Introduction
Colorectal cancer (CRC) is one of the most prevalent
types of gastrointestinal tumors worldwide, ranking third in cancer
incidence and second in mortality rate. CRC led to 191,000 deaths
in 2015 in China (1), and in 2018,
it was estimated that there were more than 180,0000 new cases of
CRC and 881,000 cancer-related deaths (2). The main treatment methods for CRC
include surgical resection, chemoradiotherapy, and chemotherapy,
but the 5-year survival of patients remains low (3–5).
Moreover, with the repeated use of chemotherapeutic drugs, CRC
tumors are prone to develop drug resistance, ultimately resulting
in failed chemotherapeutic regimens (6,7).
Thus, it is crucial to understand the mechanism of CRC to develop
effective therapeutic strategies.
Long non-coding RNAs (lncRNAs) are a type of RNA
that is abnormally expressed in various tumors, including
hepatocellular carcinoma (8),
pancreatic cancer (9), bladder
cancer (10), and CRC (11). They are associated with many
biological processes, including cell proliferation, apoptosis,
metabolism, and drug resistance (12–14).
The lncRNA colorectal neoplasia differentially expressed (CRNDE),
which was initially found to be upregulated in colorectal adenomas
and carcinomas, is transcribed from human chromosome 16 (15). Previous findings showed that CRNDE
is involved in the occurrence and development of numerous cancers,
such as CRC (16), hepatocellular
carcinoma (17), and non-small
cell lung cancer (18), and can
act as a biomarker for specific malignant tumors (19,20).
CRNDE promotes the proliferation and inhibits the apoptosis of
cervical cells by regulating the PI3K/Akt pathway, resulting in
poor prognosis (21), and CRNDE
inhibition increases the chemosensitivity of medulloblastoma cells
(22). Accumulating evidence has
shown that CRNDE is upregulated in CRC and is associated with the
regulation of CRC cell apoptosis, proliferation, and drug
sensitivity (23–25). However, the specific underlying
mechanisms of CRNDE-mediated cancer processes need to be
elucidated.
The Warburg effect is one of the main metabolic
alterations in cancer that enhances tumor growth (26,27).
Previous findings have indicated that Akt/mTOR activation promotes
the Warburg effect and tumorigenesis in non-small cell lung cancer
(28). In addition, Yang and Chen
found that CRNDE enhances the progression of tongue squamous cell
carcinoma by regulating the PI3K/Akt/mTOR signaling pathway
(29). However, whether CRNDE
affects the Warburg effect in CRC cells by regulating Akt/mTOR
signaling, thereby influencing associated biological processes of
tumorigenesis, needs to be clarified. To address this, CRNDE was
overexpressed or downregulated in the human colorectal carcinoma
cell line HCT-116, via transfection to evaluate its effect on cell
apoptosis, cisplatin sensitivity, the Warburg effect, and Akt/mTOR
activity. The aim of the present study was to clarify the mechanism
of CRNDE, provide new targets for CRC treatment, reduce the
occurrence of tumor cell drug resistance in CRC treatment, improve
the effectiveness rate of CRC treatment and patient quality of
life.
Materials and methods
Cell culture
Human colorectal carcinoma cell lines, HCT-116 and
LOVO, and the human normal colonic epithelial cell line NCM460 were
purchased from the Shanghai Institutes for Biological Sciences,
Chinese Academy of Science. HCT-116 and NCM460 cells were cultured
in McCoy's 5A medium (Gibco, Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher
Scientific, Inc.). LOVO cells were cultured in F12K medium
supplemented with 10% FBS. All cells were incubated at 37°C with 5%
CO2. After confluence reached 90%, the cells were
harvested and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) was performed to detect the expression of CRNDE
and experiments were performed in triplicate.
Cell transfection and treatment
The CRNDE overexpression vector
pCDH-CMV-MCS-EF1-CopGFP-T2A-Puro (CRNDE sequence: forward,
5-GCTCTAGATGTTGGCTGAAATTCAT-3, reverse,
5-CGGGATCCTTATAGTCTATAAACAGG-3) and interference vector pSICOR
(short hairpin RNA sequence: 5-GGTGTTAAGTGTGATGCTTCC-3) were
supplied by Addgene (Cambridge). HCT-116 cells were seeded into
24-well plates at a density of 5×105 cells/well. When
the confluence reached 70-80%, the cells were transfected with
pCDH-CRNDE (CRNDE overexpression, CRNDE-OE), empty
pCDH-CMV-MCS-EF1-CopGFP-T2A-Puro vectors (CRNDE-OE negative
control, CRNDE-NC), pSICOR-shCRNDE (CRNDE interference, sh-CRNDE),
or empty pSICOR vectors (sh-CRNDE negative control, sh-NC) using
Lipofectamine™ 2000 (Invitrogen, Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Non-transfected
HCT-116 cells served as the control. After 24 h of transfection,
the transfection rate and effect of CRNDE on HCT-116 cell
proliferation, apoptosis, the Warburg effect, and Akt/mTOR complex
1 (mTORC1) pathway activity were evaluated.
After 24 h of transfection, HCT-116 cells were
treated with cisplatin (Sigma-Aldrich) at 5 or 10 µM for 12, 24,
48, 72 and 144 h. Cell counting kit-8 (CCK-8) assays were performed
to evaluate the cisplatin sensitivity of cells. Activity of the
Akt/mTORC1 pathway in HCT-116 cells was evaluated after 24 h of
treatment with 10 µM cisplatin. To verify the influence of the
Akt/mTORC1 pathway on the regulatory effect of CRNDE on the Warburg
effect, HCT-116 cells were treated with the Akt inhibitor AZD5363
(500 nM; MedChemExpress) and the mTOR inhibitor AZD2014 (5 mM;
MedChemExpress) for 6 h. Subsequent to 24-h transfection, ATP
production, glucose uptake, and lactic acid levels were also
measured.
RT-qPCR
Total RNA was extracted from HCT-116, LOVO, NCM460,
and HCT-116 cells after 24 h of transfection using TRIzol (Ambion;
Thermo Fisher Scientific, Inc.) and reverse-transcribed into cDNA.
The collected cDNA was amplified by PCR using the following primer
sequences: Forward, 5-ATTCATCCCAAGGCTG-3, reverse
5-CAAAGACCAACGGCTG-3; GAPDH forward, 5-CCACTCCTCCACCTTTG-3, reverse
5-CACCACCCTGTTGCTGT-3. GAPDH was used as an internal
control. Data were analyzed using the 2−ΔΔCq method
(30).
