Introduction
Breast cancer is the most common type of cancer and
the leading cause of cancer-related mortality in women worldwide,
with an estimated 2.26 million cases and 680,000 deaths worldwide
in 2020 (1). In recent years, the
survival rates have improved due to extensive research being
conducted on the biological behavior of breast cancer cells
(2,3). However, once patients fail to respond
to the traditional treatment, their quality of life and survival
rate is significantly decreased (4). Therefore, it is important to identify
novel tumor markers and reliable prognostic indicators to guide
decision making during the treatment of breast cancer to improve
disease outcomes and survival.
Lactate dehydrogenase C4 (LDH-C4) is a tumor
testis-associated antigen, which has been discovered to play an
important role in the development of numerous different types of
tumor, such as lung cancer, breast cancer and renal cell carcinoma.
A previous study found that LDH-C4 expression was associated with
tumorigenesis and metastasis in several cancer types, including
breast cancer (5). The LDH-C4 gene
is located on chromosome 11, and is the secondary replication
product of the LDH-A gene (6).
At present, research into the regulation of the
LDH-A gene is relatively mature (7), but the specific transcriptional
regulatory mechanism of LDH-C4 remains unclear, to the best of our
knowledge. It was previously reported that the integrity of the GC
box and Cre site, SP and cAMP response element binding protein 1
transcription factors, and CpG methylation were involved in the
activation and expression of the LDH-C4 gene (8,9). DNA
methylation is an epigenetic marker involved in the process of gene
expression regulation; it is functionally equivalent to changes at
the genetic level and reverses gene silencing. Abnormal DNA
methylation plays an important role in the clinical diagnosis and
prognosis of various tumor types, such as lung, breast and prostate
cancer (10). Although numerous
studies have reported the biological functions of LDH-C4 in healthy
and cancerous tissues, to the best of our knowledge, no previous
study has investigated the methylation status of LDH-C4 in breast
cancer. DNA methylation is catalyzed by DNA methyltransferases
(DNMTs) (11). Results of a
previous study demonstrated that the expression levels of DNMTs
were significantly upregulated in a number of tumor types (11), which often preceded the abnormal
methylation pattern. The increase in DNMT activity promotes the
deamination of methylated cytosine, increases the rate of cytosine
to thymine transitions, and promotes the occurrence of point
mutations in the DNA (11). It
also promotes the hypermethylation of tumor suppressor genes,
silences their expression and inhibits their tumor-suppressive
effects (12). It was previously
demonstrated that DNMTs were abnormally expressed in breast cancer
(13). However, little research
has been conducted on the association between DNMTs and the
methylation of LDH-C4.
The present study aimed to determine the potential
of LDH-C4 gene methylation and DNMT expression to act as biomarkers
for the prognostic evaluation of breast cancer. In total, 136
breast cancer and adjacent tissues were collected, and
methylation-specific PCR (MSP) was used to detect the methylation
level of LDH-C4. Immunohistochemical (IHC) analysis was performed
to measure the protein expression levels of DNMTs. The correlation
between the methylation status of the LDH-C4 promoter and the
expression of DNMTs was analyzed, and the association between
patient clinicopathological parameters and prognosis was
explored.
Materials and methods
Patient studies
Primary breast cancer and corresponding adjacent
healthy tissues were obtained from 136 women patients with breast
cancer (age range,42–75 years old) following surgical resection at
The Third Clinical Hospital of Hebei Medical University
(Shijiazhuang, China) between July 2019 and December 2020. Data on
the clinicopathological characteristics of each patient were
obtained from the hospital records and pathological diagnosis, The
patients did not receive chemotherapy, radiotherapy, endocrine
therapy or other associated treatment before operation. The present
study was approved by the Ethics Committee of The Third Clinical
Hospital of Hebei Medical University (approval no. 2021-034-5) and
all patients provided written informed consent prior to
participation.
MSP
MSP was performed to detect the methylation status
of LDH-C4. Briefly, the genomic DNA was extracted from tissues
using the phenol chloroform method. The tissues were put into the
EP tube and lysis buffer β-mercaptoethanol and protease K were
added in a 56°C water bath. DNA extract was added, mixed well and
centrifuged (11,433 × g; 7 min; 4°C). The supernatant was collected
and added to double the volume of isopropanol at- 20°C for 10 min,
and centrifuged again (11,433 × g; 7 min; 4°C). The supernatant was
removed and 70% ethanol was added into the precipitation, mixed
well and centrifuged (11,433 × g; 7 min; 4°C). The precipitation
was collected and dried, ddH2O was added and the concentration was
measured using ultra-micro ultraviolet spectrophotometer (Shanghai
Ruiyue Experimental Equipment Co., Ltd.). Subsequently, the DNA was
modified and purified using a hydrogen sulfite modification kit
containing a DNA polymerase (Shanghai Yubo Biological Technology
Co., Ltd.). Subsequently, a total of 3 µl modified DNA was used as
a template for PCR amplification, which was performed using the
following reaction conditions: Initial denaturation at 95°C for 4
min, followed by 35 cycles of denaturation at 95°C for 45 sec,
annealing at 60°C for 30 sec and extension at 72°C for 30 sec, and
a final extension step at 72°C for 10 min. The PCR products were
analyzed using 2% agarose gel electrophoresis and by UV gel
electrophoresis imaging and a gel image analyzer. All PCR reactions
were repeated in triplicate. The methylated (M) and unmethylated
(U) primers of LDH-C4 were as follows: (M) forward,
5′-TCTGGGGTGTAGCGGTCGTC-3′ and reverse,
5′-GCCCACATACTAAATCACGCG-3′; and (U) forward,
5′-GTAGTTTGGGGTTGAGTGGTTGTT-3′ and reverse,
5′-CACCCCCAATACATAATCACAACA-3′. Normal placental DNA modified by
CpG methyltransferase was used as the positive control for MSP
amplification. Deionized water was used as the blank control.
