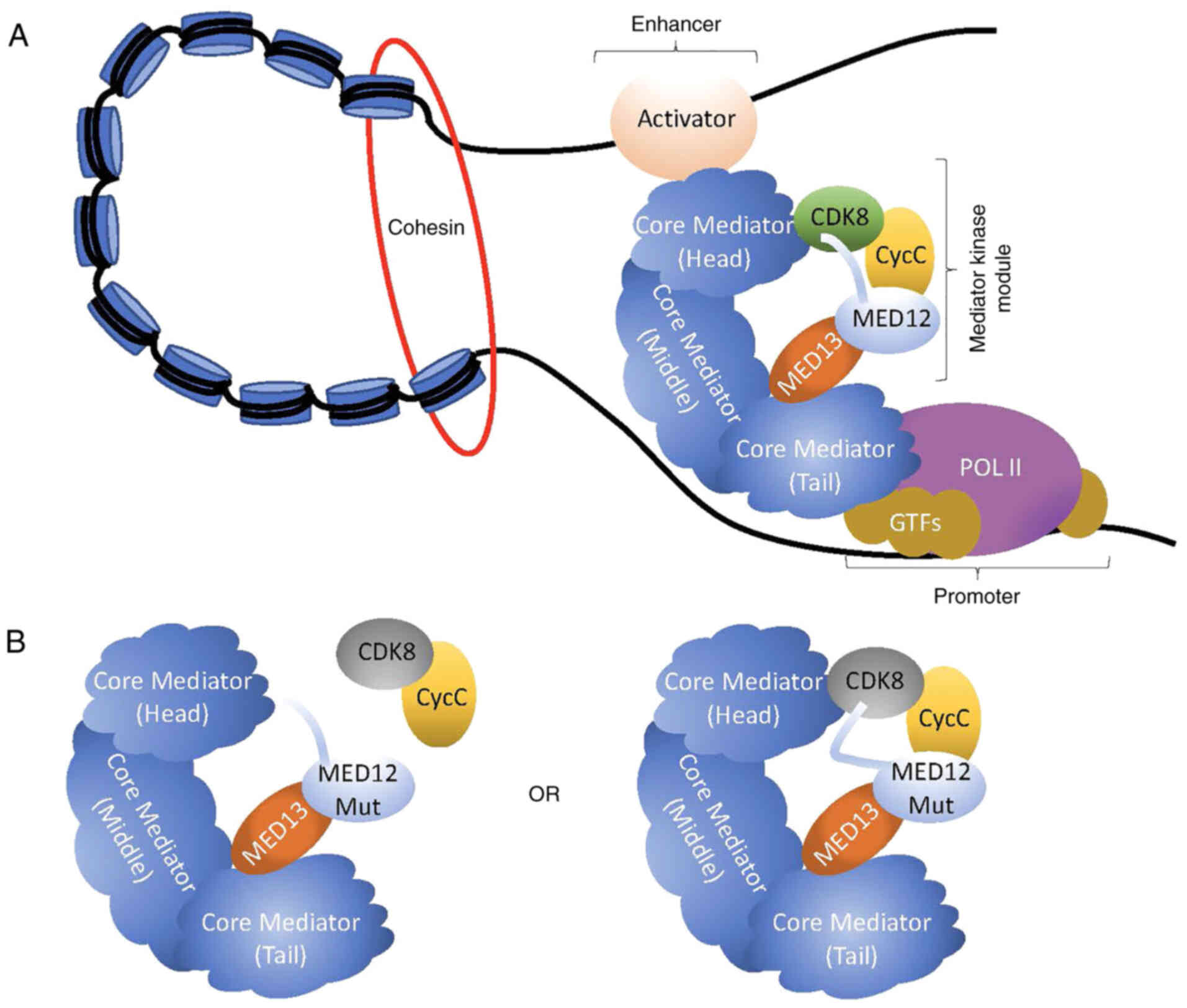
Med12 is an X-linked gene that provides instructions
for making a protein called Mediator complex subunit 12 (MED12).
Mediator is a large multi-subunit complex that is critical for gene
regulation. MED12 plays an important role in Mediator function by
activating and repressing transcription through regulation of RNA
polymerase II (Fig. 1A) (1,2). The
structure of Mediator includes a dissociable kinase module,
comprised of four subunits: MED13, MED12, CyclinC (CycC) and CDK8.
Association or dissociation of the kinase module is an important
indicator of transcriptional regulation with activation or
repression occurring in a context-dependent manner (3,4). The
exact reasons behind context-dependent activation vs. repression
are currently unclear, but appear to involve induced conformational
changes within core Mediator that ultimately affect the ability of
Mediator to interact with Polymerase II (5–9).
Earlier studies have indicated that MED12 functions as a link
between MED13 and CycC-CDK8, with MED12 making a direct connection
to CycC (8,10) (Fig.
1A). Furthermore, it has been shown that the exact
configuration of the kinase module has important implications for
CDK8 kinase activity as CDK8 has been reported to only be fully
activated when it is bound to CycC and MED12 (8,10,11).
Specifically, CycC contains a specified surface groove that acts as
the docking site for MED12 (8).
Mutagenic disruption of the MED12-CycC interface uncouples
CycC-CDK8 from Mediator, which in turn greatly diminishes CDK8
kinase activity; thus indicating that the MED12-CycC interface
plays a critical role in regulating CDK8 kinase activity (8) (Fig.
1B). However, these studies have been complicated as recent
findings determined that the N-terminal domain of MED12 directly
binds to CDK8 thereby positioning an activation helix close to the
T-loop in CDK8 (12,13) (Fig.
1A). Furthermore, these studies reported that MED12 mutations
do not disrupt the MED12-CDK8 interaction, but rather alter the
positioning of the activation helix thereby diminishing CDK8 kinase
activity (12,13) (Fig.
