Introduction
Neoadjuvant chemotherapy (NAC) is the standard of
care for patients with locally advanced breast cancer (1). However, breast cancer continues to
progress during taxane-based NAC in 3–6% of patients (2,3).
These patients have poorer survival than patients whose breast
cancer responds well to NAC (2).
Taxanes are important breast cancer drugs used
worldwide for both adjuvant therapy and treatment after recurrence.
They arrest proliferation and cause the death of cancer cells
(4). The main target of taxanes is
β-tubulin. After binding to β-tubulin, taxanes stabilize the
microtubules and disrupt their dynamics, thus interrupting
microtubule-associated functions in mitotic cells (4). Mechanisms of taxane resistance have
been widely studied. One mechanism involves microtubule
alterations, whereby mutations of the TUBA1A and TUBB
genes and alterations in microtubule-associated proteins affect
taxane binding and microtubule dynamics (5). Another mechanism involves ATP-binding
cassette (ABC) transporters. ABC transporters work as drug efflux
pumps that mediate the transmembrane transport of intracellular
substrates, such as taxanes (6).
Overexpression of the ABC transporter, ABCB1, and upregulation of
ABC transporters have been reported to affect taxane resistance
(7). Although many other
mechanisms (e.g., DNA repair and reactive oxygen species
metabolism) have been reported (4), the mechanism of taxane resistance
remains unclear.
The development of next-generation sequencing (NGS)
has allowed extensive genomic information on individual tumors to
be obtained in an extremely short period of time. Specific
mutations identified by NGS may affect therapeutic efficacy and
induce resistance to some treatments (8). For example, NGS analysis reportedly
indicated that PIK3CA mutations reduced sensitivity to
anthracycline-taxane-based NAC (9).
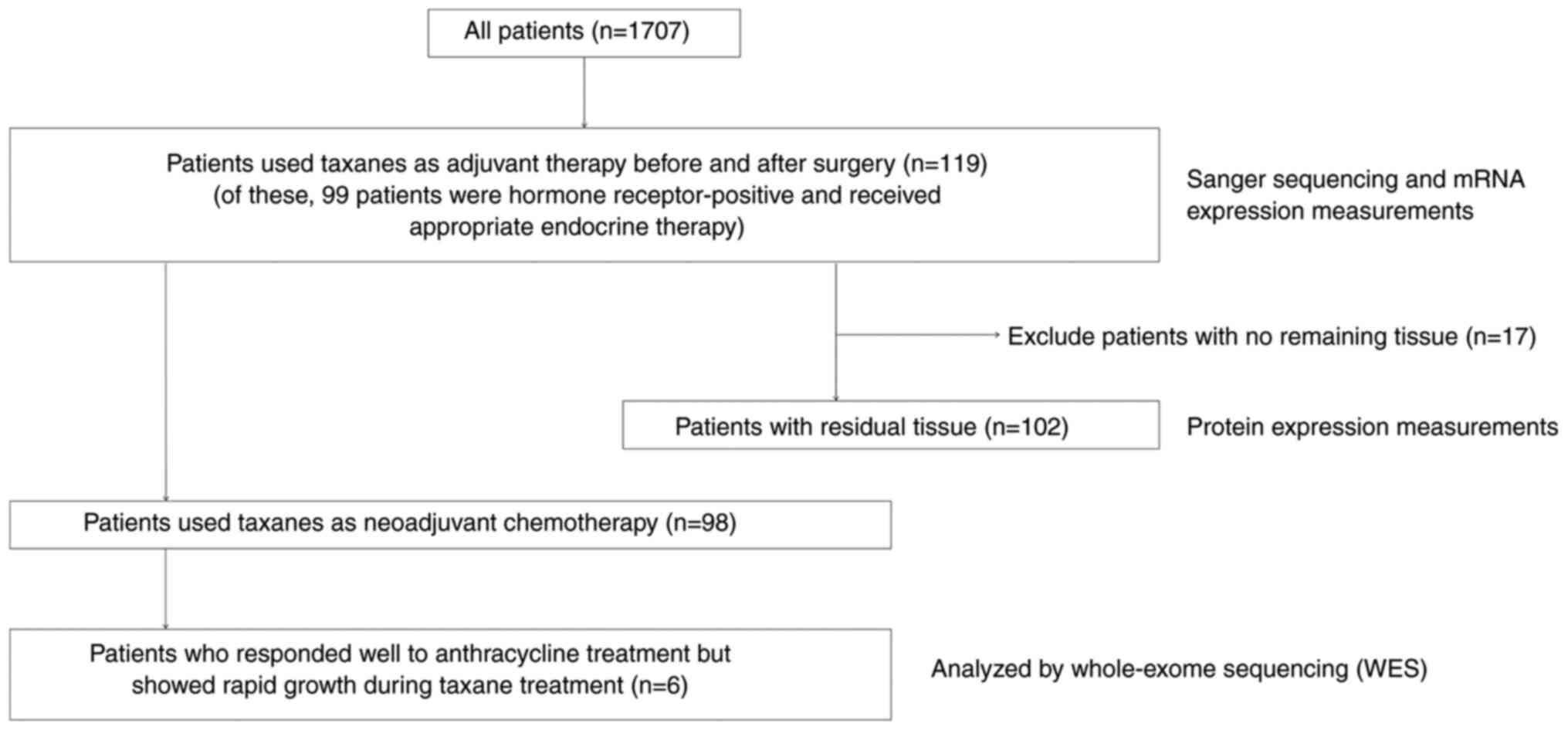
The present study included six patients with breast
cancer whose tumors responded well to anthracycline treatment but
showed rapid growth during taxane treatment with NAC. Therefore,
the hypothesis was that these six tumors may express common
mutations involved in taxane resistance. In the present study, the
novel mechanisms of taxane resistance in breast cancer using NGS
were examined.
Materials and methods
Patients and breast cancer
tissues
A total of 1,707 patients with primary breast cancer
were treated at the Department of Breast Surgery, Nagoya City
University Hospital, between January 2001 and December 2012,
including 98 patients with primary invasive breast cancer who were
treated with anthracycline-taxane-based NAC. Of these 98 patients,
six had tumors that initially responded well to anthracycline
treatment but showed rapid growth during taxane treatment. Breast
cancer tissue samples from these six patients were collected and
analyzed by WES (Fig. 1). The
inclusion criterion was patients with primary breast cancer who had
undergone surgical treatment and the exclusion criterion was stage
IV breast cancer.
Of the 1,707 patients, 119 patients used taxanes as
adjuvant therapy before and after surgery. Sanger sequencing and
mRNA expression measurements were performed using the 119 samples.
The measured genes were V-type proton ATPase catalytic subunit A
(ATP6V1A) and apolipoprotein B mRNA editing enzyme catalytic
subunit 3F (APOBEC3F). Samples of breast cancer tissues from
102 of these 119 patients were also included to evaluate ATP6V1A
and APOBEC3F protein expression levels. All 1,707 patients
underwent surgical treatment. Concerning treatment, the 119
patients who underwent taxane treatment also received 12 cycles of
paclitaxel (80 mg/m2 once weekly) or four cycles of
docetaxel (75 mg/m2 once every 3 weeks) for adjuvant
therapy. Patients with hormone receptor-positive breast cancer
(n=99) received appropriate endocrine therapy (Fig. 1). Computed tomography scans were
performed to determine the tumor diameter and efficacy of each NAC
regimen.
The study protocol was approved by the Institutional
Review Board of Nagoya City University (approval no. 70-00-0172;
Nagoya, Japan) and conformed to the guidelines of the 1996
Declaration of Helsinki. Written informed consent for the use of
resected tumor tissues was provided by all patients before
treatment.
DNA extraction and WES
DNA was extracted from approximately 500 mg of
frozen breast cancer tissue using a salt precipitation method, as
described previously (10). DNA
samples were enriched for WES using a SureSelect XT Human All Exon
v.5 Kit (Agilent Technologies). Sequencing was performed on an
Illumina HiSeq system (Illumina) with 100-bp paired-end runs,
following the manufacture's protocols. The primer sequences used
were: Read1, ACACTCTTTCCCTACACGACGCTCTTCCGATCT; Read2,
GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT. Image analysis and base calling
were performed using Genome Analyzer Pipeline version 1.5 (SeqNova™
CSmapping, Hokkaido System Science Co., Ltd.) with the default
parameters. Sequence reads were aligned to human genome assembly
hg19 (GRCh37) and variants were identified using DNAnexus software
(Palo Alto) using default parameters.
Structural modeling of ATP6V1A and
APOBEC3F proteins
ATP6V1A protein conformations from wild-type and
mutant genes were simulated using PDFAMS software (In-Silico
Sciences Inc.) with reference to the protein data bank (PDB) ID
5BN4_A protein. ATP6V1A octamer conformations were simulated with
reference to PDB ID 3VR6_ABCDEFGH protein. The APOBEC3F N-terminal
and C-terminal conformations from wild-type and mutant genes were
simulated with reference to PDB ID 5K83_A and ID 5K83_D proteins,
respectively. Each terminal model was connected to construct the
complete predicted model of APOBEC3F.
