Introduction
Lung cancer is one of the most commonly diagnosed
malignancies, and accounts for 1.6 million deaths annually
worldwide; in addition, non-small cell lung cancer (NSCLC) makes up
the majority of lung cancer cases (1,2). The
prognosis of patients with NSCLC is generally poor, with a 5-year
overall survival rate of <18% (3), and even lower for patients in
advanced stages (0–10%) (4).
Different subtypes of NSCLC are associated with different molecular
biological characteristics (5).
The main histological subtypes of NSCLC include lung adenocarcinoma
(LUAD) and lung squamous cell carcinoma (LUSC) (6). There is a significant difference in
the clinical treatment between the LUAD and LUSC subtypes. For
instance, patients with LUSC experience a longer overall survival
time than those with LUAD after treatment with ipilimumab (7). In addition, treatment with gefitinib,
which targets EGFR kinase mutated cases, is more suitable for
patients with LUAD (8). Therefore,
the development of effective molecular diagnostic biomarkers and
potential molecules for the identification and treatment of the
LUAD and LUSC subtypes is of significant interest.
Long non-coding RNA (lncRNA) is a type of non-coding
transcript >200 nucleotides in length (9). lncRNA molecules have emerged as key
regulators in tumor development and progression (10–12).
For example, Wei et al (13) suggested that BCAR4 contributed to
glioma progression by enhancing cell proliferation and activating
the EGFR/PI3K/AKT pathway, which may represent a new target for the
treatment and prognosis of patients with glioma. Upregulation of
the lncRNA HOTTIP has also been recognized as a marker of poor
survival in patients with renal cell carcinoma (14).
Long intergenic non-protein coding RNA 628
(LINC00628) is an lncRNA that has been reported to suppress the
growth and metastasis of breast cancer and promote the apoptosis of
breast cancer cells in previous studies (15,16).
Zhang et al (17) also
suggested that downregulation of LINC00628 aggravated the
progression of colorectal cancer by inhibiting p57 expression.
Recent studies have described the aberrant expression of mRNAs,
circRNAs and lncRNAs that could distinguish between LUAD and LUSC
(18–20), indicating the importance of
analyzing functional molecules in different subtypes of NSCLC. Xu
et al (20) demonstrated
that the expression levels of LINC00628 were upregulated in LUAD
tissue, which promoted LUAD cell proliferation, migration and
invasion. However, the expression levels and clinical significance
of LINC00628 in other subtypes of NSCLC remain poorly
understood.
The aim of the present study was to analyze the
expression levels of LINC00628 in NSCLC and normal tissue, as well
as in the LUAD and LUSC subtypes. Moreover, the ability of
LINC00628 to discriminate between patients with LUAD and those with
LUSC was also evaluated. Lastly, the clinical significance of
LINC00628 in patient prognosis was also analyzed in patients with
NSCLC.
Materials and methods
Patients and tissue collection
Tissue samples were collected from 128 patients with
NSCLC, including 73 patients with LUAD and 55 patients with LUSC,
who underwent curative resection between February 2015 and October
2019 at Weifang People's Hospital (Weifang, Shandong, China). NSCLC
and adjacent normal tissue samples (located 3 cm from the edge of
the tumors) were collected from the patients. All tissue samples
were verified by histopathological examination and stored in liquid
nitrogen for further use. The inclusion criteria were as follows:
i) The patients were diagnosed with NSCLC by pathological
examination; ii) had not received any tumor therapy before surgery;
and iii) had a complete clinicopathological record. The following
patients were excluded from the study: i) Patients <18 years or
>80 years; ii) pregnant or lactating patients; iii) patients
with autoimmune diseases or other malignancies; iv) patients who
had received any anti-tumor treatment. Follow-up surveys after the
surgery were conducted by telephone or via outpatient visits.
Survival was recorded for all patients. The protocols used for
tissue collection and analysis were approved by The Ethics
Committee of Weifang People's Hospital (approval no. 0014647) and
adhered with its guidelines. Written informed consent was obtained
from the patients or their family members (for patients who had no
ability to read and/or write) prior to sample collection.
Bioinformatics analysis
In the present study, starBase v3.0 (http://starbase.sysu.edu.cn/index.php)
was used to analyze The Cancer Genome Atlas (TCGA) datasets in
order to compare the expression levels of LINC00628 in LUAD, LUSC
and matched normal tissues from patients with cancer. In addition,
Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/index.html)
was used to generate the survival curves of patients with LUAD and
LUSC based on the expression of LINC00628, as well as to evaluate
the association between LINC00628 and overall survival
prognosis.
RNA extraction
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from tissue
samples. NanoDrop® 2000 (Thermo Fisher Scientific, Inc.)
was used to measure the purity and concentration of the extracted
RNA. cDNA was then reverse-transcribed from RNA using the RevertAid
First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.) at
42°C for 30 min followed by 85°C for 5 sec.
Reverse transcription-quantitative PCR
(RT-qPCR)
The expression levels of LINC00628 were measured
using RT-qPCR, which was carried out using a SYBR Green PCR Kit
(Bio-Rad laboratories, Inc.) on an Applied Biosystems 7900
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were: i) Initial denaturation
for 10 min at 95°C; ii) 40 cycles of denaturation for 30 sec at
95°C, annealing for 20 sec at 58°C and extension for 30 sec at
72°C; and iii) final extension for 10 min at 72°C. GAPDH was used
as an endogenous control for LINC00628. The primer sequences were
as follows: LINC00628 forward, 5′-ACTCCGCCTGGATGGGAATA-3′ and
reverse, 5′-CAGGACTTGGCCCACCTATC-3′; GAPDH forward,
5′-CAAGGTCATCCATGACAACTTTG-3′ and reverse,
5′-GTCCACCACCCTGTTGCTGTAG-3′. All protocols were performed
following the manufacturer's instructions. The expression levels of
LINC00268 were calculated using the 2−ΔΔCq method
(21) and normalized to those of
GAPDH.