IHC analysis
Paraffin-embedded tissue sections (4 µm) were
deparaffinized in xylene and rehydrated using a graded alcohol
series. The sections were subsequently washed with PBS (pH 7.2) for
5 min and heated in a microwave oven for 5 min at 65°C in 10 mmol/l
sodium citrate buffer (pH 6.0) for antigen retrieval. The sections
were subsequently washed with PBS and immersed in 0.3% hydrogen
peroxide in methanol for 20 min at room temperature. After washing
with PBS, the sections were incubated in 1:10 normal goat serum
(5%; Shanghai Yanjin Biotechnology Co., Ltd.) at room temperature
in a humidified chamber for 45 min to prevent non-specific
immunoglobulin binding. The sections were subsequently incubated
with rabbit anti-DNMT1 (cat. no. ab19905), anti-DNMT3a (cat. no.
ab226261) and anti-DNMT3b (cat. no. ab2851) polyclonal antibodies
(all 1:500; Abcam) at 4°C overnight; the primary antibody was
replaced with normal IgG to serve as the control. The sections were
then thoroughly washed with PBS three times, and incubated with
goat anti-rabbit polyclonal antibody (1:100; cat. no. ZB-2010;
Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.) at 37°C
for 30 min, followed by washing 3 times with PBS. Then, the
horseradish peroxidase (HRP)-conjugated streptavidin working
solution was added and incubated C for 30 min. A
streptavidin-biotinylated HRP-based detection system (Shanghai
Hexing Biological Technology Co., Ltd.) was used to determine the
specific binding of each antibody. Sections were subsequently
counterstained with hematoxylin at room temperature for 5 min and
prepared for visualization using a light microscope in high power
field.
The expression was determined by the intensity of
the positive cells using ImageJ software (v1.8.0; National
Institutes of Health). Briefly, the area of positive staining was
scored using the following scale:0–2, negative expression;3–7,
positive staining (of those,3–4, weak positive expression; and 5–7,
strong positive expression). The staining intensity was graded as
follows: 0, no staining; 1, mild staining; 2, moderate staining;
and 3, intense staining. The staining intensity and positive
staining scores were multiplied together, and scores <4
indicated negative expression, while scores ≥4 indicated positive
expression.
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.). Enumeration data were expressed using rate.
The association between DNMT expression or the methylation status
of the promoter region of LDH-C4 and clinicopathological risk
factors was statistically evaluated using a χ2 or
Fisher's exact test. The correlation between the methylation status
of the LDH-C4 promoter and the expression levels of DNMTs was
detected using Spearman's rank correlation analysis. The overall
survival of patients was estimated using the Kaplan-Meier method,
and statistical differences between the groups were determined
using a log-rank test. Univariate and multivariate analyses of
overall survival according to prognostic factors were analyzed
using Cox regression analysis. P<0.05 was considered to indicate
a statistically significant difference.
Results
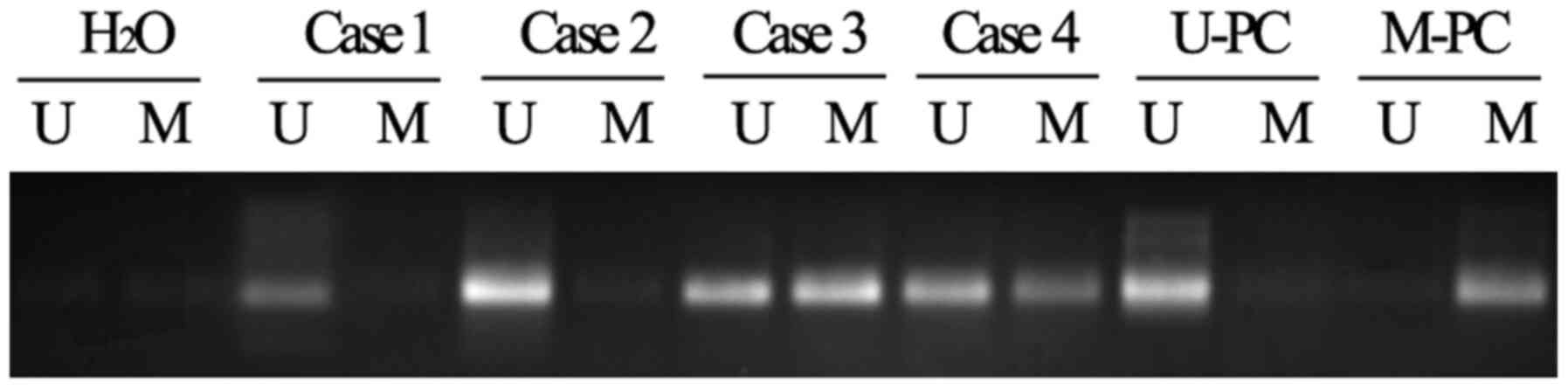
Methylation status of the promoter
region of LDH-C4 in breast cancer tissues
Using the MSP method and primers designed to amplify
the promoter region of the LDH-C4 gene, it was revealed that a
marked proportion of breast cancer tissues had low levels of
methylation compared with adjacent healthy tissues. In total, 59 of
the 136 (43.38%) breast cancer tissues samples contained
methylation patterns in the promoter region of the LDH-C4 gene,
while 121 of the 136 (88.97%) corresponding adjacent healthy tissue
samples contained methylation in the promoter region of the LDH-C4
gene (Fig. 1).
