Introduction
Liver cancer was the seventh most common type of
cancer and the second leading cause of cancer-associated mortality
worldwide in 2020 (1). Among
patients with liver cancer, hepatocellular carcinoma (HCC) accounts
for >90% of cases. Patients with HCC typically demonstrate a
poor prognosis, which can be attributed to a diagnosis at an
advanced stage (2). Previous
studies have reported that immunotherapy can serve an important
role in the treatment of HCC (3,4).
Based on these previous studies, it is necessary to evaluate the
functional molecules associated with HCC in order to determine a
potential therapeutic target in the HCC immune
microenvironment.
Tumor-associated macrophages (TAMs) are a type of
primary tumor-infiltrating immune cells (5) that have been demonstrated to promote
tumor growth, metastasis and invasion (6,7).
TAMs can be divided into two main types as follows: The classically
activated M1 phenotype and the alternatively activated M2 phenotype
(8). M1 macrophages are polarized
by the activation of interferon gamma (IFN-γ), promoting the
T-helper (Th)1 immune response and antitumor activity (9,10).
M2 macrophages are polarized by interleukin (IL)-4/IL-13 and are
associated with the anti-inflammatory Th2 immune response, which
demonstrates a pro-tumor activity (11). Previous studies have revealed that
most TAMs are of the M2 phenotype, which promote tumor growth and
progression and therefore worsen the prognosis of patients with HCC
(12,13). To further validate the results of
these studies, it is necessary to explore and block the key stages
of macrophage M2 polarization in order to inhibit HCC
progression.
The signaling lymphocyte activation molecule (SLAM)
family comprises the following nine members: CD150 (SLAMF1), CD48
(SLAMF2), CD229 (SLAMF3), CD244 (SLAMF4), CD84 (SLAMF5), CD352
(SLAMF6), CD319 (SLAMF7), CD353 (SLAMF8) and CD84-H1 (SLAMF9)
(14). These are surface molecules
that are expressed on hematopoietic cells. They are also a type of
immune molecule that can regulate innate and adaptive immunity by
activating associated immune cells (14,15).
Among these nine molecules, SLAMF6 (Ly108 in mice) is highly
expressed in activated T and B cells. Its expression can also be
increased in dendritic cells and macrophages via inflammatory
signals (16), functioning as both
a positive and negative regulator of the immune response (17). SLAMF6 can inhibit the secretion of
T-dependent and T-independent antibodies (18). It has also been reported that
SLAMF6 can activate natural killer cells to induce cytotoxicity and
influence IFN-γ production (19,20).
SLAMF3 is involved in the inhibition of HCC progression and cell
proliferation (21,22). In addition to SLAMF6, there are
several SLAMF family members that are expressed on the macrophage
surface, including SLAMF3, SLAMF4, SLAMF5 and SLAMF7 (23). SLAMFs have been demonstrated to
negatively influence the function of macrophages by reducing IL-6
production and increasing IL-13 secretion (24).
As a membrane protein that is expressed on immune
cells, SLAMF6 has been proven to serve a role in numerous diseases
(25–28). For example, although the expression
of SLAMF6 on peripheral T cells is not markedly different in
patients with systemic lupus erythematosus, it has been determined
that the co-stimulatory function of SLAMF6 is lacking (25). In addition, SLAMF6 can promote Th17
differentiation (26). SLAMF6 can
also stimulate the interaction between colonic innate immune cells
and Gram-negative bacteria, thus reducing mucosal protection and
enhancing inflammation, resulting in lethal colitis in mice
(27). The administration of
anti-SLAMF6 antibodies combined with ibrutinib, which is a Bruton
tyrosine kinase inhibitor that serves as an anticancer targeted
drug, can effectively eliminate tumor cells in the spleen, bone
marrow, liver and peritoneal cavity (28). SLAMF6 can affect the function of
Th2 by binding to downstream proteins and transcription factors,
such as nuclear factor-κB (NF-κB) (29), which serves a role in macrophage
polarization (30).
The present study investigated the expression of
SLAMF6 in the peripheral CD14+ monocytes of patients
with HCC to determine whether this expression was associated with
the clinicopathological characteristics of these patients.
Furthermore, SLAMF6 expression on different phenotype macrophages
was detected in order to explore the relationship between this and
HCC progression. This may provide a novel therapeutic target for
HCC treatment.
Materials and methods
Blood sampling and mononuclear cell
isolation
Blood samples were obtained from 34 patients with
HCC and 33 healthy individuals at the Shandong Cancer Hospital and
Institute (Jinan, China). Patients with HCC did not receive any
form of antitumor therapy and were pathologically diagnosed with
HCC prior to sampling. None of the healthy donors were positive for
hepatitis B virus (HBV), hepatitis C virus or human
immunodeficiency virus, consumed excessive alcohol or received any
chemotherapy prior to sampling. Human peripheral mononuclear cell
isolation was performed by centrifuging (800 × g; 15 min; 25°C)
blood samples with an EZ-Sep™ lymphocyte separation tube (Dakewe
Biotech Co., Ltd.). The mono-macrophages were marked with
corresponding markers immediately after the isolation. The present
study was approved by the Medical Ethics Committee of Shandong
Cancer Hospital and Institute and informed consent was obtained
from each participant. The clinicopathological characteristics of
the healthy donors and patients with HCC are summarized in Table I.
 | Table I.Clinicopathological characteristics
of patients with hepatocellular carcinoma and healthy donors. |
Table I.
Clinicopathological characteristics
of patients with hepatocellular carcinoma and healthy donors.
| Variable | HCC patients
(n=34) | Healthy donors
(n=33) |
|---|
| Sex |
|
|
| Male, n
(%) | 26 (76.47) | 25 (75.76) |
| Female,
n (%) | 8 (23.53) | 8 (24.24) |
| Age, mean years
(range) | 60 (41–80) | 44 (21–62) |
| HBV-DNA, n (+,
%) | 12 (35.29) | 0 |
| AFP, n (>100,
%) | 14 (41.18) | 0 |
| Single tumor, n
(%) | 13 (38.24) | 0 |
| Lymphatic
metastasis, n (%) | 10 (29.41) | 0 |
| Distant metastasis,
n (%) | 7 (20.59) | 0 |
| TNM stage I–II, n
(%) | 13 (38.24) | 0 |
Mouse HCC model establishment and
macrophage isolation
C57BL/6 mice were purchased from Beijing
Weitonglihua Laboratory Animal Co., Ltd. and maintained in a
controlled room (specific pathogen free; 12-h light/dark cycle;
free access to food and water, temperature, 20–26°C; relative
humidity, 40–70%). All experiments involving mice were approved by
the Animal Care Committee of Shandong Cancer Hospital and Institute
and performed according to the Animal Management Rules of the
Chinese Ministry of Health. To establish a murine model of HCC, 100
µl PBS solution containing 1×106 H22 cells (cat. no.
CL-0341; American Type Culture Collection) was subcutaneously
injected into eight C57BL/6 male mice (8–10 weeks old; average
weight, 22.32±0.19 g). After 14 days, the mice were euthanized via
intraperitoneal injection of 100 mg/kg sodium pentobarbital
followed by cervical dislocation (average weight, 24.17±0.62 g).
The subsequent experiments were conducted immediately after the
tissues were collected. The infiltrating lymphocytes from each type
of tissue were isolated by centrifugation (800 × g; 20 min; 25°C)
over 40% Percoll solution (GE Healthcare) (31), after which the expression level of
SLAMF6 was evaluated by flow cytometry.
Flow cytometry
Flow cytometry was performed to detect the
expression of SLAMF6 and various surface markers in macrophages.
