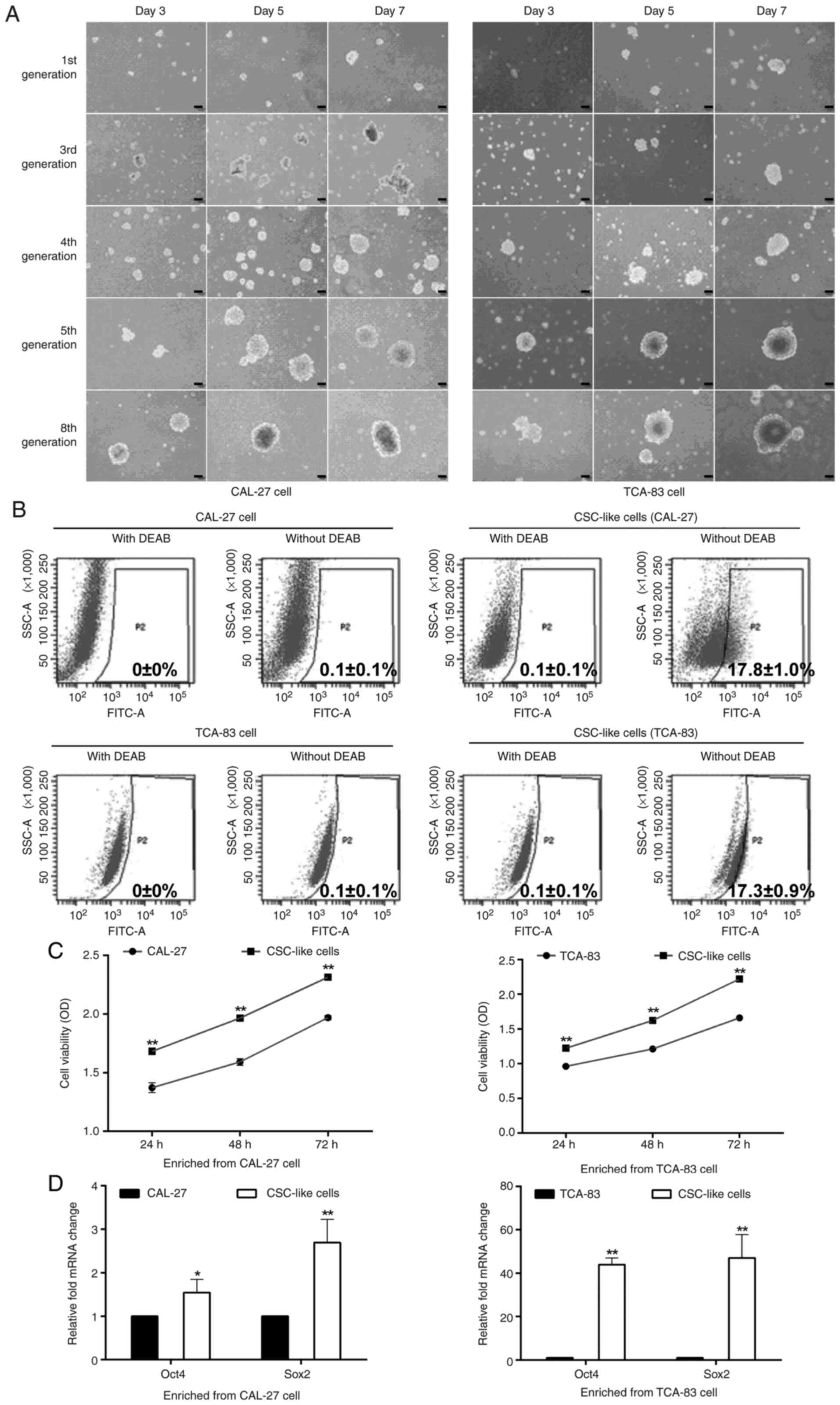 | Figure 1.Identification of CSC-like cells. (A)
Tumor sphere formation. At the same culture time points, tumor
sphere formation was enhanced after several cycles of passage.
Scale bar, 100 µm. (B) ALDH activity. ALDH+ cells (P2)
were analyzed using flow cytometry. CSC-like cells exhibited a high
ALDH activity. A specific inhibitor of ALDH, DEAB, was used to
control for background fluorescence. CAL-27 cells and TCA-83 cells
were the parental cells. (C) Viability of the parental cells and
CSC-like cells. (D) mRNA expression levels of Oct4 and Sox2.