Polymerase chain reaction (PCR) and
Sanger sequencing
DNA was extracted from the breast cancer tissues of
119 taxane-treated breast cancer patients. PCR was performed once
using 1 µg DNA from each sample. The PCR reactions were performed
using an LA-Taq Kit (Takara Bio Inc.) in a 50-µl reaction volume,
as described previously (10). The
primer sequences used were: ATP6V1A: (forward)
5′-ACTCTGGTTAAGTAGTTGGTC-3′, (reverse)
5′-ATCGTTTGAACCCAGGAGGCAGAG-3′; APOBEC3F: (forward)
5′-AGAAATGTGCTTCCTCTCTTGGTTC-3′, (reverse)
5′-ATCTTTCATGCTGTTCCTCCCGCTC-3′. The cycling conditions were as
follows: initial denaturation at 94°C for 10 min, and 40 cycles of
94°C for 30 sec, 53°C for 30 sec and 72°C for 30 sec for the
melting, annealing, and elongation phases of the reaction,
respectively. The products were purified using a Qiagen PCR
purification kit (Qiagen, Inc.). Mutations in the two genes were
analyzed by direct sequencing using an ABI Prism 3100 analyzer
(Thermo Fisher Scientific, Inc.). The mutations were then analyzed
by BLAST, and chromatograms were analyzed by manual review in both
forward and reverse sequences (10).
RNA extraction and reverse
transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from frozen breast cancer
tissue sections using an RNeasy Mini Kit (Qiagen) according to the
manufacturer's protocol, as described previously. RT was performed
using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol
(11). The High-Capacity cDNA
Reverse Transcription Kit included RT Buffer, RT Random Primers,
dNTP Mix and MultiScribe Reverse Transcriptase. TaqMan Gene
Expression assays (Thermo Fisher Scientific, Inc.) were used to
measure mRNA expression levels of ATP6V1A, APOBEC3F, and the
housekeeping gene ACTB. RT-qPCR was performed using a 7500
Fast Real-Time PCR System (Thermo Fisher Scientific, Inc.). Samples
were amplified independently in duplicate. The results were
converted into relative concentrations using an in-run standard
curve, and then normalized to the mean values of ACTB to
account for variations in the amount of input cDNA (11). The assay numbers were Hs01097169_m1
for ATP6V1A, Hs01665324_m1 for APOBEC3F, and 4333762T
for ACTB (Thermo Fisher Scientific, Inc.).
Immunohistochemistry (IHC)
Tissues were fixed by immersion in 10% neutral
buffered formalin at room temperature for 24–48 h and then embedded
in paraffin to prepare sections. A section (4 µm) from each
paraffin-embedded specimen was first stained with hematoxylin and
eosin to ascertain whether it included enough invasive ductal
carcinoma cells and if the fixation quality was adequate for IHC
analysis, as described previously (11). Serial sections were then prepared
from suitable tissue blocks and float-mounted on adhesive-coated
glass slides for the staining of estrogen receptor α (ERα),
progesterone receptor (PgR), and human epidermal growth factor
receptor 2 (HER2) using the following primary antibodies: mouse
monoclonal anti-human antibodies against ERα (1:100, 1D5; Dako),
PgR (1:100, PgR636; Dako), and rabbit anti-human c-erbB2
oncoprotein antibody (1:200; Dako) to stain for HER2, as well as
rabbit polyclonal anti-human ATP6V1A antibody (1:100, ab103445;
Abcam) and rabbit polyclonal anti-human APOBEC3D+APOBEC3F antibody
(1:50, ab74205; Abcam). The Dako EnVision system (Dako) was used to
detect ERα, PgR, and HER2. Expression levels of ERα and PgR were
scored by assigning proportion and intensity scores, according to
Allred's procedure (12). HER2
immunostaining was evaluated using the HercepTest (Dako) and
fluorescence in situ hybridization (13). For the IHC analysis of ATP6V1A and
APOBEC3F, tissue microarrays were prepared on 2 mm diameter slides
after confirming whether an appropriate number of invasive ductal
carcinoma cells were present and whether the fixation quality was
suitable for IHC analysis. The ATP6V1A and APOBEC3F protein
expression levels were evaluated according to the cytoplasmic
H-score, which was calculated by assessing the entire slide using
the Aperio Image Scope system (Leica Biosystems). Cytoplasmic
staining intensity (0, 1+, 2+, or 3+) was determined for each
cancer cell. An H-score was assigned using the formula [1 × (%
cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)] (14,15).
Prediction methods
The damaging effects of mutations were predicted
using the web-based algorithms PolyPhen2 v2.2.2 (http://genetics.bwh.harvard.edu/pph2/),
PROVEAN v1.1 (http://provean.jcvi.org/index.php), SIFT v5.0.3
(http://sift.bii.a-star.edu.sg/), FATHMM
v2.3 (http://fathmm.biocompute.org.uk/) and Mutation Taster
(https://www.mutationtaster.org/).
Polyphen2, PROVEAN and SIFT classification tools were used to
extract mutations that are most likely to affect protein function.
Two additional online tools (FATHMM and Mutation Taster) were used
to study the effect on the protein function. FATHMM and Mutation
Taster were selected because they are widely used and score the
pathological effects of mutations that have been extracted on the
basis of the protein function. The relationship between the genes
was examined using GeneMANIA v3.6.0 (https://genemania.org/).
Statistical analyses
mRNA expression levels and IHC results were analyzed
by researchers blinded to the clinical data. Statistical
calculations were performed using JMP 13 software (SAS Institute,
Inc.). Data are presented as mean ± standard deviation. Receiver
operating characteristic (ROC) analyses were used to determine the
optimal cut-off level for mRNA and protein expressions, which were
assessed using Youden's index. Disease-free survival (DFS) was
censored at the date of last follow-up if patients were still
relapse-free and alive, and overall survival (OS) was censored at
the time when patients were alive. DFS and OS were evaluated using
the Kaplan-Meier method and differences between survival curves
were assessed with the log-rank test. Cox's proportional hazards
model was used for univariate and multivariate analyses of
prognostic factors (16).
P<0.05 was considered significant. The associations between mRNA
and protein expression were assessed by Student's t-test. This
study complied with the reporting recommendations for tumor marker
prognostic studies (REMARK) criteria (17,18).
Results
Identification of common relevant
somatic mutations
Six patients whose tumors responded well to
anthracycline treatment but showed rapid growth during NAC taxane
treatment were analyzed in the present study (Table I). Computed tomography scans were
performed to determine the tumor diameter and efficacy of each NAC
regimen. The response criteria were the Response Evaluation
Criteria in Solid Tumors (RECIST) criteria (version 1.1) (19). Details are shown in Table SI. WES of the tumor samples was
also performed using Illumina HiSeq to investigate the mechanisms
responsible for taxane resistance, and in three available patient
blood samples to exclude germline variants.
 | Table I.Characteristics of breast cancer
tumors intrinsically resistant to taxanes (n=6). |
Table I.
Characteristics of breast cancer
tumors intrinsically resistant to taxanes (n=6).
