Introduction
Colorectal cancer (CRC) is one of the commonest
cancers worldwide, next to the cancers of lung and breast, while it
continues to be among the top 3 cancers both in incidence and
mortality in Japan (1,2). The incidence of CRC is in an
increasing trend with the Westernized lifestyle getting more and
more prevalent in Japan, making it one of the most important public
health burdens to countermeasure along with other issues such as
infectious diseases (3).
Accumulated clinical evidence shows that the life expectancy of CRC
patients is favorable when their CRCs are found in early stage with
5-year overall survival (OAS) rate of more than 84%, whereas the
OAS of advanced CRC patients are much lower, 60.0% for those with
clinical stage IIIb and 18.8% for those with clinical stage IV,
suggesting the importance of early detection of CRCs (4).
MicroRNAs (miRNAs) are small non-coding RNAs 20–25
nucleotides in length that play roles in transcriptional and
post-transcriptional gene regulation in normal cells. MiRNAs have
also been shown to play essential roles in the development and
progression of various types of cancers (5). Recently, medical researchers
worldwide made substantial efforts to clarify the roles of miRNAs
in and their potential applicability to early detection of cancers,
which have led to the accumulation of useful evidence for future
development of clinical tests based on blood miRNA measurements in
practical medicine (6–8).
Although there are a considerable number of studies
that investigated the possible usefulness of miRNA expression data
for early detection of CRCs (9–11),
there is still little evidence that clarified the detectability of
CRC onset by the miRNA expression analysis using the samples from
cohort study participants who were before clinical diagnoses.
Accordingly, we set out to examine the early
detectability of CRCs by serum miRNAs using the preserved serum
samples of the cohort participants who were affected with CRC
within 2 years after study enrollments.
Materials and methods
Study subjects
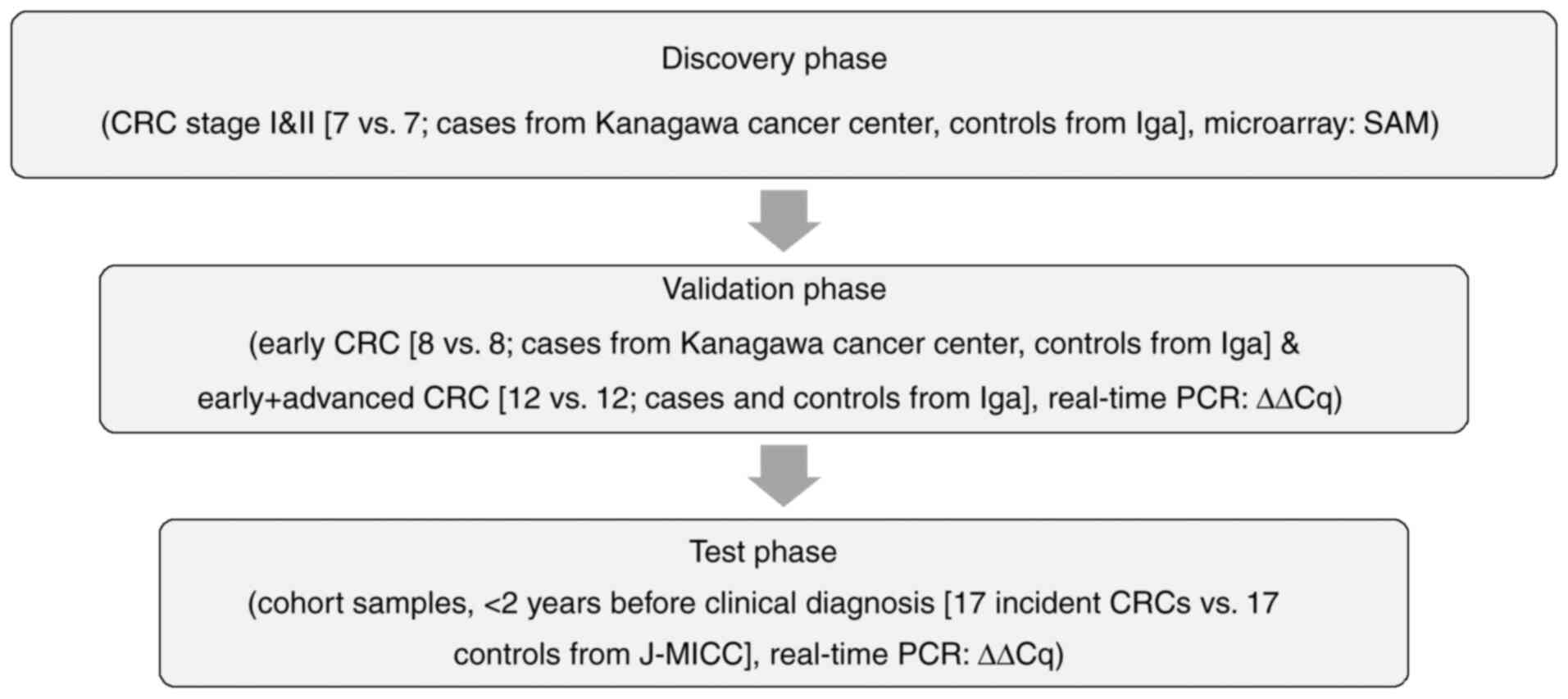
For the discovery phase, the discovery cohort
included seven CRC patients who underwent surgery at Kanagawa
Cancer Center Hospital between years of 2019 and 2020. In the first
validation phase, the case subjects were 8 CRC patients who
underwent surgery at Kanagawa Cancer Center Hospital between years
of 2019 and 2020. In the second validation phase, the case subjects
were 12 CRC patients who visited or were admitted to Iga City
General Hospital between years of 2014 and 2016. In the discovery
and validation phases, the control subjects were age- and
gender-matched cancer-free subjects from Iga City Cohort Study who
participated between years of 2013 and 2014 without any history of
cancer and without any cancer incidence at least within a year
after participation, with one-to-one matching.