Human peripheral blood mononuclear cells (CD14+) were
stained with anti-human SLAMF6-PE [cat. no. 317208; 3 µl stock
solution/test (1×106 cells); BioLegend, Inc.] and
anti-human CD14-FITC [cat. no. 301804; 5 µl stock solution/test
(1×106 cells); BioLegend, Inc.] antibodies for 30 min at
4°C. Furthermore, murine macrophages (CD11b+) were
stained with anti-mouse SLAMF6-PE [cat. no. 2124557; 3 µl stock
solution/test (1×106 cells); eBioscience; Thermo Fisher
Scientific, Inc.] and anti-mouse CD11b-PE-cy7 [cat. no.
E07514-1633; 2 µl stock solution/test (1×106 cells);
eBioscience; Thermo Fisher Scientific, Inc.] antibodies for 30 min.
A total of ~10,000 cells were analyzed using BD FACSAria II (BD
Biosciences) equipment and cells were gated according to their
forward and side scatter properties.
Cell culture
Murine HCC cell lines, including H22 [cultured in
RMPI-1640 containing 10% FBS and 1% penicillin/streptomycin mixture
(cat. no. 15140-122; Gibco; Thermo Fisher Scientific, Inc.)] and
Hepa1–6 (cat. no. CL-0105; cultured DMEM containing 10% FBS and 1%
penicillin/streptomycin mixture), were obtained from the American
Type Culture Collection. Murine peripheral macrophages (PMs) and
bone marrow-derived macrophages (BMDMs) were cultured in DMEM
containing 10% FBS and 1% penicillin/streptomycin mixture and
placed at 37°C in a humidified incubator containing 5%
CO2. The human monocyte cell line THP-1 (cat. no.
BNCC100407; BeNa Culture Collection; Beijing Beina Chunglian
Institute of Biotechnology) was cultured in RMPI-1640 supplemented
with 10% FBS and 1% penicillin/streptomycin mixture. DMEM (cat. no.
C11995500CP) was obtained from Gibco; Thermo Fisher Scientific,
Inc., RMPI-1640 (cat. no. SH30809.01) was purchased from HyClone;
Cytiva and FBS (cat. no. WXBD5226V) was obtained from
Sigma-Aldrich; Merck KGaA.
Macrophage polarization
Mouse peritonitis was induced by intraperitoneal
injection of 1 ml sterile 6% starch solution in six 8–10 week old
male C57BL/6 mice. After 72 h, mice were euthanized via
intraperitoneal injection of 100 mg/kg sodium pentobarbital
followed by cervical dislocation. The peritoneal cells were
harvested by irrigating the abdominal cavity and the PMs were
enriched by quick adhesion. Model establishment, harvesting of
cells and quick adhesion were described previously (32). PMs were polarized into M1
phenotypes using 100 ng/ml lipopolysaccharide (LPS) for 24 h at
37°C, while M2 macrophages were polarized using 20 ng/ml IL-4 for
24 h at 37°C. PBS was used as a control. Furthermore, six 4–6 week
old male C57BL/6 mice were euthanized via intraperitoneal injection
of 100 mg/kg sodium pentobarbital followed by cervical dislocation.
The BMDMs were collected by flushing the femurs and tibias
(32). BMDMs were polarized into
the M1 phenotype using 20 ng/ml granulocyte macrophage
colony-stimulating factor (cat. no. AF-315-03-20; PeproTech, Inc.)
treatment for 5 days at 37°C, while the M2 phenotype was obtained
using 100 ng/ml macrophage colony-stimulating factor (cat. no.
AF-315-02-10; PeproTech, Inc.) treatment for 5 days at 37°C.
Samples were then stimulated using 100 ng/ml LPS for 24 h to induce
M1 or 20 ng/ml IL-4 for 24 h to induce M2.
HCC conditioned medium (HCM)
After culturing H22 cells for 24 h, serum-free DMEM
was collected for use as HCM. Newly isolated PMs (2×105
per well) seeded into 12-well-plates were exposed to 200 µl HCM,
producing a final volume of 1,000 µl with complete medium (DMEM
containing 10% FBS). The control group was treated with 200 µl
FBS-free DMEM added to 800 µl complete medium. The final percentage
of HCM was 20%.
SLAMF6 interference
SLAMF6 expression was inhibited in murine PMs via
transfection with SLAMF6 small interfering RNA (siRNA). Three
siRNAs were tested and all sequences were working. The sequences of
the siRNAs are presented in Table
II. SLAMF6 siRNA (120 pmol) or non-targeting negative control
siRNA (120 pmol) (Biosune Biotechnology Co., Ltd.) were transfected
into the PMs in each of the 12 wells using lipofectamine RNAiMAX
(cat. no. 13778-150; Invitrogen; Thermo Fisher Scientific, Inc.)
for 24 h at 37°C. After 24 h, the PMs, which had been transfected
with siRNAs, were treated with Bay11-7082 (10 µM; cat. no.
HY-13453; MedChemExpress), an inhibitor of NF-κB, for 2 h before
IL-4 (20 ng/ml) was added for 24 h. SLAMF6-silenced PMs, negative
control PMs and their culture supernatants were collected
(immediately or after storage at −80°C in an ultra-low temperature
freezer within 1 month) to examine the effect of SLAMF6 on cell
proliferation and invasion, since macrophages exert their function
on tumor cells via secretion of cytokines.
 | Table II.Sequence of Ly108 small interfering
RNAs used for transfection and of primers used for RT-qPCR. |
Table II.
Sequence of Ly108 small interfering
RNAs used for transfection and of primers used for RT-qPCR.
| Primer | Sequence |
|---|
| Small interfering
RNA |
|
|
Control, sense |
UUCUCCGAACGUGUCACGUTT |
|
Ly108-366, sense |
GCUAAUGAAUGGCGUUCUATTUAGAACGCCAUUCAUUAGCTT |
|
Ly108-567, sense |
CCUGCAAAUCAGCAACCUUTTAAGGUUGCUGAUUUGCAGGTT |
|
Ly108-1190, sense |
CCAUGACAAUUUACUCCAUTTAUGGAGUAAAUUGUCAUGGTT |
| RT-qPCR |
|
|
Homo-β-actin, forward |
CATGTACGTTGCTATCCAGGC |
|
Homo-β-actin, reverse |
CTCCTTAATGTCACGCACGAT |
|
Homo-SLAMF6, forward |
GGCCCAGGGAATGTAGTTTCA |
|
Homo-SLAMF6, reverse |
ACTGACTCCCCCAGAATCCC |
|
Homo-TNF-α, forward |
GAGGCCAAGCCCTGGTATG |
|
Homo-TNF-α, reverse |
CGGGCCGATTGATCTCAGC |
|
Mouse-β-actin, forward |
CGGGCCGATTGATCTCAGC |
|
Mouse-β-actin, reverse |
TACAGCCCGGGGAGCATCGT |
|
Mouse-ly108, forward |
CCTTCAGGGTAATGGGTTGGTT |
|
Mouse-ly108, reverse |
CCTTCAGGGTAATGGGTTGGTT |
|
Mouse-arg-1, forward |
TGTCCCTAATGACAGCTCCTT |
|
Mouse-arg-1, reverse |
GCATCCACCCAAATGACACAT |
|
Mouse-TNF-α, forward |
GCCACCACGCTCTTCTGTCT |
|
Mouse-TNF-α, reverse |
TGAGGGTCTGGGCCATAGAAC |
|
Mouse-iNOS, forward |
GCCACCAACAATGGCAACAT |
|
Mouse-iNOS, reverse |
TCGATGCACAACTGGGTGAA |
|
Mouse-IL-1β, forward |
ACCTTCCAGGATGAGGACATGA |
|
Mouse-IL-1β, reverse |
AACGTCACACACCAGCAGGTTA |
Analysis of tumor cell migration,
invasion and proliferation, and in vivo assessment
The culture supernatant of SLAMF6-silenced PMs and
control PMs was used to culture Hepa1–6 cells. Transwell assay cell
migratory and invasive abilities were determined using 24-well
transwell chambers with an 8-µm pore polycarbonate membrane insert
(Corning, Inc.; cat. no. 3422). For invasion assay only, the
membrane was pre-coated with 50 µl Matrigel (cat. no. 356234; BD
Biosciences) for half an hour. Then, ~2.5×104 Hepa1–6
cells suspended in 200 µl serum-free DMEM were loaded onto the
upper chamber, while 600 µl medium collected from corresponding
macrophages, as aforementioned in the SLAMF6 interference section,
was added to the lower chamber. After 16 h (migration) or 24 h
(invasion), the cells on the lower chamber were stained with 0.1%
crystal violet for 10 min at room temperature. The number of
invaded cells were counted under a light microscope (magnification,
×200).