| Patients | Age (years) | Histology | Nuclear grade | ER (Allred
score) | PgR (Allred
score) | HER2 IHC | HER2 FISH | Blood sample | Year of
surgery |
|---|
| 1 | 31 | Invasive ductal
carcinoma | 2 | 2 | 0 | 1+ |
| NA | 2006 |
| 2 | 36 | Invasive ductal
carcinoma | 3 | 0 | 0 | 0 |
| (+) | 2008 |
| 3 | 50 | Invasive ductal
carcinoma | 3 | 5 | 0 | 0 |
| (+) | 2009 |
| 4 | 61 | Invasive ductal
carcinoma | 3 | 4 | 0 | 1+ |
| NA | 2010 |
| 5 | 52 | Invasive ductal
carcinoma | 1 | 3 | 0 | 2+ | Negative | (+) | 2012 |
| 6 | 38 | Invasive ductal
carcinoma | 3 | 2 | 0 | 1+ |
| NA | 2012 |
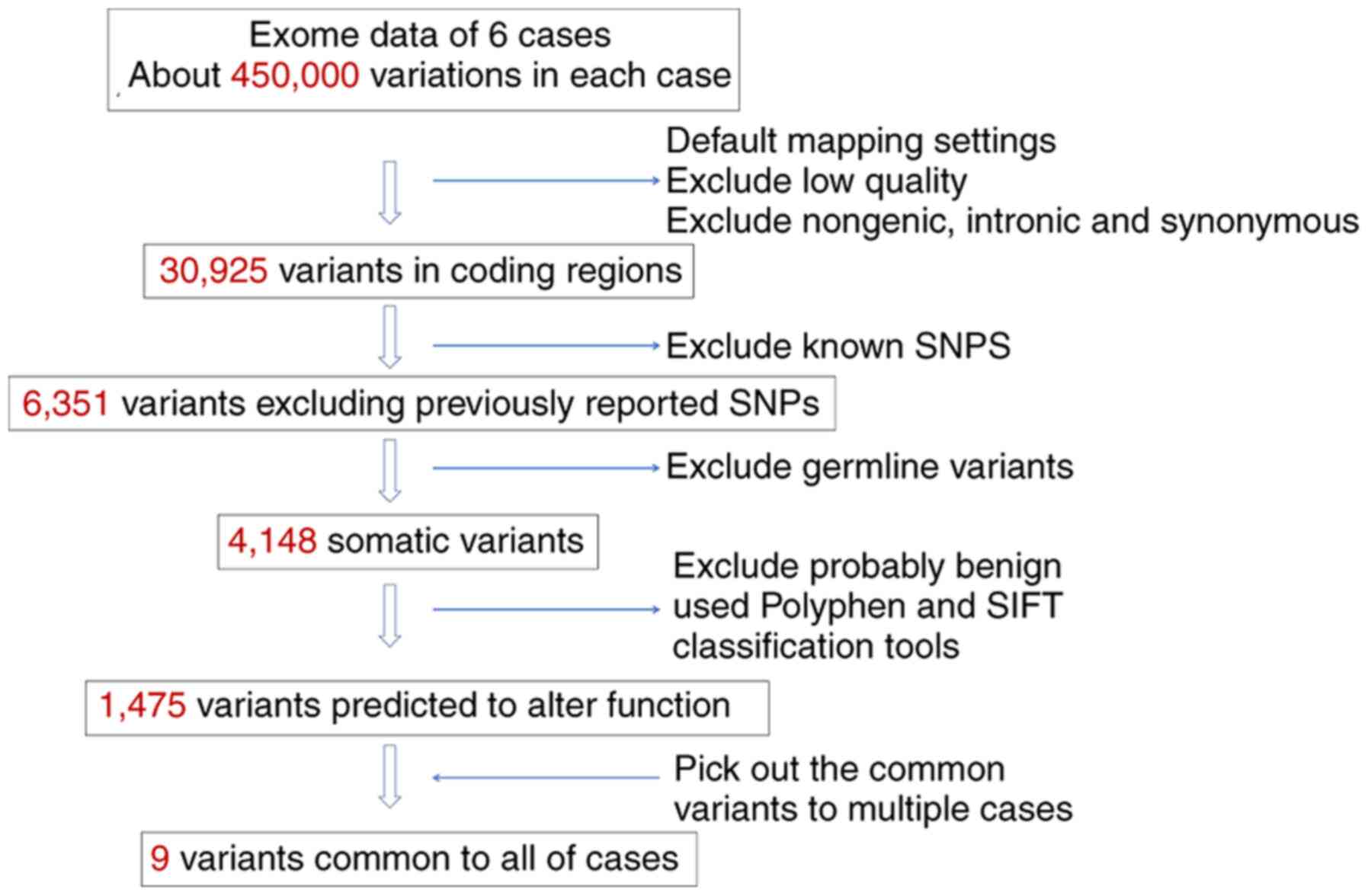
Approximately 450,000 genetic variations per
individual were identified (Fig.
2). Low-quality mapping, and all non-genomic, intronic, and
synonymous variants were excluded. Finally, 30,925 variants were
identified in coding regions. The variants were further filtered by
excluding all known variants through comparison with the dbSNP
database v130, and 6,351 variants were selected. After excluding
germline variants on the basis of exome data from the blood
samples, 4,148 somatic variants were selected. Not all mutations
are likely to be pathologically relevant, and thus those likely to
affect protein function or those that affected highly conserved
amino acids, making them functionally important, were selected. The
functional consequences of amino acid changes using the Polyphen2,
PROVEAN and SIFT classification tools were predicted, and 1,475
genes had at least one potentially destructive mutation.
Subsequently, variants common to multiple cases were sought, and
nine variants of eight different genes common to all six cases were
identified.
Protein structure visualization of
ATP6V1A and APOBEC3F somatic mutations
The nine variants were narrowed down to two
variations on two different genes: the R552P mutation of ATP6V1A
[variant allele frequency (VAF): 0.44] and the T114P mutation of
APOBEC3F (VAF: 0.45). These variations were chosen by focusing on
the role of the V-ATPase, the functional structure of ATP6V1A, in
cancer metastasis (20), and
because only T114P in APOBEC3F was predicted to damage the protein
structure by all three online prediction tools utilized in the
present study.
The structural changes caused by the R552P mutation
of ATP6V1A are shown in Fig.
3A. ATP6V1A encodes subunit A of the V1 domain of the
heteromultimeric vacuolar H+-ATPase (V-ATPase) complex.
The V1 domain forms an A3B3DF catalytic
octamer with B, D, and F subunits. We considered that the
ATP6V1A R552P mutation may cause structural changes because
it was located at the border of subunit A (Fig. 3B). The pathological prediction
score was calculated using two additional online prediction tools:
the FATHMM score was 0.99 (pathogenic) and the Mutation Taster
score was 103 (disease-causing).
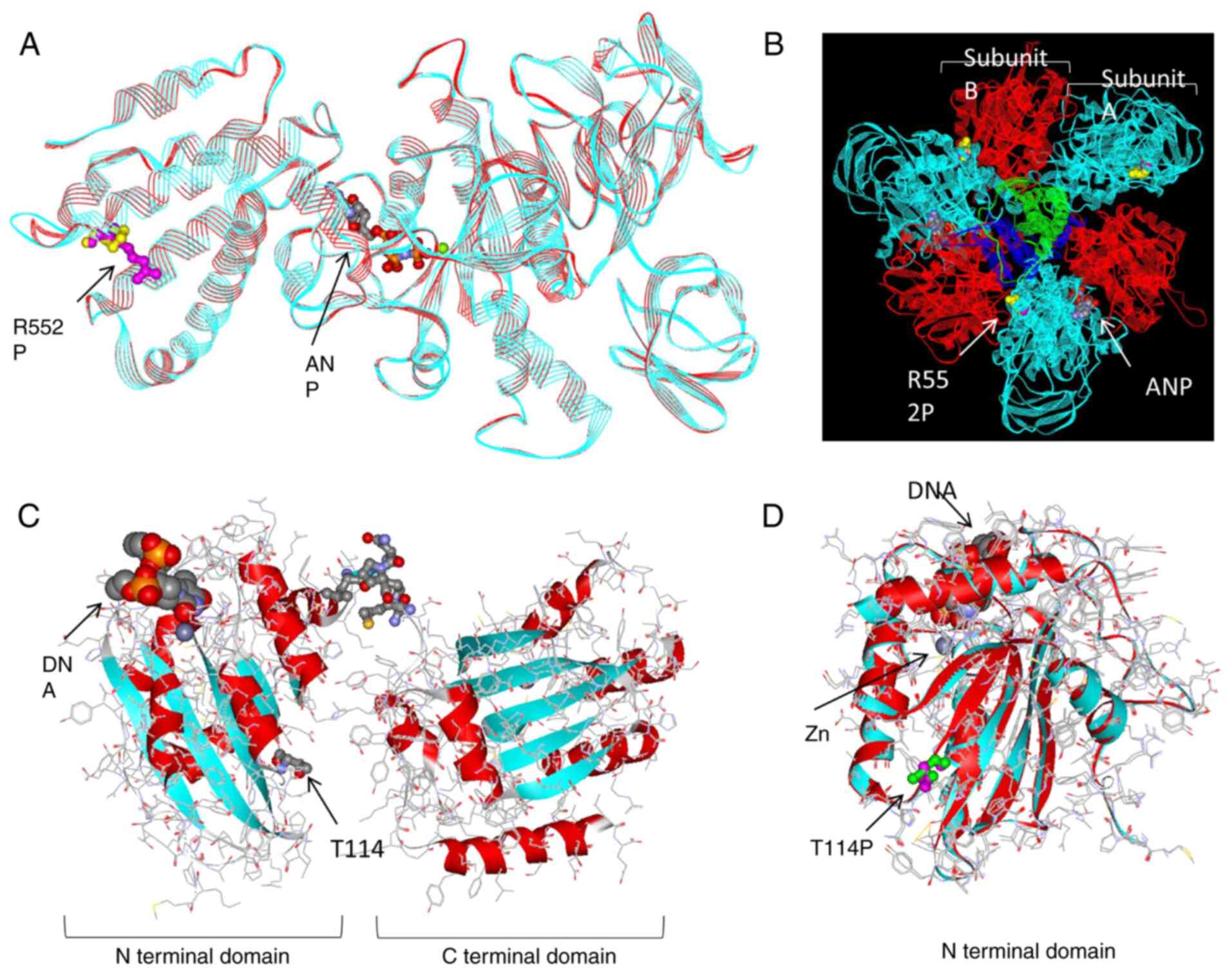 | Figure 3.Protein structure visualization of
ATP6V1A and APOBEC3F somatic mutations. (A)
Superimposed modeling of ATP6V1A. Turquoise ribbons, main wild-type
chains; red ribbons, main mutant chains; magenta spheres, R552
(wild-type); yellow spheres, P552 (mutant-type). Gray and red balls
in the center show ANP, an analog of ATP. (B) Octamer modeling of
V-ATPase V1 domain. Turquoise ribbons: subunit A (encoded by
ATP6V1A), red ribbons: subunit B, green ribbon: subunit D and blue
ribbon: subunit F; magenta and yellow spheres: R552P; gray and red
spheres: ANP. (C) Predicted model of wild-type APOBEC3F. Turquoise
and red ribbons: main wild-type chains; small gray and red spheres
in upper center: undetermined area; small gray and red spheres in
lower center: T114 (wild-type); large gray, red, and orange spheres
model: DNA. (D) Superimposed model of N-terminal domain of
APOBEC3F. Turquoise ribbons: main wild-type chains; red ribbons:
main mutant chains; green spheres: T114 (wild-type); magenta
spheres: P114 (mutant-type). ANP, phospho-aminophosphonic
acid-adenylate ester; ATP, adenosine triphosphate. |
APOBEC3F can deaminate DNA cytosine to uracil in a
zinc-dependent manner. APOBEC3F has two zinc-binding sites: one in
the N-terminal domain and one in the C-terminal domain (Fig. 3C). The results showed that the
T114P mutation may also cause some structural changes because of
its location near a zinc-binding site, which is the
deaminase-active region (Fig. 3D).