The test cohort included patients from the Japan
Multi-Institutional Collaborative Cohort Study (J-MICC Study), one
of the largest genome cohort studies in Japan with 92,610
participants who were recruited nationwide from 14 areas of Japan
between 2005 and 2014 (12). We
obtained informed consent from the participants in the J-MICC
Study. We collected blood samples using a 7 ml vacuum tube for
serum and 7 ml EDTA-2Na containing vacuum tube for plasma and buffy
coat. We also conducted a survey using self-administered
questionnaires to the participants that included items on drinking,
smoking, physical activities, sleep, food consumption, medical
history, and reproductive history.
For the test phase for early detectability of CRCs
before clinical diagnosis, cases were the 17 participants of J-MICC
Shizuoka and Daiko Studies and Iga City Cohort Study, all of which
are the sub-cohorts of the J-MICC Study, who developed CRCs within
2 years after participation, and the control subjects were age- and
gender-matched cancer-free subjects from the corresponding cohorts
(Shizuoka, Daiko and Iga City Cohort Studies) without any history
of cancer and without any cancer incidence at least within a year
after participation, with one-to-one matching. We first discovered
the significantly up/down-regulated miRNAs using the clinical CRC
samples (7 early CRCs and 7 controls in the 1st stage, discovery
phase) by microarray analysis based on SAM (significance analysis
of microarrays) (13). We next
verified their replicability by real-time PCR (8 early CRCs and 8
controls, together with 12 early and advanced CRCs and 12 controls
in the 2nd stage, validation phase). Finally, we tested the early
detectability using the cohort samples of J-MICC Shizuoka &
Daiko Studies (14,15) who developed CRC within 2 years
after participation (3rd stage, test phase) (Fig. 1). Written informed consent was
obtained from all participants. The protocol of this study was
approved by the Ethics Review Committee of the Nagoya University
Graduate School of Medicine (approval no. 2019-0314-8), Aichi
Cancer Center, Kanagawa Cancer Center and all the participated
institutions.
Follow-up
In the J-MICC Study, we followed up the cancer
incidence by the survey of regional cancer registries, where
cancers were coded based on ICD-O-3T as C00-C80 (and CRCs were
coded as C18-C20). For the follow-up of death, we conducted the
survey on death certificates and classified the underlying causes
of deaths based on ICD-10, where cancers were coded as C00-C97 (and
CRCs were coded as C18-C20). Participants who moved out of the
catchment areas were considered as dropouts, who were excluded from
the follow-up survey after the timepoints of move-outs and treated
as truncated.
MiRNA expression analysis with
microarray
MiRNAs were extracted from 400 µl of serum samples
using the miRNeasy extraction kit (Qiagen). Prior to microarray
analyses, the integrity of total RNA samples was checked by
detecting the peak from small RNA fragments using the Agilent 2100
Bioanalyzer (Agilent Technologies). In the discovery phase, miRNA
expression measurements were conducted using a 3D-Gene miRNA
Labeling kit and a 3D-Gene Human miRNA Oligo Chip (Toray Industries
Inc.) for the detection of the expressions of microRNA sequences
from 271 organisms: 38,589 hairpin precursors and 48,860 mature
microRNAs registered in the miRBase release 22 (www.mirbase.org). The raw data of each spot was
normalized by substitution with a mean intensity of the background
signal determined by all blank spots' signal intensities of 95%
confidence intervals. Measurements of spots with the signal
intensities greater than 2 standard deviations (SD) of the
background signal intensity were considered to be valid. A relative
expression level of a given miRNA was calculated by comparing the
signal intensities of the valid spots throughout the microarray
experiments. The normalized data were normalized per array, such
that the 75th percentile of the signal intensity was adjusted to
1.
Quantitative RT-PCR analysis
To validate the replicability of the miRNAs detected
by microarray analyses, we conducted the quantitative RT-PCR
(qRT-PCR) analyses. miRNAs were extracted from 200 µl of serum
samples using the miRNeasy Mini Kit (Qiagen). The miRNAs were then
reverse transcribed using the miRCURY LNA RT Kit (Qiagen). The
obtained cDNA was quantified by qRT-PCR in the ABI-PRISM 7900
Sequence Detection System and the StepOnePlus System (Applied
Biosystems, Foster City, CA) using the miRCURY LNA miRNA PCR Kit
(Qiagen) according to manufacturer's instructions (details of the
primers used are shown in Table
S1). The experiment was performed in duplicate and the average
cycle threshold (Cq) value was obtained. Quantification of miRNA
expression levels was conducted based on the comparative cycle
threshold (ΔΔCq) method, where the expression levels of each miRNA
were normalized based on the expression levels of miR-16-5p as an
internal control (16). Two-tailed
P-values are calculated based on a Student's t-test of the
2−ΔΔCq values for each gene comparing cases and
controls. In the test phase for early detectability of CRCs before
clinical diagnosis, experiments were conducted with blinding to
examiners (whose names are in the Acknowledgements).