CCK-8 assay
The effect of CRNDE on the proliferation and
cisplatin sensitivity of HCT-116 cells was investigated using the
CCK-8 assay. Harvested HCT-116 cells were seeded into 96-well
plates at a density of 5×103 cells/well and cultured
overnight at 37°C with 5% CO2. After treatment, the
cells were incubated with 10 ml of CCK-8 solution (Bioswamp; Wuhan
Bienle Biotechnology Co., Ltd.) for 4 h at 37°C with 5%
CO2. The optical density of wells was measured at 450 nm
using a Multiskan FC apparatus (Thermo Fisher Scientific,
Inc.).
Flow cytometry
Flow cytometry was performed to evaluate cell
apoptosis using an Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) apoptosis detection kit (BD
Biosciences). Harvested HCT-116 cells (5×105) were
centrifuged at 1,000 × g at 4°C for 5 min and resuspended in 1 ml
of phosphate-buffered saline (PBS; Bioswamp), followed by
centrifugation at 1,000 × g at 4°C for 5 min. The cells were then
resuspended in 200 ml of binding buffer (Bioswamp) and stained with
10 µl of Annexin V-FITC and 10 µl of PI in the dark at 4°C for 30
min. After adding 300 µl of binding buffer, the cells were
subjected to flow cytometry (Beckman Coulter, Brea, CA, USA) and
analyzed using CytExpert software (Beckman Coulter, Inc.; version
2.0).
Hoechst-33258 staining
Hoechst 33258 staining was performed to evaluate
apoptosis. After treatment, HCT-116 cells were fixed in 4%
paraformaldehyde at 4°C for 10 min. After two washes with PBS for 3
min, the cells were stained with 2 ml of Hoechst 33258 staining
solution (10 µg/ml) at room temperature for 5 min. Finally, the
cells were photographed using an inverted fluorescence microscope
(Leica).
Western blot analysis
Total proteins were extracted from HCT-116 cells
using radioimmunoprecipitation assay lysis buffer (Bioswamp) and
quantified using a bicinchoninic acid assay kit (Bioswamp).
SDS-PAGE (12%) was performed and the proteins (20 µg) were
separated and transferred onto polyvinylidene fluoride membranes
(MilliporeSigma). After blocking with 5% skimmed milk powder at
37°C for 1 h, the membranes were incubated for 1 h at room
temperature with primary antibodies against B-cell lymphoma-2
(Bcl-2, 1:1,000, PAB30041), glucose transporter type 1 (GLUT1,
1:1,000, PAB30526), hexokinase 2 (HK2, 1:1,000, PAB30052),
phosphorylated (p)-Akt (1:1,000, PAB30040), Akt (1:10,000,
PAB30042), p-mTOR (1:1,000, PAB43425-P), mTOR (1:1,000, PAB46102),
and GAPDH (1:1,000, PAB36269) purchased from Bioswamp, as well as
those targeting cytochrome-C (cyt-c, 1:500, ab216971),
cleaved-caspase 3 (1:500, ab32042), lactate dehydrogenase A (LDHA,
1:5,000, ab101562), pyruvate kinase M2 (PKM2, 1:1,000, ab137852),
p-eukaryotic translation initiation factor 4E-binding protein 1
(p-4EBP-1, 1:2,000, ab60529), 4EBP-1 (1:5,000, ab2606), eukaryotic
translation initiation factor 4E (EIF-4E, 1:5,000, ab32024), p-S6K
(1:1,000, ab60948), S6K (1:5,000, ab32529), p-S6 (1:10,000,
ab184551), and S6 (1:1,000, ab32132), purchased from Abcam
(Cambridge, UK), followed by incubation with goat anti-rabbit IgG
secondary antibody (1:20,000, Bioswamp, PAB160011) for 1 h at 4 C.
An enhanced chemiluminescence kit (Pierce; Thermo Fisher
Scientific, Inc.) was used to detect the specific bands and
analyzed using Tanon GIS software (version 4.2; Tanon Science and
Technology Co., Ltd.) using GAPDH as a control.
Biochemical assay
Biochemical assays were performed to evaluate ATP
production (A095, Nanjing Jiancheng Bioengineering Institute),
glucose uptake (KA4086, Abnova), and lactic acid level (A019-2,
Nanjing Jiancheng Bioengineering Institute) in HCT-116 cells using
corresponding commercial kits according to the manufacturer's
instructions.
Statistical analysis
Data are presented as means ± standard deviation.
Differences in data were analyzed using one-way analysis of
variance followed by Tukey's post-hoc test. Data were analyzed
using SPSS software (v19.0; IBM Corp.). P<0.05 was considered to
indicate a statistically significant difference.