To prepare murine hepatoma homografts,
2×105 H22 cells in 100 µl PBS were subcutaneously
injected into the groins of 14 C57BL/6 male mice (8–10 week old;
average weight, 22.72±0.17 g), which were anesthetized by
intraperitoneal injection with 50 mg/kg sodium pentobarbital. At 8
days after H22 cell injection, SLAMF6-silenced PMs and control PMs
were injected into the tumor site, after which time the size of
tumors was measured. Tumor volume was calculated using the
following formula: 1/2ab2 (a, long diameter; b, short
diameter). The procedure was repeated every other day. On the 18th
day, all mice were euthanized via intraperitoneal injection of 100
mg/kg sodium pentobarbital followed by cervical dislocation
(average weight, 25.03±0.71 g), after which tumors were collected
and their size and weight were determined. The maximum tumor volume
was as follows: 1/2×17.0×16.5×16.5=2,314.125 mm3.
RNA isolation and reverse
transcription quantitative (RT-q)PCR
According to the manufacturer's protocol, RNA was
extracted from THP-1 cells and PMs using the Fastagen RNA isolation
kit (cat. no. 220011; Fastagen Biotech). Subsequently, RNA was
reverse transcribed into cDNA using the Takara reverse
transcription kit (cat. no. RR037A; Takara Biotechnology Co.,
Ltd.). The PCR thermocycling conditions (Bio-Rad Laboratories,
Inc.) were as follows: 95°C for 30 sec, followed by 40 cycles of
95°C for 5 sec. The relative expression levels were normalized to
endogenous control and were expressed as 2−ΔΔCq
(33). The primers used for PCR
are listed in Table II.
Western blotting
PMs were transfected with negative control or Ly108
siRNA and lysed with RIPA lysis buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology) at 4°C. Proteins (5 µg/lane), the
concentration of which was determined by the BCA protein
determination method (cat. no. A53225; Thermo Fisher Scientific,
Inc.), were separated by 10% SDS-PAGE and transferred onto PVDF
membranes. After blocking membranes with 5% bovine serum albumin
(cat. no. A8010; Beijing Solarbio Science & Technology Co.,
Ltd.) for 2 h at room temperature, membranes were incubated with
primary antibodies against phosphorylated (p)-NF-κB p65 (Cell
Signaling Technology, Inc.; cat. no. 3033; 1:1,000); NF-κB (Cell
Signaling Technology, Inc.; cat. no. 8242; 1:1,000) and β-actin
(Cell Signaling Technology, Inc.; cat. no. 3700; 1:1,000) at 4°C
overnight. The secondary antibodies (anti-mouse IgG, HRP-linked,
cat. no. 7076S, 1:2,000, Cell Signaling Technology, Inc.;
anti-rabbit IgG (H+L) antibody conjugated to biotin, cat. no.
14708S, 1:2,000, Cell Signaling Technology, Inc.) were incubated
with the membranes at 25°C for 1 h. A western blot Imaging System
(Tanon Science and Technology Co., Ltd.) was used to detect protein
bands and obtain images using the Alldoc-ECL 2019 software (Tanon
Science and Technology Co., Ltd.). The data were analyzed via
densitometry and normalized to expression of the internal control
(β-actin).
Statistical analysis
All data were analyzed using GraphPad Prism 8
software (GraphPad Software Inc.). A Student's t-test was used to
determine differences between two groups. One-way ANOVA followed
the Tukey's post hoc test was used to determine the differences
between more than two groups. Data are presented as the mean ±
standard error of the mean. P<0.05 was considered to indicate a
statistically significant difference.
Results
Enhanced SLAMF6 expression in TAMs is
related to the severity of HCC
SLAMF6 expression level was detected in the
peripheral CD14+ monocytes of patients with HCC and
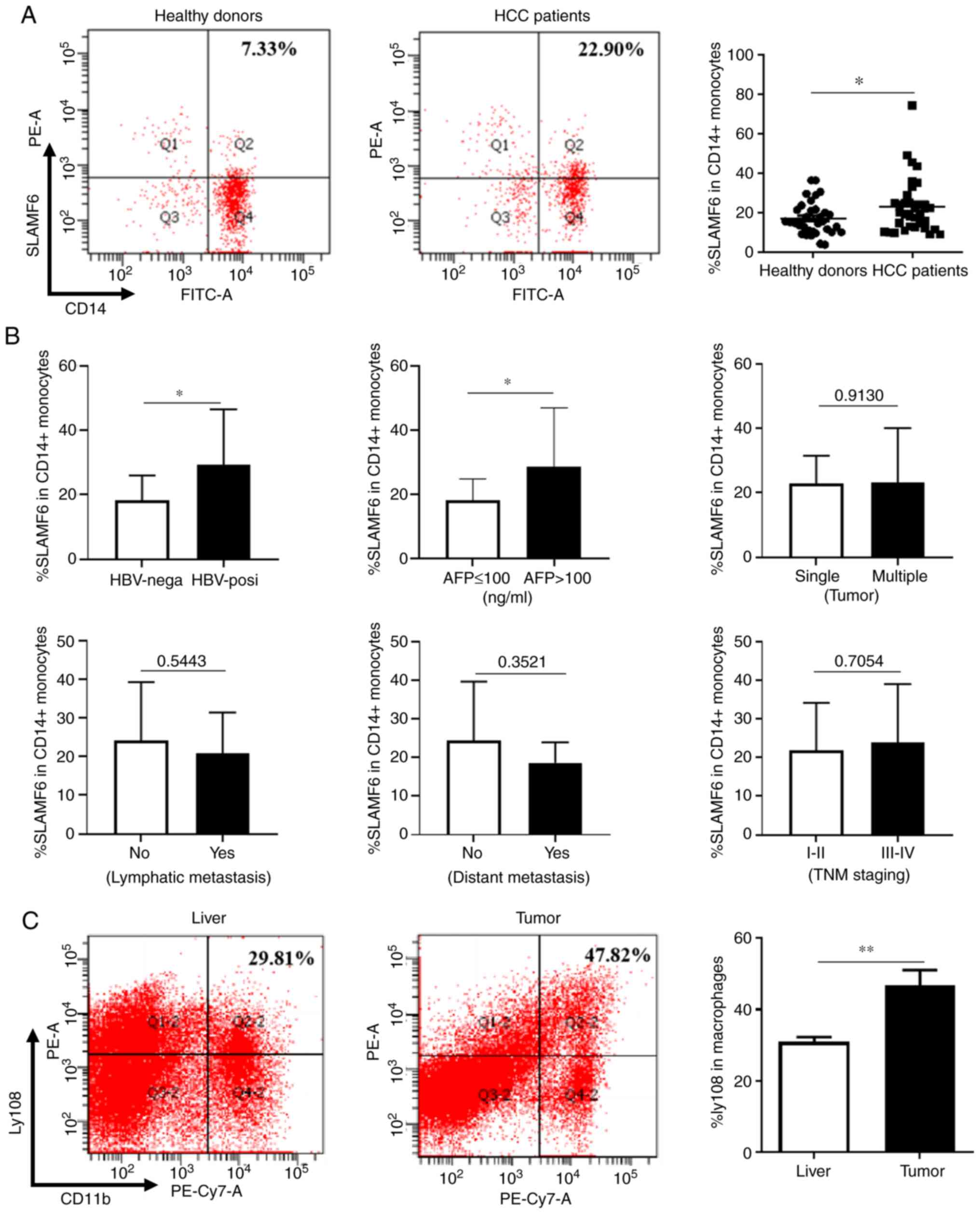
healthy donors. As presented in Fig.