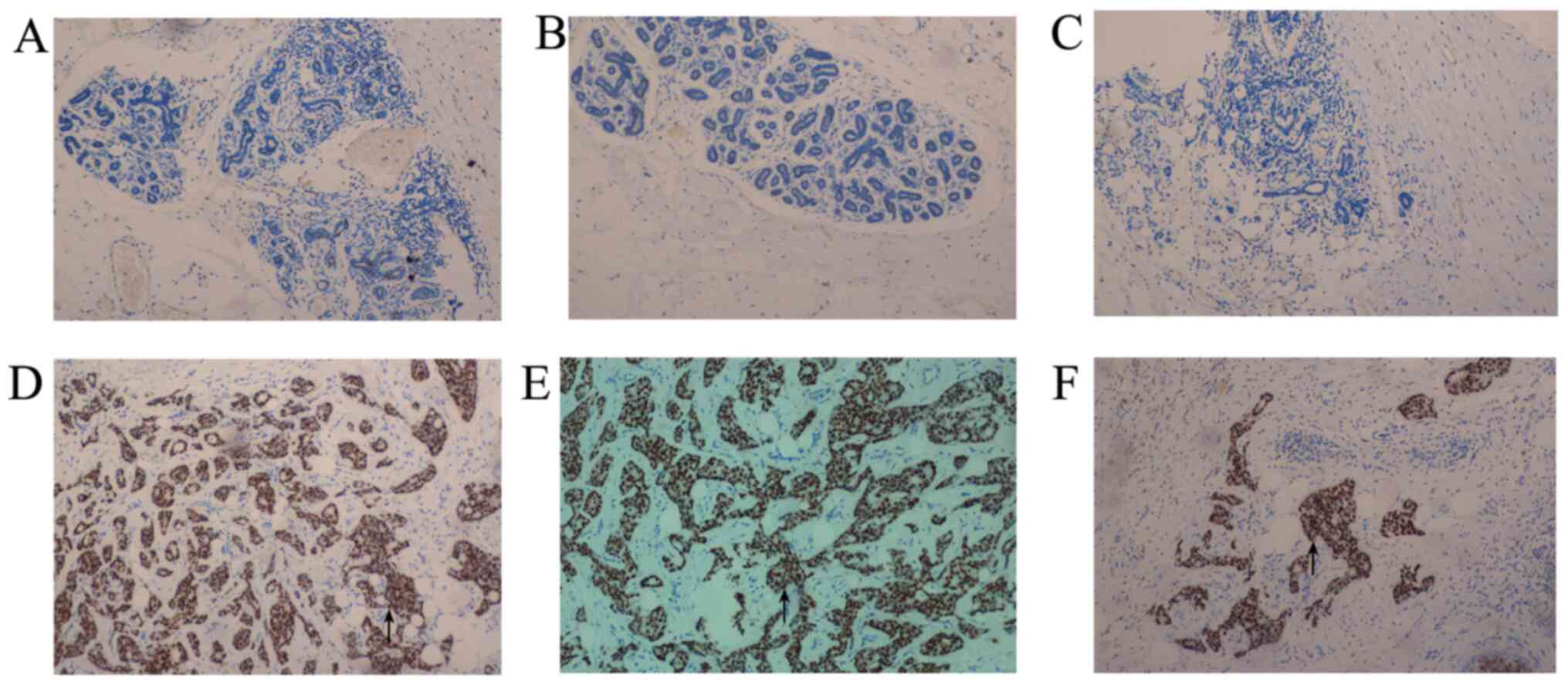
Expression of DNMT1, DNMT3a and DNMT3b
in breast cancer tissues
To determine whether the methylation status of the
LDH-C4 promoter region was correlated with the protein expression
levels of DNMTs, their expression levels were analyzed in breast
cancer and corresponding adjacent healthy tissues using IHC
analysis. As shown in Fig. 2,
DNMT1, DNMT3a and DNMT3b expression levels were observed in the
nuclei of breast cancer cells. DNMT1, DNMT3a and DNMT3b were
positively expressed in 11.76, 9.56 and 16.18% of adjacent healthy
tissues, respectively, while DNMT1, DNMT3a and DNMT3b positive
expression was found in 35.29, 41.91 and 39.71% of breast cancer
tissues, respectively.