The T114P of APOBEC3F was predicted to damage the protein in three
online prediction tools at the selection stage, but when calculated
using two additional online prediction tools, the FATHMM score was
0.04 (neutral) and the Mutation Taster score was 38
(polymorphism).
No mutations were identified by Sanger
sequencing
A search for R552P in ATP6V1A and T114P in APOBEC3F
in 119 breast cancers treated with taxanes was conducted. None of
the mutations described above were found in Sanger sequencing.
mRNA expression analysis in breast
cancer patients
The functions of ATP6V1A and APOBEC3F
may be important and predicted that overexpression of
ATP6V1A or APOBEC3F may affect taxane resistance.
Thus, the correlation between the mRNA expression levels of the two
genes and patient prognosis in 119 breast cancers treated with
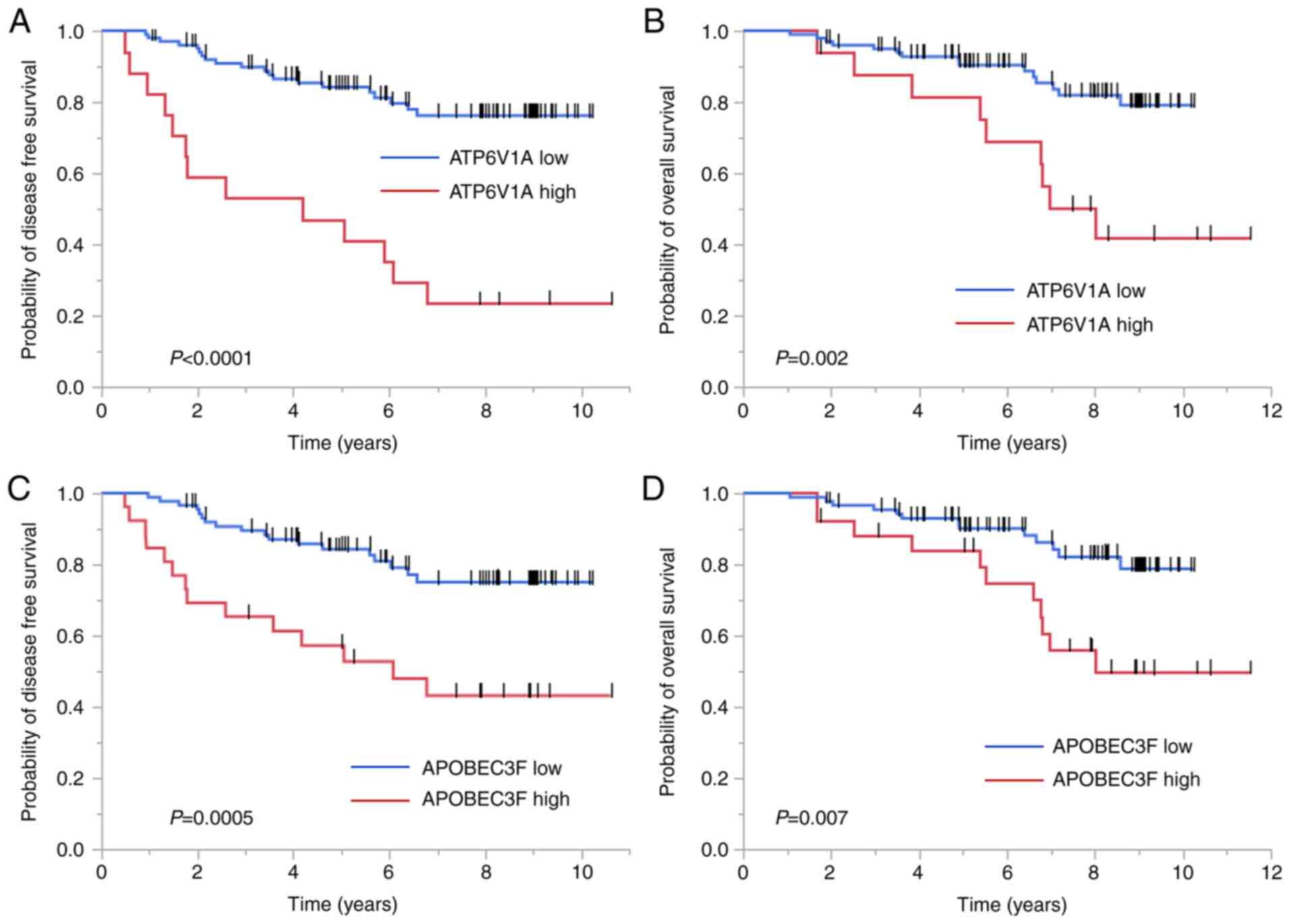
taxanes at our institute was examined (Table II). The Kaplan-Meier analysis
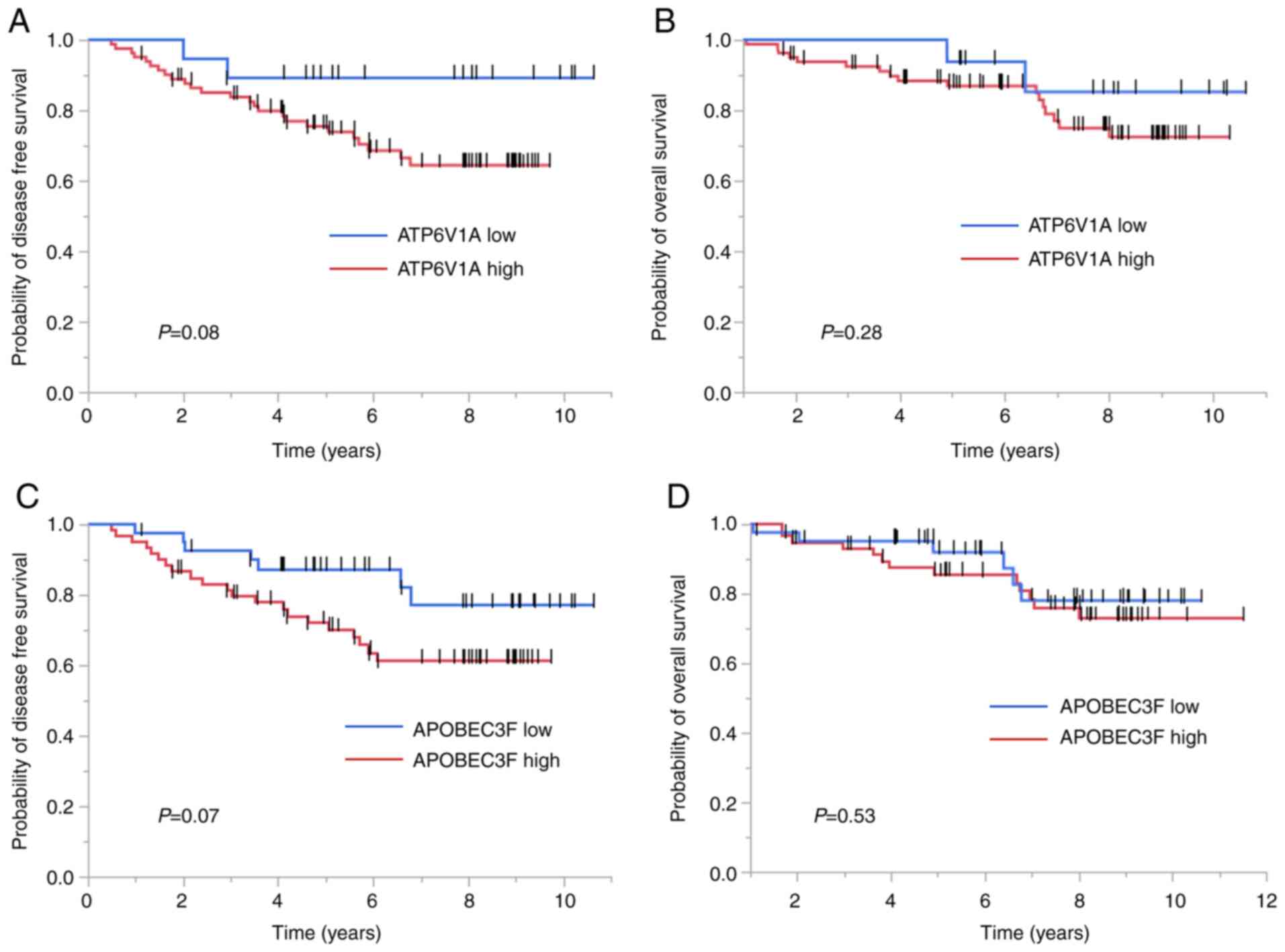
revealed that higher ATP6V1A expression was inversely
correlated with DFS (P<0.0001) and OS (P=0.002) (Fig. 4A and B). Inverse correlations were
also found between APOBEC3F expression and DFS (P=0.0005)
and OS (P=0.007) (Fig. 4C and
D).
