Introduction
Cervical carcinoma is one of the most common
malignancies in women from Mexico, representing the second most
common female neoplasm, in 2020 the incidence rate was 6.5% and the
mortality rate was 7.7% (1).
Epidemiological and molecular data indicate that persistent
infection with high-risk human papillomavirus (HPV) is a risk
factor for cervical cancer development and has been associated with
other pathologies, such as head and neck, and anal cancers
(2,3). HPV DNA is present in virtually all
cervical cancer and precursor lesions (4). The expression of the EGFR protein has
been associated with HPV infection, as evidenced by its increasing
expression level as the grade of intraepithelial neoplasia
increases (5). The HPV16 E5
oncogene cannot transform keratinocytes by itself, but it increases
the efficiency of cell immortalization by E6/E7 (6). The E5 protein inhibits the
downregulation of the EGFR receptor in the presence of the ligand,
increasing the steady-state level of EGFR on the cell surface
(7).
The EGFR is part of the ErbB family of tyrosine
kinases receptors; the other members are HER2/ErbB2, HER3/ErbB3 and
HER4/ErbB4. The activation of the EGFR leads to autophosphorylation
and this change initiates a cascade of downstream signaling
pathways. The activity of the EGFR initiates an allosteric
interaction between the kinase domain, in an asymmetric dimer, and
the helix αC, which is an important element in kinase regulation.
The conformation of the helix αC defines an inactive conformation
that is characteristic of numerous kinases. In the active
conformation, the helix αC has a conserved glutamate, which is able
to form a salt bridge with a lysine that coordinates ATP (8–11).
A significant change in the EGFR results from the
presence of mutations in the intracellular tyrosine kinase domain.
These mutations are restricted to the first four (exons 18-21) of
the seven exons that encode the tyrosine kinase domain. Previous
studies have demonstrated that these small mutations prolong the
activity of ligand-activated receptors (12–17).
In addition, previous studies have also identified somatic
mutations or evidence of gene amplification (13,17);
however, activating mutations in exons 19-21, from the EGFR, have
not been identified in cervical cancer (18).
Our previous research demonstrated that the EGFR was
not phosphorylated in the CALO and INBL cervical cancer cell lines
(19). Since the receptor was not
phosphorylated we hypothesized that it was a dead kinase due to the
presence of inactivating mutations. Therefore, the aim of this
study was to analyze the catalytic activity of the isolated EGFR as
well as the presence of mutations in the control region αC of the
catalytic domain.
Materials and methods
Cell culture conditions
The HPV-associated cervical cancer cell lines, CALO,
INBL (cell lines established in the Cell Differentiation and Cancer
Research Unit, FES Zaragoza, National University of Mexico, Mexico
City, Mexico) (20), HeLa [cat.
no. American Type Culture Collection (ATCC)-CCL-2] and CasKi (cat.
no. CRM-CRL-1550), as well as the THP1 cell line (cat. no. ATCC
TIB-202) (all purchased from ATCC), were cultured in RPMI-1640
(Microlab Industrial) medium supplemented with 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.). All the cell lines were incubated
at 37°C in a humidified incubator with 5% CO2. The cells
were treated with 20 ng/ml EGF for 5 min.
Cell lysis
The cell lines were lysed with ice-cold lysis buffer
[1% Triton X-100, 5 mM EDTA, 140 mM NaCl, 50 mM Tris (pH 7.4), 1 mM
phenylmethylsulfonylfluroide, 1 mM NaF, 1% aprotinin, 1 µM
leupeptin, 1 µM pepstatin and 100 µM Na3VO4]
for 15 min. The lysates were then centrifuged at 1,363 × g at 4°C
for 15 min and the supernatants were then collected.
Immunoprecipitation and
immunoblotting
For immuno-precipitation, treated or untreated cells
were lysed as aforementioned. The total protein content of the
lysates was determined using the Bio-Rad protein assay (Bio-Rad
Laboratories, Inc.) and 150 µg protein was incubated with protein
A-agarose beads (Invitrogen; Thermo Fisher Scientific, Inc.)
previously coupled with 0.3 µg anti-EGFR (cat. no. sc-53274) or
anti-HER2 (cat. no. sc-284) antibodies (Santa Cruz Biotechnology,
Inc.) for 3 h at 4°C. The immunoprecipitated proteins were washed
five times with ice-cold lysis buffer, resolved with 10% SDS-PAGE
and transferred to nitrocellulose membranes (Trans-Blot; Bio-Rad
Laboratories, Inc.). The membranes were blocked in TBS with 0.1%
Tween-20 and 3% bovine serum albumin (Santa Cruz Biotechnology,
Inc.) overnight at 4°C.
The membranes were analyzed using both mouse
anti-phosphotyrosine antibodies [pY20 (cat. no. sc-508) and pY99
(cat. no. sc-7020); Santa Cruz Biotechnology, Inc.] for 2 h,
followed by incubation for 45 min with HRP-conjugated rabbit
anti-mouse antibody (cat. no. 31450; Thermo Fisher Scientific,
Inc.) at room temperature (dilutions, 1:10,000). The proteins were
visualized using an enhanced chemiluminescence detection system
(Super Signal; Pierce; Thermo Fisher Scientific, Inc.). To
determine the presence of EGFR and HER2 in the same membrane, the
anti-phosphotyrosine antibodies were stripped. After stripping the
membrane with 0.1 M glycine (pH 2.5) and blocking with 5% bovine
serum albumin overnight, the same membranes were incubated with the
anti-EGFR (1:1,000) or anti-HER2 (1:1,000) antibodies (Santa Cruz
Biotechnology, Inc.) for 2 h at room temperature, then treated as
mentioned above. All figures show representative results from at
least three independent experiments.