Results
CRNDE silencing inhibits proliferation
and promotes apoptosis and cisplatin sensitivity of HCT-116
cells
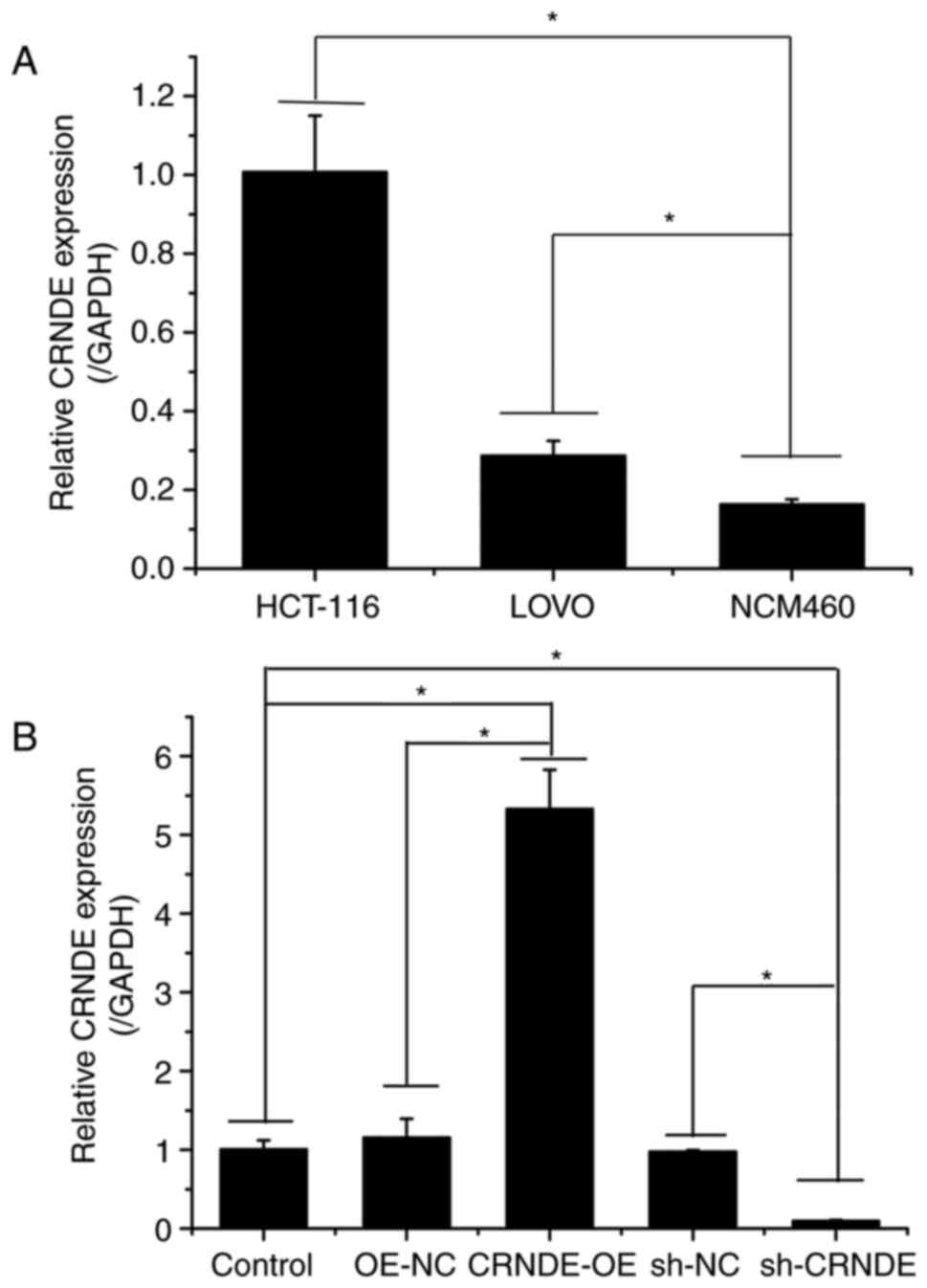
As shown in Fig.
1A, the expression of CRNDE in HCT-116 and LOVO cells was
higher than that in NCM460 cells. As HCT-116 cells exhibited the
highest expression of CRNDE, they were selected for subsequent
experiments.
To investigate the effect of CRNDE on CRC cells,
CRNDE was overexpressed or inhibited in HCT-116 cells via
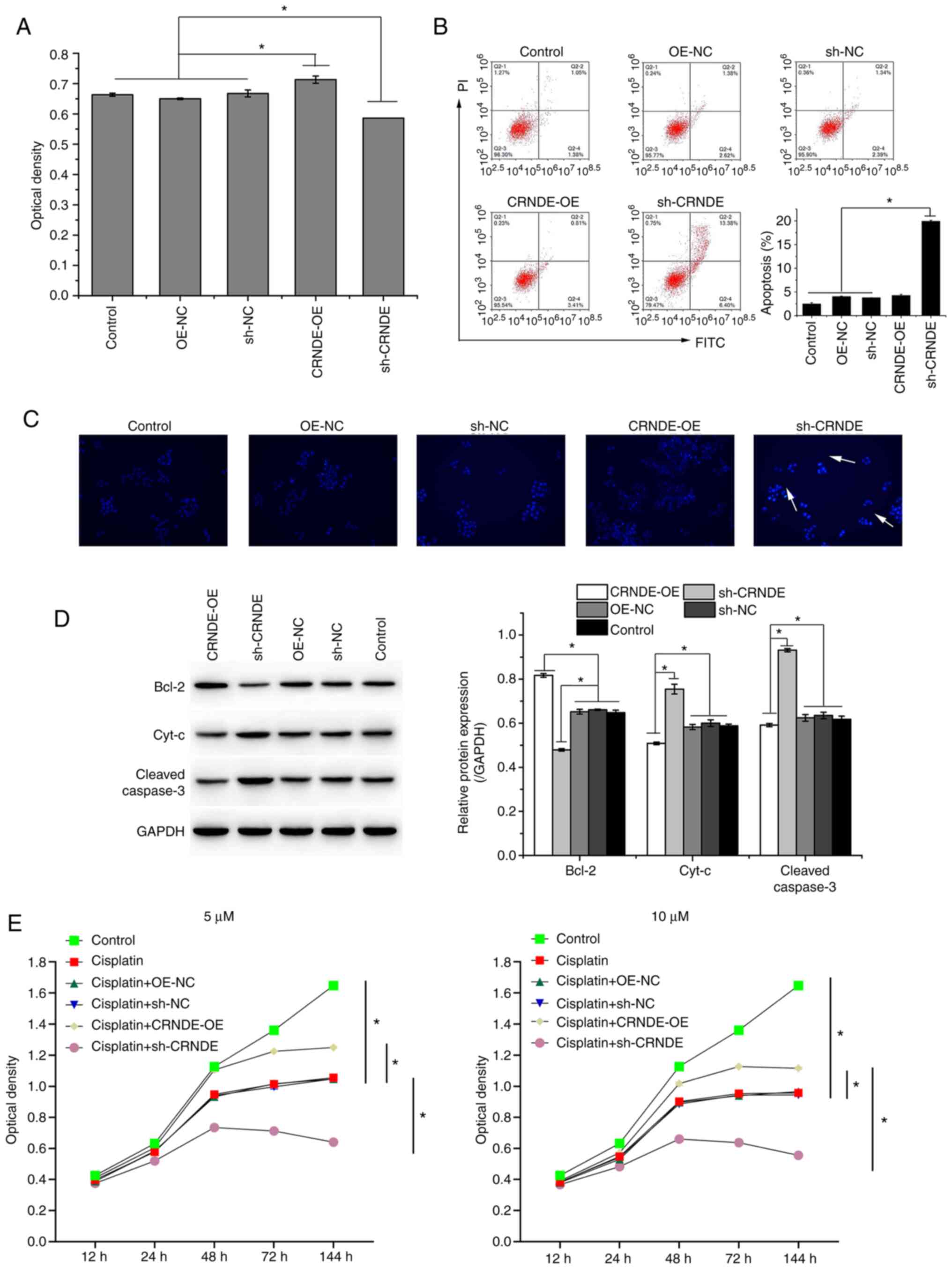
transfection (Fig. 1B). The CCK-8
assay demonstrated that CRNDE inhibition suppressed the viability
of HCT-116 cells, as indicated by the decrease in optical density
(Fig. 2A). In addition, CRNDE
inhibition (sh-CRNDE) promoted the apoptosis of HCT-116 cells, as
verified by the increase in the apoptosis rate, detected by flow
cytometry (Fig. 2B), and the
bright blue cells (white arrows), detected by Hoechst-33258
staining (Fig. 2C). The expression
of apoptosis-related proteins was detected using western blotting.