Statistical analysis
All data are shown as the mean ± SD. SPSS 22.0 (IBM
Corp.) and GraphPad Prism 7.0 software (GraphPad Software, Inc.)
were used to perform the statistical analysis. Paired Student's
t-tests were used to compare the differences in LINC00628
expression between tumor tissue samples (NSCLC, LUAD or LUSC) and
adjacent healthy tissue controls. Unpaired Student's t-test was
used to analyze the differences in LINC00628 expression between
LUAD and LUSC cases. The χ2 test was used to assess the
relationship between LINC00628 and the clinicopathological features
of patients with NSCLC. The ability of LINC00628 to discriminate
between patients with LUAD and LUSC was assessed using a receiver
operating characteristic (ROC) curve. In this analysis, the
expression of LINC00628 in LUAD and LUSC was used to construct ROC
curve, and the area under the curve (AUC) was calculated to
indicate the accuracy of LINC00628 in distinguishing LUAD and LUSC.
The diagnostic sensitivity and specificity were obtained at the
optimal cut-off value point, which was the value when the sum of
the sensitivity and specificity of the ROC curve was the highest.
The relationship between LINC00628 expression and the overall
survival of patients with NSCLC, LUAD or LUSC was analyzed using
Kaplan-Meier curves. The patients were divided into low- and
high-expression groups according to the median of the group.
Univariate and multivariate Cox regression analysis was used to
examine the prognostic value of LINC00628 in patients with NSCLC.
Each experiment was repeated at least three times. P<0.05 was
considered to indicate a statistically significant difference.
Results
LINC00628 expression is associated
with the overall survival of patients with NSCLC
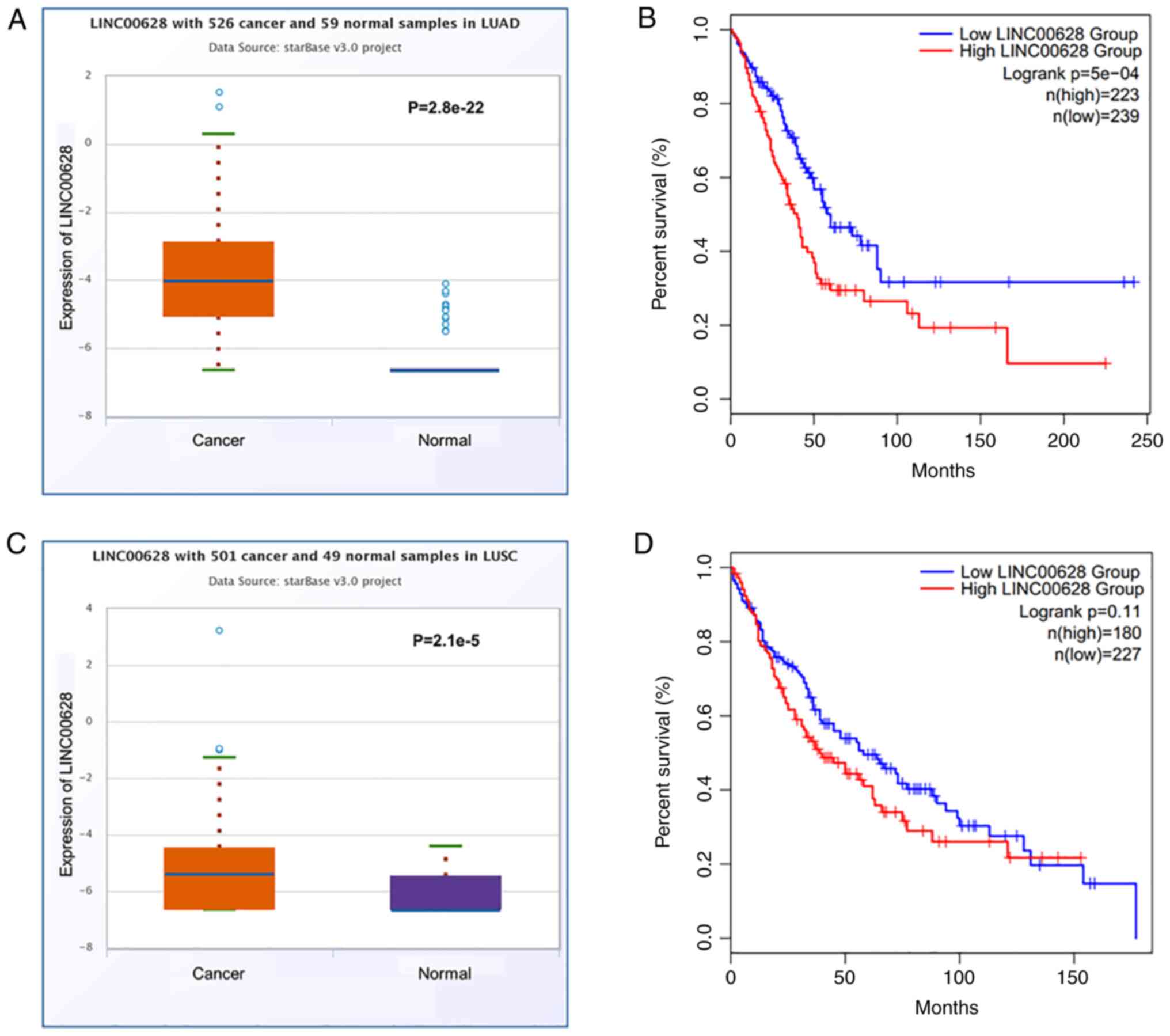
Bioinformatics analysis of TCGA datasets suggested
that the expression levels of LINC00628 were significantly
upregulated in LUAD (n=526) compared with adjacent normal tissues
(n=59) (Fig. 1A; mean, 0.11 vs.