Clinical correlation between DNMT expression and the
methylation status of the promoter region of LDH-C4 and the
clinicopathological factors of patients with breast cancer. To
further explore the possible effects of the methylation of LDH-C4
and DNMT expression in breast cancer, the correlation between the
methylation status of the promoter region of LDH-C4 and DNMT
expression, and the clinicopathological factors of patients with
breast cancer was determined. As shown in Table I, the methylation status of the
promoter region of LDH-C4 was not associated with age, menopausal
status, tumor size, tumor histology, TNM stage or antigen Ki-67
(Ki-67) expression (Ki-67 is associated with poor
clinicopathological factors (14)
in breast cancer), but was significantly associated with
histological grade, estrogen receptor (ER), progesterone receptor
(PR) and HER-2 status, and lymph node metastasis (P<0.05). The
expression of DNMT1 was not associated with age, menopausal status,
tumor size, tumor histology, TNM stage or Ki-67 expression, but was
significantly associated with ER, PR and HER-2 status, histological
grade and lymph node metastasis (P<0.05). The expression of
DNMT3a was not associated with age, menopausal status, tumor size,
tumor histology, TNM stage, and PR or HER-2 status (P>0.05), but
was significantly associated with histological grade, ER and Ki-67
expression, and lymph node metastasis (P<0.05). The expression
of DNMT3b was not associated with age, menopausal status, tumor
size, tumor histology, TNM stage, or PR, HER-2, ER or Ki-67
expression, but was significantly associated with histological
grade and lymph node metastasis (P<0.05). These data suggested
that hypomethylated profiles of the LDH-C4 promoter and DNMT
expression may indicate a poor prognosis of patients with breast
cancer.
 | Table I.Association between methylation of
LDH-C4 promoter and DNMT expression and the clinicopathological
factors of patients with breast cancer. |
Table I.
Association between methylation of
LDH-C4 promoter and DNMT expression and the clinicopathological
factors of patients with breast cancer.
|
|
| LDH-C4 | DNMT1, % | DNMT3a, % | DNMT3b, % |
|---|
|
|
|
|
|
|
|
|---|
| Clinicopathological
factor | Total (n) | U | M | P-value | High | Low | P-value | High | Low | P-value | High | Low | P-value |
|---|
| Age, years |
|
|
| 0.967 |
|
| 0.456 |
|
| 0.792 |
|
| 0.643 |
| ≤50 | 64 | 38 | 26 |
| 21 | 43 |
| 25 | 39 |
| 24 | 40 |
|
|
>50 | 72 | 40 | 32 |
| 22 | 50 |
| 29 | 43 |
| 25 | 47 |
|
| Menopausal
status |
|
|
| 0.603 |
|
| 0.397 |
|
| 0.541 |
|
| 0.784 |
| Pre- | 66 | 46 | 20 |
| 23 | 43 |
| 27 | 39 |
| 25 | 41 |
|
|
Post- | 70 | 39 | 31 |
| 24 | 46 |
| 24 | 46 |
| 24 | 73 |
|
| Tumor size, cm |
|
|
| 0.128 |
|
| 0.471 |
|
| 0.593 |
|
| 0.354 |
|
<2 | 74 | 40 | 34 |
| 26 | 48 |
| 28 | 46 |
| 31 | 43 |
|
| ≥2 | 62 | 37 | 35 |
| 22 | 40 |
| 28 | 34 |
| 21 | 41 |
|
| Tumor
histology |
|
|
| 0.087 |
|
| 0.174 |
|
| 0.237 |
|
| 0.0782 |
| Ductal
carcinoma | 126 | 90 | 36 |
| 58 | 68 |
| 34 | 92 |
| 48 | 78 |
|
| Lobular
carcinoma | 9 | 4 | 5 |
| 2 | 7 |
| 5 | 4 |
| 3 | 6 |
|
|
Other | 1 | 1 | 0 |
| 0 | 1 |
| 0 | 1 |
| 0 | 1 |
|
| TNM stage |
|
|
| 0.689 |
|
| 0.258 |
|
| 0.264 |
|
| 0.741 |
| I | 42 | 30 | 12 |
| 9 | 33 |
| 10 | 32 |
| 15 | 27 |
|
| II | 63 | 32 | 31 |
| 29 | 34 |
| 37 | 26 |
| 23 | 40 |
|
|
III | 21 | 13 | 8 |
| 4 | 17 |
| 5 | 18 |
| 9 | 12 |
|
| ER |
|
|
| 0.027 |
|
| 0.031 |
|
| 0.011 |
|
| 0.073 |
| + | 90 | 51 | 39 |
| 41 | 49 |
| 38 | 52 |
| 44 | 46 |
|
| − | 46 | 21 | 15 |
| 28 | 18 |
| 24 | 22 |
| 25 | 21 |
|
| PR |
|
|
| 0.014 |
|
| 0.046 |
|
| 0.136 |
|
| 0.364 |
| + | 82 | 50 | 32 |
| 25 | 57 |
| 32 | 50 |
| 36 | 46 |
|
| − | 54 | 24 | 30 |
| 27 | 27 |
| 22 | 32 |
| 20 | 34 |
|
| Ki-67 |
|
|
| 0.734 |
|
| 0.828 |
|
| 0.044 |
|
| 0.475 |
|
≤30% | 28 | 21 | 7 |
| 9 | 18 |
| 13 | 15 |
| 11 | 17 |
|
|
>30% | 108 | 79 | 29 |
| 37 | 71 |
| 28 | 100 |
| 56 | 52 |
|
| HER-2 |
|
|
| 0.014 |
|
| 0.0201 |
|
| 0.117 |
|
| 0.602 |
| + | 41 | 35 | 6 |
| 13 | 28 |
| 16 | 25 |
| 19 | 21 |
|
| − | 95 | 60 | 35 |
| 46 | 49 |
| 38 | 57 |
| 40 | 55 |
|
| Lymph node
metastasis |
|
|
| 0.014 |
|
| 0.001 |
|
| 0.006 |
|
| 0.026 |
|
With | 71 | 35 | 36 |
| 41 | 30 |
| 38 | 33 |
| 32 | 39 |
|
|
Without | 65 | 21 | 44 |
| 15 | 50 |
| 17 | 48 |
| 18 | 47 |
|
| Histological
grade |
|
|
| 0.034 |
|
| 0.043 |
|
| 0.026 |
|
| 0.039 |
| I | 43 | 20 | 23 |
| 18 | 25 |
| 21 | 24 |
| 19 | 24 |
|
| II | 61 | 35 | 26 |
| 20 | 41 |
| 26 | 35 |
| 24 | 37 |
|
|
III | 22 | 16 | 6 |
| 9 | 13 |
| 11 | 11 |
| 8 | 14 |
|
Association between the methylation
status of the promoter region of LDH-C4 and DNMT expression in
breast cancer tissues
As shown in Table
II, χ2 analysis revealed that the unmethylated level
of LDH-C4 in breast cancer tissues was inversely correlated with
the expression of DNMT1 (r=−0.273; P=0.018), DNMT3a (r=−0.216;
P=0.032) and DNMT3b (r=−0.298; P=0.010). These results indicated
that DNMT1, DNMT3a and DNMT3b expression may play an important role
in the demethylation of LDH-C4.