CRNDE overexpression upregulated the anti-apoptotic protein Bcl-2
while inhibiting the expression of the pro-apoptotic proteins Cyt-c
and Cleaved-caspase 3, whereas CRNDE inhibition had the opposite
effect (Fig. 2D). The results of
western blotting were consistent with those of flow cytometry and
Hoechst-33258 staining. After transfection, HCT-116 cells were
treated with cisplatin at 5 or 10 µM. Compared to that with control
cells, cisplatin decreased the optical density of HCT-116 cells
(Fig. 2E). Compared to that of
cells treated with cisplatin, the optical density of cells treated
with cisplatin combined with CRNDE overexpression was increased,
whereas that of cells treated with cisplatin combined with CRNDE
inhibition was decreased (Fig.
2E), indicating that CRNDE silencing decreased HCT-116 cell
viability, and increased HCT-116 cell cisplatin sensitivity.
CRNDE silencing inhibits the Warburg
effect in HCT-116 cells
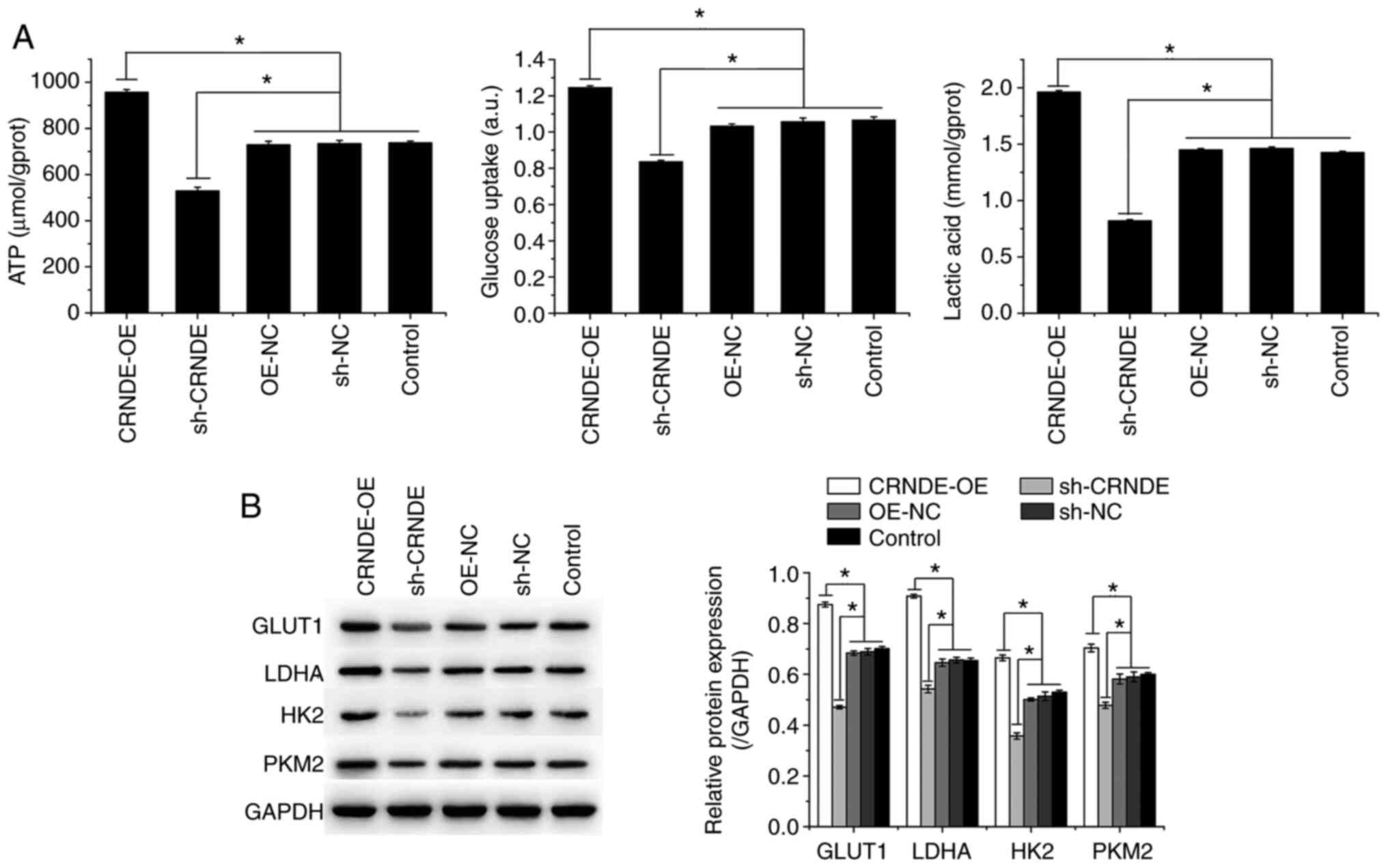
Biochemical testing showed that compared to those in
the control group, ATP and lactic acid levels and glucose uptake
were increased in the CRNDE-OE group but decreased in the sh-CRNDE
group (Fig. 3A). The expression of
GLUT1, LDHA, HK2, and PKM2 was also detected by western blotting.
CRNDE overexpression (CRNDE-OE) promoted the expression of GLUT1,
LDHA, HK2, and PKM2, which was inhibited by CRNDE interference
(sh-CRNDE) in HCT-116 cells (Fig.
3B). These results indicated that CRNDE silencing inhibited the
Warburg effect in HCT-116 cells.