In vitro kinase assay
For the in vitro kinase assay, a Universal
kinase assay kit (fluorometric) (Abcam) was used, according to the
manufacturer's instructions. Briefly, 3×106 from each
cell line, treated or untreated, were lysed with ice-cold lysis
buffer [1% Triton X-100, 5 mM EDTA, 140 mM NaCl, 50 mM Tris (pH
7.4), 1% aprotinin, 1 µM leupeptin, 1 µM pepstatin] for 15 min. The
lysates were centrifuged at 1,363 × g at 4°C for 15 min, then the
supernatants were collected and the EGFR receptor was
immunoprecipitated as aforementioned. The immunoprecipitated
proteins were washed three times with ice-cold lysis buffer, three
times with ice-cold PBS (pH 7.4), then three times with 50 mM Tris
(pH 7.4).
To each tube, 20 µl ADP assay buffer, 20 µl ADP
sensor buffer, 10 µl ADP sensor, 1 µM ATP and 0.03 µg polyGluTyr
poly amino acid (Sigma-Aldrich; Merck KGaA), as the substrate, were
added to a total ADP assay volume of 50 µl/sample. For the in
vitro kinase assay without substrate, the polyGluTyr was
omitted. The reaction mixture was incubated at room temperature for
30 min. The fluorescence intensity was measured at
excitation/emission, 540/590 nm in black plates in a Fluoroskan
Ascent Fluorometer (Thermo Fisher Scientific, Inc.).
Finally, the HeLa cell line was used as a control
for catalytic activity of EGFR and the myelomonocytic THP1 cell
line was used as a negative control.
DNA extraction and sequencing
For DNA extraction, DNAzol (Invitrogen; Thermo
Fisher Scientific, Inc.) was used, according to the manufacturer's
specifications. Briefly, 3×106 from each cell line was
lysed with 0.75 ml DNAzol, then the lysates were centrifuged
(10,000 × g at 4°C for 10 min) and the supernatant was transferred
to a new tube. Subsequently, the DNA was precipitated following the
addition of 0.5 ml 100% ethanol. The samples were centrifuged at
4,000 × g at 4°C for 2 min, then the supernatant was discarded. The
obtained DNA was washed twice with 70% ethanol, air-dried for 20
sec, then dissolved in DNase free water.
The obtained DNA was amplified using PCR. DreamTaq
DNA polymerase (cat. no. EP0702; Thermo Fisher Scientific, Inc.)
was used and the following primers were used: αC helix forward,
5′-CGTAAACGTCCCTGTGCTAGG-3′ and reverse,
5′-CCTTATCTCCCCTCCCCGTAT-3′; and GAPDH forward,
5′-GGGACGCTTTCTTTCCTTTCGC-3′ and reverse,
5′-GTCGGGTCAACGCTAGGCTG-3′ (Applied Biosystems; Thermo Fisher
Scientific, Inc.). GAPDH was used as the internal control. The
following thermocycling conditions were used: Initial denaturation
at 95°C for 5 min; 58°C for 1 min; 72°C for 1 min and 95°C for 1
min for 30 cycles. The product was visualized using agarose gel
(1.5%) electrophoresis and the DigiDoc-It transilluminator
(Analytik Jena GmbH). The bands corresponding to the expected
products were excised and purified with a Zymoclean Gel DNA
Recovery kit (Zymo Research Corp.) and sent for sequencing using
the Sanger method to the Faculty of Higher Studies (Facultad de
Estudios Superiores) Iztacala facilities at National University of
Mexico (Mexico City, Mexico) (ABI Prism 7500; Applied Biosystems;
Thermo Fisher Scientific, Inc.).
Modelling of amino acid sequence
The putative amino acid sequences for EGFR in CALO
and INBL cells, obtained from Basic Local Alignment Search Tool
(BLASTx) for proteins (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome),
were modeled using SWISS-MODEL Homology Modeling (https://swissmodel.expasy.org/interactive).
SWISS-MODEL is a fully automated protein structure
homology-modelling server. The SWISS-MODEL Workspace is a web-based
working environment, where several modelling projects can be
carried out. Protein sequence and structure databases necessary for
modelling are accessible from the workspace and are updated in
regular intervals. Tools for template selection, model building and
structure quality evaluation can be invoked from within the
workspace directly or via the web page menu. Building a homology
model comprises four steps: i) Identification of structural
template(s); ii) alignment of target sequence and template
structure(s); iii) model-building; and iv) model quality evaluation
(21–23).
Statistical analysis
All the data were obtained from three independent
experiments for statistical analysis. The data are presented as the
mean ± standard error of the mean. An unpaired Student's t-test
(two-tailed) for parametric data was used to compare treatment
groups using the GraphPad Prism v8.0.1 statistical package
(GraphPad Software, Inc.). P-values and 95% confidence intervals
were calculated. P<0.05 was considered to indicate a
statistically significant difference.
Results
Presence of the EGFR in the cervical
cancer cell lines
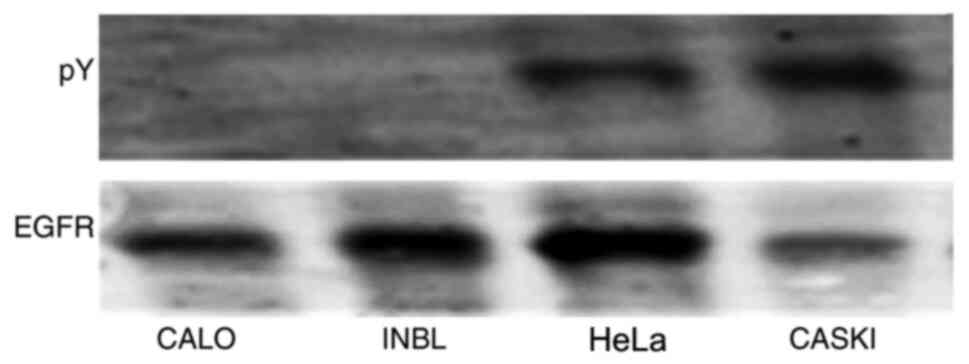
Our previous research demonstrated that the cervical
cancer cell lines, CALO and INBL expressed the EGFR; however, this
molecule was not phosphorylated (19). The CALO, INBL, HeLa and CasKi cell
lines were lysed to determine the presence of the EGFR in the
cervical cancer cells, and the proteins were immunoprecipitated
with protein-A agarose beads coupled with an anti-EGFR antibody,
resolved using SDS-PAGE and transferred to nitrocellulose
membranes. The results showed that the EGFR was present in all the
cell lines; however, it was not phosphorylated in the CALO and INBL
cell lines (Fig. 1).