1A, the percentage of peripheral CD14+ monocytes
expressing SLAMF6 in patients with HCC was significantly higher
compared with healthy donors. The relationship between SLAMF6
expression level and the severity of HCC was subsequently analyzed.
As presented in Fig. 1B, the
expression level of SLAMF6 was assessed in six groups of patients
categorized according to the following criteria: i) The HBV status
(negative or positive); ii) the α-fetoprotein (AFP) level (> or
<100 ng/ml); iii) the number of tumors (single or multiple); iv)
the lymphatic metastasis occurrence (yes or no); v) the distant
metastasis occurrence (yes or no); and vi) the different stages of
tumors. Among these groups, only two presented with differences in
SLAMF6 expression. The expression level of SLAMF6 in
CD14+ monocytes from patients with HCC who were
HBV-positive was significantly higher than in HBV-negative
individuals. Furthermore, an increased expression of SLAMF6 was
demonstrated in patients with HCC whose serum AFP level was >100
ng/ml compared with those exhibiting AFP level <100 ng/ml, which
indicated a relationship between SLAMF6 expression and HCC poor
prognosis (34). Since the age
distribution of the patients and the healthy donors was too
different for SLAMF6 expression, we made the comparison between the
low and high age groups and found no difference between the two age
groups (Fig. S1).
To determine the expression level of SLAMF6 in TAMs,
a murine tumor model of HCC was established, after which the
expression of Ly108 was determined in TAMs isolated from tumor and
liver tissues. As presented in Fig.
1C, when compared with liver tissues, the TAMs isolated from
tumor tissues expressed a higher level of Ly108. These results
demonstrated that SLAMF6 expression was increased in TAMs, which
may be associated with the poor prognosis of patients with HCC.
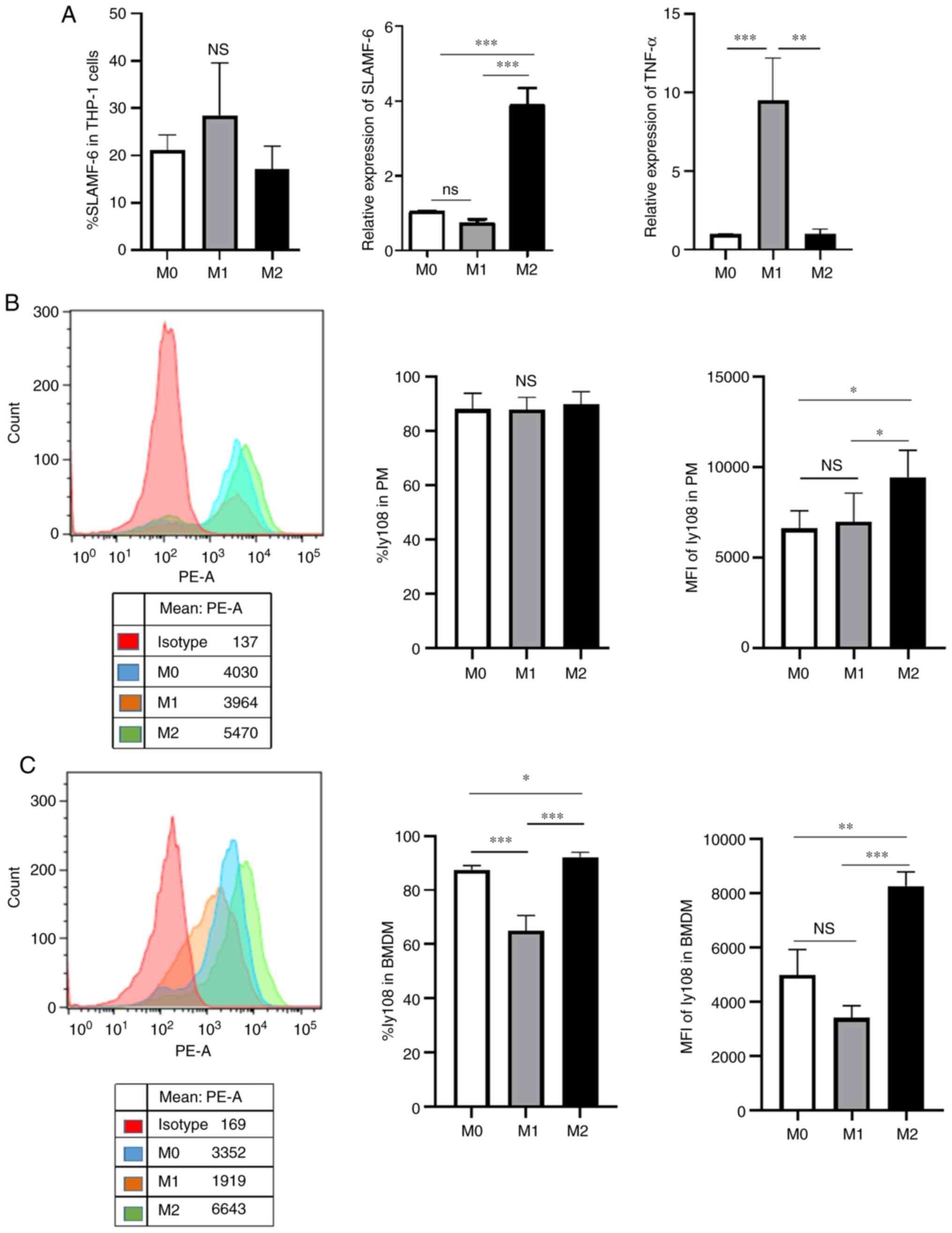
M2-like macrophages exhibit a higher
SLAMF6 expression
As previously reported, TAMs are considered to be
M2-like macrophages (35). The
present study therefore compared the expression level of SLAMF6 in
different types of macrophages. To achieve this, human THP-1,
murine PMs and murine BMDMs were polarized into M1- and M2-like
macrophages. As presented in Fig.
2A, different expression levels of TNF-α, a M1 marker,
indicated that polarization was successful. In addition, the
results from RT-qPCR demonstrated that the human M2-like phenotype
exhibited higher SLAMF6 expression level compared with the M1-like
phenotype. However, the results from flow cytometry analysis
revealed that the human M1-like phenotype expressed a higher
percentage of SLAMF6 compared with the M2-like phenotype.
For murine PMs, the mean fluorescence intensity
(MFI) of Ly108 expression in the M2-like phenotype was higher than
that in the M1-like phenotype. However, the percentage expression
of Ly108 did not significantly differ between the two groups
according to flow cytometry analysis (Fig. 2B), which may be due to the fact
that the PMs we collected were induced by the starch and already
expressed ~90% Ly108. Regarding murine BMDMs, the percentage of
expression and MFI of Ly108 in M2-like macrophages were higher than
in M1-like macrophages (Fig. 2C).