CRNDE silencing inhibits activation of
the Akt/mTORC1 pathway in HCT-116 cells
Western blotting demonstrated that compared to that
in the control cells, the protein expression of p-Akt, p-S6K, p-S6,
and p-mTOR was increased in the CRNDE-OE group, whereas that of
p-4EBP-1 and EIF-4E was decreased (Fig. 4). Conversely, the protein
expression of p-Akt, p-S6K, p-S6, and p-mTOR was decreased in the
sh-CRNDE group, whereas that of p-4EBP-1 and EIF-4E was increased
(Fig. 4). These results indicate
that CRNDE silencing inhibited the activation of the Akt/mTORC1
pathway in HCT-116 cells.
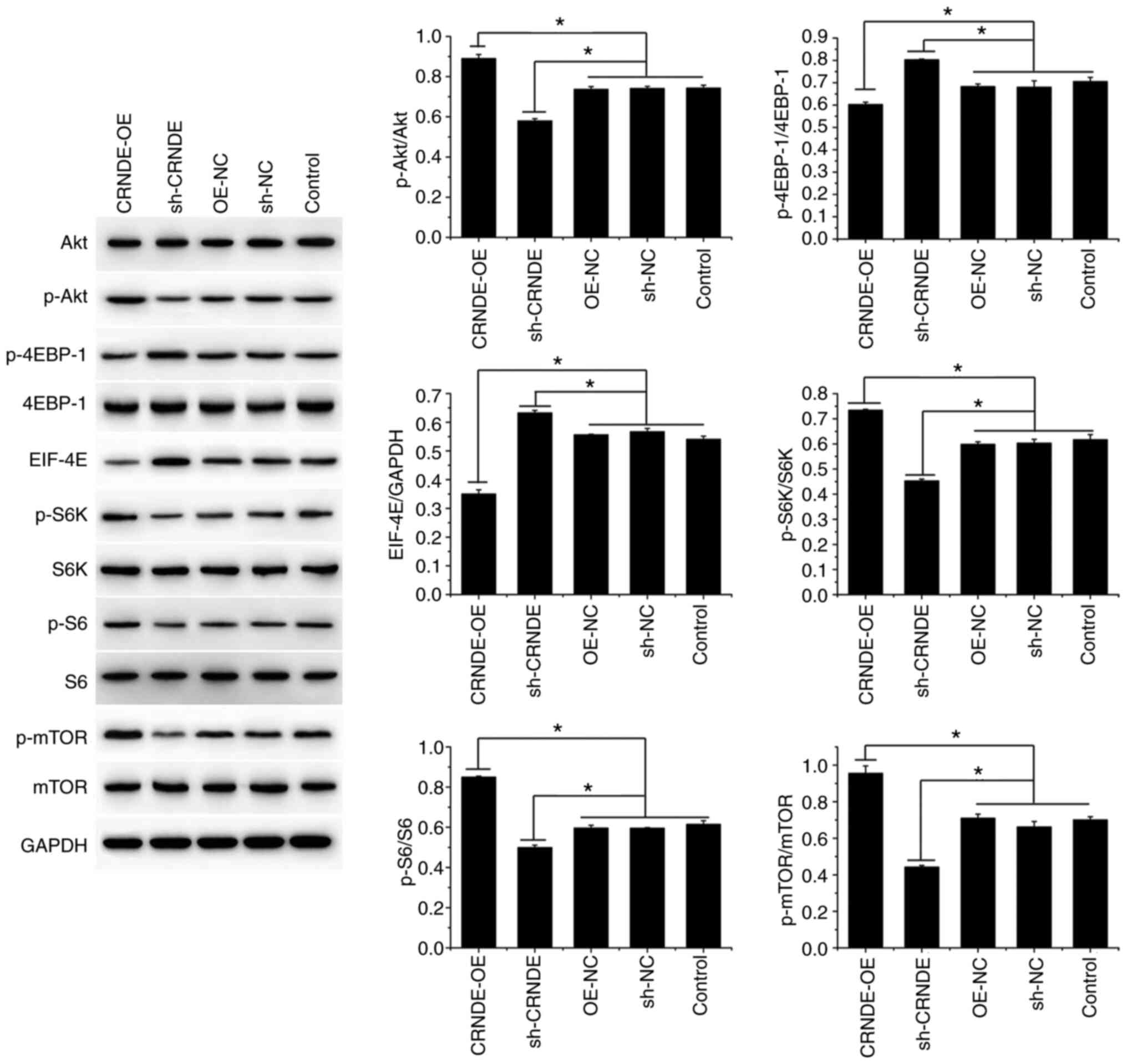 | Figure 4.CRNDE silencing inhibits Akt/mTORC1
activation in HCT-116 cells. The protein expression of Akt, p-Akt,
4EBP-1, p-4EBP-1, EIF-4E, S6K, p-S6K, S6, p-S6, mTOR, and p-mTOR in
transfected HCT-116 cells was detected by western blotting. Data
are presented as the mean ± SD, n=3, *P<0.05. |
CRNDE silencing suppresses the Warburg
effect in HCT-116 cells by inhibiting Akt/mTOR activation
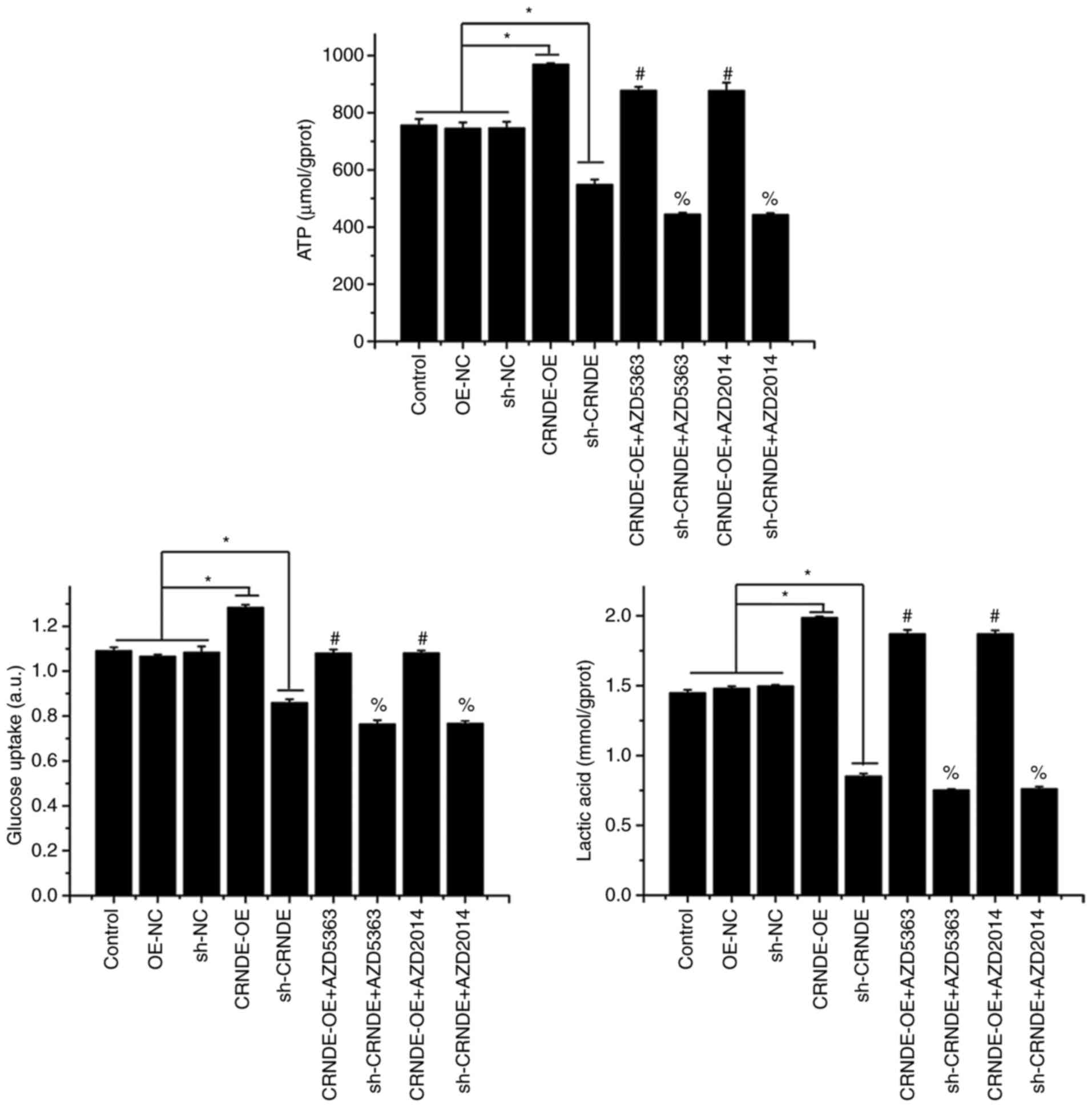
To verify the influence of the Akt/mTOR pathway on
the regulatory effect of CRNDE on the Warburg effect in HCT-116
cells, Akt and mTOR were suppressed by corresponding inhibitors in
transfected HCT-116 cells. As shown in Fig. 5, compared with the CRNDE-OE group,
ATP production, lactic acid levels, and glucose uptake decreased in
the CRNDE-OE+AZD5363 and CRNDE-OE+AZD2014 groups. In addition, the
decrease in ATP production, lactic acid levels, and glucose uptake
in the sh-CRNDE group were further decreased by Akt and mTOR
inhibition. These results indicated that the effect of CRNDE on the
Warburg effect in HCT-116 cells may be mediated by the Akt/mTOR
pathway.
Discussion
In general, normal cells use mitochondria to oxidize
glucose molecules as their main energy metabolism pathway. Glucose
forms pyruvate through multi-step glycolysis. Pyruvic acid enters
the mitochondria and is oxidized through the tricarboxylic acid
cycle to produce ATP to meet the energy needs of cells (31). However, in tumor cells, pyruvic
acid produced by glycolysis does not enter the mitochondria but is
reduced to lactic acid by LDH, even in the presence of sufficient
oxygen (31). This phenomenon,
known as the Warburg effect, is a form of metabolic reprogramming