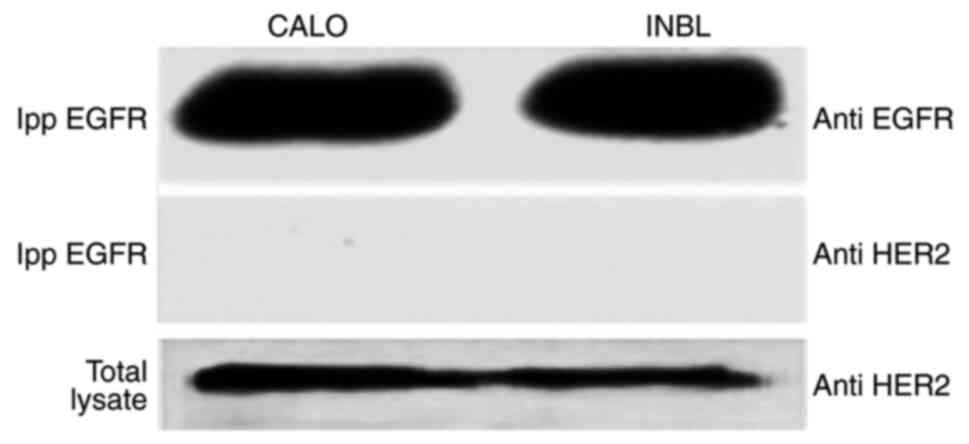
EGFR is present in the CALO and INBL
cervical cancer cell lines, but does not co-precipitate with
HER2
As HER2 was also found to be present in the CALO and
INBL cell lines (19), the ability
of the EGFR to interact with HER2 was analyzed using a
co-precipitation assay. The results showed that HER2 does not
co-precipitate with the EGFR or vice-versa (Fig. 2).
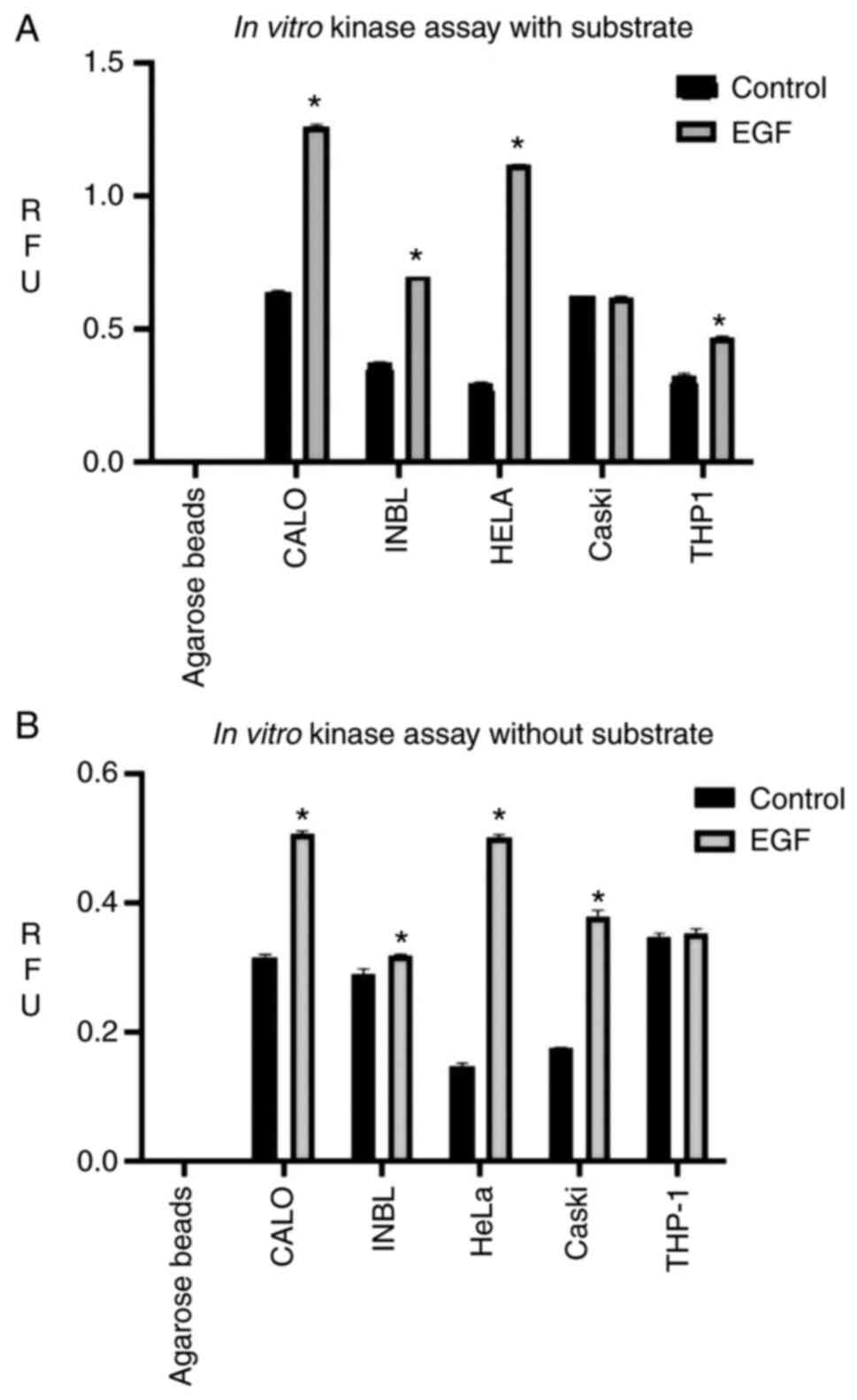
Non-phosphorylated EGFR is present in
the CALO and INBL cell lines and is catalytically active
Our previous research demonstrated that the EGFR was
not phosphorylated in the CALO and INBL cell lines (19), thus to determine the catalytic
activity of the EGFR, the CALO, INBL, HeLa, CasKi and THP1 cell
lines were stimulated with EGF. After 5 min, the cells were lysed,
and the proteins were immunoprecipitated with protein A-agarose
beads coupled with an anti-EGFR antibody, then the catalytic
activity was determined using a Universal kinase assay kit.