 | Table II.Association between promoter
methylation of LDH-C4 and expression levels of DNMT1, DNMT3a or
DNMT3b in breast cancer. |
Table II.
Association between promoter
methylation of LDH-C4 and expression levels of DNMT1, DNMT3a or
DNMT3b in breast cancer.
| Relative expression
level | LDH-C4 |
|
|
|
|---|
|
|
|
|
|---|
| U | M | r | χ2 | P-value |
|---|
| DNMT1 |
|
| −0.273 | 6.524 | 0.018 |
| High | 37 | 11 |
|
|
|
| Low | 40 | 48 |
|
|
|
| DNMT3a |
|
| −0.216 | 4.730 | 0.032 |
| High | 40 | 17 |
|
|
|
| Low | 37 | 42 |
|
|
|
| DNMT3b |
|
| −0.298 | 7.163 | 0.010 |
| High | 29 | 25 |
|
|
|
| Low | 48 | 34 |
|
|
|
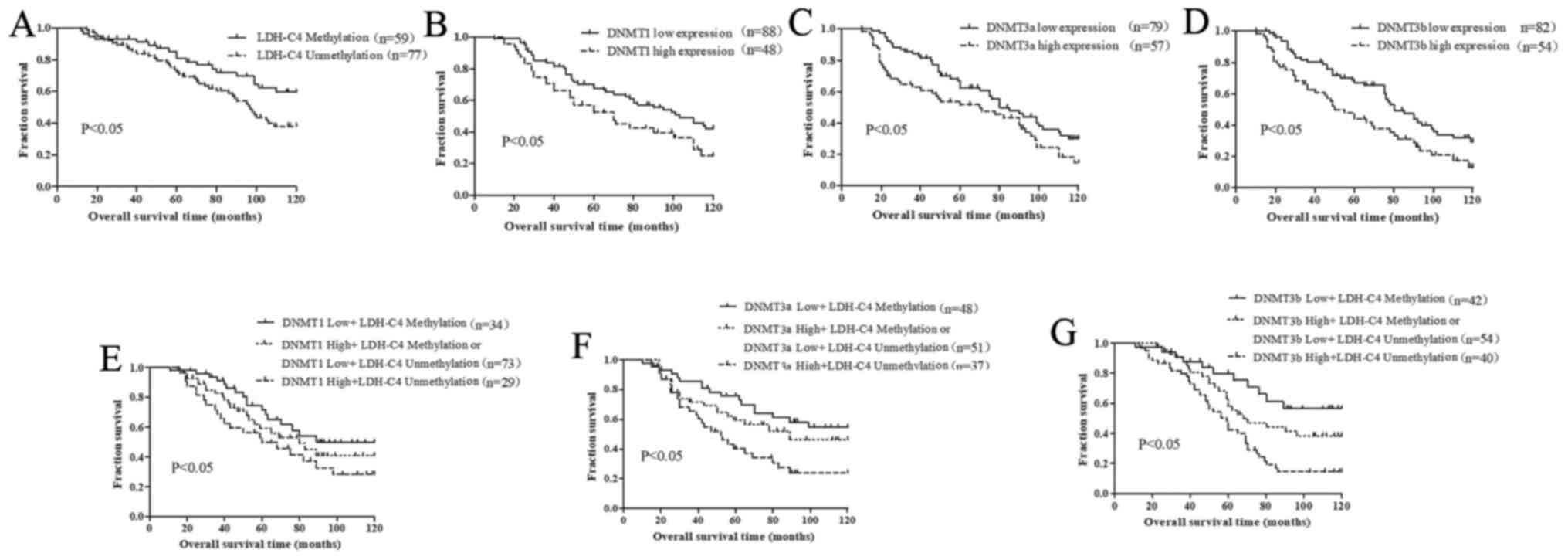
Methylation of the LDH-C4 promoter and
DNMT expression are associated with the poor prognosis of patients
with breast cancer
To determine the association between DNMT expression
and patient survival, or the methylation status of the promoter
region of LDH-C4 and patient survival, Kaplan-Meier curve analysis
for overall survival was performed. Kaplan-Meier analysis revealed
that patients with a demethylated LDH-C4 promoter exhibited a
shorter survival time and a poorer prognosis compared with those
patients with a methylated LDH-C4 promoter (P<0.05; Fig. 3A). Patients with high expression
levels of DNMT1, DNMT3a and DNMT3b exhibited a shorter survival
time and worse prognosis compared with those with low expression
levels of DNMTs (Fig. 3B-D).