Database search
We referred to the databases of miRCancer
(www.mircancer.ecu.edu) (17) to search for the previously reported
miRNAs whose expressions were associated with the development or
progression of CRCs.
Statistical analysis
The comparisons between the miRNA expression data of
CRC patients and control subjects based on microarray measurements
were conducted using SAM, using the ‘samr’ package of R ver. 3.6.3,
where q-value is used as a means to control the positive false
discovery rate in multiple hypothesis testing (18), after excluding the miRNAs with
percent presence of the signals in either of cases or controls was
less than 50%. ROC curve analyses were conducted using the MedCalc
ver. 20.011 (MedCalc Software Ltd.). Graphs for miRNA expression
levels were drawn using the ‘beeswarm’ package of R. Comparisons of
the two miRNA expression levels were conducted using the Student's
t-test (unpaired, 2-sided) as well as by Mann-Whitney's U test, and
those of more than three miRNA expression levels were conducted by
one-way analysis of variance (ANOVA) with post hoc analysis by
Tukey's test by using Stata (StataCorp).
Results
Study characteristics
The characteristics of the study subjects are shown
in Table I. Briefly, the discovery
set consisted of 7 early CRC cases and 7 age- and gender-matched
controls, the first validation set consisted of 8 early CRC cases
with 8 age- and gender-matched controls, and the second validation
set consisted of 12 CRC cases with mixture of early and advanced
stages together with 12 age- and gender-matched controls, whereas
the test set of cohort samples for pre-clinical early detection
consisted of 17 cohort participants who developed CRC with mixture
of early and advanced clinical stages within 2 years after
participation, together with 17 age- and gender-matched
controls.
 | Table I.Characteristics of the study
subjects. |
Table I.
Characteristics of the study
subjects.
|
| Discovery | Validation-1 | Validation-2 | Test (cohort,
J-MICC) |
|---|
|
|
|
|
|
|
|---|
| Variables | Case (n=7) | Control (n=7) | Case (n=8) | Control (n=8) | Case (n=12) | Control (n=12) | Case (n=17) | Control (n=17) |
|---|
| Male (%) | 5 (71.4%) | 5 (71.4%) | 4 (33.3%) | 4 (33.3%) | 8 (66.7%) | 8 (66.7%) | 12 (70.6%) | 12 (70.6%) |
| Age, mean ± SD | 58.4±6.9 | 58.0±5.7 | 74.5±4.2 | 64.5±1.3 | 69.0±7.7 | 63.8±4.5 | 59.7±8.0 | 59.5±7.3 |
| Cancer site |
|
|
|
|
|
|
|
|
| Colon
(C) | 0 (0.0%) | - | 4 (50.0%) | - | 2 (16.7%) | - | 4 (23.5%) | - |
| Sigmoid
(S) | 2 (28.6%) | - | 1 (12.5%) | - | 6 (50.0%) | - | 7 (41.2%) | - |
| Rectum
(R) | 5 (71.4%) | - | 3 (37.5%) | - | 4 (33.3%) | - | 4 (23.5%) | - |
|
S-R | 0 (0.0%) | - | 0 (0.0%) | - | 0 (0.0%) | - | 2 (11.8%) | - |
| Clinical stage |
|
|
|
|
|
|
|
|
| I | 6 (85.7%) | - | 3 (37.5%) | - | 1 (8.3%) | - | 3 (17.6%) | - |
| II | 1 (14.3%) | - | 5 (62.5%) | - | 4 (33.3%) | - | 7 (41.2%) | - |
|
III | 0 (0.0%) | - | 0 (0.0%) | - | 7 (58.3%) | - | 3 (17.6%) | - |
| IV | 0 (0.0%) | - | 0 (0.0%) | - | 0 (0.0%) | - | 2 (11.8%) | - |
|
(Unknown) | 0 (0.0%) | - | 0 (0.0%) | - | 0 (0.0%) | - | 2 (11.8%) | - |
Microarray miRNA expression analysis
of CRC patients and controls
We conducted the miRNA expression analyses according
to the workflow presented in Fig.