closely associated with cancer occurrence and development (32). Although less ATP is produced by
glycolysis than by oxidative phosphorylation, glycolysis
intermediates provide the carbon sources needed for rapid cell
proliferation (33). In addition,
lactic acid generated during glycolysis decreases the pH value of
the extracellular matrix (34),
whereas acidic microenvironments promote tumor invasion,
metastasis, and radiotherapy resistance (35,36).
Glucose uptake is the first step of the Warburg effect, during
which specific membrane transporters such as GLUTs are required
(37). This glucose is then
phosphorylated by HK to generate glucose-6-phosphate, which is
subsequently converted into phosphoenol pyruvate (38), which is then converted into
pyruvate by PK, and the pyruvate is finally converted to lactic
acid by LDH (39), which is the
last step of glycolysis. Inhibiting the activity of GLUTs, HK, PK,
and LDH has been shown to suppress the Warburg effect in cancer
cells (40,41), thereby modulating their apoptosis,
proliferation, migration, invasion, and drug resistance (42–45).
The current study indicated that CRNDE inhibition decreased the
levels of ATP and lactic acid, glucose uptake, and the expression
of GLUT1, LDHA, HK2, and PKM2 in HCT-116 cells. In addition, CRNDE
inhibition suppressed the proliferation of HCT-116 cells while
promoting their apoptosis and cisplatin sensitivity, which may be
associated with its inhibitory action on the Warburg effect.
Additionally, the current study indicated that CRNDE
inhibition decreases the activity of Akt/mTORC1, as demonstrated by
the decrease in the protein expression of p-Akt, p-S6K, p-S6, and
p-mTOR and the increase in the protein expression of p-4EBP-1 and
EIF-4E. The activation of mTOR is associated with a series of
proteins involved in two structurally and functionally distinct
complexes, mTORC1 and mTORC2 (46). Of note, mTORC1 is regulated by Akt
via phosphorylation at Ser 2448 (47). The Akt/mTORC1 pathway and its
downstream factors are involved in the regulation of cellular
metabolism. mTORC1, a central activator of the Warburg effect, has
been shown to induce glycolytic enzymes in tumor cells (48). p70S6K is an effector of mTORC1 that
promotes glucose metabolism from glycolysis to pentose phosphate,
which in turn enhances cancer cell growth (49,50).