*P<0.05 and **P<0.01 vs. parental cells. CSC, cancer stem
cell; ALDH, aldehyde dehydrogenase; DEAB, diethylaminobenzaldehyde;
OD, optical density; Oct4, octamer-binding transcription factor 4;
Sox2, sex-determining region Y (SRY)-box 2. |
ZNF750 decreases the percentage of
ALDH+ cells
A high ALDH activity can be used to identify
CSC-like cells characterized by a significantly higher
proliferation rate (22). In the
present study, to demonstrate the inhibitory effects of ZNF750 on
CSC maintenance, the CSC-like cells (enriched from CAL-27 cells)
were transduced with a lentiviral vector encoding the ZNF750 gene;
a significant decrease in the number of ALDH+ cells from
15.9±0.6% (oe-control group) to 11.7±0.5% (oe-ZNF750 group) in the
CSC-like cell population without DEAB was observed. Nevertheless,
the knockdown of the ZNF750 gene increased the number of
ALDH+ cells from 13.9±0.9% (sh-control group) to
23.0±2.5% (sh-ZNF750 group) in the CSC-like cell population without
DEAB (Fig. 2A and B). The similar
results were also revealed in CSC-like cells which enriched from
TCA-83 cells (Fig. 2A and C).
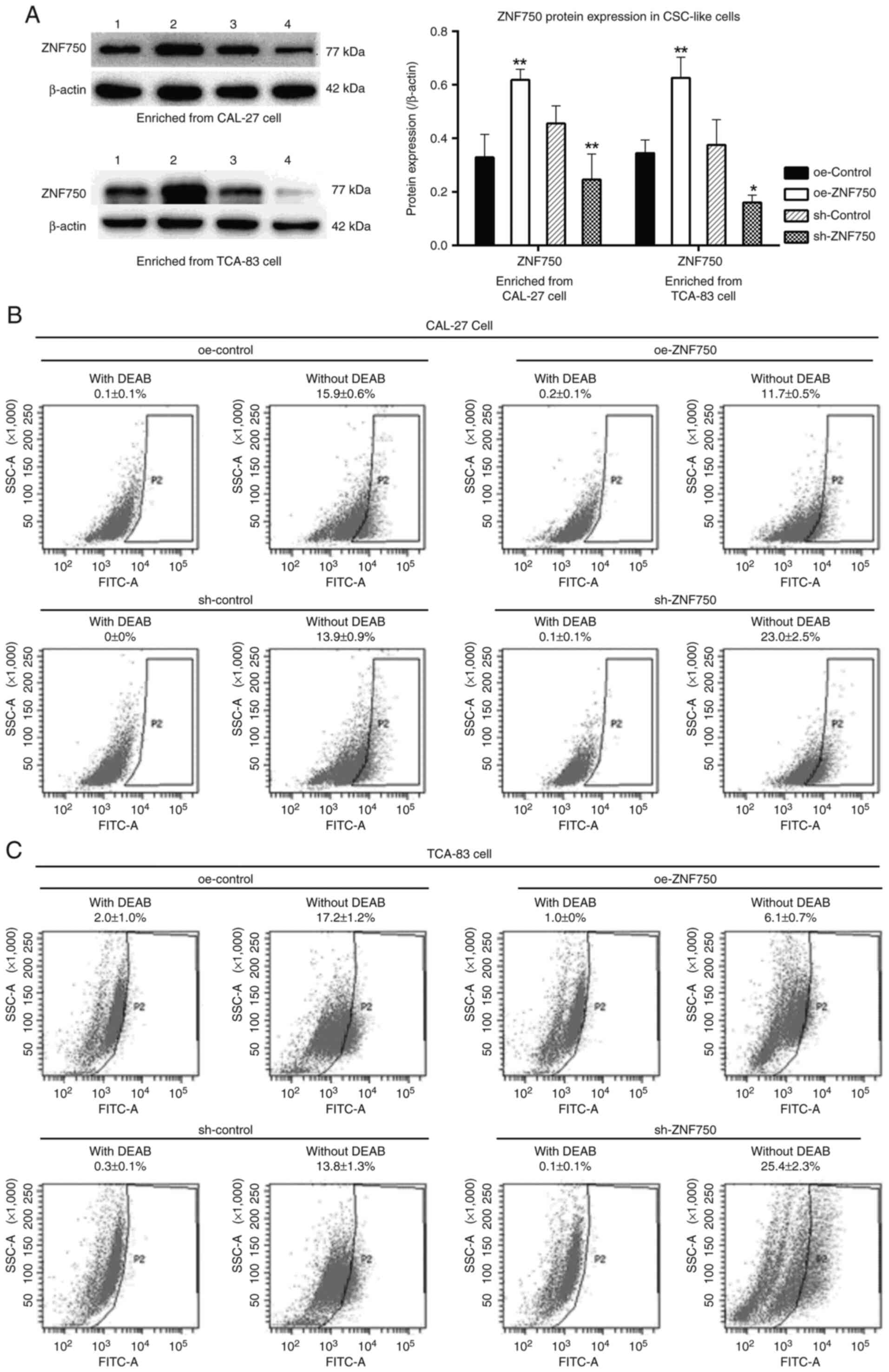 | Figure 2.Inhibitory effects of ZNF750 on the
ALDH activity of oral CSC-like cells. (A) ZNF750 protein expression
level. 1, oe-control group; 2, oe-ZNF750 group; 3, sh-control
group; 4, sh-ZNF750 group. (B) ALDH activity in CSC-like cells
(enriched from CAL-27 cells). (C) ALDH activity in CSC-like cells
(enriched from TCA-83 cells). ZNF750 effectively eliminated the CSC
specific marker, ALDH. ALDH+ cells (P2) were analyzed
using flow cytometry. ALDH specific inhibitor, DEAB, was used to
control for background fluorescence. Cells were treated with or
without DEAB. *P<0.05, **P<0.01 vs. oe-control or sh-control
group. ZNF750, zinc-finger protein 750; ALDH, aldehyde
dehydrogenase; CSC, cancer stem cell; oe-, overexpression; sh-,
short hairpin; DEAB, diethylaminobenzaldehyde. |
ZNF750 inhibits the self-renewal
ability of oral CSC-like cells
Tumor sphere- and colony-formation assays were
performed to evaluate the self-renewal ability and tumor
propagation potency of oral CSC-like cells (enriched from CAL-27
cells). Compared with the oe-control group, the tumor sphere and
colony formation in the oe-ZNF750 group was significantly
decreased, characterized by the decreased number and size of cell
spheres and colonies. However, the knockdown of the ZNF750 gene
significantly enhanced the number and size of the cell spheres and
colonies (Fig. 3A and B).