0.02, respectively; P=2.8×10−22). The Kaplan-Meier
curves generated using TCGA data also indicated that patients with
LUAD and high LINC00628 expression experienced worse overall
survival compared with those with low LINC00628 expression
[Fig. 1B; log-rank
P=5×10−4; hazard ratio (HR=1.7)]. Moreover, the
expression levels of LINC00628 were significantly increased in LUSC
(n=501) compared with adjacent normal tissues (n=59) (Fig. 1C; mean, 0.06 vs. 0.02,
respectively; P=2.1×10−5). Kaplan-Meier curves for
patients with LUSC indicated that there was no significant
difference in overall survival between patients with high or low
LINC00628 expression (Fig. 1D;
log-rank P=0.11; HR=1.33).
LINC00628 expression is dysregulated
patients with LUAD and LUSC
The expression levels of LINC00628 were
significantly upregulated NSCLC tumor compared with the adjacent
normal tissue samples (Fig. 2A;
n=128; P<0.001). In addition, the expression of LINC00628 was
also significantly increased in LUAD (n=73) and LUSC (n=55) tumor
tissue compared with the adjacent normal tissue controls (Fig. 2B, P<0.001). Moreover, LINC00628
expression was significantly increased in patients with LUAD
compared with those with LUSC (Fig.
2C, P<0.001). The expression of LINC00628 in LUAD and LUSC
was used to construct an ROC curve, and the AUC was calculated as a
measure of the accuracy of LINC00628 in discriminating between LUAD
and LUSC. The ROC curve results revealed that the AUC of the curve
was 0.861, indicating that LINC00628 could accurately distinguish
LUAD from LUSC. At the optimal cut-off value of 0.955, the
sensitivity was 83.64%, and the specificity was 89.04% (Fig. 2D).
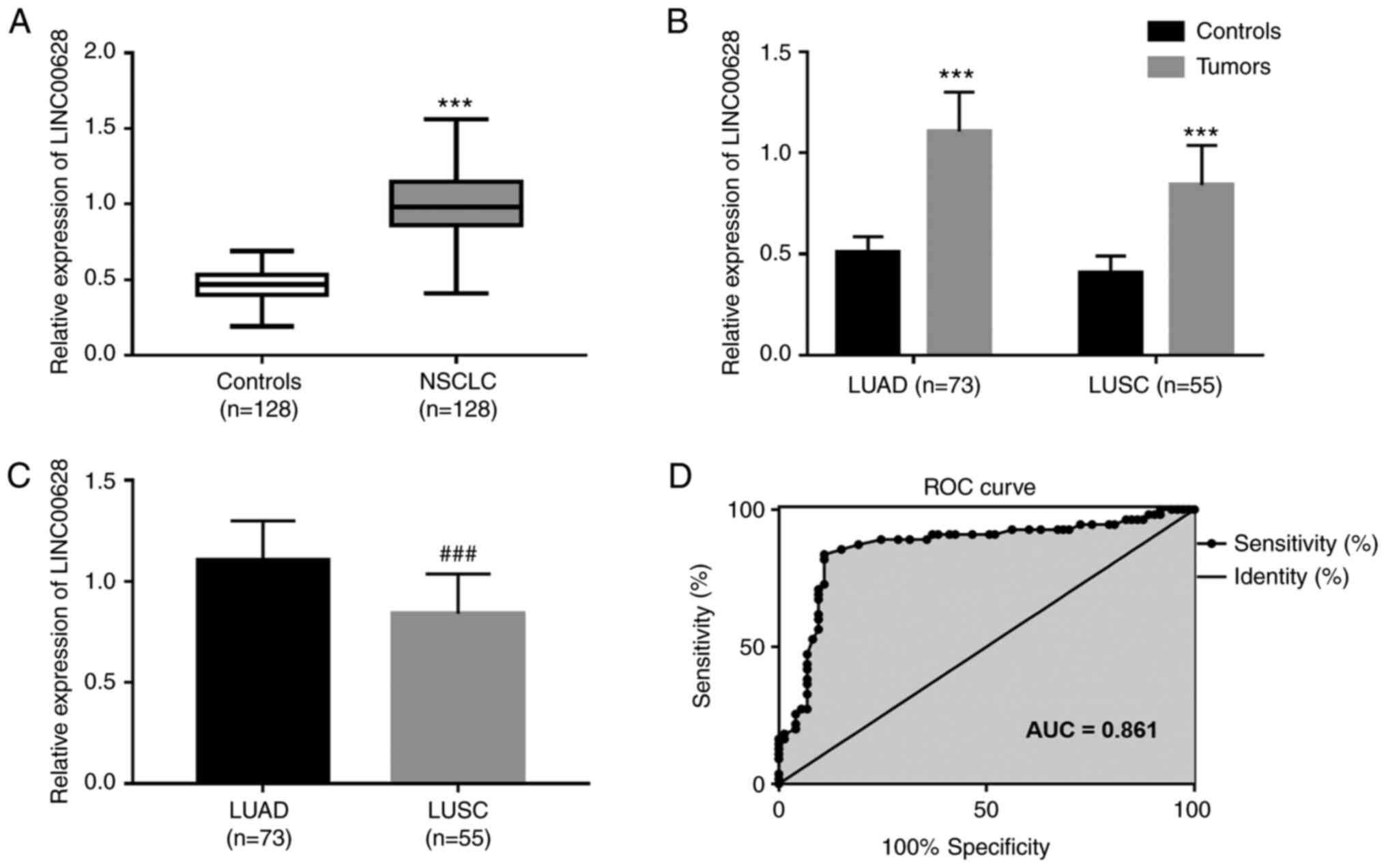 | Figure 2.Upregulation of LINC00628 in LUAD and
LUSC tumor tissue. (A) Relative expression of LINC00628 in NSCLC
tumor and adjacent normal tissue samples (mean, 0.99 vs. 0.46,
respectively; ***P<0.001 vs. Controls). (B) Relative expression
of LINC00628 in LUAD and LUSC tumors, compared with adjacent normal
tissue samples (LUAD, mean, 1.11 vs. 0.51, respectively,
***P<0.001 vs. Controls; LUSC, mean, 0.84 vs. 0.41,
respectively, ***P<0.001 vs. Controls). (C) Relative expression
of LINC00628 in patients with LUAD compared with patients with LUSC
(mean, 1.11 vs. 0.84, respectively; P<0.001).
###P<0. vs. LUAD. (D) LINC00628 expression can
discriminate between LUAD and LUSC (AUC=0.861). LINC00628, long
intergenic non-protein coding RNA 628; LUAD, lung adenocarcinoma;