1B). Importantly, in contrast to previous studies, it has been
shown that disruption of the MED12-CycC interface has no effect on
MED12-dependent CDK8 activation (12). Rather, upon MED12 mutation, the
entire kinase module becomes destabilized, which ultimately leads
to diminished CDK8 kinase activity (12). Nonetheless, all of these studies
demonstrate that mutations within MED12 cause conformational
changes in the Mediator kinase module that ultimately disrupt CDK8
kinase activity. Consequently, altered CDK8 kinase activity, due to
site-specific mutations within MED12, has important implications in
gene regulation (3,4,14).
Importantly, a number of the genes that show altered expression
upon MED12 mutations are associated with several different types of
human cancers, thus showing a link between MED12 and tumor
formation (15). Therefore, the
present review aimed to discuss tumorigenesis as a result of
somatic mutations in MED12. Furthermore, the review also discussed
drug resistance mechanisms and potential treatment options for
MED12-related disorders.
Uterine leiomyomas, also commonly known as fibroids,
are a classification of muscle tumors that are benign. Extensive
research has pointed out that most uterine leiomyoma-associated
MED12 mutations are located within exon 2, with only some
exceptions occurring within exon 1 (15–35)
(Table I). Based on the current
literature there are two plausible mechanisms behind MED12
mutant-uterine leiomyoma tumorigenesis. First, it is postulated
that this mutation disrupts the interaction between MED12 and
CyclinC-CDK8 in the kinase module of Mediator (36) (Fig.
1B). Extensive studies have shown that complete CDK8 activation
depends on the interaction between CycC and MED12, with this
interaction specifically occurring within the first two exons of
MED12 (8,22,24,37–39).
Therefore, mutations occurring within MED12 exon 1 or exon 2 often
lead to disruption of the interaction between CycC and MED12, which
subsequently leads to reduced CDK8 kinase activity (Fig. 1B). Secondly, recent studies have
now postulated that MED12 mutations found in patients with uterine
leiomyoma do not affect the MED12-CycC interaction, but rather
abolish CDK8 kinase activity through displacement of a
MED12-mediated activation loop on CDK8 (12,13)
(Fig. 1B). Nonetheless, one common
theme among these studies is that reduced CDK8 kinase activity, as
a result of MED12 mutations, causes cell cycle dysregulation
thereby resulting in abnormal cell growth and ultimately tumor
formation (40–42). Further studies will need to be
completed to gain a full understanding of the mechanistic basis
behind tumorigenesis in uterine leiomyomas as a result of MED12
mutations.
Phyllodes tumors are fibroepithelial tumors found in
the breast, which can include benign, borderline and malignant
tumors with ~1 in every 4 tumors being malignant. MED12 mutations
have been identified in up to 80% of phyllodes tumors with little
variance of frequency amongst malignant (77%), benign (83%) and
borderline (80%) tumors (15,39)
(Table I). These findings indicate
that MED12 mutations play an important role in the development of
phyllodes tumors, but do not appear to be involved in the
progression from benign grade to malignant grade tumors. Of note,
in another study, patients with MED12-mutant phyllodes tumors
showed improved disease-free survival when compared with patients
with phyllodes tumors lacking MED12 mutations (43). The reason for this is still
unknown, but it may provide insight into the role of MED12
mutations in phyllodes tumor formation and progression. The
majority of MED12 mutations found in phyllodes tumors reside within
exon 2, which would lead to impairment of CDK8 kinase activity as
seen in uterine leiomyomas. However, further studies are required
to investigate the mechanistic basis behind MED12 mutant phyllodes
tumorigenesis.
Fibroadenomas are a benign breast fibroepithelial
tumor commonly found in women <30 years old, which occur more
frequently than phyllodes tumors. The MED12 mutation patterns of
both phyllodes tumors and fibroadenomas are very similar, with
missense and deletion-insertion mutations occurring mainly in exon
2 (and a small number in exon 1), with codon 44 being the most
commonly mutated (86%) (15,38,44)
(Table I). Codons 36 and 43 are
also affected, but at a much less frequent rate (16). Similar to uterine leiomyomas and
phyllodes tumors, exon 2 mutations disrupt CDK8 kinase activity
(44). In addition to MED12
mutations, MED12 suppression in fibroadenomas, as shown by RNA
interference, enhances the activation of transforming growth
factor-b (TGF-b) signaling, which causes activation of
mitogen-activated protein kinase/extracellular signal-regulated
kinase, ultimately causing resistance to cancer drugs that target
this specific pathway (45). These
observations thus provide insight into drug resistance mechanisms
involving tumors containing MED12 alterations.
Chronic lymphatic leukemia (CLL) is a hematological
disease that results from the accumulation of mature B lymphocytes
in peripheral blood, bone marrow and lymph nodes. The extent of
this disease observed in patients varies from being highly
indolent, which may never require therapy, to very aggressive and
requiring extensive treatment (46). It has been found that ~5-9% of the
mutations found in patients with lymphatic leukemia correspond to
mutations in the N-terminus of MED12, specifically within exon 1
and exon 2 (37,46–48)
(Table I). CLL-associated MED12
mutations have been found to affect NOTCH signaling through
disruption of the MED12-CycC interface (46). NOTCH signaling can act in an
oncogenic or tumor suppressive manner depending on cellular context
(49–52). CLL-associated MED12 mutations have
been shown to increase NOTCH as a direct result of MED12-CycC
dissociation and subsequent loss of CDK8 kinase activity (8,17,37,53).
Consequently, enhanced NOTCH signaling in these patients increases
apoptotic resistance to ultimately promote tumor formation
(46,53).
Prostate cancer is a hormone-driven cancer regulated
through interactions of circulating androgens to the androgen
receptor (54). Of note, a link
has been established between MED12 overexpression and TGF-β
signaling in castration-resistant prostate cancer (CRPC), a highly
advanced and aggressive type of prostate cancer that is
unresponsive to hormone treatment (55). Specifically, MED12 overexpression,
as assayed by immunohistochemistry on tissue microarrays, was
observed in 70 distance metastatic and 90 local recurrent CRPC
samples (55). MED12
overexpression in these samples was significantly correlated to
activated TGF-β signaling (55).