Furthermore, cell viability was decreased by ~20% in the oe-ZNF750
group at 24, 48 and 72 h compared with the oe-control group;
however, it was increased by 1.24-, 1.31- and 1.30-fold at 24, 48
and 72 h, respectively, in the sh-ZNF750 group compared with the
sh-control group (Fig. 3C). The
increased cell viability observed in the sh-ZNF750 group suggested
that ZNF750 exerts inhibitory effects on the self-renewal ability
of oral CSC-like cells.
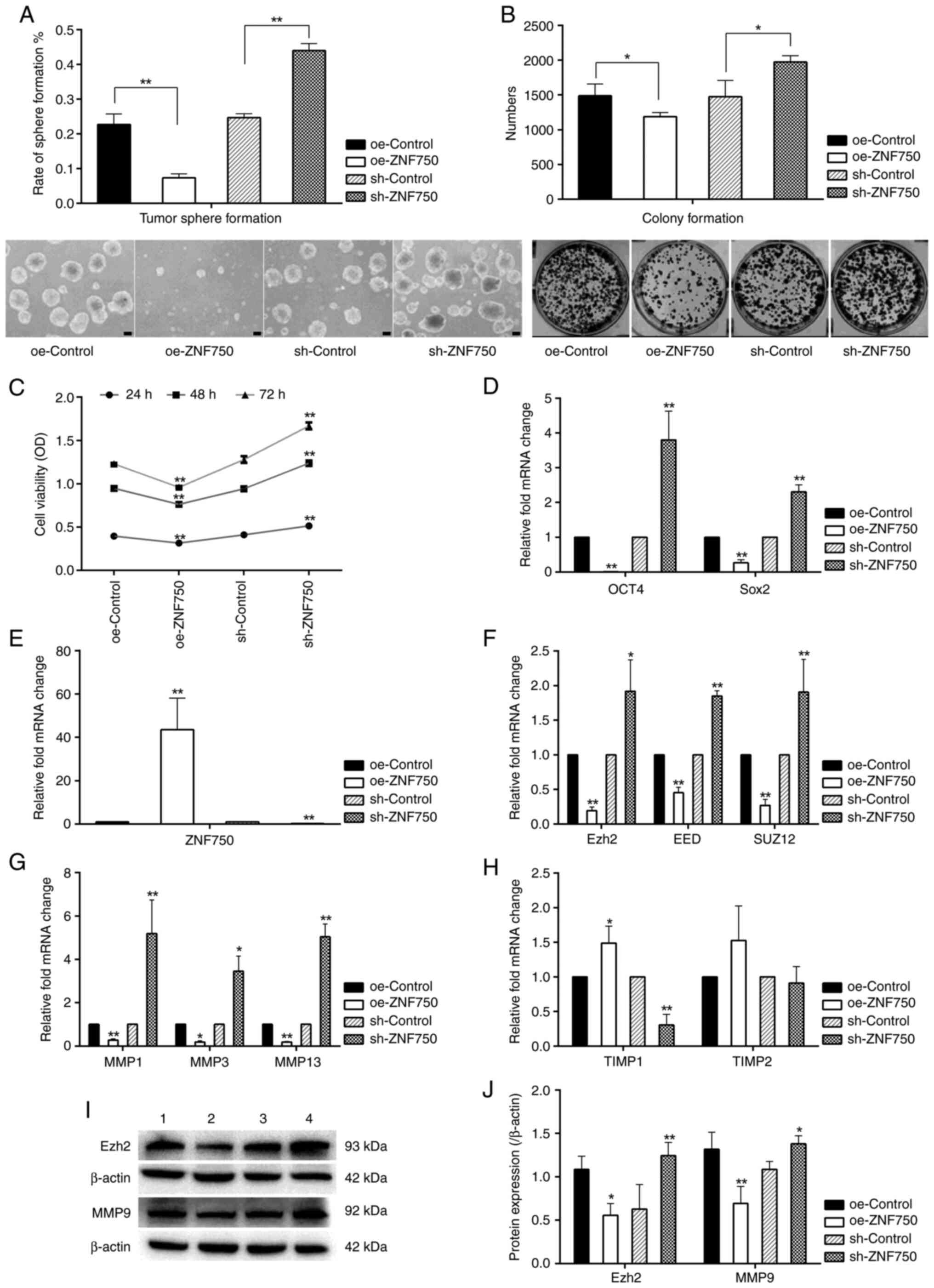 | Figure 3.Regulatory effects of ZNF750 on stem
cell properties, Ezh2, SUZ12 and EED and metastasis-related genes.