LUSC, lung squamous cell carcinoma; NSCLC, non-small cell lung
cancer; AUC, area under the curve. |
Relationship between LINC00628 and the
clinicopathological features of patients with NSCLC
As shown in Table
I, the expression of LINC00628 was significantly associated
with tumor size (P=0.013), histological type (P=0.009), lymph node
metastasis (P=0.021) and TNM stage (P=0.008). However, there were
no significant associations between LINC00628 expression and age,
sex and smoking status (all P>0.05). These findings suggested
that LINC00628 may be involved in the development of NSCLC.
 | Table I.Relationship between LINC00628
expression and the clinicopathological characteristics of patients
with non-small cell lung cancer. |
Table I.
Relationship between LINC00628
expression and the clinicopathological characteristics of patients
with non-small cell lung cancer.
|
|
| LINC00628
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Total (n=128) | Low (n=62) | High (n=66) | P-valuea |
|---|
| Age, years |
|
|
| 0.656 |
| ≤60 | 45 | 23 | 22 |
|
|
>60 | 83 | 39 | 44 |
|
| Sex |
|
|
| 0.809 |
|
Female | 53 | 25 | 28 |
|
|
Male | 75 | 37 | 38 |
|
| Smoking |
|
|
| 0.633 |
| No | 53 | 27 | 26 |
|
|
Yes | 75 | 35 | 40 |
|
| Tumor size, cm |
|
|
| 0.013 |
| ≤3 | 66 | 39 | 27 |
|
|
>3 | 62 | 23 | 39 |
|
| Histological
type |
|
|
| 0.009 |
|
LUAD | 73 | 28 | 45 |
|
|
LUSC | 55 | 34 | 21 |
|
| Lymph node
metastasis |
|
|
| 0.021 |
|
Negative | 65 | 38 | 27 |
|
|
Positive | 63 | 24 | 39 |
|
| TNM stage |
|
|
| 0.008 |
|
I–II | 59 | 36 | 23 |
|
|
III–IV | 69 | 26 | 43 |
|
High LINC00628 is associated poor
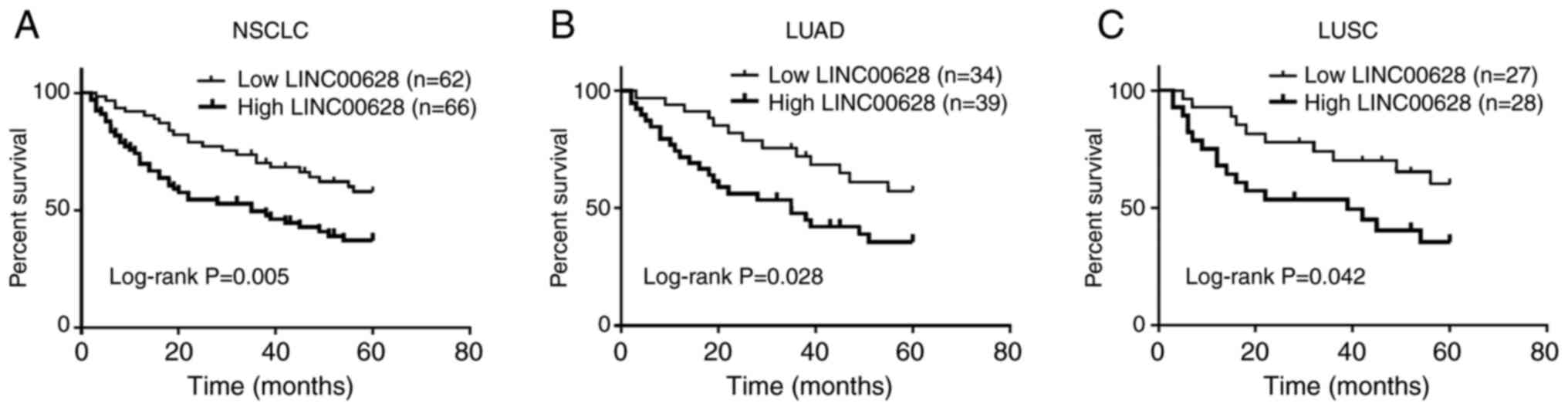
overall survival in patients with NSCLC
Kaplan-Meier survival curves and multivariate Cox
regression analyses were used to evaluate the prognostic value of
LINC00628 for patients with NSCLC. As shown in Fig. 3A, high LINC00628 expression was
associated with worse overall survival in patients with NSCLC
(log-rank P=0.005). Cox regression analysis indicated that lymph
node metastasis (HR=1.508; 95% CI, 1.085-2.167; P=0.047), TNM stage
(HR=2.541; 95% CI, 1.883-3.496; P=0.003) and LINC00628 (HR=2.887;
95% CI, 1.973-4.243; P=0.002) were associated with survival of
patients with NSCLC, and TNM stage (HR=2.139; 95% CI, 1.527-2.896;
P=0.008) and LINC00628 expression (HR=2.161, 95% CI, 1.452-3.051;
P=0.005) were confirmed as two independent prognostic factors
(Table II).