Moreover, a demethylated LDH-C4 promoter and high DNMT expression
levels predicted an unfavorable prognosis (Fig. 3E-G).
To further investigate the prognostic factors for
poor breast cancer outcomes, univariate and multivariate analyses
were performed. The results of the univariate analysis revealed
that overall survival was significantly associated with the
methylation of the LDH-C4 promoter, and DNMT1, DNMT3a and DNMT3b
expression, histological grade, ER, PR and HER-2 status, and lymph
node metastasis (Table II).
Multivariate analysis revealed that a demethylated LDH-C4 promoter,
high expression of DNMT3a and DNMT1, histological grade and lymph
node metastasis were independent prognostic factors for patients
with breast cancer (Table
III).
 | Table III.Univariate and multivariate analyses
of prognostic factors in breast cancer for overall survival. |
Table III.
Univariate and multivariate analyses
of prognostic factors in breast cancer for overall survival.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | P-value | 95% CI | HR | P-value | 95% CI |
|---|
| LDH-C4 methylation
status | 2.304 | 0.015 | 1.115-3.457 | 1.784 | 0.007 | 0.874-2.477 |
|
Methylation vs.
unmethylation |
|
|
|
|
|
|
| Expression of
DNMT1 | 3.654 | 0.036 | 2.654-4.759 | 2.114 | 0.001 | 0.998-3.327 |
| High
vs. low |
| 0.021 |
|
|
|
|
| Expression of
DNMT3a | 2.881 |
| 1.497-4.776 | 1.964 | 0.003 | 0.716-2.997 |
| High
vs. low |
| 0.047 |
|
|
|
|
| Expression of
DNMT3b | 3.244 |
| 2.229-5.436 | 1.374 | 0.645 | 1.185-3.249 |
| High
vs. low |
|
|
|
|
|
|
| Age, years | 1.742 | 0.357 | 0.860-2.374 |
|
|
|
| <50
vs. ≥50 |
|
|
|
|
|
|
| Menopausal
status | 1.476 | 0.184 | 0.461-1.127 |
|
|
|
| Pre-
vs.post- |
|
|
|
|
|
|
| Tumor size, cm | 1.655 | 0.31 | 1.417-2.652 |
|
|
|
| <2
vs. ≥2 |
|
|
|
|
|
|
| Tumor
histology | 0.87 | 0.112 | 0.650-1.117 |
|
|
|
| Ductal
vs. lobular |
|
|
|
|
|
|
| TNM stage | 1.336 | 0.155 | 0.897-1.968 |
|
|
|
| I and
II vs. III |
|
|
|
|
|
|
| ER | 1.774 | 0.041 | 1.156-2.427 | 2.347 | 0.06 | 0.671-4.623 |
| + vs.
- |
|
|
|
|
|
|
| PR | 2.668 | 0.001 | 1.497-4.634 | 1.562 | 0.332 | 0.784-3.116 |
| + vs.
- |
|
|
|
|
|
|
| Ki-67 | 1.352 | 0.357 | 0.874-2.328 |
|
|
|
| ≤30%
vs. >30% |
|
|
|
|
|
|
| HER-2 | 1.94 | 0.041 | 1.118-2.564 | 2.338 | 0.075 | 0.657-4.014 |
| + vs.
- |
|
|
|
|
|
|
| Histological
grade | 4.124 | 0.001 | 2.321-6.374 | 2.887 | 0.001 | 1.986-4.015 |
| I and
II vs. III |
|
|
|
|
|
|
| Lymph node
metastasis | 6.327 | 0.001 | 4.365-8.457 | 3.016 | 0.001 | 0.1.336-5.417 |
| Yes vs.
no |
|
|
|
|
|
|
Discussion
Due to the increasing prevalence of breast cancer,
early diagnosis and treatment are crucial for the effective
prevention and control of the disease, in addition to improving the
prognosis of patients (15).
However, highly sensitive and specific tumor markers for the
auxiliary diagnosis and prognosis prediction of breast cancer are
currently limited. Therefore, it remains important to identify
novel tumor markers for breast cancer (16). LDH-C4 is a nicotinamide adenine
dinucleotide-dependent kinase, which exists in mammalian sperm and
other germ cells (17). LDH-C4 is
also a tumor testis-associated antigen, that has strong
immunogenicity (18) and therefore
may represent a novel target for tumor immunotherapy. Previous
studies have found that LDH-C4 is highly expressed in lung cancer
(19,20), breast cancer (21) and renal cell carcinoma (22). It has been shown to have important
clinical value as a specific molecular marker of breast cancer
(21). However, the specific
expression profile of LDH-C4 and its underlying molecular
regulatory mechanisms remain unclear; thus, further investigations
are required.