1. We selected the candidate miRNA based on the combined
strategy of theoretical selection with best statistical
significance and the empirical selection by database search
(17) and our previous reports
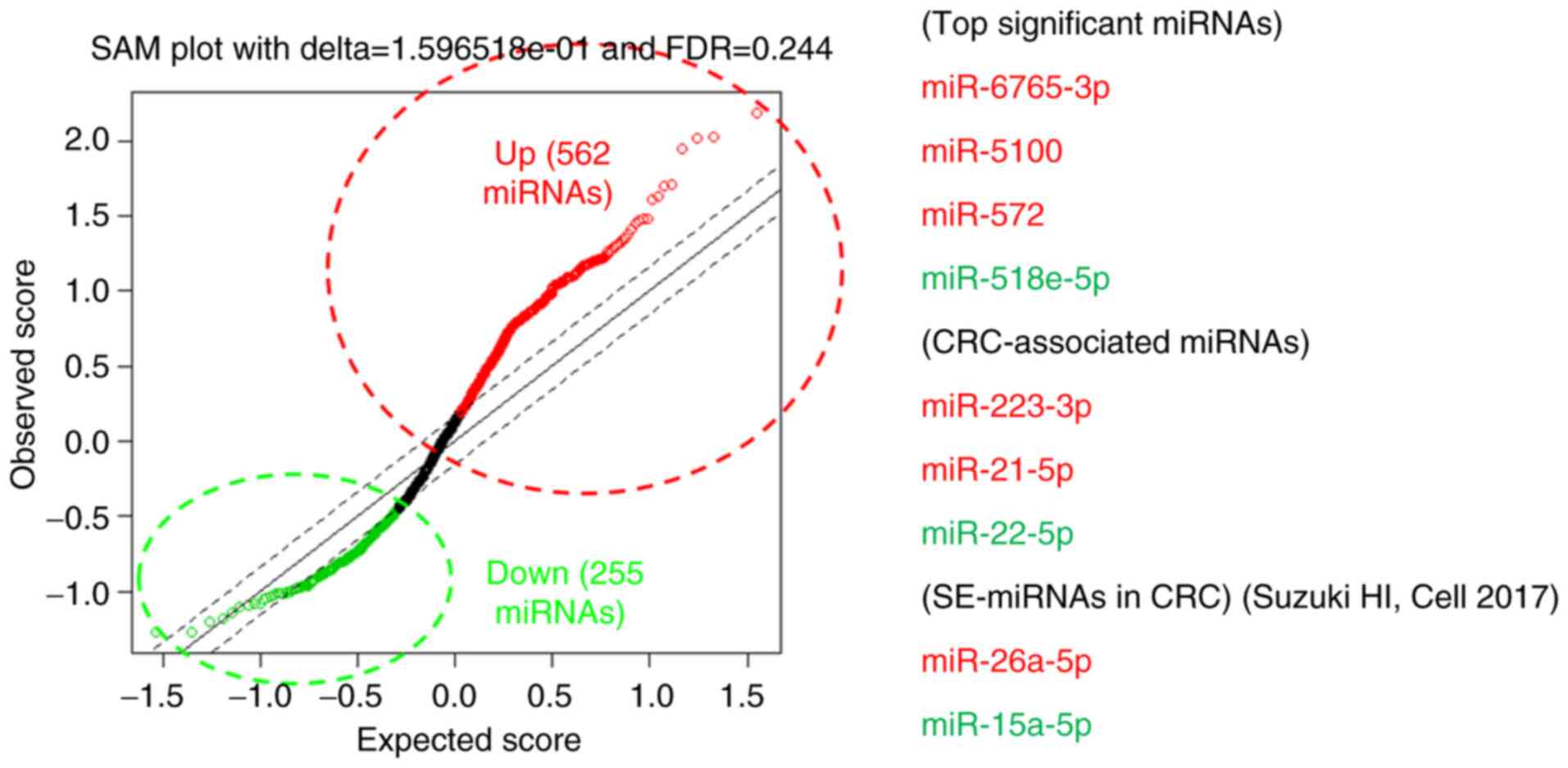
(9,10). The comprehensive expression
measurements of miRNAs by microarrays and subsequent SAM analysis
revealed miR-6765, miR-5100, miR-572 (up-regulated) and miR-518e-5p
(down-regulated) as the miRNAs with best discrimination abilities.
In addition, we searched for the miRNAs with smallest q-values
which were previously reported in association with CRCs among the
miRNAs that fulfilled the criteria of FDR <0.25 (562
up-regulated miRNAs and 255 down-regulated miRNAs), which led us to
adopt miR-223-3p, miR-26a-5p and miR-21-5p as the up-regulated,
miR-22-5p and miR-15a-5p as the down-regulated candidate miRNAs
(Table II and Fig. 2). We also confirmed that there was
no statistically significant difference in expression levels of
miR-16-5p (the internal control) observed between cases and
controls (Fig. S1).
 | Table II.Selected miRNAs in the discovery
phase. |
Table II.
Selected miRNAs in the discovery
phase.