 | Table II.Cox regression analysis in patients
with non-small cell lung cancer. |
Table II.
Cox regression analysis in patients
with non-small cell lung cancer.
| A, Univariate
analysis |
|---|
|
|---|
| Variables | HR | 95% CI | P-value |
|---|
| Age | 1.139 | 0.831-1.556 | 0.293 |
| Sex | 1.214 | 0.794-1.809 | 0.302 |
| Smoking | 1.148 | 0.851-1.667 | 0.227 |
| Tumor size | 1.409 | 0.965-1.973 | 0.089 |
| Histological
type | 1.132 | 0.921-1.480 | 0.298 |
| Lymph node
metastasis | 1.508 | 1.085-2.167 | 0.047 |
| TNM stage | 2.541 | 1.883-3.496 | 0.003 |
| LINC00628
expression | 2.887 | 1.973-4.243 | 0.002 |
|
| B, Multivariate
analysis |
|
|
Variables | HR | 95% CI | P-value |
|
| Age | 1.115 | 0.821-1.435 | 0.334 |
| Sex | 1.261 | 0.854-1.896 | 0.289 |
| Smoking | 1.124 | 0.864-1.593 | 0.255 |
| Tumor size | 1.313 | 0.942-1.574 | 0.192 |
| Histological
type | 1.102 | 0.929-1.382 | 0.283 |
| Lymph node
metastasis | 1.415 | 0.993-1.991 | 0.066 |
| TNM stage | 2.139 | 1.527-2.896 | 0.008 |
| LINC00628
expression | 2.161 | 1.452-3.051 | 0.005 |
Furthermore, the prognostic value of LINC00628 in
patients with LUAD and LUSC were also analyzed. In LUAD tumor
tissue samples, high LINC00628 was associated with worse overall
survival (Fig. 3B; log-rank
P=0.028). Univariate Cox regression analysis results demonstrated
the relationship of lymph node metastasis, TNM stage and LINC00628
expression with overall survival of patients with LUAD (P<0.05),
and the results from multivariate Cox regression analysis also
indicated that LINC00628 expression (HR=2.437; 95% CI=1.551-3.496;
P=0.001), lymph node metastasis (HR=1.532; 95% CI, 1.035-2.159;
P=0.047), TNM stage (HR=2.222; 95% CI, 1.533-2.935; P=0.006) were
independently associated with the survival prognosis of patients
with LUAD (Table III). However,
in LUSC, although high LINC00628 expression was associated with
worse overall survival (Fig. 3C;
log-rank P=0.042), the results of multivariate Cox regression
analysis suggested that there was no statistical significance
between LINC00628 expression and the prognosis of patients with
LUSC (HR=1.896; 95% CI, 0.952-2.886; P=0.088). Nevertheless, TNM
stage (HR=2.201; 95% CI, 1.417-3.071; P=0.013) was an independent
factor for the prognosis of patients with LUSC (Table IV).
 | Table III.Cox regression analysis for patients
with lung adenocarcinoma. |
Table III.
Cox regression analysis for patients
with lung adenocarcinoma.
| A, Univariate
analysis |
|---|
|
|---|
| Variables | HR | 95% CI | P-value |
|---|
| Age | 1.331 | 0.727-1.992 | 0.398 |
| Sex | 1.454 | 0.713-2.348 | 0.551 |
| Smoking | 1.278 | 0.749-1.873 | 0.412 |
| Tumor size | 1.496 | 0.888-2.256 | 0.072 |
| Lymph node
metastasis | 1.583 | 1.185-2.354 | 0.037 |
| TNM stage | 2.639 | 1.798-4.551 | 0.002 |
| LINC00628
expression | 2.949 | 1.982-5.173 | <0.001 |
|
| B, Multivariate
analysis |
|
|
Variables | HR | 95% CI | P-value |
|
| Age | 1.302 | 0.716-1.647 | 0.549 |
| Sex | 1.465 | 0.730-2.232 | 0.672 |
| Smoking | 1.272 | 0.746-1.808 | 0.456 |
| Tumor size | 1.464 | 0.871-2.193 | 0.108 |
| Lymph node
metastasis | 1.532 | 1.035-2.159 | 0.047 |
| TNM stage | 2.222 | 1.533-2.935 | 0.006 |
| LINC00628
expression | 2.437 | 1.551-3.496 | 0.001 |
 | Table IV.Cox regression analysis for patients
with lung squamous cell carcinoma. |
Table IV.