Surprisingly, it was found that the EGFR receptor
isolated from the CALO and INBL cell lines showed high catalytic
activity (Fig. 3A). The EGFR,
present in the CALO cell line, had similar catalytic activity to
the receptor in the HeLa cell line; however, the INBL cell line had
a reduced EGFR catalytic activity. The EGFR present in the CasKi
cell line had the same catalytic activity with or without EGF
treatment. The myelomonocytic THP1 cell line was used as a negative
control; however, the receptor was present in this cell line, and a
slight increase in the activity of EGFR was observed when treated
with EGF (Fig. 3A).
To analyze the autophosphorylation activity of the
receptor an in vitro kinase assay without substrate was
designed (Fig. 3B). The catalytic
activity of the EGFR in all the cell lines was lower than the
activity observed in the presence of the substrate. The EGFR showed
similar catalytic activity in the CALO, HeLa and CasKi cell lines;
however, the basal activity of the EGFR in the CALO cell line was
higher compared with the HeLa cell line. When the cells were
stimulated with EGF, the catalytic activity of the receptor in the
CALO and HeLa cell lines increased at the same level; however, in
the CasKi cell line, the receptor showed an increase in catalytic
activity (75%) compared with that in the HeLa cell line (Fig. 3B). In the assay without substrate,
only THP1 cell line did not have an increase in the catalytic
activity of the EGFR protein with or without EGF treatment.
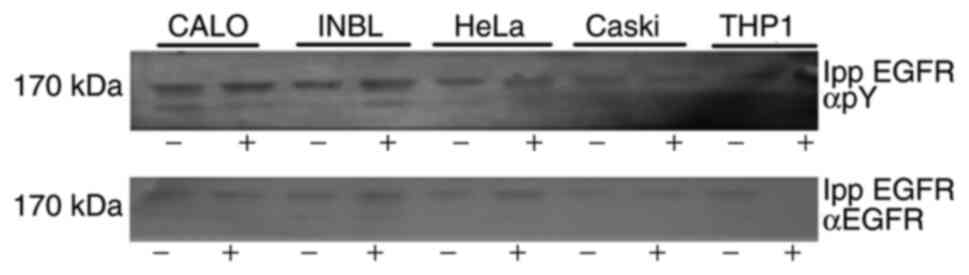
The immunoprecipitated EGFR, used to determine the
enzymatic activity without a substrate, was separated using
SDS-PAGE to analyze its autophosphorylation ability (Fig. 4). The results showed the presence
of the phosphorylated receptor in all the cell lines (Fig. 4, upper panel), indicating that the
isolated EGFR had catalytic activity and it could phosphorylate its
tyrosine residues. After the membrane was stripped, it was
re-probed with anti-EGFR antibodies, which confirmed the presence
of EGFR (Fig. 4, bottom
panel).
Sequence determination of the αC
domain in the EGFR isolated from the CALO and INBL cell lines
Our previous research demonstrated that the EGFR was
not phosphorylated in the CALO and INBL cell lines (19), and a possible explanation for the
lack of tyrosine phosphorylation of the EGFR in the CALO and INBL
cell lines is the presence of mutations. Therefore, possible
mutations in the catalytic domain of the receptor were
investigated. DNA from the CALO and INBL cell lines was isolated
and PCR was used to amplify the exons corresponding to the αC helix
in the EGFR. The PCR products were visualized using agarose gel
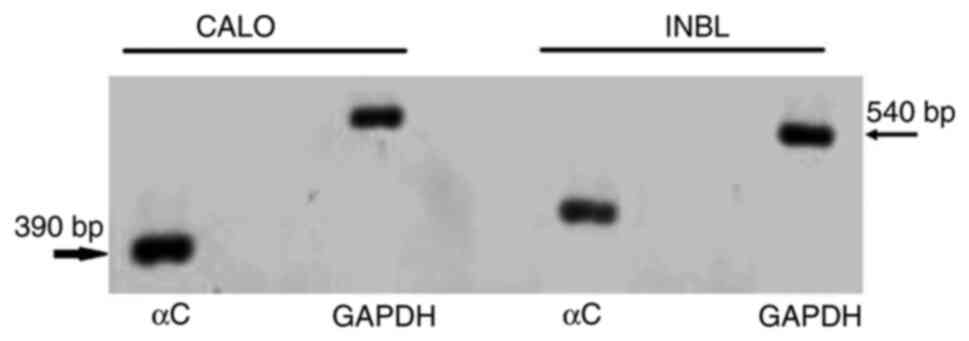
electrophoresis (Fig. 5). The
pattern of expression of the αC helix (390 bp) was different in
both cell lines (Fig. 5).