Another characterized downstream factor of mTORC1 is 4EBP1, which
attenuates the initiation of protein translation by binding and
inactivating EIF-4E. Previous findings have shown that 4EBP1
inhibition, regulated by mTOR activation, promotes glucose uptake
and glycolysis (51). The
suppression of Akt/mTORC1 activation downregulates the expression
of GLUT1 and HK2 and inhibits glucose uptake, subsequently
impairing glucose metabolism (52). In addition, the current study found
that the increase in ATP and lactic acid levels and glucose uptake
in HCT-116 cells induced by CRNDE overexpression was counteracted
by Akt and mTOR inhibition, indicating that the effect of CRNDE on
the Warburg effect in HCT-116 cells may be mediated by the
Akt/mTORC1 pathway.
In conclusion, the present study provides evidence
demonstrating that CRNDE inhibition suppresses the Warburg effect
in HCT-116 cells, in turn inhibiting their proliferation and
promoting their apoptosis and cisplatin sensitivity. The mechanism
underlying the action of CRNDE silencing may be the inactivation of
Akt/mTORC1 signaling. In vivo experiments are to be designed
in follow-up studies to verify the findings of the current
study.
Acknowledgements
Not applicable.
Funding
The present study was supported by the PhD-initiated funding
project of Hubei University of Chinese Medicine (grant no.
10110428).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WY and XY contributed to conception of the study.
WY, YW and XY designed and performed the experiments, analyzed the
data and drafted the manuscript. CT, YL and SC analyzed the data
and provided technical support. All authors have read and approved
the final manuscript. WY, YW, CT, YL, SC and XY confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Binefa G, Rodriguez-Moranta F, Teule A and
Medina-Hayas M: Colorectal cancer: From prevention to personalized
medicine. World J Gastroenterol. 20:6786–6808. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim SE, Paik HY, Yoon H, Lee JE, Kim N and
Sung MK: Sex- and gender-specific disparities in colorectal cancer
risk. World J Gastroenterol. 21:5167–5175. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Woolston A, Khan K, Spain G, Barber LJ,
Griffiths B, Gonzalez-Exposito R, Hornsteiner L, Punta M, Patil Y,
Newey A, et al: Genomic and transcriptomic determinants of therapy
resistance and immune landscape evolution during Anti-EGFR
treatment in colorectal cancer. Cancer Cell. 36:35–50, e9. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marjaneh RM, Khazaei M, Ferns GA, Avan A
and Aghaee-Bakhtiari SH: The role of microRNAs in 5-FU resistance
of colorectal cancer: Possible mechanisms. J Cell Physiol.
234:2306–2316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wei L, Wang X, Lv L, Liu J, Xing H, Song
Y, Xie M, Lei T, Zhang N and Yang M: The emerging role of microRNAs
and long noncoding RNAs in drug resistance of hepatocellular
carcinoma. Mol Cancer. 18:1472019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiong G, Liu C, Yang G, Feng M, Xu J, Zhao
F, You L, Zhou L, Zheng L, Hu Y, et al: Long noncoding RNA GSTM3TV2
upregulates LAT2 and OLR1 by competitively sponging let-7 to
promote gemcitabine resistance in pancreatic cancer. J Hematol
Oncol. 12:972019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martens-Uzunova ES, Bottcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou
A, Liu J, Che L and Li J: Long noncoding RNA GAS5 inhibits
progression of colorectal cancer by interacting with and triggering
YAP phosphorylation and degradation and is negatively regulated by
the m6A reader YTHDF3. Mol Cancer. 18:1432019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu D, Lu C, Qu X, Li P, Chen K, Shan L and
Zhu X: LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and
drug resistance via the miR-134-5p/MBTD1 axis. Aging (Albany NY).
11:8374–8385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang P, Xu J, Wang Y and Cao X: An
interferon-independent lncRNA promotes viral replication by
modulating cellular metabolism. Science. 358:1051–1055. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Yin M, Peng G and Zhao Y: CRNDE:
An important oncogenic long non-coding RNA in human cancers. Cell
Prolif. 51:e124402018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu
XY, Yu ZW, Jia YH, Bai XF, Li L, et al: The lncRNA CRNDE promotes
colorectal cancer cell proliferation and chemoresistance via
miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol
Cancer. 16:92017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ji D, Jiang C, Zhang L, Liang N, Jiang T,
Yang B and Liang H: LncRNA CRNDE promotes hepatocellular carcinoma
cell proliferation, invasion, and migration through regulating
miR-203/BCAT1 axis. J Cell Physiol. 234:6548–6560. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jing H, Xia H, Qian M and Lv X: Long
noncoding RNA CRNDE promotes non-small cell lung cancer progression
via sponging microRNA-338-3p. Biomed Pharmacother. 110:825–833.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang C, Zhang B, Ge H, Xu Y, Li G and Wu
J: Long non-coding RNA CRNDE as a potential prognostic biomarker in
solid tumors: A meta-analysis. Clin Chim Acta. 481:99–107. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding C, Han F, Xiang H, Xia X, Wang Y, Dou
M, Zheng J, Li Y, Xue W, Ding X and Tian P: LncRNA CRNDE is a
biomarker for clinical progression and poor prognosis in clear cell
renal cell carcinoma. J Cell Biochem. 119:10406–10414. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang HY, Huang CP, Cao MM, Wang YF and Liu
Y: Long non-coding RNA CRNDE may be associated with poor prognosis
by promoting proliferation and inhibiting apoptosis of cervical
cancer cells through targeting PI3K/AKT. Neoplasma. 65:872–880.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun XH, Fan WJ, An ZJ and Sun Y:
Inhibition of long noncoding RNA CRNDE increases chemosensitivity
of medulloblastoma cells by targeting miR-29c-3p. Oncol Res.
28:95–102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding J, Li J, Wang H, Tian Y, Xie M, He X,
Ji H, Ma Z, Hui B, Wang K and Ji G: Long noncoding RNA CRNDE
promotes colorectal cancer cell proliferation via epigenetically
silencing DUSP5/CDKN1A expression. Cell Death Dis. 8:e29972017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun F, Liang W and Qian J: The
identification of CRNDE, H19, UCA1 and HOTAIR as the key lncRNAs
involved in oxaliplatin or irinotecan resistance in the
chemotherapy of colorectal cancer based on integrative
bioinformatics analysis. Mol Med Rep. 20:3583–3596. 2019.PubMed/NCBI
|
|
25
|
Gao H, Song X, Kang T, Yan B, Feng L, Gao
L, Ai L, Liu X, Yu J and Li H: Long noncoding RNA CRNDE functions
as a competing endogenous RNA to promote metastasis and oxaliplatin
resistance by sponging miR-136 in colorectal cancer. Onco Targets
Ther. 10:205–216. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wiese EK and Hitosugi T: Tyrosine Kinase
signaling in cancer metabolism: PKM2 Paradox in the Warburg Effect.