These results suggested that the M2-like phenotype exhibited higher
SLAMF6 expression level compared with the M1-like phenotype.
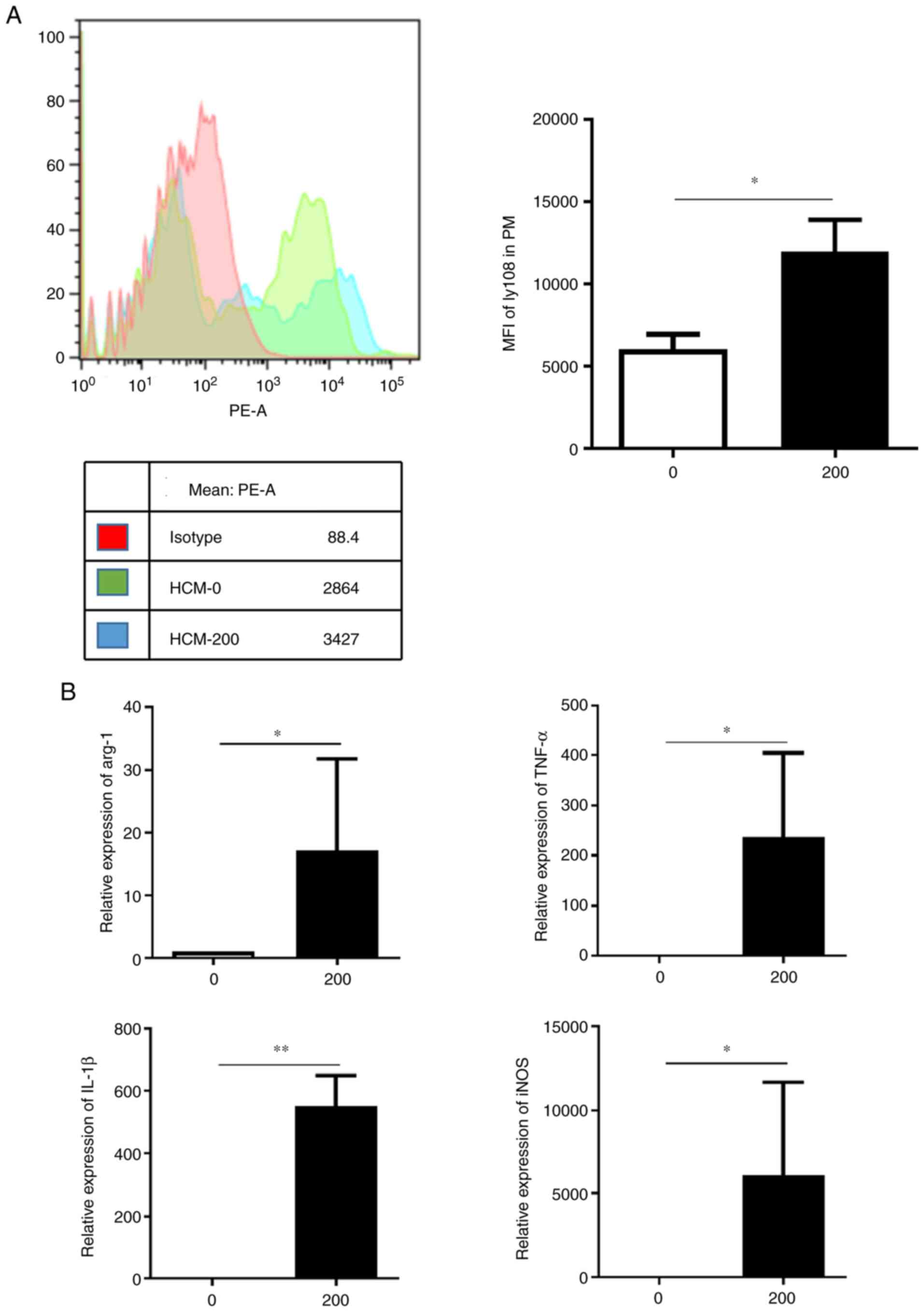
Macrophage SLAMF6 expression and
polarization is promoted in the tumor microenvironment
The aforementioned results indicated that the
M2-like phenotype exhibited increased SLAMF6 expression level.
Thus, the present study aimed to determine the reason behind this
increase. The effect of HCC microenvironment on PMs was therefore
assessed by using HCM. As presented in Fig. 3A, 200 µl HCM significantly promoted
Ly108 expression in PMs and induced two positive peaks rather than
one positive peak following the negative peak. This result may be
due to the fact that HCM enhanced Ly108 expression in macrophages
that already expressed a certain amount of Ly108, and HCM might
trigger the Ly108 expression in macrophages that did express the
molecule at a lower level. In addition, the expression levels of M2
marker arginase-1 (arg-1) and of the M1 markers TNF-α, IL-1β and
inducible nitric oxide synthase (iNOS) were increased in the HCM
group compared with the control group (Fig. 3B). It was therefore hypothesized
that the tumor microenvironment may enhance SLAMF6 expression and
that an unknown mechanism may promote M2 polarization in these
conditions.
Ly108 silencing reverses M2
polarization and inhibits HCC progression
Previous studies have revealed that M2-like
macrophages can promote HCC growth and invasion (6,8,12).
Ly108 siRNA was therefore used to investigate the role of Ly108 in
macrophage polarization and HCC progression. As presented in
Fig. 4A, Ly108 expression was
successfully decreased following siRNA transfection. The results
also demonstrated that following Ly108 silencing, the expression of
the M2 marker arg-1 was decreased, while the expression of certain
M1 markers, including TNF-α, IL-1β and iNOS, was increased. The
culture supernatant of macrophages treated with Ly108 or control
siRNA was subsequently co-cultured with Hepa1–6 cells (Fig. 4B). Following co-culture, the
proliferation, migration and invasion of Hepa1–6 cells was
significantly decreased. These results suggested that Ly108 could
inhibit the migratory and invasive abilities of Hepa1–6 cells.