The bands were then isolated using a Zymoclean Gel
DNA Recovery kit and sequenced using an ABI Prism sequencer. The
analysis of the obtained DNA sequences were then compared to the
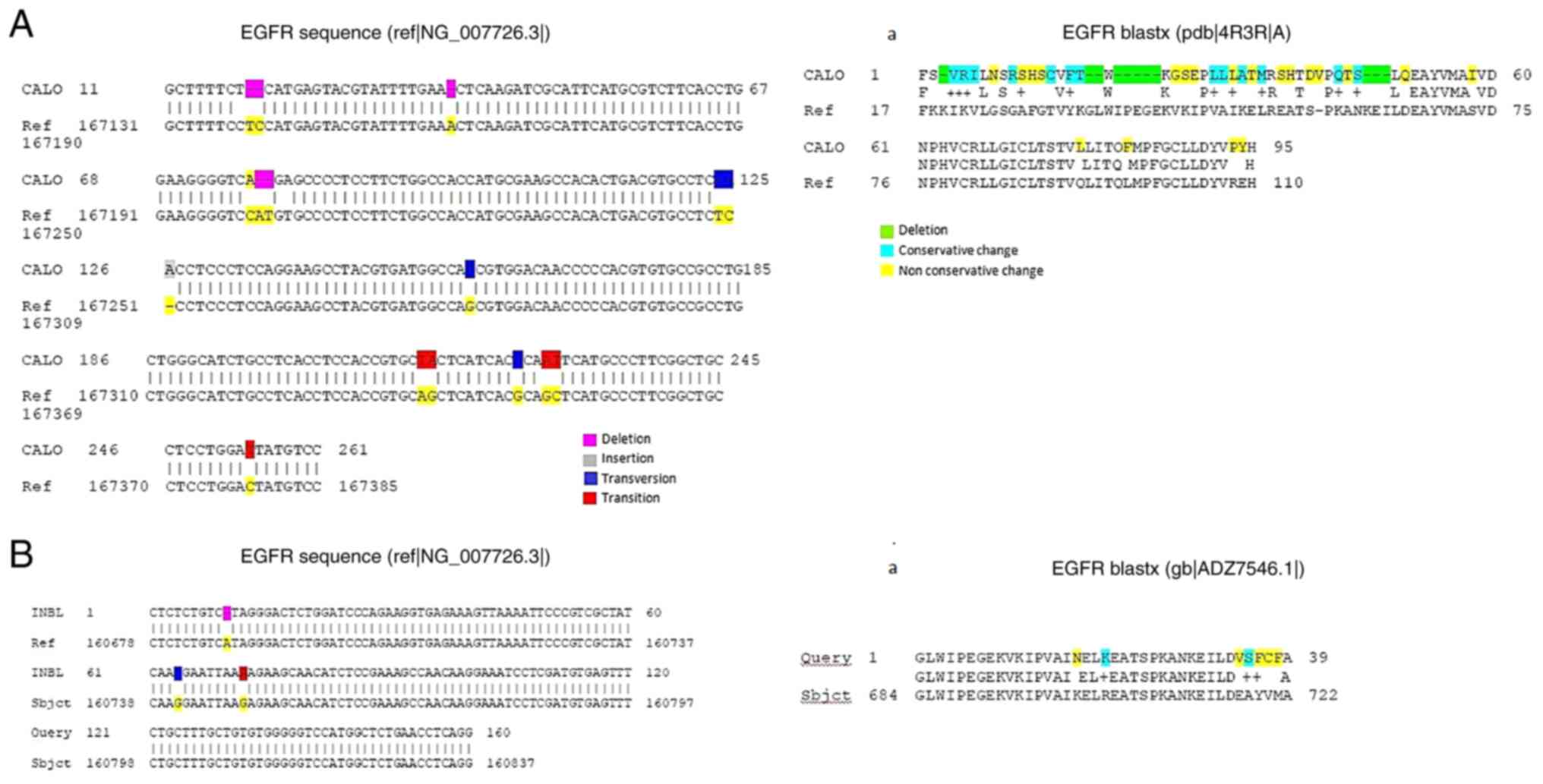
normal EGFR sequence reported and is shown in Fig. 6. The obtained sequence for the EGFR
present in the CALO cell line is shown in Fig. 6A. The presence of multiple
deletions (positions 19, 20, 40, 78 and 79), one insertion
(position 126), transversions (124, 157, 214 and 224) and
transitions (215, 227, 228 and 254) was detected. Using the BLASTx
program, the DNA sequence was translated into the putative amino
acid sequence. The putative amino acid sequence of the EGFR present
in the CALO cell line showed multiple differences as compared with
that in the normal EGFR amino acid sequence. For example, deletions
of amino acids (positions 3, 18, 19, 21-25 and 47-49), conservative
changes (positions 4-6, 11, 16, 31, 33, 36, 44 and 46) and
non-conservative changes (positions 2, 8, 10, 12-14, 17, 27-29, 32,
34, 35, 38, 39, 41, 42, 45, 51, 58, 77, 82, 93 and 94) (Fig. 6A-a).
With respect to the INBL cervical cancer cell line
(Fig. 6B), the DNA sequence of the
EGFR showed only three changes, such as a deletion (position 10), a
transversion (position 64) and a transition (position 72). The
translation of the DNA sequence into the putative amino acid
sequence, using the BLASTx program, is shown in Fig. 6B-a. The comparison with the EGFR
reference sequence indicated some differences: Non-conservative
(positions 17, 34, 36, 37 and 38) and conservative differences
(positions 20, and 36).
The DNA sequences were processed by BLASTx and the
putative amino acid sequences were obtained. The results showed
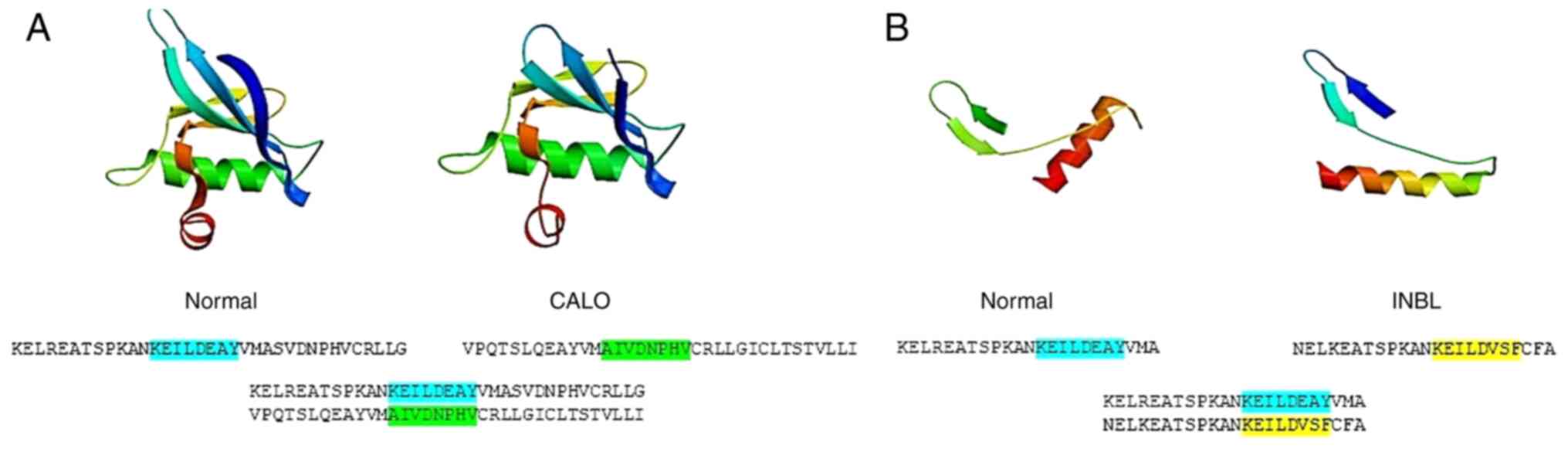
that in the CALO cervical cancer cell line the conserved glutamic
acid residue at position 738 in the normal EGFR was replaced by
asparagine, and in the INBL cell line, the glutamic acid residue
was replaced by valine. Other residues found in the reference
sequence of the helix αC in the normal EGFR were also different in
the EGFR present in the cervical cancer cell lines. In the CALO
cell line, a sequence of amino acids contained multiples changes.