(A) Tumor sphere and (B) colony formation ability. ZNF750
significantly suppressed the ability of tumor sphere (Scale bar,
100 µm) and colony formation (magnification, ×1). (C) Effect of
ZNF750 on cell viability. (D) mRNA expression profiles of stemness
factors (Oct4 and Sox2). (E) Verification of ZNF750 mRNA expression
in the oe-ZNF750 and sh-ZNF750 groups. mRNA expression profiles of
(F) epigenetic regulatory genes (Ezh2, SUZ12 and EED), and
metastasis-related genes (G) MMP1, MMP3, MMP13 and (H) TIMP1 and
TIMP2. (I and J) Ezh2 and MMP9 protein expression. 1, oe-control
groups; 2, oe-ZNF750 groups; 3, sh-control groups; 4, sh-ZNF750
groups. *P<0.05 and **P<0.01 vs. oe-control or sh-control
group. ZNF750, zinc-finger protein 750; Ezh2, enhancer of zeste
homolog 2; SUZ12, SUZ12 polycomb repressive complex 2 subunit; EED,
embryonic ectoderm development; Oct4, octamer-binding transcription
factor 4; Sox2, sex-determining region Y (SRY)-box 2; oe-,
overexpression; sh-, short hairpin; TIMP, tissue inhibitors of
matrix metalloproteinases. |
Effect of ZNF750 on stemness-related
transcription factors
To determine the effects of ZNF750 on cancer
stemness, the transcriptional activities of the genes encoding Oct4
and Sox2 were investigated using RT-qPCR. The expression levels of
Oct4 and Sox2 were significantly increased in the sh-ZNF750 group
by ~3.8- and 2.3-fold, respectively, compared with the sh-control
group, although it was significantly reduced in the oe-ZNF750 group
compared with the oe-control group (Fig. 3D).
ZNF750 decreases the expression of
genes associated with epigenetic regulation and metastasis
The present study revealed that ZNF750 mRNA and
protein was effectively overexpressed or silenced in the oe-ZNF750
and sh-ZNF750 groups, respectively (Figs. 2A and 3E). The mRNA expression levels of
epigenetic regulatory genes (Ezh2, EED and SUZ12) were
significantly decreased in the oe-ZNF750 group compared with the
oe-Control. However, this was significantly reversed by the
knockdown of the ZNF750 gene compared with the sh-Control group.
CSCs are considered to be involved in tumor metastasis (23). The expression levels of the
metastasis-related genes, MMP1, MMP3 and MMP13, were significantly
downregulated compared with the oe-Control groups; whereas that of
TIMP1 was significantly increased by ZNF750 overexpression. The
opposite effect was observed in the sh-ZNF750 group (Fig. 3F-H). With regards to TIMP2
expression, no significant differences were observed between the
groups. As the aberrant expression of Ezh2 is a frequent occurrence
in tumors (14), and TIMP1 exerts
potent inhibitory effects against MMP9 (24), Ezh2 and MMP9 protein expression
levels were investigated. The present study found that Ezh2 and
MMP9 protein expression levels were elevated in the sh-ZNF750 group
compared with the sh-control group, however, they were decreased in
the oe-ZNF750 group compared with the oe-control group (Fig. 3I and J).