 | Table II.Clinicopathological characteristics
of patients. |
Table II.
Clinicopathological characteristics
of patients.
|
Characteristics | Samples for mRNA
analysis (%) | Samples for
immunohisto-chemistry (%) |
|---|
| No. of
patients | 119 | 102 |
| Age (years) |
|
|
| Mean ±
SD | 49.0±9.4 | 48.8±8.9 |
|
Range | 26-70 | 26-75 |
| Histology |
|
|
|
Invasive ductal carcinoma | 103 (87) | 90 (88) |
|
Invasive lobular
carcinoma | 5 (4) | 5 (5) |
| Ductal
carcinoma in situ | 0 (0) | 0 (0) |
| Other
special types | 11 (9) | 7 (7) |
| Tumor size |
|
|
|
≤2.0 | 54 (45) | 51 (50) |
|
2.1-5.0 | 54 (45) | 43 (42) |
|
>5.0 | 8 (7) | 7 (7) |
|
Unknown | 3 (3) | 1 (1) |
| No. of positive
lymph nodes |
|
|
| 0 | 35 (29) | 32 (31) |
|
1-3 | 60 (50) | 49 (48) |
|
4-9 | 14 (12) | 14 (14) |
|
≥10 | 9 (8) | 6 (6) |
|
Unknown | 1 (1) | 1 (1) |
| Nuclear grade |
|
|
| 1 | 33 (28) | 29 (28) |
| 2 | 27 (23) | 23 (23) |
| 3 | 55 (46) | 49 (48) |
|
Unknown | 4 (3) | 1 (1) |
| ER (Allred
score) |
|
|
|
Positive (3–8) | 99 (83) | 88 (86) |
|
Negative (0–2) | 19 (16) | 14 (14) |
|
Unknown | 1 (1) | 0 (0) |
| PgR (Allred
score) |
|
|
|
Positive (3–8) | 82 (69) | 72 (70) |
|
Negative (0–2) | 33 (28) | 29 (29) |
|
Unknown | 4 (3) | 1 (1) |
| HER2 (Hercep
test) |
|
|
|
0,1+ | 92 (77) | 81 (80) |
| 2+ | 8 (7) | 7 (6) |
| 3+ | 16 (13) | 13 (12) |
|
Unknown | 4 (3) | 2 (2) |
| Adjuvant
therapy |
|
|
|
Paclitaxel | 71 (60) | 58 (57) |
|
Docetaxel | 48 (40) | 44 (43) |
The multivariate analysis revealed a high
ATP6V1A mRNA expression as an independent prognostic factor
for poor DFS (hazard ratio: 3.85; 95% confidence interval:
1.43-9.89; P=0.009; Table III).
A ROC curve analysis was carried out to determine the cut-off
values of ATP6V1A and APOBEC3F mRNA using Youden's
index (sensitivity + specificity - 1). Cut-off levels for relative
ATP6V1A and APOBEC3F mRNA expression were 3.5 and
2.1, respectively. ROC area under the curve (AUC) was 0.56 for DFS
according to ATP6V1A mRNA expression, and the ROC AUC was
0.67 for DFS according to the APOBEC3F mRNA expression.
 | Table III.Univariate and multivariate analyses
of factors predicting disease-free survival (n=119). |
Table III.
Univariate and multivariate analyses
of factors predicting disease-free survival (n=119).
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Tumor size
(cm) | 1.11 |
0.91-1.30 |
0.28 |
|
|
|
| Lymph nodes
(Positive/Negative) | 1.05 |
0.51-2.31 |
0.90 |
|
|
|
| Tumor grade | 1.60 |
1.03-2.59 | 0.03a | 1.37 | 0.81-2.44 |
0.24 |
| ER
(Positive/Negative) | 0.41 |
0.20-0.93 | 0.03a |
|
|
|
| PgR
(Positive/Negative) | 0.32 |
0.16-0.66 | 0.002a | 0.32 | 0.14-0.75 | 0.008a |
| HER2
(Positive/Negative) | 1.49 |
0.63-3.16 |
0.35 |
|
|
|
| ATP6V1A
(High/Low) | 5.29 | 2.56-10.53 |
<0.0001a | 3.85 | 1.43-9.89 | 0.009a |
| APOBEC3F
(High/Low) | 3.23 |
1.58-6.48 | 0.002a | 1.88 | 0.66-4.82 |
0.21 |
Protein expression of ATP6V1A and
APOBEC3F in breast cancer
IHC was performed to analyze the protein expression
levels in samples from 102 breast cancers treated with taxanes at
our institute (Table II).
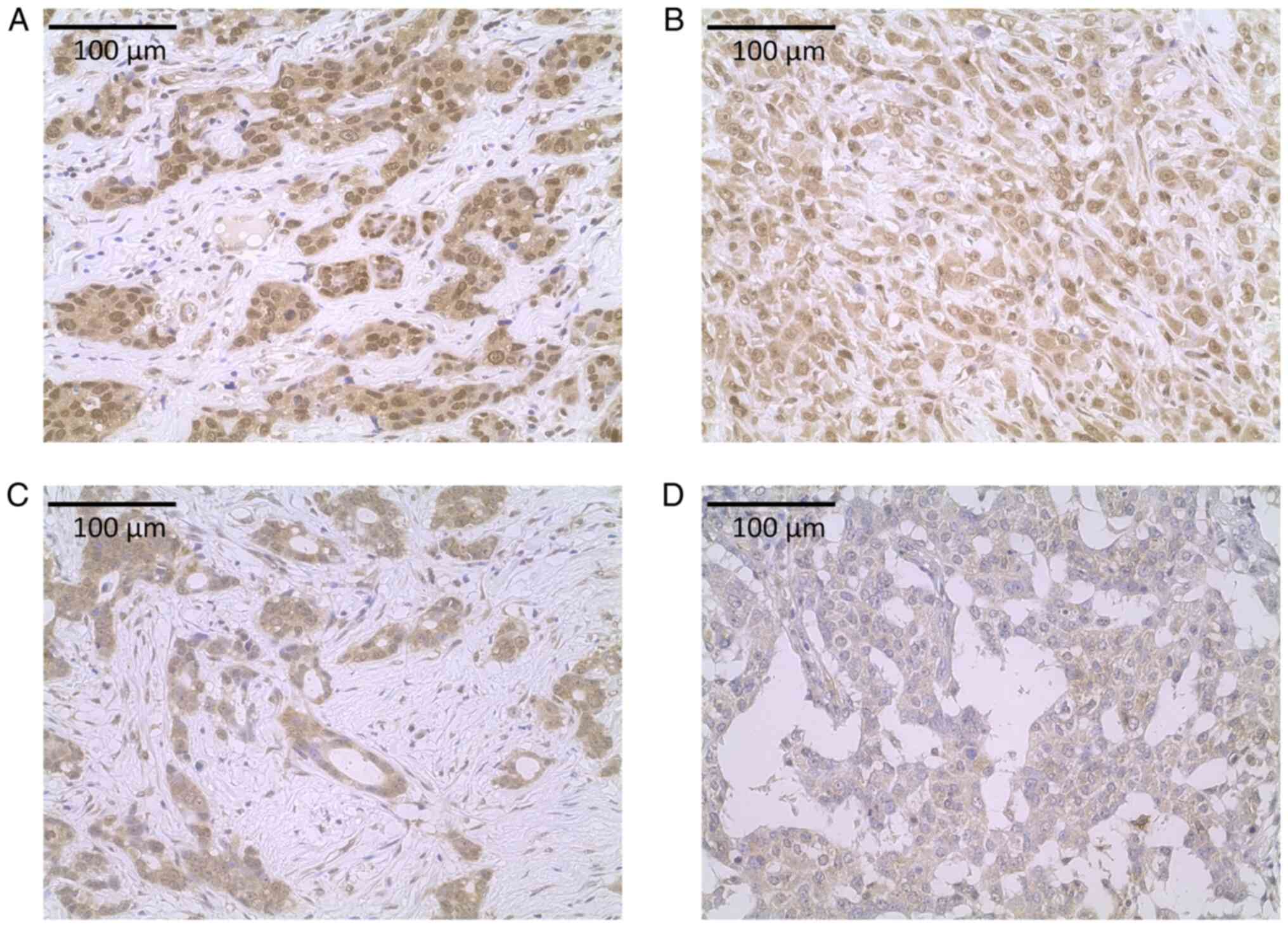
Representative images of ATP6V1A and APOBEC3F protein expression
are shown in Fig. 5. High and low
expression of ATP6V1A (Fig. 5A and
B) as well as high and low expression of APOBEC3F (Fig. 5C and D) are shown, respectively.