Front Cell Dev Biol. 6:792018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li L, Liang Y, Kang L, Liu Y, Gao S, Chen
S, Li Y, You W, Dong Q, Hong T, et al: Transcriptional regulation
of the warburg effect in cancer by SIX1. Cancer Cell.
33:368–385.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li J, Cheng D, Zhu M, Yu H, Pan Z, Liu L,
Geng Q, Pan H, Yan M and Yao M: OTUB2 stabilizes U2AF2 to promote
the Warburg effect and tumorigenesis via the AKT/mTOR signaling
pathway in non-small cell lung cancer. Theranostics. 9:179–195.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Z and Chen W: Long non-coding RNA
CRNDE promote the progression of tongue squamous cell carcinoma
through regulating the PI3K/AKT/mTOR signaling pathway. RSC Adv.
9:21381–21390. 2019. View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lunt SY and Vander Heiden MG: Aerobic
glycolysis: Meeting the metabolic requirements of cell
proliferation. Annu Rev Cell Dev Biol. 27:441–464. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vaupel P, Schmidberger H and Mayer A: The
Warburg effect: Essential part of metabolic reprogramming and
central contributor to cancer progression. Int J Radiat Biol.
95:912–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brooks GA: Cell-cell and intracellular
lactate shuttles. J Physiol. 587((Pt 23)): 5591–5600. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Held-Warmkessel J and Dell DD: Lactic
acidosis in patients with cancer. Clin J Oncol Nurs. 18:592–594.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Peppicelli S, Bianchini F and Calorini L:
Extracellular acidity, a ‘reappreciated’ trait of tumor environment
driving malignancy: Perspectives in diagnosis and therapy. Cancer
Metastasis Rev. 33:823–832. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shiraishi T, Verdone JE, Huang J, Kahlert
UD, Hernandez JR, Torga G, Zarif JC, Epstein T, Gatenby R,
McCartney A, et al: Glycolysis is the primary bioenergetic pathway
for cell motility and cytoskeletal remodeling in human prostate and
breast cancer cells. Oncotarget. 6:130–143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Holman GD: Chemical biology probes of
mammalian GLUT structure and function. Biochem J. 475:3511–3534.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Al-Azzam N: Sirtuin 6 and metabolic genes
interplay in Warburg effect in cancers. J Clin Biochem Nutr.
66:169–175. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hong X, Zhong L, Xie Y, Zheng K, Pang J,
Li Y, Yang Y, Xu X, Mi P, Cao H, et al: Matrine reverses the
warburg effect and suppresses colon cancer cell growth via
negatively regulating HIF-1α. Front Pharmacol. 10:14372019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu W, Yang Z, Huang R, Min Z and Ye M:
SIRT6 promotes the Warburg effect of papillary thyroid cancer cell
BCPAP through reactive oxygen species. Onco Targets Ther.
12:2861–2868. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lu Z, Guo Y, Zhang X, Li J, Li L, Zhang S
and Shan C: ORY-1001 Suppresses cell growth and induces apoptosis
in lung cancer through triggering HK2 mediated warburg effect.
Front Pharmacol. 9:14112018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lin G, Wu Y, Cai F, Li Z, Su S, Wang J,
Cao J and Ma L: Matrine promotes human myeloid leukemia cells
apoptosis through warburg effect mediated by hexokinase 2. Front
Pharmacol. 10:10692019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang S, Zhang Y, Cai Q, Ma M, Jin LY, Weng
M, Zhou D, Tang Z, Wang JD and Quan Z: Circular RNA FOXP1 promotes
tumor progression and Warburg effect in gallbladder cancer by
regulating PKLR expression. Mol Cancer. 18:1452019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Icard P, Shulman S, Farhat D, Steyaert JM,
Alifano M and Lincet H: How the Warburg effect supports
aggressiveness and drug resistance of cancer cells? Drug Resist
Updat. 38:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lu Z, Shi X, Gong F, Li S, Wang Y, Ren Y,
Zhang M, Yu B, Li Y, Zhao W, et al: RICTOR/mTORC2 affects
tumorigenesis and therapeutic efficacy of mTOR inhibitors in
esophageal squamous cell carcinoma. Acta Pharm Sin B. 10:1004–1019.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen GQ, Tang CF, Shi XK, Lin CY, Fatima
S, Pan XH, Yang DJ, Zhang G, Lu AP, Lin SH and Bian ZX:
Halofuginone inhibits colorectal cancer growth through suppression
of Akt/mTORC1 signaling and glucose metabolism. Oncotarget.
6:24148–24162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sun Q, Chen X, Ma J, Peng H, Wang F, Zha
X, Wang Y, Jing Y, Yang H, Chen R, et al: Mammalian target of
rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is
critical for aerobic glycolysis and tumor growth. Proc Natl Acad
Sci USA. 108:4129–4134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Duvel K, Yecies JL, Menon S, Raman P,
Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S,
et al: Activation of a metabolic gene regulatory network downstream
of mTOR complex 1. Mol Cell. 39:171–183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yalcin S, Marinkovic D, Mungamuri SK,
Zhang X, Tong W, Sellers R and Ghaffari S: ROS-mediated
amplification of AKT/mTOR signalling pathway leads to
myeloproliferative syndrome in Foxo3(−/-) mice. EMBO J.
29:4118–4131. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Maiese K, Chong ZZ, Shang YC and Wang S:
mTOR: On target for novel therapeutic strategies in the nervous
system. Trends Mol Med. 19:51–60. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Siska PJ, van der Windt GJ, Kishton RJ,
Cohen S, Eisner W, MacIver NJ, Kater AP, Weinberg JB and Rathmell
JC: Suppression of glut1 and glucose metabolism by decreased
Akt/mTORC1 signaling drives T cell impairment in B cell leukemia. J
Immunol. 197:2532–2540. 2016. View Article : Google Scholar : PubMed/NCBI
|