Results of a previous study demonstrated that the
CpG methylation frequency in the promoter region of sperm
expressing the LDH-C gene was low, while the methylation frequency
of CpG in liver cells without the LDH-C gene was high (23). These results suggested that
methylation status may affect the activity of the LDH-C gene
promoter, thus regulating the expression of the gene. Tang and
Goldberg (8) previously reported
that methylation plays an important role in the transcription of
the LDH-C gene in prostate cancer tissues. Results of the present
study revealed that the demethylation rate of the LDH-C4 gene in
breast cancer tissues was significantly higher compared with that
of adjacent tissues, suggesting that the demethylation of LDH-C4
may be closely associated with the occurrence and development of
breast cancer. In addition, a correlation was identified between
the methylation status of LDH-C4 and the histological grade, ER, PR
and HER-2 status, and lymph node metastasis. These aforementioned
pathological indexes were also closely associated with the
prognosis of the disease.
DNMTs play an important role in DNA methylation
(24), and the main members of the
family include DNMT1, DNMT3a and DNMT3b. The expression of DNMTs is
associated with aberrant methylation in tumor tissue (25), and they participate in the
occurrence and development of tumors to varying degrees (26,27).
Previous studies have revealed that DNMTs interact with
transcription factors, histone methyltransferases and microRNA to
regulate a variety of tumor-related genes (28). However, to the best of our
knowledge, it remains unclear whether LDH-C4 demethylation is
affected by DNMTs in breast cancer. Results of the present study
demonstrated that DNMT1, DNMT3a and DNMT3b proteins were expressed
to differing degrees in breast cancer tissues. A previous study
also indicated that DNMT expression was upregulated in breast
cancer (29). Findings of the
present study also indicated that the expression levels of DNMT1
were correlated with ER, PR and HER-2 expression, histological
grade and lymph node metastasis. In addition, the expression levels
of DNMT3a were found to be associated with histological grade, ER
and Ki-67 expression, and lymph node metastasis, while the
expression levels of DNMT3b were associated with histological grade
and lymph node metastasis. Analysis to identify a correlation
between LDH-C4 demethylation and DNMT expression revealed that the
protein expression levels of DNMT were reduced in line with the
demethylation of LDH-C4 in breast cancer tissues, indicating that
the low expression of DNMTs may be an important biological event
promoting the occurrence of LDH-C4 demethylation. The 5-year
overall survival of patients with breast cancer with demethylated
LDH-C4 was also found to be significantly reduced compared with
methylated LDH-C4. In addition, the prognosis of patients with
LDH-C4 demethylation and high DNMT expression was poor compared
with methylated LDH-C4 and low DNMT expression, respectively.
Multiple Cox regression analyses also revealed that LDH-C4 gene
demethylation, and DNMT3a and DNMT1 expression were independent
prognostic factors of breast cancer. However, DNMT3b expression was
not found to be an independent risk factor for breast cancer.
In conclusion, the findings of the present study
revealed that the expression levels of DNMTs were closely
associated with the methylation level of LDH-C4 in breast cancer.
The observed low expression levels of DNMT3a and DNMT3b may be an
important molecular event of LDH-C4 gene demethylation, which
suggests their potential use for the diagnosis and treatment of
early-stage breast cancer. In addition, these markers may also have
potential as comprehensive indicators to assist clinical prognosis
evaluation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Health Department of
Hebei Province (grant no. 20210337).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JZ and XW confirm the authenticity all the raw data.
JZ and XW designed the study. FXZ and FQZ collected the
characteristics of the patients and performed the experiments. HW
and BZ analyzed the data. JZ wrote the manuscript. All authors
agreed to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work was
appropriately investigated and resolved. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Third Clinical Hospital of Hebei Medical
University (approval no.2021-034-5) and all patients provided
written informed consent prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reis-Filho JS and Pusztai L: Gene
expression profiling in breast cancer: Classification,
prognostication, and prediction. Lancet. 378:1812–1823. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lnning PE: Breast cancer prognostication
and prediction: Are we making progress. Ann Oncol. 18 (Suppl
8):viii3–7. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shao Y, Sun X, He Y, Liu C and Liu H:
Elevated Levels of Serum Tumor Markers CEA and CA15-3 Are
Prognostic Parameters for Different Molecular Subtypes of Breast
Cancer. PLoS One. 10:e01338302015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kong L, Du W, Cui Z, Wang L, Yang Z, Zhang
H and Lin D: Expression of lactate dehydrogenase C in MDA MB 231
cells and its role in tumor invasion and migration. Mol Med Rep.