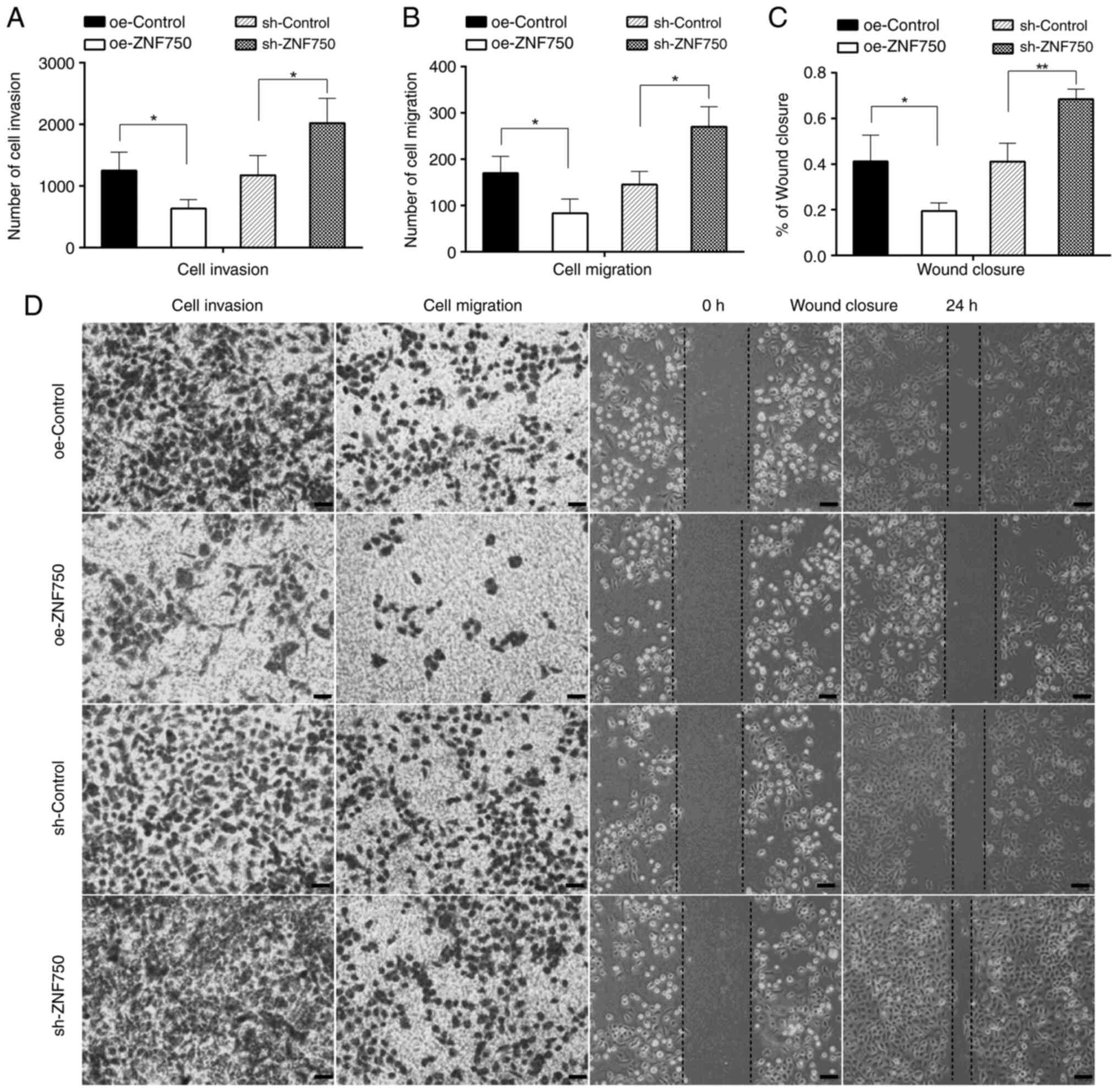
ZNF750 inhibits the invasive and
migratory abilities of CSC-like cells
In order to examine the effects of ZNF750 on the
metastatic ability of CSCs, the cell invasive and migratory
abilities were analyzed. The results revealed that the numbers of
cells that had invaded or migrated to the lower chamber of the
Transwell in the oe-ZNF750 group were significantly lower compared
with those in the oe-control group. However, the invasive and
migratory abilities were increased by 1.72- and 1.86-fold,
respectively, in the sh-ZNF750 group compared with the sh-control
group. Moreover, the wound closure ability of the CSC-like cells
was significantly reduced in the oe-ZNF750 group (19.50±0.04%)
compared with the oe-control group (41.25±0.11%). However,
knockdown of the ZNF750 gene led to a more rapid wound closure; the
wound had almost healed in the sh-ZNF750 group, in which the cells
exhibited high migration, covering 68.46±0.04% of the scratch,
which was significantly higher compared with the coverage observed
in the sh-control group (41.16±0.08%) at 24 h (Fig. 4A-D). The aforementioned findings
indicated that the ZNF750 gene significantly regulated the invasive
and migratory capacity of CSC-like cells.
Discussion
CSCs are hypothesized to be involved in tumor
metastasis and recurrence, thus contributing to the high mortality
rate of patients with cancer. Therefore, they have become a
potential target for anticancer therapy (5). The functional relevance of ZNF750 in
oral CSC-like cells has not yet been studied, at least to the best
of our knowledge. In the present study, it was revealed that ZNF750
inhibited the cancer stem cell-like behaviors of oral CSC-like
cells enriched from tumor spheres. The overexpression of ZNF750 was
efficient in repressing the ALDH-positive cell population and
impairing the self-renewal capability of CSC-like cells. It also
inhibited the expression levels of stemness factors (Oct4, Sox2),
epigenetic regulatory genes (Ezh2, SUZ12 and EED) and cell invasion
and metastasis related-genes, whereas it increased the expression
of TIMP1.