The most significant was the lack of the KEILDEAY motif in the CALO
cell line. The sequence, AIVDNPHV, lacked tyrosine and other
necessary residues (only conserved D) to form the interphase
between the dimers. The INBL cell line contained the sequence,
KEILDVSF, which is similar to the reference EGFR sequence; however,
it included three different amino acids. Both cell lines contained
the loss of the phosphorylated tyrosine present in the KEILDEAY
motif (Fig. 7A and B). These
putative amino acid sequences were modelled using the SWISS-MODEL
Workspace and the 3D structure showed differences in the amino acid
spatial arrangement, for example, in the CALO cell line, the
β-sheet was shorter and the torsion was slightly lower; in the INBL
cell line the most evident change was the torsion, which was bigger
(Fig. 7A and B).
Discussion
The understanding of cancer biology has led to the
identification of numerous receptors with tyrosine kinase activity
(RTK) (12,24) involved in malignant transformation.
The EGFR is the most common RTK that is mutated and overexpressed
in epithelial cancers (25–28).
Our previous research showed that the EGFR was present in the CALO
and INBL cervical cancer cell lines, but was not phosphorylated
(19); therefore, we hypothesized
that it was a dead kinase. To investigate the hypothesis, the
present study was performed to determine if it had null catalytic
activity due to a mutation in the catalytic domain. The
amplification of the EGFR gene is common in cancer and mutations in
the EGFR tyrosine kinase domain have been demonstrated to occur in
cervical cancer; these mutations are mainly somatic (13–17).
The present study provides evidence for in vitro catalytic
activity of this molecule, despite the presence of some mutations
in the catalytic domain in both cell lines. In addition, it was
found that the immunoprecipitated EGFR, from the activated CALO and
INBL cell lines was not phosphorylated compared with that in the
HeLa and CasKi cell lines (Fig.
1). This is unusual as typically this receptor is
hyperphosphorylated. Our previous research demonstrated that the
CALO and INBL cell lines expressed EGFR, HER2 (19) and JAK3 (24), and only the EGFR was found not to
be constitutively phosphorylated. However, when the EGFR was
isolated and analyzed using an in vitro kinase assay
(Fig. 2), it was found that the
receptor had catalytic and autophosphorylating activity. It could
be possible that in the CALO and INBL cervical cancer cell lines,
the ERRFI1 molecule could block the autocatalytic activity of EGFR,
which is responsible for the inhibition observed (29,30).
In the present study, a differential behavior of the catalytic
activity of the receptor was found. It is proposed that when the
isolated receptor is washed under stringent conditions,
non-covalent interactions are broken. Therefore, the regulator
molecule might be separated, enabling the catalytic activity of
EGFR.
On the other hand, EGFR is part of the ErbB family
of tyrosine kinases receptors. The activation of EGFR by its ligand
leads to autophosphorylation; this change initiates a cascade of
downstream signaling pathways regulating cellular proliferation,
differentiation and survival (8,26,30).
With respect to HER3, this kinase lacks several residues that are
important for catalysis. Since the activation of EGFR family
members is switched by an allosteric interaction between the kinase
domain in an asymmetric kinase domain dimer, HER3 is specialized to
serve as an activator of other EGFR family members (8–11).
The helix αC is an essential element in kinase regulation. The
conformation of this domain defines an inactive conformation that
is characteristic of numerous kinases, such as CDKs (31), Src kinases (32), Zap70 (33) and EGFR (34). In the active conformation, the
helix αC has a conserved glutamic acid residue (position 738 in
EGFR), which is able to form a salt bridge with a conserved lysine
(position 721 in EGFR), which coordinates ATP (8). In HER3, the glutamic acid 738 residue
is replaced by histidine, an amino acid with a positive charge. It
has been reported that the recombinant HER3 does not have a
detectable kinase activity (10)
or has minimal activity when immunoprecipitated (11,35).
Due to these characteristics, HER3 has been designated as a
pseudokinase (8,9,36).
Similarly, in the CALO cervical cancer cell line, in the putative
sequence of helix αC, in the catalytic domain, the conserved
glutamic acid 738 residue is replaced by asparagine, and in the
INBL cell line it is replaced by valine. Therefore, both amino
acids lack a negative charge and cannot form a saline bridge with
the conserved residue lysine 721. There were also differences in
the helix αC from the isolated EGFR in comparison to the EGFR
reference sequence. In the CALO cell line, one notable difference
was found; the lack of the KEILDEAY motif, which changed to
AIVDNPHV. It lacked the tyrosine residue and other residues
required to form the interphase between the dimers. Both cell lines
lacked the tyrosine residue, that can be phosphorylated, present in
the KEILDEAY motif; therefore, the activity of the receptor may
change.
The activation of the EGFR family involves the
formation of asymmetric dimers (homo- or heterodimers) between
their kinase domains, in which one kinase domain (activator) acts
as an allosteric activator of the other (receiver) (9,37).
In this mechanism, the helix αC plays an essential role in the EGFR
family by forming part of a docking site on the receiver domain for
the activator kinase domain (9).