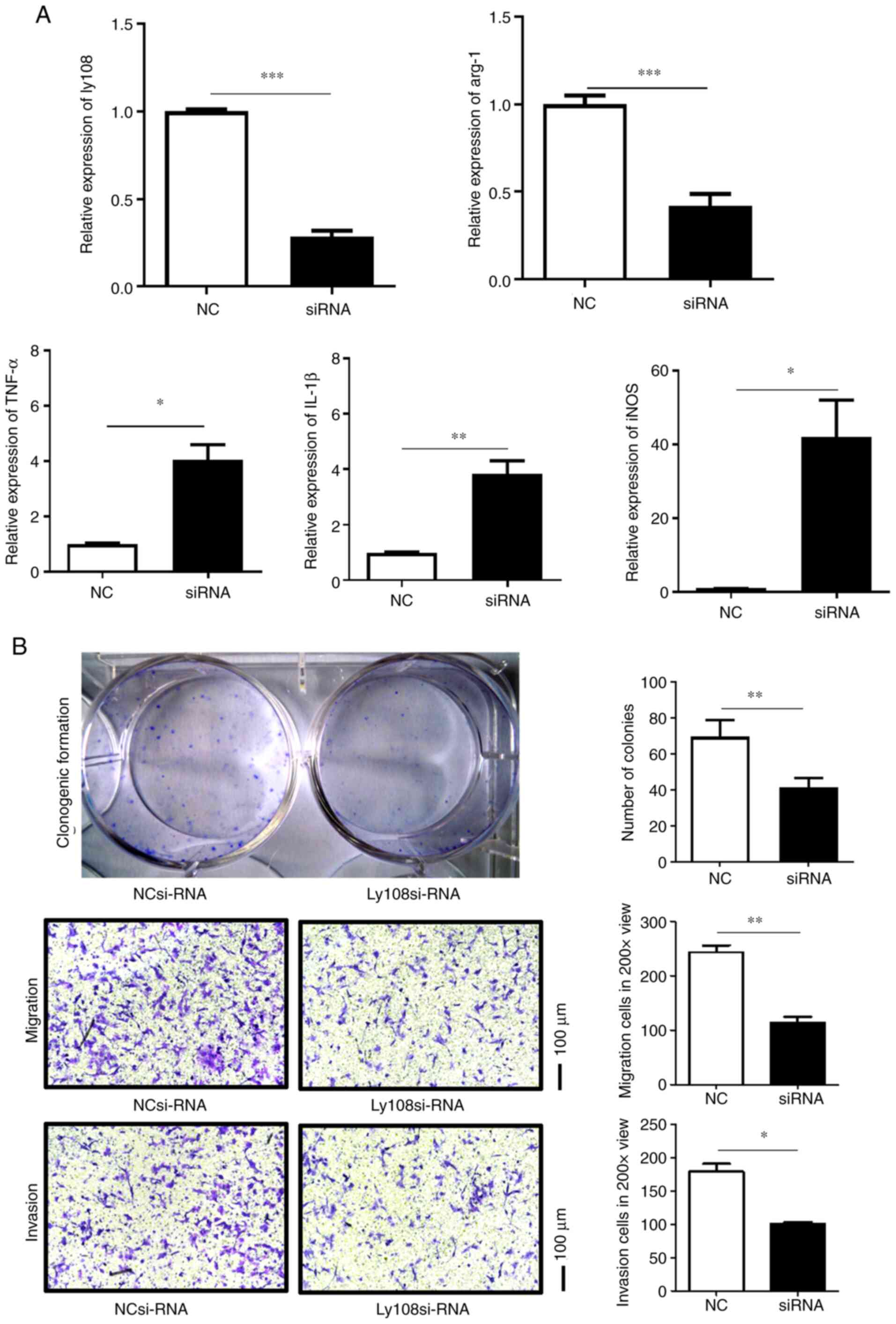 | Figure 4.Ly108 promotes M2 polarization and
hepatocellular carcinoma cell migration and invasion. (A) Reverse
transcription quantitative PCR was performed to determine the
relative expression of Ly108, arg-1, TNF-α, IL-1β and iNOS in PMs,
which were transfected with SLAMF6 siRNA or negative control siRNA.
(B) Clonogenic formation and Transwell assays were performed to
evaluate the proliferation, migration and invasion of Hepa1–6
cells. *P<0.05, **P<0.01 and ***P<0.001. NC, negative
control; si, small interfering; Arg-1, arginase-1; IL-1β,
interleukin 1β; iNOS, inducible nitric oxide synthase; TNF-α, tumor
necrosis factor-α. |
Macrophage Ly108 promotes HCC growth
in vivo
To improve our understanding of the effect of TAM
Ly108 on HCC progression in vivo, a murine model of HCC was
established. Ly108 siRNA or control siRNA was transfected into
M2-like macrophages, which were subsequently injected into C57BL/6
mice. The results demonstrated that, compared with control M2-like
macrophages, Ly108 siRNA-transfected M2-like macrophages inhibited
the growth of H22 tumors in terms of growth rate, gross tumor
volume and tumor weight (Fig.
5A).
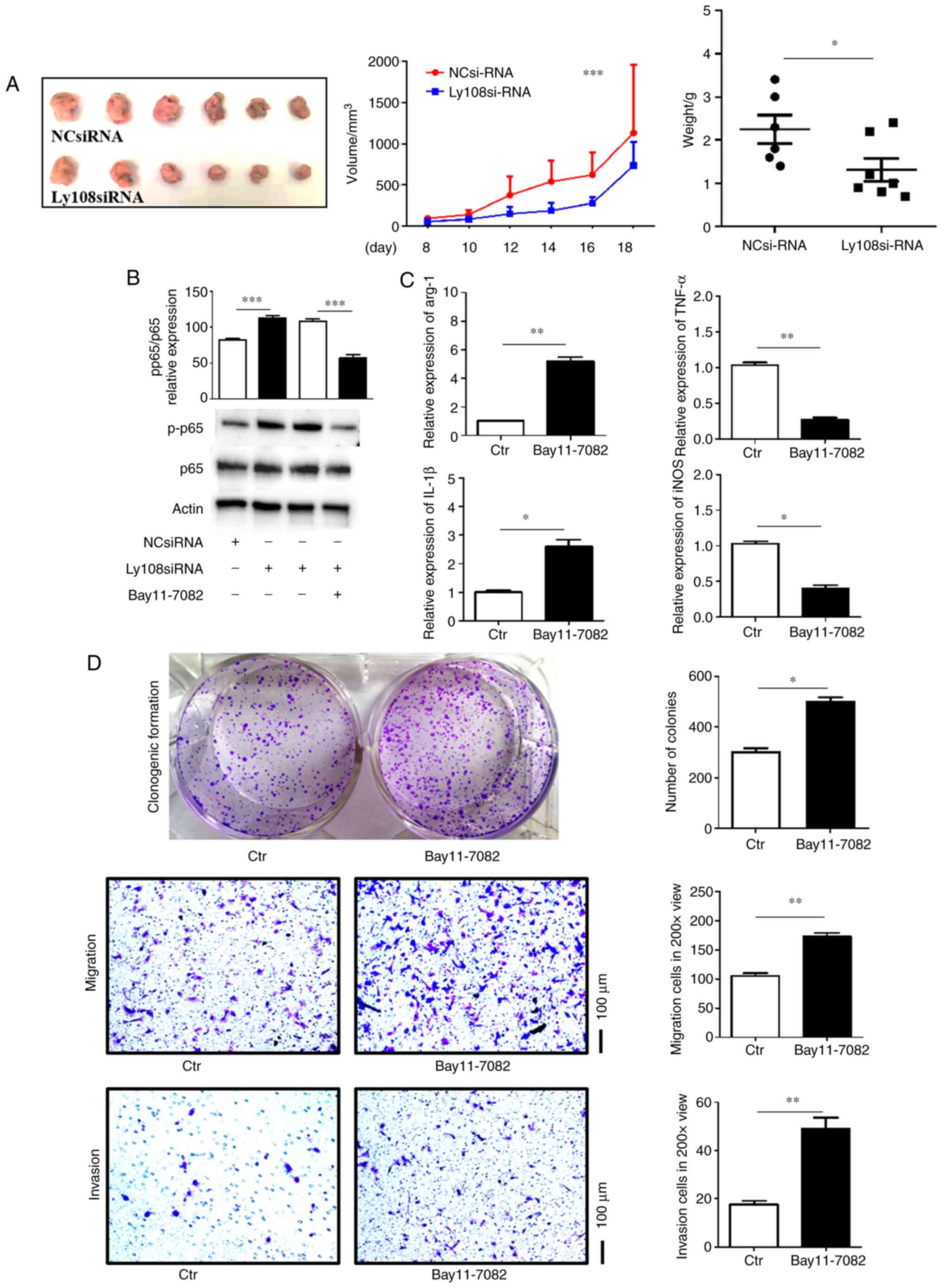 | Figure 5.Ly108 promotes hepatocellular
carcinoma growth by inhibiting the NF-κB signaling pathway. (A) A
murine HCC model was prepared (n=6) and the resulting tumors were
weighed at the time of sacrifice. Tumor images are presented in the
left panel, while summary data are presented in the right panel.