In addition to MED12 overexpression, MED12 is also mutated in ~5%
of prostate cancers (56–58) (Table
I). Unlike the previously discussed tumors, MED12 mutations in
prostate cancer do not reside within the N-terminus, with the most
common mutation found in exon 26 (57). Further studies will need to be
performed to determine the mechanistic basis behind prostate
cancer-associated MED12 mutations. Of note, however, most prostate
cancer-associated MED12 mutations are in close proximity to MED12
mutations found in the X-linked intellectual disability (XLID)
disorders, FG, Lujan and Ohdo syndromes (33,59–61).
XLID-associated MED12 mutations have previously been shown to
impair recruitment of CycC-CDK8 into Mediator, which ultimately
promotes hyperactivated Sonic Hedgehog signaling (62–64).
It is thus plausible that MED12 mutations in prostate cancer may
promote tumorigenesis through a similar mechanism.
Breast and ovarian cancer are two additional
examples of cancer subtypes that are primarily driven by hormones;
with breast cancer being driven by estrogen and progesterone, and
ovarian cancer by estrogen. Recently, MED12 mutations have been
found in breast (up to 33.3%) and ovarian (~3%) cancer, however
insight into the mechanistic basis behind these mutations is still
lacking (65–68) (Table
I). One noteworthy finding in ovarian cancer is that MED12
mutations correlate with high tumor mutational burden (TMB-H) as
mutations were exclusively found in tumors classified as TMB-H
(68). Of note, TMB-H has been
linked to the generation of neoantigens and clinical response to
immunotherapies in various cancer subtypes (68). Thus, MED12 mutations could serve as
a potential biomarker for immunotherapy in ovarian cancer. In
breast cancer, MED12 expression levels have been correlated to the
regulation of angiotensin converting enzyme 2 (ACE2) levels and
subsequent severe acute respiratory syndrome-related coronavirus 2
(SARS-CoV-2) infection (69). The
details of this study will be further discussed in the therapeutics
section of the current review.
Several studies have found that loss of genomic and
non-genomic functions of MED12 are associated with the development
of resistance to chemotherapeutics. A specific example of this is
seen in non-small cell lung cancer, where loss of MED12, as assayed
by RNA interference, causes drug resistance, by increasing the
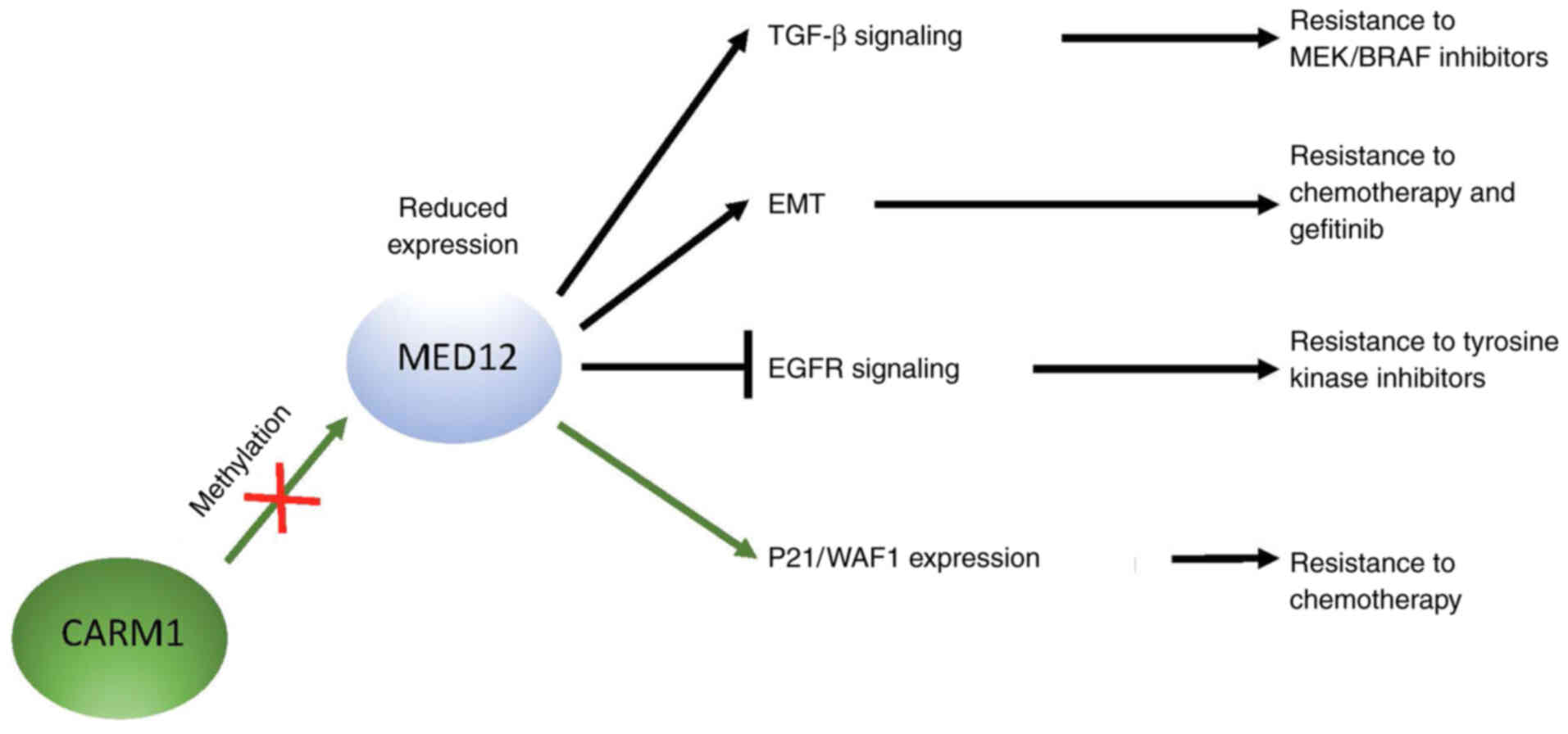
level of TGF-β receptor II (R2) (45,70).