This activation mechanism of the EGFR family resembles the
activator kinase when it plays a similar role to that of the
activation of a cyclin-dependent kinase (38).
With respect to HER3, the kinase domain lacks
several residues that are necessary for catalysis. The activation
of the EGFR family members is initiated by an allosteric
interaction between the kinase domain in an asymmetric dimer. Thus,
HER3 may have acquired a specialization to serve as an activator of
the EGFR family members (9).
Similarly, the EGFR present in the CALO and INBL cervical cancer
cell lines did not show catalytic activity in the presence of other
molecules in the cellular environment in the present study. In
addition, it contained differences in the amino acids located in
the catalytic domain (Figs. 6 and
7). Thus, it is hypothesized these
changes in the amino acid sequence and the spatial arrangement may
change the EGFR conformation and confer a different activity in
these cell lines, such as an activator of asymmetric dimers
emulating the function of HER3.
The weak kinase activity of EGFR in the CALO and
INBL cervical cancer cell lines may resemble the ability of the
HER3 kinase to drive resistance to agents which inhibit other
family members, for example EGFR or HER2 (39). Therefore, the EGFR without kinase
activity in the cellular environment in both the CALO and INBL cell
lines may serve to activate HER2 (which is constitutively active)
(19) and, similar to HER3, also
plays a critical role in the ability of HER2 to escape growth
inhibition by some inhibitors, such as lapatinib (40). It was demonstrated that the EGFR
protein does not interact with HER2 in the CALO and INBL cell
lines; however, it is necessary to determine if this interaction is
possible in other cervical cell lines, such as HeLa and CasKi, to
have a complete understanding of this function of EGFR protein. In
addition, further studies are being conducted to prove the
interaction between the EGFR protein and ERRF1 and its
participation in cervical carcinogenesis, as this molecule has an
intrinsic oncogenic potential.
Acknowledgements
The authors would like to thank Mr. José
Chavarría-Mancilla (Laboratory of Molecular Oncology, FES Zaragoza,
National University of Mexico, Mexico City, Mexico) for technical
assistance.
Funding
This study was supported by grants from Programa de Apoyos a
Proyectos de Investigación e Innovación Tecnológica (PAPIIT) (grant
nos. IN221512 and IN222218) from the Dirección General de Asuntos
del Personal Académico (DGAPA) Universidad Nacional Autónoma de
México (UNAM).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AVM and ISC were involved in the conception and
design of the study. AVM, RBR, OZC and VDM performed the
experiments. AVM, RBR and OZC analyzed the data. AVM, RBR, OZC,
VDM, AGH, BWS and ISC interpreted the results of the experiments.
AVM and VDM performed analysis of the sequencing data. AVM, RBR and
OZC prepared the figures. AVM, VDM, AGH and ISC drafted the
manuscript. AVM, VDM, AGH, BWS and ISC edited and revised the
manuscript. AVM, VDM, AGH, BWS and ISC approve the final version of
the manuscript. All authors have read and approved the final
manuscript. AVM, OZC and ISC confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bouvard V, Baan R, Straif K, Grosse Y,
Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C,
Galichet L, et al: A review of human carcinogens-Part B: Biological
agents. Lancet Oncol. 10:321–322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zur Hausen H: Papillomaviruses and cancer:
From basic studies to clinical application. Nat Rev Cancer.
2:342–350. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Munoz N, Bosch FX, de Sanjose S, Herrero
R, Castellsaque X, Shah KV, Snijders PJ and Meijer CJ;
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group, : Epidemiologic classification of human
papillomavirus types associated with cervical cancer. N Engl J Med.
348:518–527. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Soonthornthum T, Arias-Pulido H, Joste N,
Lomo L, Muller C, Rutledge T and Verschraegen C: Epidermal growth
factor receptor as a biomarker for cervical cancer. Ann Oncol.
22:2166–2178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stöppler MC, Straight SW, Tsao G, Schlegel
R and Mccance DJ: The E5 gene of HPV16 enhances keratinocyte
immortalization by full-length DNA. Virology. 223:251–254. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tomakidi P, Cheng H, Kohl A, Komposch G
and Alonso A: Modulation of the epidermal growth factor receptor by
the human papillomavirus type 16 E5 protein in raft cultures of
human keratinocytes. Eur J Cell Biol. 79:407–412. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jura N, Shan Y, Cao X, Shaw DE and Kuriyan
J: Structural analysis of the catalytically inactive kinase domain
of the human EGF receptor 3. Proc Natl Acad Sci USA.
106:21608–21613. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sierke SL, Cheng K, Kim HH and Koland JG:
Biochemical characterization of the protein tyrosine kinase
homology domain of the ErbB3 (HER3) receptor protein. Biochem J.
322:757–763. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wallasch C, Weiss FU, Niederfellner G,
Jallal BA, Issing W and Ullrich A: Heregulin-dependent regulation
of HER2/neu oncogenic signaling by heterodimerization with HER3.
EMBO J. 14:4267–4275. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi F, Telesco SE, Liu Y, Radhakrishnan R
and Lemmon MA: ErbB3/HER3 intracellular domain is competent to bind
ATP and catalyze autophosphorylation. Proc Natl Acad Sci USA.
107:7692–7697. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor underlying responsiveness of non-small-cell
lung cancer to gefitinib. N Engl J Med. 350:2129–2139. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Iida K, Nakayama K, Rahman MT, Rahman M,
Ishikawa M, Katagiri A, Yeasmin S, Otsuki Y, Kobayashi H, Nakayama
S and Miyazaki K: EGFR gene amplification is related to adverse
clinical outcomes in cervical squamous cell carcinoma, making the
EGFR pathway a novel therapeutic target. Br J Cancer. 105:420–427.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wolber L, Hess S, Bohlken H, Tennstedt P,
Eulenburg C, Simon R, Gieseking F, Jaenicke F, Mahner S and
Choschzinck M: EGFR gene copy number increase in vulvar carcinoma
is linked with poor clinical outcome. J Clin Pathol. 65:133–139.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Q, Tang Y, Cheng X, Ji J, Zhang J and
Zhou X: EGFR protein expression and gene amplification in squamous
intraepithelial lesions and squamous cell carcinomas of the cervix.