Tumor growth was measured via tumor size, as presented in the
middle panel. (B) PMs were transfected with negative control siRNA
or Ly108 siRNA, with or without Bay11-7082 prior to treatment with
20 ng/ml IL-4. Western blotting was performed to determine p65 and
p-p65 expression. (C) Ly108 siRNA transfection and Bay11-7082 or
control DMSO stimulation prior to IL-4 induction was performed on
murine PMs. Reverse transcription quantitative PCR was performed to
determine the relative expression of arg-1, TNF-α, IL-1β and iNOS.
(D) The macrophage culture supernatants in Figure 5C were collected and co-cultured
with Hepa1–6 cells for the transwell assays. Clonogenic formation
assays were then performed. *P<0.05, **P<0.01 and
***P<0.001. HCC, hepatocellular carcinoma; NC, negative control;
si, small interfering; Arg-1, arginase-1; IL-1β, interleukin 1β;
iNOS, inducible nitric oxide synthase; TNF-α, tumor necrosis
factor-α; NF-κB, nuclear factor-κB; IL-4, interleukin 4; Ctr,
control; p, phosphorylated; PMs, peritoneal macrophages. |
Ly108 promotes HCC growth via the
NF-κB pathway
The present study aimed to determine the potential
underlying mechanism of Ly108. As presented in Fig. 5B, the Ly108 siRNA-transfected group
presented higher expression of p65 phosphorylation. Furthermore,
treatment with Bay11-7082, an inhibitor of NF-κB, inhibited the
phosphorylation of p65. In addition, when Ly108 siRNA-transfected
macrophages were treated with Bay11-7082, they were able to
re-polarize to the M2 phenotype. This result was determined
according to higher Arg-1 and IL-1β levels and lower TNF-α and iNOS
levels (Fig. 5C). Macrophages of
which phenotype was reversed by Bay11-7082 treatment also promoted
the proliferation, migration and invasion of Hepa1–6 cells
(Fig. 5D). These results indicated
that Ly108 may promote HCC progression by inhibiting the NF-κB
pathway.
Discussion
SLAMF6 is an immune regulator that is expressed at
the surface of hematopoietic cells. Previous studies have revealed
that SLAMF6 is involved in numerous diseases, including systemic
lupus erythematosus and lethal colitis (25,27).
However, the role of macrophages SLAMF6 expression in HCC
progression remains unknown. The present study reported increased
SLAMF6 expression levels in the HCC microenvironment, which
facilitated M2-polarization and promoted HCC cell migration,
invasion and proliferation.
The results from the present study suggested that
there was a relationship between SLAMF6 and TAM-related
tumor-promoting inflammation. SLAMF6 expression level was higher in
the peripheral CD14+ monocytes of patients with HCC
compared with healthy donors and was associated with certain
clinicopathological characteristics of patients. Furthermore, TAMs,
also known as M2-like macrophages, exhibited higher SLAMF6 (Ly108
in mice) expression level compared with M1-like macrophages. In
addition, the tumor microenvironment was determined to be the cause
of high SLAMF6 expression and could also promote macrophage
polarization. SLAMF6 expression in TAMs was able to promote HCC
cell proliferation, migration and invasion, indicating the
important role of SLAMF6 in HCC development.
After demonstrating that SLAMF6 was upregulated in
the peripheral CD14+ monocytes of patients with HCC, our
study aimed to elucidate the relationship between SLAMF6 expression
level and certain clinicopathological characteristics of patients
with HCC, including HBV status, AFP level, the number of tumors,
the lymphatic metastasis occurrence, the distant metastasis
occurrence and the different stages of tumors. Only SLAMF6
expression level in the CD14+ monocytes from patients
with HCC that were HBV positive or whose serum AFP levels was
>100 ng/ml was significantly higher compared with the control
group. This may be due to the diversity of the patients from whom
blood samples were obtained. Upregulated SLAMF6 levels were also
observed in the murine HCC model, which verifies this
conclusion.
Macrophages are classified into two phenotypes: The
classically activated M1 phenotype and the alternatively activated
M2 phenotype (8). Among these, the
M2-like phenotype is considered to be a TAM, serving an important
role in the promotion of tumor growth and progression and
accelerating HCC cell invasion and migration, thereby worsening
patient prognosis (12,13). The results from the present study
revealed that TAMs had higher SLAMF6 expression levels compared
with M1-like macrophages. Furthermore, these findings were in
accordance with those obtained in human blood samples, indicating
that the monocytes from patients with HCC may have a tendency
towards the M2 phenotype and may therefore exhibit higher SLAMF6
expression level. The present study also demonstrated that the
tumor microenvironment may facilitate macrophage SLAMF6 expression
and polarization. The tumor microenvironment is extremely complex
and its components could promote the expression of SLAMF6. However,
this conclusion is difficult to verify. The results from the
present study indicated that SLAMF6 silencing significantly
inhibited the proliferation, migration and invasion of Hepa1–6
cells. Furthermore, SLAMF6 silencing could inhibit tumor growth in
the murine HCC model. Taken together, these finding suggested a
potential role of SLAMF6 in TAM-mediated HCC progression.
NF-κB is a signaling pathway that serves an
important role in macrophage polarization (30). It has been reported that SLAMF6
regulates the function of Th2 cells via the NF-κB pathway (29). The results from the present study
demonstrated that SLAMF6 accelerated HCC growth by inhibiting the
NF-κB signaling pathway.
The findings from our study suggested that increased
SLAMF6 expression levels in patients with HCC may be caused by the
tumor microenvironment, which promoted HCC development via the
NF-κB signaling pathway. However, further investigation is required
to determine the relationship between SLAMF6 expression and the
clinicopathological characteristics of patients with HCC.
Furthermore, determining why SLAMF6 expression level was
upregulated in M2 macrophages and elucidating the mechanisms
underlying the interactions between SLAMF6 and NF-κB should be the
aim of future studies.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Shandong Provincial Natural
Science Foundation, China (grant no. ZR2020QH177) and the Key
Research & Development Plan of Shandong Province (grant no.
2019GSF108013).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article and are available from the
corresponding author on reasonable request.
Authors' contributions
QM, XD and QY were responsible for the flow
cytometry and molecular biology experiments. DX, ZL, YL, QJ, FG and
SJ were responsible for the human sample collection. ZW, XC and WY
were responsible for the mouse model of hepatocellular carcinoma.
PS designed the research and drafted the manuscript. QM and PS
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Shandong Cancer Hospital and Institute. Blood
collection methods were carried out in accordance with Guidelines
for the Collection of Venous Blood Specimens of China
(WS/T661-2020). Informed consent was obtained from all
participants. Animal experimental protocols were approved by the
Animal Ethics Committee of Shandong Cancer Hospital and Institute.