MED12 physically interacts with TGF-βR2 in the cytoplasm, which
results in a reduction of TGF-β signaling (45). Consequently, reduced MED12
expression promotes TGF-β signaling that has been linked directly
to drug resistance, including resistance to MEK and BRAF
inhibitors, downstream effectors in the TGF-β signaling pathway
(45) (Fig. 2). Furthermore, reduced MED12
expression is also associated with an
epithelial-to-mesenchymal-like phenotype that has been correlated
to chemotherapy resistance in colon cancer and gefitinib resistance
in lung cancer (45) (Fig. 2). Since MED12 mutations abrogate
CDK8 kinase activity, either due to a disruption of MED12-CycC or a
displacement of the activation loop of CDK8, it can be argued that
loss of MED12 expression mimics a MED12 mutant setting. Thus, these
results indicate that inhibition of TGF-β signaling in cancers with
either reduced MED12 expression or MED12 mutations could prove to
be a beneficial therapeutic strategy.
In another study, MED12 overexpression was
correlated to improved disease prognosis in human breast cancer
(71). Specifically, this was
shown to involve the interaction of MED12 with
coactivator-associated arginine methyltransferase 1 (CARM 1) for
which MED12 is known to be a substrate (71). CARM1 and MED12 expression show a
positive correlation, and high expression of both predicts improved
prognosis in patients with breast cancer after chemotherapy
(71). CARM1 methylates MED12 at
residues R1862 and R1912, and mutations within these sites promote
MED12 downregulation, which results in resistance to chemotherapy
drugs (71,72) (Fig.
2). The drug response mechanism shown in these cells is
distinct from the mechanism involving TGF-β signaling activation as
methylated MED12 represses p21/WAF1 transcription, which ultimately
promotes cell cycle progression and cell proliferation (71). Overall, this study showed that
MED12 expression levels, as a result of methylation status, may
serve as a potential biomarker to determine severity of disease and
potential drug resistance.
Finally, in a recent study, a genome-wide clustered
regularly interspaced short palindromic repeats (CRISPR) knockout
screen was employed in HeLa and MCF10A (non-malignant breast
epithelial) cells to identify mechanisms that regulate resistance
to ataxia telangiectasia and Rad3 related inhibitors (ATRi)
(73). ATR plays a pivotal role in
the DNA damage response pathway thereby making it an attractive
target for chemotherapeutics. For example, ATRi have been shown to
be highly efficient in targeting cancer cells that have a large
amount of DNA damage and are deficient in ATM/p53 (74–76).
Through the CRISPR knockout screen, MED12 was identified as playing
a crucial role in ATRi resistance (73). Specifically, MED12 knockout or
knockdown was shown to increase the CldU-to-IdU ratio, thus
indicating that loss of MED12 promotes restoration of replication
fork speed in the presence of ATRi (73). Overall, this study provides further
insight into the pivotal role of MED12 in cancer drug resistance
pathways.
Patients with cancer have been shown to be at an
increased risk for severe symptoms from SARS-CoV-2 infection,
although the precise reasons behind this are unclear (69). Of note, a recent study has found
that chemotherapy treatment gradually decreases MED12 levels in
breast cancer cells (CAL-51 cells), which subsequently promotes an
increase in ACE2 expression (69).
Since ACE2 plays a critical role in viral entry of SARS-CoV-2, this
finding demonstrates a link between chemotherapy treatment, MED12
expression and the likelihood of viral infection. Furthermore, upon
further investigation, it was found that CRISPR-mediated knockout
of MED12 affects the expression of 31 different viral-dependent
host factors, including centromere protein F and numerous
fibroblast growth factors (69).
Thus, MED12 plays a role in blocking SARS-CoV-2 infection through
the repression of ACE2 expression to control viral entry and by
potentiating broad anti-viral defense pathways in the host.
EGFR signaling regulates growth, proliferation,
survival and differentiation in cells. This transmembrane tyrosine
kinase receptor is commonly upregulated or mutated in various
cancers, resulting in the activation of several oncogenic pathways.
Of note, MED12 can bind to the EGFR promoter through
Mediator-polymerase II complex to stimulate EGFR transcription and
thus subsequently promote tumor proliferation (77). MED12 and EGFR expression levels are
directly related to MED12 knockdown, causing decreased EGFR
signaling, which in turn causes tumor dormancy (77). Specifically, in ovarian cancer
cells, depletion of MED12 was enough to cause tumor dormancy by
causing the cell cycle to arrest in the G0-G1
phase (77). This finding is of
importance in cancer therapeutics as several studies have suggested
that tumor relapse may be prevented by inhibiting the conversion of
tumor dormancy to tumor proliferation (77,78).
Thus, it may be beneficial to keep tumor cells in the dormant state
and prevent the relapse of cancer through the use of MED12
inhibitors. However, further studies must be conducted to confirm
this strategy as a therapeutic option.
DNA sequencing is key in modern cancer therapeutics
as it allows for the generation of personalized treatment options.
In one study, researchers investigated the efficacy of genetic
sequencing on patients with non-small cell lung cancer who
developed resistance to EGFR tyrosine kinase inhibitors (Fig. 2) (79). Of note, a direct correlation was
found between reduced MED12 expression and drug resistance in these
patients (79). The researchers
found that deep sequencing analysis of tumors was efficient in
identifying drug response and resistance biomarkers as well as
clonal and subclonal genetic populations (79). The identification of biomarkers
allows for serial biomarker analysis of samples gathered from each
patient to guide the initial and subsequent treatments to prevent
or overcome drug resistance. It is important that this genotyping
is enriched with additional biomarkers to predict signaling
pathways that crosstalk and are involved in adaptive resistance to
treatment. Targeting these signaling pathways, and adapting the
treatment based on serial genetic analysis, might be a more
effective alternative to EGFR tyrosine kinase inhibitor monotherapy
(79). This study thus provides
important insight into the efficacy and use of genotyping in cancer
therapeutics as well as the role of MED12 in potential drug
resistance.