| A, miRNAs
upregulated in CRCs |
|---|
|
|---|
| miRNAs | Fold-change | q-value (%) |
|---|
|
|---|
| Smallest
q-value |
|
|
|
miR-6765-3p | 3.356 | 0 |
|
miR-5100 | 3.114 | 0 |
|
miR-572 | 3.152 | 0 |
| CRC associated,
smallest |
|
|
| q-value |
|
|
|
miR-223-3p | 2.867 | 0 |
|
miR-26a-5p | 2.862 | 4.091 |
| CRC associated,
reported by our group (9,10) |
|
|
|
miR-21-5p | 1.766 | 9.431 |
|
| B, miRNAs
downregulated in CRCs |
|
| miRNAs |
Fold-change | q-value
(%) |
|
| Smallest
q-value |
|
|
|
miR-518e-5p | 0.381 | 15.197 |
| CRC associated,
smallest |
|
|
| q-value |
|
|
|
miR-22-5p | 0.578 | 15.197 |
|
miR-15a-5p | 0.474 | 15.197 |
Validation with quantitative RT-PCR
analysis with independent samples
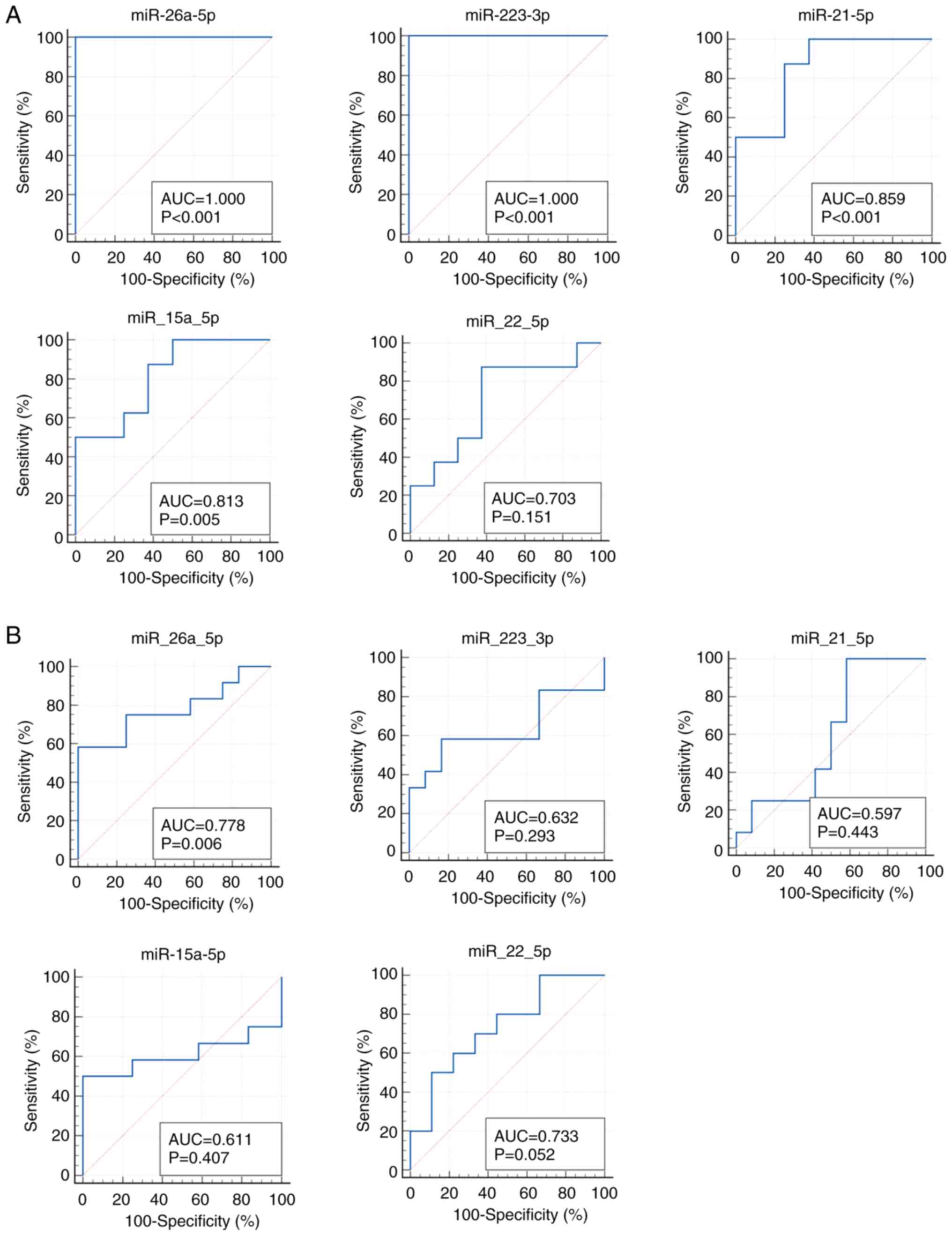
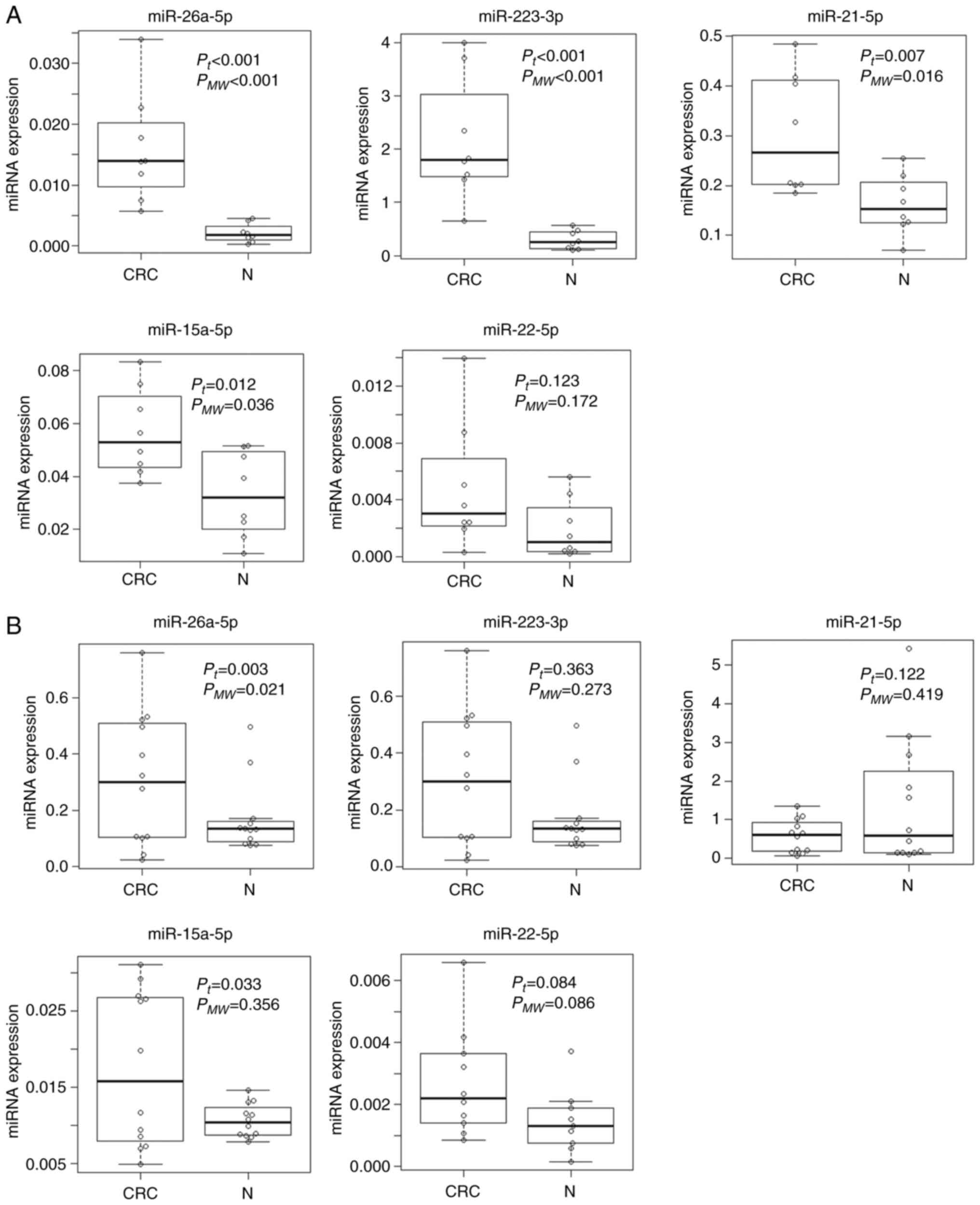
We next verified the replicability of microarray
discovered miRNAs by qRT-PCR using independent samples. In the
first validation set with 8 CRC samples with early clinical stage
from Kanagawa Cancer Center and 8 age- and gender-matched controls,
miR-26a-5p and miR-223-3p demonstrated the highest diagnostic
accuracy of AUC=1.000 (sensitivity 100.0% and specificity 100.0%),
followed by miR-21-5p with AUC of 0.859 (sensitivity 100.0% and
specificity 62.5%) (Fig. 3A). In
the second validation set with the independent set with 12 advanced
CRCs and 12 controls from Iga City General Hospital, most of the
selected miRs demonstrated moderate accuracy with AUCs of 0.6-0.8
(Fig. 3B). The expressions of the
top 3 significant up-regulated miRNAs (miR-6765, miR-5100 and
miR-572) and the top significant down-regulated miRNA (miR-518e-5p)
were not detected in the qRT-PCR of the validation phase. The