The present study revealed that ZNF750 efficiently
suppressed the stemness characteristics of CSC-like cells, as
evidenced by the reduced numbers of ALDH-positive cells. ALDH is an
intracellular enzyme involved in detoxification and drug resistance
via the oxidation of aldehydes; its activity has been widely used
as a functional stem cell marker in various cell lines, including
oral cancer cells (25). In human
head and neck squamous cell carcinoma, ALDH-positive cells have
been reported to exhibit typical CSC behavior and an increased
tumorigenic ability (26). The
findings of the present study demonstrated that ALDH activity in
CSC-like cells was reduced by the overexpression of ZNF750, whereas
it was increased following the knockdown of the ZNF750 gene. This
indicated that ZNF750 had the ability to suppress the stemness
characteristics of oral CSC-like cells.
Furthermore, both sphere and colony formation assays
have been used to evaluate the proliferative ability of CSCs by
measuring the ability of single CSCs to form spheres over multiple
passages (27). Consistent with
these findings, the present study demonstrated the potent
anti-proliferative effects of ZNF750 on oral CSC-like cells,
manifested by the inhibition of cell viability, and the tumor
sphere and colony forming abilities of the cells in the oe-ZNF750
group compared with the control group. By contrast, the
self-renewal ability of the CSC-like cells was enhanced by the
knockdown of the ZNF750 gene. This was accompanied by the loss of
their differentiated phenotype, as assessed by the increased
expression of Oct4 and Sox2, two human embryonic stem cell
pluripotency markers (28).
However, the expression levels of the stemness factors, Oct4 and
Sox2, were downregulated by ZNF750 overexpression. The expression
levels of the Oct4 and Sox2 transcription factors have been
demonstrated to be associated with metastasis and a poor prognosis
in OSCC (29). As recently
demonstrated, Sox2 expression in tumors is associated with
increased tumor invasive, metastatic and proliferative activities
in cancer, and contributes to drug resistance, tumor aggressiveness
and a poor prognosis (30). Sox2
can enhance the expression of key pluripotent factors, such as
Oct4, driving cancer cells toward a stem-like state (31). Therefore, these results indicated
that ZNF750 exerted an inhibitory effect on the stemness
characteristics of oral CSC-like cells.
Multiple molecular abnormalities may contribute to
aggressive behavior, possibly via the epigenetic regulation of CSCs
(32). The present study
demonstrated that the expression levels of Ezh2, SUZ12 and EED,
which are components of the polycomb-repressive complex 2 (PRC2),
were decreased by ZNF750 overexpression. Ezh2 is a catalytic
subunit of PRC2, and functions as an epigenetic silencer of several
tumor suppressor genes through histone 3 lysine 27 trimethylation
(H3K27me3) (33). Histone lysine
methyltransferases have been linked to the pathogenesis of cancer
(34). The aberrant expression of
Ezh2 is a frequent occurrence in multiple tumors, and there is
evidence to suggest that the upregulation of Ezh2 promotes cancer
cell proliferation, migration, invasion and metastasis, as well as
CSC self-renewal (14). The
present study revealed that oral CSC-like cells expressed higher
levels of Ezh2 in the sh-ZNF750 compared with the sh-control group.
However, the overexpression of ZNF750 decreased the Ezh2 protein
expression in oral CSC-like cells. Moreover, SUZ12 and EED mRNA
expression levels were also decreased by ZNF750 overexpression,
whereas they were increased in the sh-ZNF750 group. Therefore, the
increased PRC2 components (Ezh2, SUZ12 and EED) may trimethylate
H3K27me3, hence increasing the repression of transcription and
leading to tumor-suppressor gene inactivation in the sh-ZNF750
group. This consequently resulted in an increased self-renewal,
migratory and invasive abilities of the CSC-like cells. The
aforementioned results suggested that ZNF750 may downregulate the
expression of the epigenetic silencer, Ezh2, to suppress the
self-renewal ability of oral CSC-like cells.
The self-renewal and pluripotency properties of CSCs
are considered to enable primary tumors to metastasize (35). Activated MMPs can degrade the
highly rigid extracellular matrix and basement membrane components,
thus mediating matrix degradation and leading to tumor cell
invasion (36). All MMPs are
specifically inhibited by TIMPs, a family of proteins that includes
TIMP1, TIMP2, TIMP3 and TIMP4. The balance between MMPs and their
inhibitors is an important factor that affects tumor invasion and
metastasis (37). It has been
reported that TIMPs have the ability to both stimulate and suppress
tumor growth (24). TIMP1 exerts
potent inhibitory effects against MMP9. With regards to TIMP2, its
low expression can lead to the activation of MMP2, whereas its high
expression inhibits MMP2 activity (24). Consistent with these aforementioned
studies, the present study demonstrated that the expression levels
of the matrix remodeling genes, MMP1, MMP3, MMP9 and MMP13, were
decreased, whereas that of TIMP1 was elevated by the overexpression
of ZNF750. The present study revealed that TIMP2 expression was not
altered by ZNF750. Additionally, the CSC-like behaviors were also
inhibited by the overexpression of ZNF750, and cell invasion and
migration were attenuated in the oe-ZNF750 group. By contrast,
these effects were reversed by the knockdown of ZNF750. These
observations provided further evidence of the suppressive effects
of ZNF750 on oral CSC-like cell invasion.