MED12 functions in regulating cell growth signals,
early development and differentiation. As a result of these
important physiological functions, mutations within MED12 are
commonly seen in several different cancers and benign tumors,
including uterine leiomyomas, phyllodes tumors, breast
fibroadenomas, lymphatic leukemia, prostate, breast and ovarian
cancer. One theme that is common to most of these tumors, with the
exception of prostate cancer, is that the mutations occur within
exon 1 or exon 2, which leads to a reduction of CDK8 kinase
activity; though it is currently unclear if this reduction is due
to disruption of MED12-CycC or a displacement of the activation
loop on CDK8 (12,13,16–32,
34,35,39,44,46–48).
The resulting mechanistic basis behind tumorigenesis as a result of
loss of CDK8 kinase activity varies between the different tumor
subtypes. At least one study has shown that loss of CDK8 kinase
activity leads to dysregulated NOTCH signaling to promote lymphatic
leukemia (46). It is of
importance to note that other studies have investigated the
relationship between CDK8 kinase activity and tumorigenesis in a
MED12-independent context in a variety of different cancers
(80,81). Nonetheless, further studies are
required to study the mechanisms behind MED12-mediated loss of CDK8
kinase activity and subsequent tumorigenesis.
One theme that is noteworthy is the fact that the
exon 1/exon 2 MED12 mutations are restricted primarily, with the
exception of lymphatic leukemia, to hormone-driven tumors with
mesenchymal stem cell origins. The reasons underlying this finding
are currently unclear, but they do suggest that MED12 plays an
important role in the development and/or differentiation of this
progenitor lineage. Of note, the only tumor subtype that has
provided clues as to the mechanistic basis behind exon1/exon2
MED12-mutation driven tumorigenesis stems from studies in lymphatic
leukemia, the only non-hormone-driven tumor. Future studies are
warranted to determine whether the mechanism underlying
hormone-driven tumorigenesis is similar or distinct to that
observed in lymphatic leukemia. Furthermore, the relationship
between exon1/exon2 mutations and the exon 26 mutations found in
prostate cancer is currently unclear. Prostate cancer and
exon1/exon2-mutant tumors are both hormone-driven, so it is unclear
as to why mutations in these tumors are found in very distinct
regions of MED12.
Although information regarding the mechanistic basis
behind MED12-mutant tumorigenesis is still lacking, there are
several studies that have begun to investigate the role of MED12 in
potential cancer therapeutics. One important finding is that
reduced MED12 expression can lead to drug resistance, including
resistance to MEK, BRAF and tyrosine kinase inhibitors; thereby,
providing important insight into drug treatment for patients who
display reduced MED12 expression (45,79).
Congruently, potential has been demonstrated in the importance of
genetic testing, specifically in determining the mutant or
expression landscape of MED12. Not only will this provide clues as
to potential drug resistance mechanisms, but it will also provide
insight into potential treatment strategies. Ultimately, future
studies will need to be performed to determine whether MED12
itself, the MED-CycC interface or the MED12-CDK8 connection can
serve as potential therapeutic targets for cancer treatment.
Not applicable.
Funding: No funding was received.
Not applicable.
SA and CGG wrote the manuscript. MB provided
guidance and performed all editing and formatting. Data
authentication is not applicable. All authors read and approved the
final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Allen BL and Taatjes DJ: The mediator
complex: A central integrator of transcription. Nat Rev Mol Cell
Biol. 16:155–166. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kornberg RD: Mediator and the mechanism of
transcriptional activation. Trends Biochem Sci. 30:235–239. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mo X, Kowenz-Leutz E, Xu H and Leutz A:
Ras induces mediator complex exchange on C/EBP beta. Mol Cell.
13:241–250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pavri R, Lewis B, Kim TK, Dilworth FJ,
Erdjument-Bromage H, Tempst P, de Murcia G, Evans R, Chambon P and
Reinberg D: PARP-1 determines specificity in a retinoid signaling
pathway via direct modulation of mediator. Mol Cell. 18:83–96.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bernecky C, Grob P, Ebmeier CC, Nogales E
and Taatjes DJ: Molecular architecture of the human Mediator-RNA
polymerase II-TFIIF assembly. PLoS Biol. 9:e10006032011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Elmlund H, Baraznenok V, Lindahl M,
Samuelsen CO, Koeck PJ, Holmberg S, Hebert H and Gustafsson CM: The
cyclin-dependent kinase 8 module sterically blocks Mediator
interactions with RNA polymerase II. Proc Natl Acad Sci USA.
103:15788–15793. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Knuesel MT, Meyer KD, Bernecky C and
Taatjes DJ: The human CDK8 subcomplex is a molecular switch that
controls Mediator coactivator function. Genes Dev. 23:439–451.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Turunen M, Spaeth JM, Keskitalo S, Park
MJ, Kivioja T, Clark AD, Mäkinen N, Gao F, Palin K, Nurkkala H, et
al: Uterine leiomyoma-linked MED12 mutations disrupt
mediator-associated CDK activity. Cell Rep. 7:654–660. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Sun Q, Ding Z, Ji J, Wang J, Kong
X, Yang J and Cai G: Redefining the modular organization of the
core Mediator complex. Cell Res. 24:796–808. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Knuesel MT, Meyer KD, Donner AJ, Espinosa
JM and Taatjes DJ: The human CDK8 subcomplex is a histone kinase
that requires Med12 for activity and can function independently of
mediator. Mol Cell Biol. 29:650–661. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu W and Ji JY: Dysregulation of CDK8 and
Cyclin C in tumorigenesis. J Genetics. 38:439–452. 2011.PubMed/NCBI
|
|
12
|
Klatt F, Leitner A, Kim IV, Ho-Xuan H,
Schneider EV, Langhammer F, Weinmann R, Müller MR, Huber R, Meister
G and Kuhn CD: A precisely positioned MED12 activation helix
stimulates CDK8 kinase activity. Proc Natl Acad Sci USA.