The correlations between ATP6V1A and APOBEC3F protein expression
levels and patient prognosis were then investigated. In the
Kaplan-Meier analyses, high ATP6V1A or APOBEC3F protein expression
showed a tendency to be associated with poor DFS. However, no
significant difference was observed (Fig. 6A and C). In addition, the
Kaplan-Meier analyses revealed no stratification of OS according to
ATP6V1A or APOBEC3F protein expression levels (Fig. 6B and D). The cut-off levels for
relative ATP6V1A and APOBEC3F H-score were 85.0 and 49.6,
respectively, while the ROC AUC for DFS was 0.49 and 0.58,
respectively. As an AUC of 0.49 or 0.58 does not indicate good
discriminatory power, the cut-off values for both mRNA levels
should be re-evaluated using a different dataset in the future.
Next, the association between mRNA and protein
expression was examined. There was no significant association
between ATP6V1A mRNA and protein expression. However, a significant
positive association was found between APOBEC3F mRNA and protein
expression (P=0.01) (data not shown).
Discussion
In this study, we identified two different mutations
in two genes that may affect taxane resistance in breast cancer
patients using WES: R552P in ATP6V1A and T114P in
APOBEC3F. Results of the present study suggested that these
mutations caused structural changes in their respective proteins.
Furthermore, high mRNA expression levels of these genes were
correlated with poor prognoses in breast cancer patients treated
with taxanes.
ATP6V1A encodes subunit A of V-ATPase, which
has a total of 14 different subunits arranged in two functional
domains (21). V-ATPase is a
proton pump that is present in intracellular and plasma membranes
and regulates pH homeostasis in all eukaryotic cells (22,23).
A literature search revealed no reports of the R552P mutation in
ATP6V1A with respect to taxane resistance. The reason for
this may be that the frequency of this mutation itself is rare.
However, this mutation was common to all six patients in the
current study, whose breast cancers grew rapidly during
taxane-based NAC. This mutation may cause structural, and therefore
functional, changes in the ATP6V1A protein, although the
significance of this mutation remains unclear.
V-ATPase is present in many types of cancer cells
(20) and its dysregulation has
been reported to affect drug resistance through pH dysregulation of
the cytoplasm and extracellular environment (22,24).
V-ATPase hyperactivity has been demonstrated to induce drug
resistance, whereas V-ATPase inhibition could overcome drug
resistance (25,26). Another study reported that a lower
extracellular pH increased the migratory capacity and resistance of
MCF7 cells to docetaxel and paclitaxel (27). Abnormalities in V-ATPase may thus
cause pH dysregulation, leading to taxane resistance. In the
present study, we identified a novel mutation, R552P in
ATP6V1A, which was common to all intrinsically
taxane-resistant cases in our cohort following NGS. The R552P
mutation in ATP6V1A was determined to be a pathogenic mutation by
four online prediction tools. This mutation introduces a proline
into the alpha-helix. Out of 20 essential amino acids, proline is
the only amino acid that contains a cyclized substituted α-amino
group (is formally an imino acid) and has an important role in the
protein folding process because of its structural properties
(28,29). Proline is recognized as a
helix-breaking amino acid, and it has been reported that
replacement of residues in the alpha-helix with proline can
destabilize the protein structure (29). Thus, the R552P mutation in ATP6V1A
may cause structural changes in ATP6V1A protein. A high
ATP6V1A mRNA expression was also significantly associated
with a poorer prognosis in patients who received taxane treatment
for breast cancer. These results suggest that structural changes
and the overexpression of ATP6V1A may cause V-ATPase hyperactivity
and taxane resistance in breast cancer. However, previous evidence
suggested that changes in cytosolic pH resulting from a higher
activity of V-ATPase and the proton transporters can affect the
packing of lipids and decrease the movement of doxorubicin, an
anthracycline (30,31). The present analysis is based on
cases in which taxane anticancer drugs were used. Since
anthracyclines are also used in many cases, their influence cannot
be ignored.
Dysregulation of the V-ATPase subunit was also
linked to poor cancer outcome (24). Liu et al (32) reported that ATP6V1A protein
expression levels were higher in gastric cancer compared with
normal tissues and were correlated with worse survival. V-ATPase
and its subunits have been reported to be more highly expressed in
many types of cancer tissues, including breast cancer, compared
with normal tissues, and its high expression was also reportedly
associated with a poor prognosis (24,33,34).
In the current study, high ATP6V1A mRNA expression was
significantly associated with a poor prognosis and was an
independent predictor of poor prognosis in breast cancer patients
receiving taxane treatment. Although, to the best of our knowledge,
the current study was the first to report the correlation between
mRNA expression levels of ATP6V1A in breast cancer and
patient outcomes, a high expression of the V-ATPase subunit
component gene was also identified as a poor prognostic factor in
previous studies (24), suggesting
that a high ATP6V1A mRNA expression and increased ATP6V1A
function may be associated with a poor prognosis in breast
cancer.
APOBEC3F is a member of the cytidine
deaminase gene family, which encodes proteins that are structurally
and functionally related to C to U RNA editing (35). However, the correlation between
APOBEC3F somatic mutations and cancer has not been widely
investigated. In the current study, we identified the T114P
mutation as a novel APOBEC3F gene variant by NGS. The T114P
mutation of APOBEC3F was predicted to be a pathogenic mutation by
three of five online prediction tools. This mutation also produced
a change to proline but was not located inside the alpha-helix.
Although its significance is unclear, this mutation was common to
all intrinsically taxane-resistant cases in the current study and
was predicted to cause structural changes in the APOBEC3F
protein.
DNA de-amination activity of APOBEC3 family proteins
has been identified as a major contributor to mutagenesis in
various types of cancer, including breast cancer (36,37).
APOBEC3B, another APOBEC3 family member, is involved in genetic
changes in breast cancer (38),
and APOBEC3B mRNA expression was associated with poorer
survival in ER-positive breast cancer (39,40).
However, the role of APOBEC3F in breast cancer has not been widely
studied. APOBEC3F is endogenously expressed in human T cells, where
it mediates antiviral immunity by catalyzing mutations in the viral
genome (36,37). Yang et al (41,42)
demonstrated that the overexpression of APOBEC3F was a risk factor
for poor survival and tumor aggressiveness in hepatocellular
carcinoma (HCC). In the current study, a higher APOBEC3F expression
was associated with poor breast cancer prognosis. Although the
study by Yang et al (42)
involved different cancer types, their findings regarding APOBEC3F
were similar to those of the present study. Given that the
overexpression of APOBEC3F reduced the immune function against
breast cancer, as in HCC, APOBEC3F is a potential therapeutic
target in these forms of cancer.