13:3533–3538. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goldberg E, Eddy EM, Duan C and Odet F:
LDHC: the ultimate testis-specific gene. J Androl. 31:86–94. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Woodford MR, Chen VZ, Backe SJ,
Bratslavsky G and Mollapour M: Structural and functional regulation
of lactate dehydrogenase-A in cancer. Future Med Chem. 12:439–455.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang H and Goldberg E: Homo sapiens
lactate dehydrogenase c (Ldhc) gene expression in cancer cells is
regulated by transcription factor Sp1, CREB, and CpG island
methylation. J Androl. 30:157–167. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang H, Kung A and Goldberg E: Regulation
of murine lactate dehydrogenase C (Ldhc) gene expression. Biol
Reprod. 78:455–461. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding W, Chen G and Shi T: Integrative
analysis identifies potential DNA methylation biomarkers for
pan-cancer diagnosis and prognosis. Epigenetics. 14:67–80. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin B and Robertson KD: DNA
methyltransferases, DNA damage repair, and cancer. Adv Exp Med
Biol. 754:3–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lam TW, Tong JH, To KF, Chan A, Liew CT,
Lai PB and Wong N: Correlative Analysis of DNA Methyltransferase
Expression and Promoter Hypermethylation of Tumor Suppressor Genes
in Hepatocellular Carcinoma. Cancer Genomics Proteomics. 3:271–277.
2006.PubMed/NCBI
|
|
13
|
Mirza S, Sharma G, Parshad R, Gupta SD,
Pandya P and Ralhan R: Expression of DNA methyltransferases in
breast cancer patients and to analyze the effect of natural
compounds on DNA methyltransferases and associated proteins. J
Breast Cancer. 16:23–31. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aman NA, Doukoure B, Koffi KD, Koui BS,
Traore ZC, Kouyate M, Toure I and Effi AB: Immunohistochemical
Evaluation of Ki-67 and Comparison with Clinicopathologic Factors
in Breast Carcinomas. Asian Pac J Cancer Prev. 20:73–79. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Parambil NA, Philip S, Tripathy JP, Philip
PM, Duraisamy K and Balasubramanian S: Community engaged breast
cancer screening program in Kannur District, Kerala, India: A ray
of hope for early diagnosis and treatment. Indian J Cancer.
56:222–227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Banin Hirata BK, Oda JM, Losi Guembarovski
R, Ariza CB, de Oliveira CE and Watanabe MA: Molecular markers for
breast cancer: prediction on tumor behavior. Dis Markers.
2014.5131582014. PubMed/NCBI
|
|
17
|
Duan C and Goldberg E: Inhibition of
lactate dehydrogenase C4 (LDH-C4) blocks capacitation of mouse
sperm in vitro. Cytogenet Genome Res. 103:352–359. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hogrefe HH, Kaumaya PT and Goldberg E:
Immunogenicity of synthetic peptides corresponding to flexible and
antibody-accessible segments of mouse lactate dehydrogenase
(LDH)-C4. J Biol. 264:10513–10519. 1989.PubMed/NCBI
|
|
19
|
Grunwald C, Koslowski M, Arsiray T, Dhaene
K, Praet M, Victor A, Morresi-Hauf A, Lindner M, Passlick B, Lehr
HA, et al: Expression of multiple epigenetically regulated
cancer/germline genes in non-small cell lung cancer. Int J Cancer.
118:2522–2528. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen L, Wu Q, Xu X, Yang C, You J, Chen F
and Zeng Y: Cancer/testis antigen LDHC promotes proliferation and
metastasis by activating the PI3K/Akt/GSK-3β-signaling pathway and
the in lung adenocarcinoma. Exp Cell Res. 398:1124142021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui Z and Chen Y, Hu M, Lin Y, Zhang S,
Kong L and Chen Y: Diagnostic and prognostic value of the
cancer-testis antigen lactate dehydrogenase C4 in breast cancer.
Clin Chim Acta. 503:203–209. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hua Y, Liang C, Zhu J, Miao C, Yu Y, Xu A,
Zhang J, Li P, Li S, Bao M, et al: Expression of lactate
dehydrogenase C correlates with poor prognosis in renal cell
carcinoma. Tumour Biol. 39:10104283176959682017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kroft TL, Jethanandani P, McLean DJ and
Goldberg E: Methylation of CpG dinucleotides alters binding and
silences testis-specific transcription directed by the mouse
lactate dehydrogenase C promoter. Biol Reprod. 65:1522–1527. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hervouet E, Peixoto P, Delage-Mourroux R,
Boyer-Guittaut M and Cartron PF: Specific or not specific
recruitment of DNMTs for DNA methylation, an epigenetic dilemma.
Clin Epigenetics. 10:172018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He M, Fan J, Jiang R, Tang WX and Wang ZW:
Expression of DNMTs and genomic DNA methylation in gastric signet
ring cell carcinoma. Mol Med Rep. 8:942–948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Supic G, Kozomara R, Zeljic K, Jovic N and
Magic Z: Prognostic value of the DNMTs mRNA expression and genetic
polymorphisms on the clinical outcome in oral cancer patients. Clin
Oral Investig. 21:173–182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu J, Shuang Z, Zhao J, Tang H, Liu P,
Zhang L, Xie X and Xiao X: Linc00152 promotes tumorigenesis by
regulating DNMTs in triple-negative breast cancer. Biomed
Pharmacother. 97:1275–1281. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Turek-Plewa J and Jagodziński PP: The role
of mammalian DNA methyltransferases in the regulation of gene
expression. Cell Mol Biol Lett. 10:631–647. 2005.PubMed/NCBI
|
|
29
|
Jahangiri R, Jamialahmadi K, Gharib M,
Emami Razavi A and Mosaffa F: Expression and clinicopathological
significance of DNA methyltransferase 1, 3A and 3B in
tamoxifen-treated breast cancer patients. Gene. 685:24–31. 2019.
View Article : Google Scholar : PubMed/NCBI
|