expression levels of replicated miRNAs (miRNAs with AUC of more
than 0.6 and the same directions of correlations as those in the
discovery phase) as well as those of the rest of the miRNAs that
failed to replicate are shown in Fig.
4.
Examination of the predictability of
CRC incidence using pre-clinical cohort samples
We next examined the predictabilities of CRC
incidence by the replicated miRNAs that demonstrated the AUC of
more than 0.6 in both of the two validation phases with the same
direction of correlations between expression levels of miRNAs and
CRC risk as those in the discovery phase 17 samples from patients
from the J-MICC Shizuoka and Daiko Studies (14,15)
who developed CRCs within 2 years after participation and age- and
gender-matched controls.
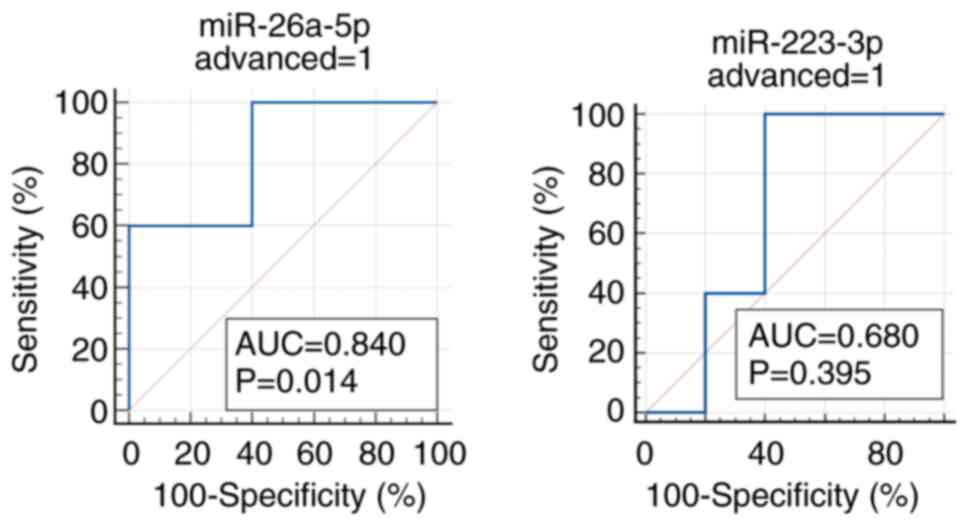
Among the two miRNAs that fulfilled the criteria
(miR-26a-5p and miR-223-3p), miR-26a-5p demonstrated the good
predictability of advanced CRC incidence (5 incident CRCs and 5
age- and gender-matched controls) with the AUC of 0.840
(sensitivity 100.0% and specificity 60.0%), whereas no other miRs
fulfilled the criteria of AUC more than 0.6, and in the same
direction of correlations as those in the discovery and validation
sets (Figs. 5 and S2). The predictability of early CRC by
miR-26a-5p and miR-223-3p did not reach the AUC of 0.6 (Fig. S3). The expression levels of
miR-26a-5p and miR-223-3p in the pre-clinical phase are shown in
Supplementary Fig. S2. In
addition, we compared the expression levels of miR-26a-5p between
early CRC, advanced CRC, and non-CRC subjects, which revealed
statistically significant difference (P=0.049, one-way ANOVA)
(Fig. S4).