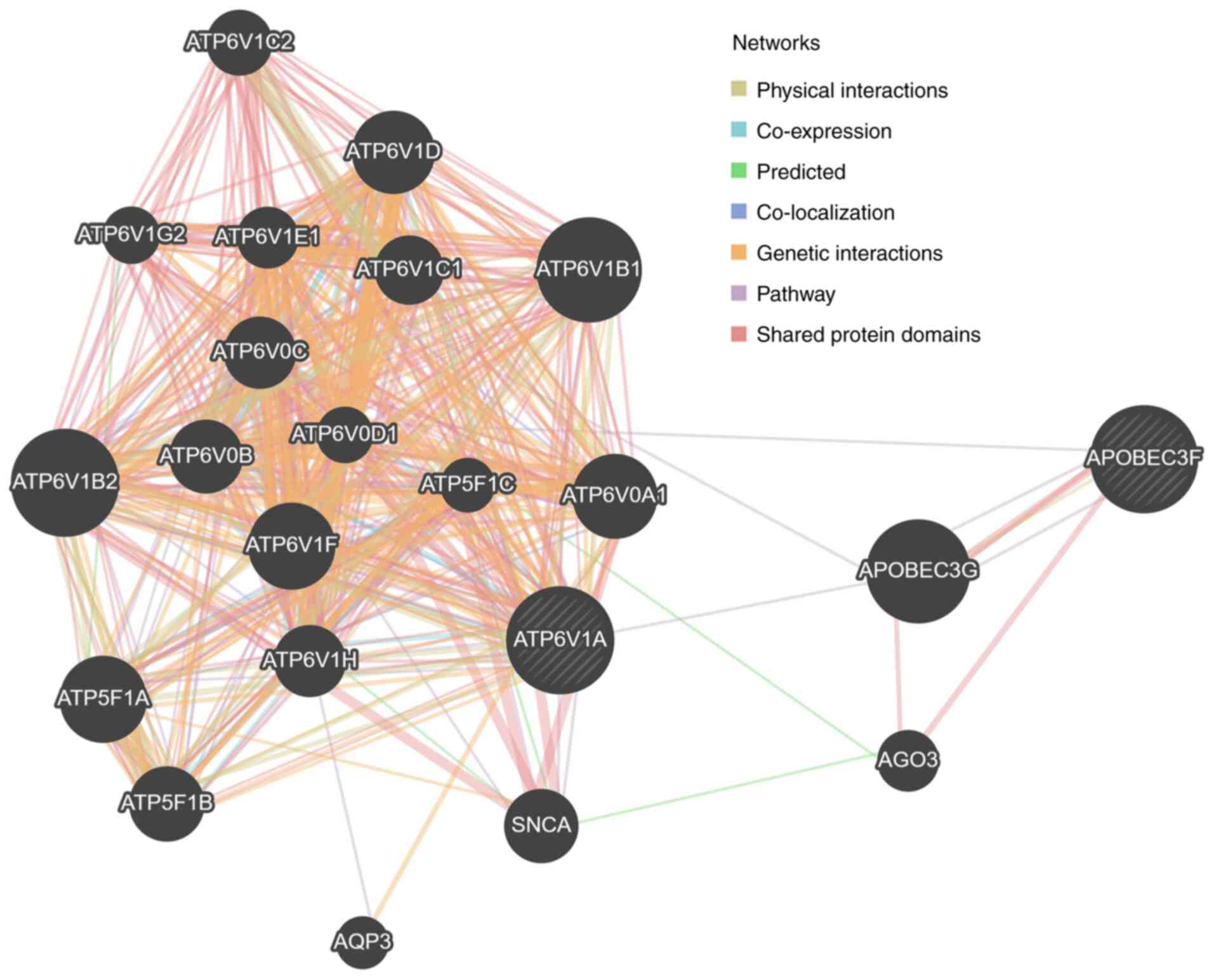
The interaction between ATP6V1A and APOBEC3F is
shown in Fig. 7 (created by
GeneMANIA), based on previous reports (43). According to this analysis, ATP6V1A
is predicted to be co-expressed with APOBEC3F via APOBEC3G.
However, no other tumors with the same mutations
were identified. Mutations common to taxane-resistant tumors are
rare. However, the current six cases with mutations identified by
NGS were not clearly identified by Sanger sequencing, suggesting
that the detection rate of Sanger sequencing should be
reviewed.
The present study had some limitations. The
prognosis evaluations were based on retrospective analyses of
archived materials from a single institute. In addition, we
examined only cases in which taxanes were used and did not include
a control group. For this reason, patients with a good prognosis
who could avoid anti-cancer drugs were not included. Although we
used this group of patients for an exploratory study, there is a
possibility of bias due to this factor. As shown in Table SII, a comparison of the
clinicopathological factors of all patients who were treated for
breast cancer at the same time and in those who had their mRNA
expression measured revealed significant differences in histology,
number of lymph node metastases, nuclear grade, and HER2
expression. A high ATP6V1A and APOBEC3F protein expression showed a
tendency to be associated with poor prognosis. However, no
significant difference was observed. There are three possible
reasons for the lack of significant differences in the correlation
between prognosis and protein expression. First, the number of
cases was insufficient. Second, the IHC methodology for ATP6V1A and
APOBEC3F protein evaluation has not been well established. Third,
because long-term follow-up tissues were used in this study, the
rate of positive staining may have changed because of tissue
deterioration over time.
This study attempted to identify novel mutations
associated with taxane resistance. ATP6V1A is a therapeutic target,
and drugs targeting ATP6V1A and inhibiting V-ATPase-dependent
growth signaling have been reported (44). An APOBEC3F-targeting drug that
inhibits cell proliferation and migration has also been reported
for the treatment of HCC (41).
Future research is to focus on investigating the mechanism of
ATP6V1A and APOBEC3F against taxane resistance in in vitro
studies and analyzing the specific function of each gene in
addition to taxane resistance.
Although attempts at cell culture studies are
currently underway, it remains difficult to include the data owing
to the restrictions resulting from the COVID-19 pandemic regarding
the use of cell laboratories.
In conclusion, using NGS, we identified two specific
mutations, R552P in ATP6V1A and T114P in APOBEC3F,
which were common to all intrinsically taxane-resistant breast
cancer patients in this study. The two mutations appeared to change
their respective protein structures. The results also showed that,
among patients who receive taxane treatment for breast cancer,
those with high ATP6V1A or APOBEC3F expression levels
are likely to have shorter survival.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Mrs. Shinobu Makino
(Department of Breast Surgery, Nagoya City University Graduate
School of Medical Sciences, Nagoya, Japan) for her technical
assistance. The authors would also like to thank Dr Marla Brunker,
Dr Susan Furness and Dr H. Nikki March for editing drafts of this
manuscript.
Funding
This work was supported by a Grant-in-Aid for Scientific
Research from the Ministry of Education, Culture, Sports, Science,
and Technology of Japan (MEXT) KAKENHI (grant no. 19K09080).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the Japanese Genotype-Phenotype
Archive (JGA) database repository on reasonable request. The
accession numbers are Study: JGAS000370, Dataset: JGAD000484.
(https://www.ddbj.nig.ac.jp/jga/index-e.html).
Authors' contributions
YWE conceived and designed the study and evaluated
the results. YWE and YD conducted mRNA expression and
immunostaining analyses. TF, TA, TH, YU, SN, YK, AK, MT and HS
provided tissue samples and assessed patient data. KO, HK and ST
assisted in evaluation of the results. NK and TT contributed to
theoretical organization of the study, the research design, and
revising and editing of the manuscript and confirmed the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional Review
Board of Nagoya City University (approval no. 70-00-0172; Nagoya,
Japan). All tissue samples were provided by a biobank maintained by
the Department of Breast Surgery, Nagoya City University, which
conforms to the guidelines of the Declaration of Helsinki. Written
informed consent for the comprehensive research use of clinical
samples was obtained from all 1,707 patients involved in this
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
WES
|
whole-exome sequencing
|
|
ATP6V1A
|
V-type proton ATPase catalytic subunit
A
|
|
APOBEC3F
|
apolipoprotein B mRNA editing enzyme
catalytic subunit 3F
|
|
DFS
|
disease-free survival
|
|
OS
|
overall survival
|
|
NAC
|
neoadjuvant chemotherapy
|
|
ABC
|
ATP-binding cassette
|
|
NGS
|
next-generation sequencing
|
|
PDB
|
protein data bank
|
|
IHC
|
immunohistochemistry
|
|
ERα
|
estrogen receptor α
|
|
PgR
|
progesterone receptor
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
VAF
|
variant allele frequency
|
|
V-ATPase
|
vacuolar H+-ATPase
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the curve
|
|
HCC
|
hepatocellular carcinoma
|
References
|
1
|
Mougalian SS, Soulos PR, Killelea BK,
Lannin DR, Abu-Khalaf MM, DiGiovanna MP, Sanft TB, Pusztai L, Gross
CP and Chagpar AB: Use of neoadjuvant chemotherapy for patients
with stage I to III breast cancer in the United States. Cancer.
121:2544–2552. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bear HD, Anderson S, Brown A, Smith R,
Mamounas EP, Fisher B, Margolese R, Theoret H, Soran A, Wickerham
DL, et al: The effect on tumor response of adding sequential
preoperative docetaxel to preoperative doxorubicin and
cyclophosphamide: Preliminary results from national surgical
adjuvant breast and bowel project protocol B-27. J Clin Oncol.
21:4165–4174. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Caudle AS, Gonzalez-Angulo AM, Hunt KK,
Liu P, Pusztai L, Symmans WF, Kuerer HM, Mittendorf EA, Hortobagyi
GN and Meric-Bernstam F: Predictors of tumor progression during
neoadjuvant chemotherapy in breast cancer. J Clin Oncol.
28:1821–1828. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pucci P, Rescigno P, Sumanasuriya S, de
Bono J and Crea F: Hypoxia and noncoding RNAs in taxane resistance.
Trends Pharmacol Sci. 39:695–709. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yin S, Zeng C, Hari M and Cabral F:
Paclitaxel resistance by random mutagenesis of alpha-tubulin.