In conclusion, the present study demonstrated that
ZNF750 had the ability to eradicate the cancer stemness and stem
cell-like behaviors of oral CSC-like cells enriched from parental
CAL-27 cells. However, the limitation of the present study is
absence of in vitro and in vivo data considering the
mechanism of ZNF750 on epigenetic genes and metastatic genes
regulation. Future studies are required to further investigate the
suppressive effects of ZNF750 in vivo in order to fully
elucidate the role of ZNF750 in OSCC tumor development.
Acknowledgements
The authors acknowledge the FACS technical support
from Professor Jie Ding (Liaocheng People's Hospital, Liaocheng,
China), Dr Padraig Strappe (Central Queensland University,
Queensland, Australia) for providing the lentiviral packaging
plasmids and Dr Zhen Meng (Peking Beijing University School and
Hospital of Stomatology, Beijing, China) for providing the TCA-83
cells.
Funding
This work was supported by the National Natural Science
Foundation of China (grant no. 81773759) and Science Foundation of
Liaocheng People's Hospital (grant no. LYQN201906).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HYC and LP contributed to the study conception and
design. CX, HLY, YKY, HYC and LP conducted experiments and data
analysis. CX and HYC wrote the first draft of the manuscript. HYC,
CX, HLY, YKY and LP confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Irani S and Dehghan A: The expression and
functional significance of vascular endothelial-cadherin, CD44, and
vimentin in oral squamous cell carcinoma. J Int Soc Prev Community
Dent. 8:110–117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hu J, Mirshahidi S, Simental A, Lee SC, De
Andrade Filho PA, Peterson NR, Duerksen-Hughes P and Yuan X: Cancer
stem cell self-renewal as a therapeutic target in human oral
cancer. Oncogene. 38:5440–5456. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akbari-Birgani S, Paranjothy T, Zuse A,
Janikowski T, Cieślar-Pobuda A, Likus W, Urasińska E, Schweizer F,
Ghavami S, Klonisch T and Łos MJ: Cancer stem cells,
cancer-initiating cells and methods for their detection. Drug
Discov Today. 21:836–842. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Teixeira MG and Corrêa L: Quality
assessment of prognostic studies using cancer stem cell markers in
oral squamous cell carcinoma. Appl Immunohistochem Mol Morphol.
26:e61–e69. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baniebrahimi G, Mir F and Khanmohammadi R:
Cancer stem cells and oral cancer: Insights into molecular
mechanisms and therapeutic approaches. Cancer Cell Int. 20:1132020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ram R, Brasch HD, Dunne JC, Davis PF, Tan
ST and Itinteang T: The identification of three cancer stem cell
subpopulations within moderately differentiated lip squamous cell
carcinoma. Front Surg. 4:122017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lei Y, Zhang D, Yu J, Dong H, Zhang J and
Yang S: Corrigendum to ‘targeting autophagy in cancer stem cells as
an anticancer therapy’ [Canc. Lett. 393 (2017) 33-39]. Cancer Lett.
416:1492018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Annett S and Robson T: Targeting cancer
stem cells in the clinic: Current status and perspectives.
Pharmacol Ther. 187:13–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh D, Minz AP and Sahoo SK:
Nanomedicine-mediated drug targeting of cancer stem cells. Drug
Discov Today. 22:952–959. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hazawa M, Lin DC, Handral H, Xu L, Chen Y,
Jiang YY, Mayakonda A, Ding LW, Meng X, Sharma A, et al: ZNF750 is
a lineage-specific tumour suppressor in squamous cell carcinoma.
Oncogene. 36:2243–2254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan L, Yang H, Xu C, Chen S, Meng Z, Li K
and Chen H: ZNF750 inhibited the malignant progression of oral
squamous cell carcinoma by regulating tumor vascular
microenvironment. Biomed Pharmacother. 105:566–572. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Otsuka R, Akutsu Y, Sakata H, Hanari N,
Murakami K, Kano M, Toyozumi T, Takahashi M, Matsumoto Y, Sekino N,
et al: ZNF750 expression is a potential prognostic biomarker in
esophageal squamous cell carcinoma. Oncology. 94:142–148. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu X, Yang Y, Xu C, Yang H, Chen S and
Chen H: RNA sequencing analysis of the CAL-27 cell response to
over-expressed ZNF750 gene revealed an extensive regulation on cell
cycle. Biomed Pharmacother. 118:1093772019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Beca F, Kensler K, Glass B, Schnitt SJ,
Tamimi RM and Beck AH: EZH2 protein expression in normal breast
epithelium and risk of breast cancer: Results from the nurses'
health studies. Breast Cancer Res. 19:212017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salcido CD, Larochelle A, Taylor BJ,
Dunbar CE and Varticovski L: Molecular characterisation of side
population cells with cancer stem cell-like characteristics in
small-cell lung cancer. Br J Cancer. 102:1636–1644. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miao ZF, Zhao TT, Wang ZN, Xu YY, Mao XY,
Wu JH, Liu XY, Xu H, You Y and Xu HM: Influence of different
hypoxia models on metastatic potential of SGC-7901 gastric cancer
cells. Tumour Biol. 35:6801–6808. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang
J, Zhang G, Wang X, Dong Z, Chen F and Cui H: Targeting cancer stem
cell pathways for cancer therapy. Signal Transduct Target Ther.