Int J Clin Exp Pathol. 7:733–741. 2014.PubMed/NCBI
|
|
16
|
Conesa-Zamora P, Torres-Moreno D, Isaac MA
and Pérez-Guillermo M: Gene amplification and immunohistochemical
expression of ERBB2 and EGFR in cervical carcinogenesis.
Correlation with cell-cycle markers and HPV presence. Exp Mol
Pathol. 95:151–155. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Q, Cheng X, Ji J, Zhang J and Zhou X:
Gene amplification of EGFR and its clinical significance in various
cervical (lesions) lesions using cytology and FISH. Int J Clin Exp
Pathol. 7:2477–2483. 2014.PubMed/NCBI
|
|
18
|
Arias-Pulido H, Joste N, Chavez A, Muller
CY, Dai D, Smith HO and Verschraegen CF: Absence of epidermal
growth factor receptor mutations in cervical cancer. Int J Gynecol
Cancer. 18:749–754. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Soto-Cruz I, Rangel-Corona R,
Valle-Mendiola A, Moreno-Morales X, Santiago-Pérez R, Weiss-Steider
B and Cáceres-Cortés JR: The tyrphostin B42 inhibits cell
proliferation and HER-2 autophosphorylation in cervical carcinoma
cell lines. Cancer Invest. 26:136–144. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Caceres-Cortes JR, Alvarado-Moreno JA,
Waga K, Rangel-Corona R, Monroy-Garcia A, Rocha-Zavaleta L,
Urdiales-Ramos J, Weiss-Steider B, Haman A, Hugo P, et al:
Implication of tyrosine kinase receptor and steel factor in cell
density-dependent growth in cervical cancers and leukemias. Cancer
Res. 61:6281–6290. 2001.PubMed/NCBI
|
|
21
|
Biasini M, Bienert S, Waterhouse A, Arnold
K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli
L and Schwede T: SWISS-MODEL: Modeling protein tertiary and
quaternary structures using evolutionary information. Nucleic Acids
Res. 42:W252–W258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Arnold K, Bordoli L, Kopp J and Schwede T:
The SWISS-MODEL workspace: A web-based environment for protein
structure homology modeling. Bioinformatics. 22:195–201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Benkert P, Biasini M and Schwede T: Toward
the estimation of the absolute quality of individual protein
structure models. Bioinformatics. 27:343–350. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Valle-Mendiola A, Weiss-Steider B,
Rocha-Zavaleta L and Soto-Cruz I: IL-2 enhances cervical cancer
cells proliferation and JAK3/STAT5 phosphorylation at low doses,
while at high doses IL-2 has opposite effects. Cancer Invest.
32:115–125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yarden Y and Pines G: The ERBB network: At
last, cancer therapy meets systems biology. Nat Rev Cancer.
12:553–563. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
London M and Gallo E: Epidermal growth
factor receptor (EGFR) involvement in epitelial-derived cancers and
its current antibody-based immunotherapies. Cell Biol Int.
44:1267–1282. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rosell R, Moran T, Queralt C, Porta R,
Cardenal F, Camps C, Majem M, López-Vivanco G, Isla D, Provencio M,
et al: Screening for epidermal growth factor receptor mutations in
lung cancer. N Engl J Med. 361:958–977. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ayati A, Moghimi S, Salarinejad S, Safavi
M, Pouramiri B and Foroumadi A: A review on progression of
epidermal growth factor receptor (EGFR) inhibitors as an efficient
approach in cancer targeted therapy. Bioorg Chem. 99:1038112020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anastasi S, Baietti MF, Frosi Y, Alema S
and Segatto O: The evolutionarily conserved EBR module of RALT/MIG6
mediates suppression of the EGFR catalytic activity. Oncogene.
26:7833–7846. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Singh D, Attri BK, Gill RK and Bariwal J:
Review on EGFR inhibitors: Critical update. Mini Rev Med Chem.
16:1134–1166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
De Bondt HL, Rosenblatt J, Jancarik J,
Jones HD, Morgan DO and Kim SH: Crystal structure of
cyclin-dependent kinase 2. Nature. 363:595–602. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu W, Doshi A, Lei M, Eck MJ and Harrison
SC: Crystal structure of c-Src reveal features of its
autoinhibitory mechanism. Mol Cell. 3:629–638. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Deindl S, Kadlecek TA, Brdicka T, Cao X,
Weiss A and Kuriyan J: Structural basis for the inhibition of
tyrosine kinase activity of ZAP-70. Cell. 129:735–746. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wood ER, Truesdale AT, McDonald OB, Yuan
D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K,
et al: A unique structure for epidermal growth factor receptor
bound to GW572016 (Lapatinib): relationship among protein
conformation, inhibitor off-rate, and receptor activity in tumor
cells. Cancer Res. 64:6652–6659. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Manning G, Whyte DB, Martinez R, Hunter T
and Sudarsanam S: The protein kinase complement of the human
genome. Science. 298:1912–1934. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guy PM, Platko JV, Cantley LC, Cerione RA
and Carraway KL III: Insect cell-expressed p180erbB3 possesses an
impaired tyrosine kinase activity. Proc Natl Acad Sci USA.
91:8132–8136. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang X, Gureasko J, Shen K, Cole PA and
Kuriyan J: An allosteric mechanism for activation of the kinase
domain of epidermal growth factor receptor. Cell. 125:1137–1149.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jeffrey PD, Russo AA, Polyak K, Gibbs E,
Hurwitz J, Massague J and Pavletich NP: Mechanism of CDK activation
revealed by structure of a cyclinA-CDK2 complex. Nature.
376:313–320. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Baselga J and Swain SM: Novel anticancer
targets: Revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer.
9:463–475. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sergina NV, Rausch M, Wang D, Blair J,
Hann B, Shokat KM and Moasser MM: Escape from HER-family tyrosine
kinase inhibitor therapy by the kinase-inactive HER3. Nature.
445:437–441. 2007. View Article : Google Scholar : PubMed/NCBI
|