Cytoskeleton (Hoboken). 70:849–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matsunaga T, Saito H, Endo S, Iguchi K,
Soda M, El-Kabbani O, Hara A and Ikari A: Roles of aldo-keto
reductases 1B10 and 1C3 and ATP-binding cassette transporter in
docetaxel tolerance. Free Radic Res. 50:1296–1308. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vaidyanathan A, Sawers L, Gannon AL,
Chakravarty P, Scott AL, Bray SE, Ferguson MJ and Smith G: ABCB1
(MDR1) induction defines a common resistance mechanism in
paclitaxel- and olaparib-resistant ovarian cancer cells. Br J
Cancer. 115:431–441. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bose R, Kavuri SM, Searleman AC, Shen W,
Shen D, Koboldt DC, Monsey J, Goel N, Aronson AB, Li S, et al:
Activating HER2 mutations in HER2 gene amplification negative
breast cancer. Cancer Discov. 3:224–237. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang L, Ye F, Bao L, Zhou X, Wang Z, Hu P,
Ouyang N, Li X, Shi Y, Chen G, et al: Somatic alterations of TP53,
ERBB2, PIK3CA and CCND1 are associated with chemosensitivity for
breast cancers. Cancer Sci. 110:1389–1400. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Endo Y, Dong Y, Yoshimoto N, Asano T, Hato
Y, Yamashita H, Sato S, Takahashi S, Fujii Y and Toyama T: HER2
mutation status in Japanese HER2-negative breast cancer patients.
Jpn J Clin Oncol. 44:619–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wanifuchi-Endo Y, Asano T, Kondo N, Hato
Y, Dong Y, Hisada T, Nishikawa S, Kato H, Takahashi S, Okuda K, et
al: Effects of serum estradiol and progesterone on
estrogen-regulated gene expression in breast cancers of
premenopausal patients. Jpn J Clin Oncol. 49:12–21. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Allred DC, Harvey JM, Berardo M and Clark
GM: Prognostic and predictive factors in breast cancer by
immunohistochemical analysis. Mod Pathol. 11:155–168.
1998.PubMed/NCBI
|
|
13
|
Wolff AC, Hammond ME, Schwartz JN, Hagerty
KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer
A, et al: American society of clinical oncology/college of american
pathologists guideline recommendations for human epidermal growth
factor receptor 2 testing in breast cancer. J Clin Oncol.
25:118–145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirsch FR, Varella-Garcia M, Bunn PA Jr,
Maria MV, Veve R, Bremmes RM, Barón AE, Zeng C and Franklin WA:
Epidermal growth factor receptor in non-small-cell lung carcinomas:
Correlation between gene copy number and protein expression and
impact on prognosis. J Clin Oncol. 21:3798–3807. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
John T, Liu G and Tsao MS: Overview of
molecular testing in non-small-cell lung cancer: Mutational
analysis, gene copy number, protein expression and other biomarkers
of EGFR for the prediction of response to tyrosine kinase
inhibitors. Oncogene. 28 (Suppl 1):S14–S23. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Endo Y, Yamashita H, Takahashi S, Sato S,
Yoshimoto N, Asano T, Hato Y, Dong Y, Fujii Y and Toyama T:
Immunohistochemical determination of the miR-1290 target arylamine
N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast
cancer. BMC Cancer. 14:9902014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hayes DF, Ethier S and Lippman ME: New
guidelines for reporting of tumor marker studies in breast cancer
research and treatment: REMARK. Breast Cancer Res Treat.
100:237–238. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
McShane LM, Altman DG, Sauerbrei W, Taube
SE, Gion M and Clark GM; Statistics Subcommittee of the NCI-EORTC
Working Group on Cancer Diagnostics, : REporting recommendations
for tumor MARKer prognostic studies (REMARK). Breast Cancer Res
Treat. 100:229–235. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fais S, De Milito A, You H and Qin W:
Targeting vacuolar H+-ATPases as a new strategy against
cancer. Cancer Res. 67:10627–10630. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishi T and Forgac M: The vacuolar
(H+)-ATPases-nature's most versatile proton pumps. Nat
Rev Mol Cell Biol. 3:94–103. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stransky L, Cotter K and Forgac M: The
function of V-ATPases in cancer. Physiol Rev. 96:1071–1091. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun-Wada GH and Wada Y: Role of
vacuolar-type proton ATPase in signal transduction. Biochim Biophys
Acta. 1847:1166–1172. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Whitton B, Okamoto H, Packham G and Crabb
SJ: Vacuolar ATPase as a potential therapeutic target and mediator
of treatment resistance in cancer. Cancer Med. 7:3800–3811. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sasazawa Y, Futamura Y, Tashiro E and
Imoto M: Vacuolar H+-ATPase inhibitors overcome
Bcl-xL-mediated chemoresistance through restoration of a
caspase-independent apoptotic pathway. Cancer Sci. 100:1460–1467.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
von Schwarzenberg K, Lajtos T, Simon L,
Müller R, Vereb G and Vollmar AM: V-ATPase inhibition overcomes
trastuzumab resistance in breast cancer. Mol Oncol. 8:9–19. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tavares-Valente D, Baltazar F, Moreira R
and Queirós O: Cancer cell bioenergetics and pH regulation
influence breast cancer cell resistance to paclitaxel and
doxorubicin. J Bioenerg Biomembr. 45:467–475. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pakula AA and Sauer RT: Genetic analysis
of protein stability and function. Annu Rev Genet. 23:289–310.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
MacArthur MW and Thornton JM: Influence of
proline residues on protein conformation. J Mol Biol. 218:397–412.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Muley H, Fadó R, Rodriguez-Rodríguez R and
Casals N: Drug uptake-based chemoresistance in breast cancer
treatment. Biochem Pharmacol. 177:1139592020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan S, Niu Y, Tan N, Wu Z, Wang Y, You H,
Ke R, Song J, Shen Q, Wang W, et al: LASS2 enhances
chemosensitivity of breast cancer by counteracting acidic tumor
microenvironment through inhibiting activity of V-ATPase proton
pump. Oncogene. 32:1682–1690. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu P, Chen H, Han L, Zou X and Shen W:
Expression and role of V1A subunit of V-ATPases in gastric cancer
cells. Int J Clin Oncol. 20:725–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cotter K, Liberman R, Sun-Wada G, Wada Y,
Sgroi D, Naber S, Brown D, Breton S and Forgac M: The a3 isoform of
subunit a of the vacuolar ATPase localizes to the plasma membrane
of invasive breast tumor cells and is overexpressed in human breast
cancer. Oncotarget. 7:46142–46157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Katara GK, Jaiswal MK, Kulshrestha A,
Kolli B, Gilman-Sachs A and Beaman KD: Tumor-associated vacuolar
ATPase subunit promotes tumorigenic characteristics in macrophages.
Oncogene. 33:5649–5654. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jarmuz A, Chester A, Bayliss J, Gisbourne
J, Dunham I, Scott J and Navaratnam N: An anthropoid-specific locus
of orphan C to U RNA-editing enzymes on chromosome 22. Genomics.
79:285–296. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Burns MB, Temiz NA and Harris RS: Evidence
for APOBEC3B mutagenesis in multiple human cancers. Nat Genet.
45:977–983. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roberts SA, Lawrence MS, Klimczak LJ,
Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL,
Saksena G, et al: An APOBEC cytidine deaminase mutagenesis pattern
is widespread in human cancers. Nat Genet. 45:970–976. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Burns MB, Lackey L, Carpenter MA, Rathore
A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N,
Nikas JB, et al: APOBEC3B is an enzymatic source of mutation in
breast cancer. Nature. 494:366–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sieuwerts AM, Schrijver WA, Dalm SU, de
Weerd V, Moelans CB, Hoeve NT, van Diest PJ, Martens JWM and van
Deurzen CH: Progressive APOBEC3B mRNA expression in distant breast
cancer metastases. PLoS One. 12:e01713432017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sieuwerts AM, Willis S, Burns MB, Look MP,
Gelder ME, Schlicker A, Heideman MR, Jacobs H, Wessels L,
Leyland-Jones B, et al: Elevated APOBEC3B correlates with poor
outcomes for estrogen-receptor-positive breast cancers. Horm
Cancer. 5:405–413. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Z, Tao Y, Xu X, Cai F, Yu Y and Ma L:
Bufalin inhibits cell proliferation and migration of hepatocellular
carcinoma cells via APOBEC3F induced intestinal immune network for
IgA production signaling pathway. Biochem Biophys Res Commun.
503:2124–2131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Z, Zhuang L, Yu Y, Zhou W, Lu Y, Xu
Q, Tang B and Chen X: Overexpression of APOBEC3F in tumor tissues
is potentially predictive for poor recurrence-free survival from
HBV-related hepatocellular carcinoma. Discov Med. 20:349–356.
2015.PubMed/NCBI
|
|
43
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chung CY, Shin HR, Berdan CA, Ford B, Ward
CC, Olzmann JA, Zoncu R and Nomura DK: Covalent targeting of the
vacuolar H(+)-ATPase activates autophagy via mTORC1 inhibition. Nat
Chem Biol. 15:776–785. 2019. View Article : Google Scholar : PubMed/NCBI
|