Discussion
In the present study, on the basis of discovery and
validation with clinical CRC samples and testing with pre-clinical
cohort samples, we identified serum miR-26a-5p as a possible early
detection marker for CRC before clinical diagnosis.
With regard to the roles of miR-26a-5p in the
colorectal carcinogenesis, considerable amount of experimental
evidence has been accumulated. The expressions of miR-26a have been
reportedly inversely correlated with PTEN expressions in
human CRC tissue samples, where miR-26a was significantly
upregulated in tumors in comparison to normal tissues, and there
was no difference in miR-26a expression levels by CRC stages
(19). MiR-26a is also
demonstrated to downregulate retinoblastoma 1 gene
expression and also to regulate glucose metabolism by targeting
PDHX in CRCs (20).
Meanwhile, miR-223-3p is shown to promote the
proliferation, growth, migration, invasion and EMT
(epithelial-mesenchymal transition) of CRCs through the negative
regulation of PRDM1 (21).
Although the present study failed to detect the early detectability
of CRCs by miR-223-3p, the high accuracy of CRC detectability in
the validation phase may indicate its importance in the development
of CRCs. MiR-21 is also shown to be a robust diagnostic marker for
CRCs, which failed to detect CRC incidence before clinical
diagnosis. Considering that the high expression of miR-21 in serum
and CRC tissue is associated with larger tumor size, distant
metastasis and poor prognosis, as demonstrated in the previous
papers (10,22), the role of miR-21 in the early
detection of CRCs might be relatively limited.
In addition, miR-26a-5p is one of the
super-enhancer-associated miRNAs (SE-miRNAs) of CRCs, where
super-enhancers (SEs) are newly found regulatory regions on the
genome consisted of multiple enhancer-like elements occupied with
high densities of master transcription factors and mediator
complexes (23). Moreover, both
miR-15a and miR-26a-5p are marked with super-enhancers in normal
sigmoid colon and other normal tissues (23), thus the roles of these SE-miRNAs in
the normal colorectal tissues as well as in the development of CRCs
should be clarified experimentally and clinically in the future.
Further clarification of the roles of SE-miRNAs in CRC development
as well as in the early detection of CRCs should be warranted.
In the present study, the detectability of selected
miRNAs in the discovery phase was improved in the first validation
set with CRC cases of early clinical stages compared to the second
validation set with CRC cases of mixture of early and advanced
clinical stages, which was in accordance with the findings with the
previous report of miRNA diagnostic model in lung cancer, although
the difference in diagnostic accuracies between early and advanced
stage of lung cancers was only a little (24). It also remains unclear whether the
changes in miRNA expression levels reflect the presence of
diseases, i.e., the existence of cancers itself, or reflect
the alterations of surrounding tissues such as microenvironments,
and it is neither clarified whether the miRNA expression changes
initiates prior to disease onset or not (25). Further biological and
epidemiological studies are warranted.
One strength to be mentioned of the present study
would be the availability of serum samples of cohort participants
who developed CRCs after participation, which enabled us to
evaluate the early detectability of CRCs before clinical diagnosis.
Further studies that will make use of the preserved samples of
large cohort studies for early detection of cancers might be
expected.
There are several limitations to be mentioned in the
present study. Although the present study analyzed the frozen
samples preserved for several years for early detectability of CRC,
the miRNAs are shown to be highly stable for ~5 years under the
storage at −80°C (26). To obtain
more reliable results, however, further studies with improved
designs, e.g., those with more frequent follow-ups with
blood sample collections will be expected. In the present study,
miRNA expression levels in the serum were measured. Although the
miRNA expression levels in serum and plasma are in good
correlations to each other, there are some advantages and
disadvantages. For example, whereas the miRNA expression levels in
serum tends to be higher due to the RNA release during coagulation
(27), the miRNA expression levels
in plasma might also be affected by delayed sample processing,
especially in cohort studies, due to the possible release of
exosomes from the blood cells. In addition, aberrant expressions of
miR-26a-5p in blood have been reported also in acute myeloid
leukemia (AML) (28), bladder
cancer (29,30), non-small cell lung cancer (31) and breast cancer (32). Although non-specificity is
recognized as a general problem of cancer detection markers, it is
worth reporting the present study results considering miR-26a-5p
can be one of the potential markers for future clinical
applications. Further investigations should be made to identify
more specific early detection methods of CRCs. There were also
constraints due to limited research funds; we had to select the
limited number of miRNAs with high accuracy carefully, although
adopting as numerous miRNAs as possible to improve the diagnostic
accuracy of miRNA prediction models would be ideal. In addition,
some of the miRNAs detected in the discovery phase indicated poor
reproducibility of expressions themselves in the validation phase,
which was considered to be attributable to the performance of the
microarray platform used. Therefore, studies with improved ways of
expression analyses, i.e., by using the next-generation
sequencings of miRNAs should be expected. Finally, statistical
power considerations are as follows. The statistical power for the
10-fold increase in expression levels with the standard deviation
of 5 in 8 cases and 8 controls is more than 90%, whereas it is more
than 85% in 7 cases and 7 controls under the same conditions.