5:82020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kaufhold S, Garbán H and Bonavida B: Yin
Yang 1 is associated with cancer stem cell transcription factors
(SOX2, OCT4, BMI1) and clinical implication. J Exp Clin Cancer Res.
35:842016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ellis BL, Potts PR and Porteus MH:
Creating higher titer lentivirus with caffeine. Hum Gene Ther.
22:93–100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lohberger B, Rinner B, Stuendl N, Absenger
M, Liegl-Atzwanger B, Walzer SM, Windhager R and Leithner A:
Aldehyde dehydrogenase 1, a potential marker for cancer stem cells
in human sarcoma. PLoS One. 7:e436642012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoshihama R, Yamaguchi K, Imajyo I, Mine
M, Hiyake N, Akimoto N, Kobayashi Y, Chigita S, Kumamaru W,
Kiyoshima T, et al: Expression levels of SOX2, KLF4 and brachyury
transcription factors are associated with metastasis and poor
prognosis in oral squamous cell carcinoma. Oncol Lett.
11:1435–1446. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaczorowska A, Miękus N, Stefanowicz J and
Adamkiewicz-Drożyńska E: Selected matrix metalloproteinases (MMP-2,
MMP-7) and their inhibitor (TIMP-2) in adult and pediatric cancer.
Diagnostics (Basel). 10:5472020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zou B, Sun S, Qi X and Ji P: Aldehyde
dehydrogenase activity is a cancer stem cell marker of tongue
squamous cell carcinoma. Mol Med Rep. 5:1116–1120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Krishnamurthy S and Nör JE: Head and neck
cancer stem cells. J Dent Res. 91:334–340. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han J, Fujisawa T, Husain SR and Puri RK:
Identification and characterization of cancer stem cells in human
head and neck squamous cell carcinoma. BMC Cancer. 14:1732014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Swain N, Thakur M, Pathak J and Swain B:
SOX2, OCT4 and NANOG: The core embryonic stem cell pluripotency
regulators in oral carcinogenesis. J Oral Maxillofac Pathol.
24:368–373. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fu TY, Hsieh IC, Cheng JT, Tsai MH, Hou
YY, Lee JH, Liou HH, Huang SF, Chen HC, Yen LM, et al: Association
of OCT4, SOX2, and NANOG expression with oral squamous cell
carcinoma progression. J Oral Pathol Med. 45:89–95. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Basati G, Mohammadpour H and Emami Razavi
A: Association of high expression levels of SOX2, NANOG, and OCT4
in gastric cancer tumor tissues with progression and poor
prognosis. J Gastrointest Cancer. 51:41–47. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ooki A, Dinalankara W, Marchionni L, Tsay
JJ, Goparaju C, Maleki Z, Rom WN, Pass HI and Hoque MO:
Epigenetically regulated PAX6 drives cancer cells toward a
stem-like state via GLI-SOX2 signaling axis in lung adenocarcinoma.
Oncogene. 37:5967–5981. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Panaccione A, Zhang Y, Mi Y, Mitani Y, Yan
G, Prasad ML, McDonald WH, El-Naggar AK, Yarbrough WG and Ivanov
SV: Chromosomal abnormalities and molecular landscape of
metastasizing mucinous salivary adenocarcinoma. Oral Oncol.
66:38–45. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Papale M, Ferretti E, Battaglia G,
Bellavia D, Mai A and Tafani M: EZH2, HIF-1, and their inhibitors:
An overview on pediatric cancers. Front Pediatr. 6:3282018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Varier RA and Timmers HT: Histone lysine
methylation and demethylation pathways in cancer. Biochim Biophys
Acta. 1815:75–89. 2011.PubMed/NCBI
|
|
35
|
Shafiei S, Kalantari E, Saeednejad Zanjani
L, Abolhasani M, Asadi Lari MH and Madjd Z: Increased expression of
DCLK1, a novel putative CSC maker, is associated with tumor
aggressiveness and worse disease-specific survival in patients with
bladder carcinomas. Exp Mol Pathol. 108:164–172. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pickup MW, Mouw JK and Weaver VM: The
extracellular matrix modulates the hallmarks of cancer. EMBO Rep.
15:1243–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stanciu AE, Zamfir-Chiru-Anton A, Stanciu
MM, Popescu CR and Gheorghe DC: Imbalance between matrix
metalloproteinases and tissue inhibitors of metalloproteinases
promotes invasion and metastasis of head and neck squamous cell
carcinoma. Clin Lab. 63:1613–1620. 2017. View Article : Google Scholar : PubMed/NCBI
|