117:2894–2905. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li YC, Chao TC, Kim HJ, Cholko T, Chen SF,
Li G, Snyder L, Nakanishi K, Chang CE, Murakami K, et al: Structure
and noncanonical Cdk8 activation mechanism within an
Argonaute-containing Mediator kinase module. Sci Adv.
7:eabd44842021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Holstege FC, Jennings EG, Wyrick JJ, Lee
TI, Hengartner CJ, Green MR, Golub TR, Lander ES and Young RA:
Dissecting the regulatory circuitry of a eukaryotic genome. Cell.
95:717–728. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Clark AD, Oldenbroek M and Boyer TG:
Mediator kinase module and human tumorigenesis. Crit Rev Biochem
Mol Biol. 50:393–426. 2015.PubMed/NCBI
|
|
16
|
Ajabnoor GMA, Mohammed NA, Banaganapalli
B, Abdullah LS, Bondagji ON, Mansouri N, Sahly NN, Vaidyanathan V,
Bondagji N, Elango R and Shaik NA: Expanded somatic mutation
spectrum of MED12 gene in uterine leiomyomas of Saudi Arabian
Women. Front Genet. 9:5522018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Banaganapalli B, Mohammed K, Khan IA,
Al-Aama JY, Elango R and Shaik NA: A Computational protein
phenotype prediction approach to analyze the deleterious mutations
of human MED12 gene. J Cell Biochem. 117:2023–2035. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Croce S and Chibon F: MED12 and uterine
smooth muscle oncogenesis: State of the art and perspectives. Eur J
Cancer. 51:1603–1610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Graaff MA, Cleton-Jansen AM, Szuhai K
and Bovée JV: Mediator complex subunit 12 exon 2 mutation analysis
in different subtypes of smooth muscle tumors confirms genetic
heterogeneity. Hum Pathol. 44:1597–1604. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heinonen HR, Sarvilinna NS, Sjöberg J,
Kämpjärvi K, Pitkänen E, Vahteristo P, Mäkinen N and Aaltonen LA:
MED12 mutation frequency in unselected sporadic uterine leiomyomas.
Fertil Steril. 102:1137–1142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Je EM, Kim MR, Min KO, Yoo NJ and Lee SH:
Mutational analysis of MED12 exon 2 in uterine leiomyoma and other
common tumors. Int J Cancer. 131:E1044–E1047. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kämpjärvi K, Mäkinen N, Kilpivaara O,
Arola J, Heinonen HR, Böhm J, Abdel-Wahab O, Lehtonen HJ, Pelttari
LM, Mehine M, et al: Somatic MED12 mutations in uterine
leiomyosarcoma and colorectal cancer. Br J Cancer. 107:1761–1765.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li N, Fassl A, Chick J, Inuzuka H, Li X,
Mansour MR, Liu L, Wang H, King B, Shaik S, et al: Cyclin C is a
haploinsufficient tumour suppressor. Nat Cell Biol. 16:1080–1091.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mäkinen N, Heinonen HR, Moore S, Tomlinson
IP, van der Spuy ZM and Aaltonen LA: MED12 exon 2 mutations are
common in uterine leiomyomas from South African patients.
Oncotarget. 2:966–969. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mäkinen N, Mehine M, Tolvanen J, Kaasinen
E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M, et al:
MED12, the mediator complex subunit 12 gene, is mutated at high
frequency in uterine leiomyomas. Science. 334:252–255. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mäkinen N, Vahteristo P, Kämpjärvi K,
Arola J, Bützow R and Aaltonen LA: MED12 exon 2 mutations in
histopathological uterine leiomyoma variants. Eur J Hum Genet.
21:1300–1303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsubara A, Sekine S, Yoshida M, Yoshida
A, Taniguchi H, Kushima R, Tsuda H and Kanai Y: Prevalence of MED12
mutations in uterine and extrauterine smooth muscle tumours.
Histopathology. 62:657–661. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McGuire MM, Yatsenko A, Hoffner L, Jones
M, Surti U and Rajkovic A: Whole exome sequencing in a Random
Sample of North American Women with leiomyomas identifies MED12
mutations in majority of uterine leiomyomas. PLoS One.
7:e332512012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mehine M, Kaasinen E, Mäkinen N, Katainen
R, Kämpjärvi K, Pitkänen E, Heinonen HR, Bützow R, Kilpivaara O,
Kuosmanen A, et al: Characterization of uterine leiomyomas by
whole-genome sequencing. N Engl J Med. 369:43–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pérot G, Croce S, Ribeiro A, Lagarde P,
Velasco V, Neuville A, Coindre JM, Stoeckle E, Floquet A, MacGrogan
G and Chibon F: MED12 alterations in both human benign and
malignant uterine soft tissue tumors. PLoS One. 7:e400152012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ravegnini G, Mariño-Enriquez A, Slater J,
Eilers G, Wang Y, Zhu M, Nucci MR, George S, Angelini S, Raut CP
and Fletcher JA: MED12 mutations in leiomyosarcoma and extrauterine
leiomyoma. Mod Pathol. 26:743–749. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schwetye KE, Pfeifer JD and Duncavage EJ:
MED12 exon 2 mutations in uterine and extrauterine smooth muscle
tumors. Hum Pathol. 45:65–70. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang H, Shen Q, Ye LH and Ye J: MED12
mutations in human diseases. Protein Cell. 4:643–646. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang L, Hu S, Xin F, Zhao H, Li G, Ran W,
Xing X and Wang J: MED12 exon 2 mutation is uncommon in intravenous
leiomyomatosis: Clinicopathologic features and molecular study. Hum
Pathol. 99:36–42. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Q, Ubago J, Li L, Guo H, Liu Y,
Qiang W, Kim JJ, Kong B and Wei JJ: Molecular analyses of 6
different types of uterine smooth muscle tumors: Emphasis in
atypical leiomyoma. Cancer. 120:3165–3177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park MJ, Shen H, Spaeth JM, Tolvanen JH,
Failor C, Knudtson JF, McLaughlin J, Halder SK, Yang Q, Bulun SE,
et al: Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt
allosteric activation of cyclin C-CDK8/19. J Biol Chem.