Cox regression analysis for patients
with lung squamous cell carcinoma.
| A, Univariate
analysis |
|---|
|
|---|
| Variables | HR | 95% CI | P-value |
|---|
| Age | 1.158 | 0.781-1.683 | 0.342 |
| Sex | 1.504 | 0.803-2.374 | 0.261 |
| Smoking | 1.257 | 0.668-1.986 | 0.552 |
| Tumor size | 1.334 | 0.923-1.894 | 0.094 |
| Lymph node
metastasis | 1.523 | 0.982-2.180 | 0.061 |
| TNM stage | 2.473 | 1.652-3.385 | 0.008 |
| LINC00628
expression | 1.998 | 0.986-3.165 | 0.052 |
|
| B, Multivariate
analysis |
|
| Variables | HR | 95% CI | P-value |
| Age | 1.126 | 0.776-1.676 | 0.354 |
| Sex | 1.445 | 0.790-2.201 | 0.277 |
| Smoking | 1.263 | 0.675-1.955 | 0.531 |
| Tumor size | 1.265 | 0.879-1.746 | 0.179 |
| Lymph node
metastasis | 1.408 | 0.961-2.052 | 0.074 |
| TNM stage | 2.201 | 1.417-3.071 | 0.013 |
| LINC00628
expression | 1.896 | 0.952-2.886 | 0.088 |
Discussion
It has been reported that aberrant expression of
lncRNA is associated with the progression of various cancer types
(22–24). Several lncRNA molecules, such as
lncRNA PTAR (25) and LINC01234
(26), have been reported to be
abnormally expressed in NSCLC. In the present study, bioinformatics
analysis of TCGA datasets demonstrated that the expression levels
of LINC00628 were significantly upregulated in LUAD and LUSC tumor
samples compared with normal tissue samples. In addition, analysis
of our own clinical samples suggested that the expression levels of
LINC00628 were significantly increased in NSCLC, LUAD and LUSC
tumor compared with adjacent normal tissue samples. In addition,
the expression of LINC00628 was significantly associated with tumor
size, histological type, lymph node metastasis and TNM stage in
patients with NSCLC.
Furthermore, dysregulation of LINC00628 has been
implicated in other malignancies (27,28).
For example, the overexpression of LINC00628 inhibits the
proliferation, invasion and migration and promotes the apoptosis of
osteosarcoma cells (27).
LINC00628 expression is downregulated in the gastric cancer tissue
and also suppresses gastric cancer cell proliferation and migration
(28). The findings of the present
study suggested that the expression levels of LINC00628 were
significantly upregulated in LUAD and LUSC tissue samples,
indicating that LINC00628 might be involved in tumor
progression.
Increasing evidence indicates that lncRNA molecules
may serve as potential biomarkers for NSCLC prognosis (15,29).
Schmidt et al (30)
reported that MALAT-1 expression levels were associated with the
survival of patients with NSCLC, and that this lncRNA acted as an
oncogene. Additionally, Lu et al (31) validated LINC00673 as a novel
oncogenic lncRNA and demonstrated the molecular mechanism by which
it promotes NSCLC. In the present study, high LINC00628 was
associated with worse overall survival in all patients with NSCLC,
and LINC00628 expression could be used as the independent
prognostic factor in LUAD.
In a study by Navarro et al (32), the lncRNA HOTTIP was proposed as a
prognostic biomarker in early-stage NSCLC, indicating the
importance of lncRNAs for prognosis prediction in NSCLC. Although
Xu et al (20) have
suggested that LINC00628 was upregulated in LUAD tissue samples and
promoted LUAD cell proliferation, migration and invasion, its
expression and clinical significance remain unexplored in other
subtypes of NSCLC. Our present study analyzed TCGA database data,
and found that patients with LUAD and LUSC with high LINC00628 had
a worse overall survival than those with low LINC00628 levels,
which was consistent with the results obtained for patients with
NSCLC. In the present study, survival analysis was carried out for
patients with NSCLC, LUAD and LUSC separately. High LINC00628 was
associated with worse overall survival in both LUAD and LUSC tumor
tissue samples. However, multivariate Cox regression analysis
indicated that LINC00628 expression was an independent prognostic
factor for patients with LUAD, but not those with LUSC. Despite
this result, the Kaplan-Meier survival curves generated our LUSC
clinical samples indicated a significant difference in the survival
of patients with low and high LINC00628 levels, which was at odds
with the survival curves constructed using TCGA datasets. These
paradoxical results may be mainly due to the differences in sample
size and follow-up time between the two analyses. Indeed, the
number of clinical samples was smaller than that of the TCGA
datasets. Indeed, only 55 patients with LUSC were included in this
study and collecting additional LUSC tissue samples for more
precise statistical analysis may prove useful in future studies. In
addition, the follow-up time in the present study was ≤60 months,
whereas the longest follow-up time for TCGA data was >150
months. In TCGA LUSC data, the survival rate appears to differ
between the high and low-expression groups up to 100 months of
patient follow-up. However, between 100 and 150 months, the
survival of the low-expression group decreases at a faster rate
than that of the high expression group, indicating that LINC00628
may be associated with early, but not late-stage survival.