The methods used for the anesthesia and euthanasia of mice were
carried out in accordance with the American Veterinary Medical
Association Guidelines for the Euthanasia of Animals (2020). All
methods involving animals were performed in accordance with ARRIVE
(Animal Research: Reporting of In Vivo Experiments)
guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BMDM
|
bone marrow-derived macrophage
|
|
HCC
|
hepatocellular carcinoma
|
|
IFN-γ
|
interferon gamma
|
|
IL-6
|
interleukin-6
|
|
LPS
|
lipopolysaccharide
|
|
PM
|
peripheral macrophage
|
|
RNS
|
reactive nitrogen species
|
|
ROS
|
reactive oxygen species
|
|
SLAMF
|
signaling lymphocytes activation
molecule family
|
|
SLE
|
systemic lupus erythematous
|
|
TAMs
|
tumor-associated macrophages
|
|
TNF-α
|
tumor necrosis factor-α
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
European Association for The Study of The
Liver, European Organisation for Research and Treatment of Cancer,
. EASL-EORTC clinical practice guidelines: Management of
hepatocellular carcinoma. J Hepatol. 56:908–943. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pardee AD and Butterfield LH:
Immunotherapy of hepatocellular carcinoma: Unique challenges and
clinical opportunities. Oncoimmunology. 1:48–55. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsuchiya N, Sawada Y, Endo I, Uemura Y and
Nakatsura T: Potentiality of immunotherapy against hepatocellular
carcinoma. World J Gastroenterol. 21:10314–10326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hernandez-Gea V, Toffanin S, Friedman SL
and Llovet JM: Role of the microenvironment in the pathogenesis and
treatment of hepatocellular carcinoma. Gastroenterology.
144:512–527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wan S, Zhao E, Kryczek I, Vatan L,
Sadovskaya A, Ludema G, Simeone DM, Zou W and Welling TH:
Tumor-associated macrophages produce interleukin 6 and signal via
STAT3 to promote expansion of human hepatocellular carcinoma stem
cells. Gastroenterology. 147:1393–1404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ruffell B, Affara NI and Coussens LM:
Differential macrophage programming in the tumor microenvironment.
Trends Immunol. 33:119–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gordon S: Alternative activation of
macrophages. Nat Rev Immunol. 3:23–35. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Locati M, Mantovani A and Sica A:
Macrophage activation and polarization as an adaptive component of
innate immunity. Adv Immunol. 120:163–184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sica A and Mantovani A: Macrophage
plasticity and polarization: In vivo veritas. J Clin Invest.
122:787–795. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wynn TA and Vannella KM: Macrophages in
tissue repair, regeneration, and fibrosis. Immunity. 44:450–462.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li
CX, Ng KT, Forbes SJ, Guan XY, Poon RT, et al: Alternatively
activated (M2) macrophages promote tumour growth and invasiveness
in hepatocellular carcinoma. J Hepatol. 62:607–616. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yao RR, Li JH, Zhang R, Chen RX and Wang
YH: M2-polarized tumor-associated macrophages facilitated migration
and epithelial-mesenchymal transition of HCC cells via the
TLR4/STAT3 signaling pathway. World J Surg Oncol. 16:92018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cannons JL, Tangye SG and Schwartzberg PL:
SLAM family receptors and SAP adaptors in immunity. Annu Rev
Immunol. 29:665–705. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Veillette A: Immune regulation by SLAM
family receptors and SAP-related adaptors. Nat Rev Immunol.
6:56–66. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calpe S, Wang N, Romero X, Berger SB,
Lanyi A, Engel P and Terhorst C: The SLAM and SAP gene families
control innate and adaptive immune responses. Adv Immunol.
97:177–250. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang N, Keszei M, Halibozek P, Yigit B,
Engel P and Terhorst C: Slamf6 negatively regulates autoimmunity.
Clin Immunol. 173:19–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang N, Halibozek PJ, Yigit B, Zhao H,
O'Keeffe MS, Sage P, Sharpe A and Terhorst C: Negative regulation
of humoral immunity due to interplay between the SLAMF1, SLAMF5,
and SLAMF6 Receptors. Front Immunol. 6:1582015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Flaig RM, Stark S and Watzl C: Cutting
edge: NTB-A activates NK cells via homophilic interaction. J
Immunol. 172:6524–6527. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bottino C, Falco M, Parolini S, Marcenaro
E, Augugliaro R, Sivori S, Landi E, Biassoni R, Notarangelo LD,
Moretta L and Moretta A: NTB-A [correction of GNTB-A], a novel
SH2D1A-associated surface molecule contributing to the inability of
natural killer cells to kill Epstein-Barr virus-infected B cells in
X-linked lymphoproliferative disease. J Exp Med. 194:235–246. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marcq I, Nyga R, Cartier F, Amrathlal RS,
Ossart C, Ouled-Haddou H, Ghamlouch H, Galmiche A, Chatelain D,
Lamotte L, et al: Identification of SLAMF3 (CD229) as an inhibitor
of hepatocellular carcinoma cell proliferation and tumour
progression. PLoS One. 8:e829182013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bouhlal H, Ouled-Haddou H, Debuysscher V,
Singh AR, Ossart C, Reignier A, Hocini H, Fouquet G, Al Baghami M,
Eugenio MS, et al: RB/PLK1-dependent induced pathway by SLAMF3
expression inhibits mitosis and control hepatocarcinoma cell
proliferation. Oncotarget. 7:9832–9843. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang N, Satoskar A, Faubion W, Howie D,
Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH and
Terhorst C: The cell surface receptor SLAM controls T cell and
macrophage functions. J Exp Med. 199:1255–1264. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fouquet G, Marcq I, Debuysscher V, Bayry
J, Rabbind Singh A, Bengrine A, Nguyen-Khac E, Naassila M and
Bouhlal H: Signaling lymphocytic activation molecules Slam and
cancers: Friends or foes? Oncotarget. 9:16248–16262. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chatterjee M, Kis-Toth K, Thai TH,
Terhorst C and Tsokos GC: SLAMF6-driven co-stimulation of human
peripheral T cells is defective in SLE T cells. Autoimmunity.
44:211–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chatterjee M, Rauen T, Kis-Toth K,
Kyttaris VC, Hedrich CM, Terhorst C and Tsokos GC: Increased
expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus
erythematosus T lymphocytes promotes Th17 differentiation. J
Immunol. 188:1206–1212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van Driel B, Wang G, Liao G, Halibozek PJ,
Keszei M, O'Keeffe MS, Bhan AK, Wang N and Terhorst C: The cell
surface receptor Slamf6 modulates innate immune responses during
Citrobacter rodentium-induced colitis. Int Immunol. 27:447–457.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yigit B, Halibozek PJ, Chen SS, O'Keeffe
MS, Arnason J, Avigan D, Gattei V, Bhan A, Cen O, Longnecker R, et
al: A combination of an anti-SLAMF6 antibody and ibrutinib
efficiently abrogates expansion of chronic lymphocytic leukemia
cells. Oncotarget. 7:26346–26360. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cannons JL, Yu LJ, Hill B, Mijares LA,
Dombroski D, Nichols KE, Antonellis A, Koretzky GA, Gardner K,
Schwartzberg PL, et al: SAP regulates T(H)2 differentiation and
PKC-theta-mediated activation of NF-kappaB. Immunity. 21:693–706.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mussbacher M, Salzmann M, Brostjan C,
Hoesel B, Schoergenhofer C, Datler H, Hohensinner P, Basílio J,
Petzelbauer P, Assinger A and Schmid JA: Cell type-specific roles
of NF-κB linking inflammation and thrombosis. Front Immunol.
10:852019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Crowe NY, Coquet JM, Berzins SP,
Kyparissoudis K, Keating R, Pellicci DG, Hayakawa Y, Godfrey DI and
Smyth MJ: Differential antitumor immunity mediated by NKT cell
subsets in vivo. J Exp Med. 202:1279–1288. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang X, Goncalves R and Mosser DM: The
isolation and characterization of murine macrophages. Curr Protoc
Immunol. Nov 15–2008.(Epub ahead of print). doi:
10.1002/0471142735.im1401s83. View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)). Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sauzay C, Petit A, Bourgeois AM, Barbare
JC, Chauffert B, Galmiche A and Houessinon A: Alpha-foetoprotein
(AFP): A multi-purpose marker in hepatocellular carcinoma. Clin
Chim Acta. 463:39–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|