293:4870–4882. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kämpjärvi K, Järvinen TM, Heikkinen T,
Ruppert AS, Senter L, Hoag KW, Dufva O, Kontro M, Rassenti L,
Hertlein E, et al: Somatic MED12 mutations are associated with poor
prognosis markers in chronic lymphocytic leukemia. Oncotarget.
6:1884–1888. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lim WK, Ong CK, Tan J, Thike AA, Ng CC,
Rajasegaran V, Myint SS, Nagarajan S, Nasir ND, McPherson JR, et
al: Exome sequencing identifies highly recurrent MED12 somatic
mutations in breast fibroadenoma. Nat Genet. 46:877–880. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshida M, Sekine S, Ogawa R, Yoshida H,
Maeshima A, Kanai Y, Kinoshita T and Ochiai A: Frequent MED12
mutations in phyllodes tumours of the breast. Br J Cancer.
112:1703–1708. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kishi T, Ikeda A, Koyama N, Fukada J and
Nagao R: A refined two-hybrid system reveals that
SCF(Cdc4)-dependent degradation of Swi5 contributes to the
regulatory mechanism of S-phase entry. Proc Natl Acad Sci USA.
105:14497–14502. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Porter DC, Farmaki E, Altilia S, Schools
GP, West DK, Chen M, Chang BD, Puzyrev AT, Lim CU, Rokow-Kittell R,
et al: Cyclin-dependent kinase 8 mediates chemotherapy-induced
tumor-promoting paracrine activities. Proc Natl Acad Sci USA.
109:13799–13804. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu D, Li CF, Zhang X, Gong Z, Chan CH, Lee
SW, Jin G, Rezaeian AH, Han F, Wang J, et al: Skp2-macroH2A1-CDK8
axis orchestrates G2/M transition and tumorigenesis. Nat Commun.
6:66412015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ng CC, Tan J, Ong CK, Lim WK, Rajasegaran
V, Nasir ND, Lim JC, Thike AA, Salahuddin SA, Iqbal J, et al: MED12
is frequently mutated in breast phyllodes tumours: A study of 112
cases. J Clin Pathol. 68:685–691. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Darooei M, Khan F, Rehan M, Zubeda S,
Jeyashanker E, Annapurna S, Shah A, Maddali S and Hasan Q: MED12
somatic mutations encompassing exon 2 associated with benign breast
fibroadenomas and not breast carcinoma in Indian women. J Cell
Biochem. 120:182–191. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang S, Hölzel M, Knijnenburg T,
Schlicker A, Roepman P, McDermott U, Garnett M, Grernrum W, Sun C,
Prahallad A, et al: MED12 controls the response to multiple cancer
drugs through regulation of TGF-β receptor signaling. Cell.
151:937–950. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu B, Słabicki M, Sellner L, Dietrich S,
Liu X, Jethwa A, Hüllein J, Walther T, Wagner L, Huang Z, et al:
MED12 mutations and NOTCH signalling in chronic lymphocytic
leukaemia. Br J Haematol. 179:421–429. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bullerdiek J and Rommel B: Factors
targeting MED12 to drive tumorigenesis? F1000Res. 7:3592018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guièze R, Robbe P, Clifford R, de Guibert
S, Pereira B, Timbs A, Dilhuydy MS, Cabes M, Ysebaert L, Burns A,
et al: Presence of multiple recurrent mutations confers poor trial
outcome of relapsed/refractory CLL. Blood. 126:2110–2117. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Aster JC, Pear WS and Blacklow SC: The
varied roles of notch in cancer. Annu Rev Pathol. 12:245–275. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Meurette O and Mehlen P: Notch signaling
in the tumor microenvironment. Cancer Cell. 34:536–548. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nowell CS and Radtke F: Notch as a tumour
suppressor. Nat Rev Cancer. 17:145–159. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ntziachristos P, Lim JS, Sage J and
Aifantis I: From fly wings to targeted cancer therapies: A
centennial for notch signaling. Cancer Cell. 25:318–334. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rosati E, Sabatini R, Rampino G, Tabilio
A, Di Ianni M, Fettucciari K, Bartoli A, Coaccioli S, Screpanti I
and Marconi P: Constitutively activated Notch signaling is involved
in survival and apoptosis resistance of B-CLL cells. Blood.
113:856–865. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lupien M and Brown M: Cistromics of
hormone-dependent cancer. Endocr Relat Cancer. 16:381–389. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shaikhibrahim Z, Offermann A, Braun M,
Menon R, Syring I, Nowak M, Halbach R, Vogel W, Ruiz C, Zellweger
T, et al: MED12 overexpression is a frequent event in
castration-resistant prostate cancer. Endocr Relat Cancer.
21:663–675. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Baca SC, Prandi D, Lawrence MS, Mosquera
JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi
M, et al: Punctuated evolution of prostate cancer genomes. Cell.
153:666–677. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Barbieri CE, Baca SC, Lawrence MS,
Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van
Allen E, Stransky N, et al: Exome sequencing identifies recurrent
SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet.