Therefore, it may be hypothesized that high LINC00628 expression
was also associated with poor overall survival in LUAD. LINC00628
might represent a potential prognostic biomarker for patients with
LUAD.
Moreover, ROC curve analysis indicated that
LINC00628 had the ability to discriminate between LUAD and LUSC.
Previous studies have identified multiple components of the immune
system and molecules that might serve as diagnostic biomarkers for
LUAD and LUSC (33,34). For instance, Shinmura et al
(33) have suggested that chloride
channel accessory 2 might be an immunohistochemical diagnostic
biomarker for differentiating between primary LUSC and primary
LUAD. In addition, fatty acids from erythrocyte total lipids might
be used as diagnostic biomarkers of LUAD and LUSC, as demonstrated
in the study by de Castro et al (34). Similarly, LINC00628 also has the
potential to be a diagnostic biomarker for LUAD and LUSC.
There are some limitations to this study. As
aforementioned, the limited sample size, especially for the LUSC
samples (only 55 patients with LUSC), may have affected the
accuracy of the results. Thus, additional LUSC tissue samples
should be collected for more precise statistical analysis in
subsequent studies. Moreover, in vitro experiments were not
carried out, including analyses of the effects of LINC00628 on
NSCLC cell proliferation, migration, invasion, to explore the
different roles that LINC00628 might have in LUAD and LUSC based on
the differential expression and clinical significance of LINC00628
in the two subtypes found in the present study. Thus, future
studies will explore the biological function of LINC00628 using
in vitro analyses. In addition, the NSCLC study population
only contained LUAD and LUSC subtypes. and further analysis in
other NSCLC subtypes is necessary.
In conclusion, LINC00628 expression levels were
increased in NSCLC tumor tissue samples and associated with tumor
development and survival prognosis. The elevated LINC00628
expression in LUAD compared with LUSC was independently associated
with LUAD overall survival, but this relationship was not found in
LUSC. Therefore, future studies should not only explore the
biological function of LINC00628 in NSCLC, but also focus on the
functional differences of LINC00628 in different NSCLC subtypes.
The findings of the present study suggest that LINC00628 may become
a diagnostic and prognostic biomarker for different subtypes of
NSCLC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TL conceived and designed the study. SX and XL
analyzed and interpreted the data. TL and SX carried out the
experiments. All authors wrote and revised the manuscript. TL and
XL confirmed the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All experimental procedures were carried out in
accordance with the guidelines of The Ethics Committee of Weifang
People's Hospital and The Declaration of Helsinki. The present
study was approved by The Ethics Committee of Weifang People's
Hospital. Written informed consent was obtained from each
patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brown S, Banfill K, Aznar MC, Whitehurst P
and Faivre Finn C: The evolving role of radiotherapy in non-small
cell lung cancer. Br J Radiol. 92:201905242019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang Q, Tang Y, Tang C, Cong H, Wang X,
Shen X and Ju S: Diminished LINC00173 expression induced miR-182-5p
accumulation promotes cell proliferation, migration and apoptosis
inhibition via AGER/NF-κB pathway in non-small-cell lung cancer. Am
J Transl Res. 11:4248–4262. 2019.PubMed/NCBI
|
|
4
|
Duma N, Santana-Davila R and Molina JR:
Non-small cell lung cancer: Epidemiology, screening, diagnosis, and
treatment. Mayo Clin Proc. 94:1623–1640. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li N, Zhao J, Ma Y, Roy B, Liu R,
Kristiansen K and Gao Q: Dissecting the expression landscape of
mitochondrial genes in lung squamous cell carcinoma and lung
adenocarcinoma. Oncol Lett. 16:3992–4000. 2018.PubMed/NCBI
|
|
6
|
Faruki H, Mayhew GM, Serody JS, Hayes DN,
Perou CM and Lai-Goldman M: Lung adenocarcinoma and squamous cell
carcinoma gene expression subtypes demonstrate significant
differences in tumor immune landscape. J Thorac Oncol. 12:943–953.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tomasini P, Khobta N, Greillier L and
Barlesi F: Ipilimumab: Its potential in non-small cell lung cancer.
Ther Adv Med Oncol. 4:43–50. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Y, Liu H, Shi X, Yao Y, Yang W and Song
Y: The long non-coding RNA HNF1A-AS1 regulates proliferation and
metastasis in lung adenocarcinoma. Oncotarget. 6:9160–9172. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long Non-Coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tasharrofi B and Ghafouri-Fard S: Long
Non-coding RNAs as regulators of the mitogen-activated protein
kinase (MAPK) pathway in cancer. Klin Onkol. 31:95–102. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei L, Yi Z, Guo K and Long X: Long
noncoding RNA BCAR4 promotes glioma cell proliferation via
EGFR/PI3K/AKT signaling pathway. J Cell Physiol. 234:23608–23617.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Su Y, Lu J, Chen X, Liang C, Luo P, Qin C
and Zhang J: Long non-coding RNA HOTTIP affects renal cell
carcinoma progression by regulating autophagy via the
PI3K/Akt/Atg13 signaling pathway. J Cancer Res Clin Oncol.
145:573–588. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ghafouri-Fard S, Abak A, Tondro Anamag F,
Shoorei H, Majidpoor J and Taheri M: The emerging role of
non-coding RNAs in the regulation of PI3K/AKT pathway in the
carcinogenesis process. Biomed Pharmacother. 137:1112792021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen DQ, Zheng XD, Cao Y, He XD, Nian WQ,
Zeng XH and Liu XY: Long non-coding RNA LINC00628 suppresses the
growth and metastasis and promotes cell apoptosis in breast cancer.