44:685–689. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Grasso CS, Wu YM, Robinson DR, Cao X,
Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC,
et al: The mutational landscape of lethal castration-resistant
prostate cancer. Nature. 487:239–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Risheg H, Graham JM Jr, Clark RD, Rogers
RC, Opitz JM, Moeschler JB, Peiffer AP, May M, Joseph SM, Jones JR,
et al: A recurrent mutation in MED12 leading to R961W causes
Opitz-Kaveggia syndrome. Nat Genet. 39:451–453. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Schwartz CE, Tarpey PS, Lubs HA, Verloes
A, May MM, Risheg H, Friez MJ, Futreal PA, Edkins S, Teague J, et
al: The original Lujan syndrome family has a novel missense
mutation (p.N1007S) in the MED12 gene. J Med Genet. 44:472–477.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Vulto-van Silfhout AT, de Vries BB, van
Bon BW, Hoischen A, Ruiterkamp-Versteeg M, Gilissen C, Gao F, van
Zwam M, Harteveld CL, van Essen AJ, et al: Mutations in MED12 cause
X-linked Ohdo syndrome. Am J Hum Genet. 92:401–406. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen M, Carkner R and Buttyan R: The
hedgehog/Gli signaling paradigm in prostate cancer. Expert Rev
Endocrinol Metab. 6:453–467. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhou H, Kim S, Ishii S and Boyer TG:
Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol
Cell Biol. 26:8667–8682. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhou H, Spaeth JM, Kim NH, Xu X, Friez MJ,
Schwartz CE and Boyer TG: MED12 mutations link intellectual
disability syndromes with dysregulated GLI3-dependent Sonic
Hedgehog signaling. Proc Natl Acad Sci USA. 109:19763–19768. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Banerji S, Cibulskis K, Rangel-Escareno C,
Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY,
Sougnez C, Zou L, et al: Sequence analysis of mutations and
translocations across breast cancer subtypes. Nature. 486:405–409.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chao X, Tan W, Tsang JY, Tse GM, Hu J, Li
P, Hou J, Li M, He J and Sun P: Clinicopathologic and genetic
features of metaplastic breast cancer with osseous differentiation:
A series of 6 cases. Breast Cancer. 28:1100–1111. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shah SP, Roth A, Goya R, Oloumi A, Ha G,
Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al: The clonal
and mutational evolution spectrum of primary triple-negative breast
cancers. Nature. 486:395–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang Y, Shi X, Zhang J, Chen X, Zhang P,
Liu A and Zhu T: A comprehensive analysis of somatic alterations in
Chinese ovarian cancer patients. Sci Rep. 11:3872021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang S, Liu F, Halfmann P, Behrens RT,
Liu P, McIlwain SJ, Ong IM, Donahue K, Wang Y, Kawaoka Y, et al:
Mediator complex subunit 12 is a gatekeeper of SARS-CoV-2 infection
in breast cancer cells. Genes Dis. Aug 17–2021.doi:
10.1016/j.gendis.2021.08.001 (Epub ahead of print).
|
|
70
|
Zhang S, O'Regan R and Xu W: The emerging
role of mediator complex subunit 12 in tumorigenesis and response
to chemotherapeutics. Cancer. 126:939–948. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wang L, Zeng H, Wang Q, Zhao Z, Boyer TG,
Bian X and Xu W: MED12 methylation by CARM1 sensitizes human breast
cancer cells to chemotherapy drugs. Sci Adv. 1:e15004632015.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Peng BL, Li WJ, Ding JC, He YH, Ran T, Xie
BL, Wang ZR, Shen HF, Xiao RQ, Gao WW, et al: A hypermethylation
strategy utilized by enhancer-bound CARM1 to promote estrogen
receptor α-dependent transcriptional activation and breast
carcinogenesis. Theranostics. 10:3451–3473. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Schleicher EM, Dhoonmoon A, Jackson LM,
Clements KE, Stump CL, Nicolae CM and Moldovan GL: Dual genome-wide
CRISPR knockout and CRISPR activation screens identify mechanisms
that regulate the resistance to multiple ATR inhibitors. PLoS
Genet. 16:e10091762020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Reaper PM, Griffiths MR, Long JM, Charrier
JD, Maccormick S, Charlton PA, Golec JM and Pollard JR: Selective
killing of ATM- or p53-deficient cancer cells through inhibition of
ATR. Nat Chem Biol. 7:428–430. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Reinhardt HC, Aslanian AS, Lees JA and
Yaffe MB: p53-deficient cells rely on ATM- and ATR-mediated
checkpoint signaling through the p38MAPK/MK2 pathway for survival
after DNA damage. Cancer Cell. 11:175–189. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Vendetti FP, Lau A, Schamus S, Conrads TP,
O'Connor MJ and Bakkenist CJ: The orally active and bioavailable
ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of
cisplatin to resolve ATM-deficient non-small cell lung cancer in
vivo. Oncotarget. 6:44289–44305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Luo XL, Deng CC, Su XD, Wang F, Chen Z, Wu
XP, Liang SB, Liu JH and Fu LW: Loss of MED12 induces tumor
dormancy in human epithelial ovarian cancer via downregulation of
EGFR. Cancer Res. 78:3532–3543. 2018.PubMed/NCBI
|
|
78
|
Srivastava S and Kulshreshtha R: Insights
into the regulatory role and clinical relevance of mediator
subunit, MED12, in human diseases. J Cell Physiol. 236:3163–3177.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Rosell R, Bivona TG and Karachaliou N:
Genetics and biomarkers in personalisation of lung cancer
treatment. Lancet. 382:720–731. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Broude EV, Győrffy B, Chumanevich AA, Chen
M, McDermott MS, Shtutman M, Catroppo JF and Roninson IB:
Expression of CDK8 and CDK8-interacting Genes as potential
biomarkers in breast cancer. Curr Cancer Drug Targets. 15:739–749.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ma D, Chen X, Shen XB, Sheng LQ and Liu
XH: Binding patterns and structure-activity relationship of CDK8
inhibitors. Bioorg Chem. 96:1036242020. View Article : Google Scholar : PubMed/NCBI
|