Eur Rev Med Pharmacol Sci. 21:275–283. 2017.PubMed/NCBI
|
|
17
|
Zhang MC, Zhang L, Zhang MQ, Yang GX, Wang
FZ and Ding P: Downregulated LINC00628 aggravates the progression
of colorectal cancer via inhibiting p57 level. Eur Rev Med
Pharmacol Sci. 24:1763–1770. 2020.PubMed/NCBI
|
|
18
|
Tian Y, Yu M, Sun L, Liu L, Wang J, Hui K,
Nan Q, Nie X, Ren Y and Ren X: Distinct patterns of mRNA and lncRNA
expression differences between lung squamous cell carcinoma and
adenocarcinoma. J Comput Biol. 27:1067–1078. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu M, Tian Y, Wu M, Gao J, Wang Y, Liu F,
Sheng S, Huo S and Bai J: A comparison of mRNA and circRNA
expression between squamous cell carcinoma and adenocarcinoma of
the lungs. Genet Mol Biol. 43:e202000542020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu SF, Zheng Y, Zhang L, Wang P, Niu CM,
Wu T, Tian Q, Yin XB, Shi SS, Zheng L and Gao LM: Long Non-coding
RNA LINC00628 interacts epigenetically with the LAMA3 promoter and
contributes to lung adenocarcinoma. Mol Ther Nucleic Acids.
18:166–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu S, Zheng Q, Xiong J, Wu H, Wang W and
Zhou W: Long non-coding RNA MVIH promotes cell proliferation,
migration, invasion through regulating multiple cancer-related
pathways, and correlates with worse prognosis in pancreatic ductal
adenocarcinomas. Am J Transl Res. 12:2118–2135. 2020.PubMed/NCBI
|
|
23
|
Wang Y, Ding X, Hu H, He Y, Lu Z, Wu P,
Tian L, Xia T, Yin J, Yuan H, et al: Long non-coding RNA lnc-PCTST
predicts prognosis through inhibiting progression of pancreatic
cancer by downregulation of TACC-3. Int J Cancer. 143:3143–3154.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng J, Li J, Qie P, Li Z, Xu Y and Tian
Z: Long non-coding RNA (lncRNA) PGM5P4-AS1 inhibits lung cancer
progression by up-regulating leucine zipper tumor suppressor
(LZTS3) through sponging microRNA miR-1275. Bioengineered.
12:196–207. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu W, Sun Z, Yang L, Han Y, Yue L, Deng L
and Yao R: lncRNA PTAR promotes NSCLC cell proliferation, migration
and invasion by sponging microRNA-101. Mol Med Rep. 20:4168–4174.
2019.PubMed/NCBI
|
|
26
|
Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang
J, Lu B, Yuan L, Sun M and Wang Z: Integrative analysis of NSCLC
identifies LINC01234 as an oncogenic lncRNA that interacts with
HNRNPA2B1 and regulates miR-106b biogenesis. Mol Ther.
28:1479–1493. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He R, Wu JX, Zhang Y, Che H and Yang L:
LncRNA LINC00628 overexpression inhibits the growth and invasion
through regulating PI3K/Akt signaling pathway in osteosarcoma. Eur
Rev Med Pharmacol Sci. 22:5857–5866. 2018.PubMed/NCBI
|
|
28
|
Zhang ZZ, Zhao G, Zhuang C, Shen YY, Zhao
WY, Xu J, Wang M, Wang CJ, Tu L, Cao H and Zhang ZG: Long
non-coding RNA LINC00628 functions as a gastric cancer suppressor
via long-range modulating the expression of cell cycle related
genes. Sci Rep. 6:274352016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song JY, Li XP, Qin XJ, Zhang JD, Zhao JY
and Wang R: A fourteen-lncRNA risk score system for prognostic
prediction of patients with non-small cell lung cancer. Cancer
Biomark. 29:493–508. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H,
Kong J, Ding K, Shen HM, Wu H, et al: Long non-coding RNA linc00673
regulated non-small cell lung cancer proliferation, migration,
invasion and epithelial mesenchymal transition by sponging
miR-150-5p. Mol Cancer. 16:1182017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Navarro A, Moises J, Santasusagna S,
Marrades RM, Viñolas N, Castellano JJ, Canals J, Muñoz C, Ramírez
J, Molins L and Monzo M: Clinical significance of long non-coding
RNA HOTTIP in early-stage non-small-cell lung cancer. BMC Pulm Med.
19:552019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shinmura K, Igarashi H, Kato H, Kawanishi
Y, Inoue Y, Nakamura S, Ogawa H, Yamashita T, Kawase A, Funai K and
Sugimura H: CLCA2 as a novel immunohistochemical marker for
differential diagnosis of squamous cell carcinoma from
adenocarcinoma of the lung. Dis Markers. 2014:6192732014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
de Castro J, Rodríguez MC,
Martínez-Zorzano VS, Sánchez-Rodríguez P and Sánchez-Yagüe J:
Erythrocyte fatty acids as potential biomarkers in the diagnosis of
advanced lung adenocarcinoma, lung squamous cell carcinoma, and
small cell lung cancer. Am J Clin Pathol. 142:111–120. 2014.
View Article : Google Scholar : PubMed/NCBI
|