Although the sample size in the present study can detect the
difference in expression levels if the difference is large enough,
the limited sample sizes of the present study should be considered
as a limitation, which should warrant further investigations with
sufficient sample sizes in the near future.
In conclusion, the present study found serum
miR-26a-5p as a potential early detection marker for CRC before
clinical diagnosis. Further investigations with improved designs
are warranted.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Nobuyuki Hamajima
(Department of Healthcare Administration, Nagoya University
Graduate School of Medicine) and Dr Hideo Tanaka (Kishiwada
Healthcare Center) for their work for initiating and organizing the
J-MICC Study as former principal investigators. The authors would
also like to thank Ms. Yoko Mitsuda and Ms. Rie Terasawa (both
affiliated with the Department of Preventative Medicine, Nagoya
University Graduate School of Medicine) for their technical
assistance.
Funding
This study was supported by Grants-in-Aid for Scientific
Research for Priority Areas of Cancer (grant no. 17015018) and
Innovative Areas (grant no. 221S0001) and by the Japan Society for
the Promotion of Science (JSPS) KAKENHI grant from the Japanese
Ministry of Education, Culture, Sports, Science and Technology
[grant nos. 16H06277 (CoBiA) and 19K10598].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The microarray data is publicly available from GEO
(www.ncbi.nlm.nih.gov/geo/; accession
no. GSE186510).
Authors' contributions
AH designed the study, analyzed the data and
prepared the manuscript. HY and YA performed the experiments and
prepared the data. YO, MS, YM, YD, YT, YS, KoT, YK, RO, MN, TT,
KeT, AM, NH, TK and KW collected the data and samples. YM, YD, NH
and KW supervised the study. AH and HY confirmed the authenticity
of all the raw data All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All of the patients agreed to provide their genetic
and clinical data for analysis after written informed consent. The
study protocol was approved by the Ethics Review Committee of the
Nagoya University Graduate School of Medicine (approval no.
2019-0314-8), Aichi Cancer Center, Kanagawa Cancer Center and all
the participated institutions.
Patient consent for publication
All study participants approved the publication of
the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Onyoh EF, Hsu WF, Chang LC, Lee YC, Wu MS
and Chiu HM: The rise of colorectal cancer in Asia: Epidemiology,
screening, and management. Curr Gastroenterol Rep. 21:362019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Akimoto N, Ugai T, Zhong R, Hamada T,
Fujiyoshi K, Giannakis M, Wu K, Cao Y, Ng K and Ogino S: Rising
incidence of early-onset colorectal cancer-a call to action. Nat
Rev Clin Oncol. 18:230–243. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moore MA, Sobue T, Kuriki K, Tajima K,
Tokudome S and Kono S: Comparison of Japanese, American-Whites and
African-Americans-pointers to risk factors to underlying
distribution of tumours in the colorectum. Asian Pac J Cancer Prev.
6:412–419. 2005.PubMed/NCBI
|
|
4
|
Hashiguchi Y, Muro K, Saito Y, Ito Y,
Ajioka Y, Hamaguchi T, Hasegawa K, Hotta K, Ishida H, Ishiguro M,
et al: Japanese society for cancer of the colon and rectum.
Japanese society for cancer of the colon and rectum (JSCCR)
guidelines 2019 for the treatment of colorectal cancer. Int J Clin
Oncol. 25:1–42. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abe S, Matsuzaki J, Sudo K, Oda I, Katai
H, Kato K, Takizawa S, Sakamoto H, Takeshita F, Niida S, et al: A
novel combination of serum microRNAs for the detection of early
gastric cancer. Gastric Cancer. 24:835–843. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka
Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda
T, et al: Integrated extracellular microRNA profiling for ovarian
cancer screening. Nat Commun. 9:43192018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shimomura A, Shiino S, Kawauchi J,
Takizawa S, Sakamoto H, Matsuzaki J, Ono M, Takeshita F, Niida S,
Shimizu C, et al: Novel combination of serum microRNA for detecting
breast cancer in the early stage. Cancer Sci. 107:326–334. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamada A, Horimatsu T, Okugawa Y, Nishida
N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, et al: Serum
miR-21-5p, miR-29a, and miR-125b are promising biomarkers for the
early detection of colorectal neoplasia. Clin Cancer Res.
21:4234–4242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toiyama Y, Takahashi M, Hur K, Nagasaka T,
Tanaka K, Inoue Y, Kusunoki M, Boland CR and Goel A: Serum
miR-21-5p as a diagnostic and prognostic biomarker in colorectal
cancer. J Natl Cancer Inst. 105:849–859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Toiyama Y, Okugawa Y, Fleshman J, Boland
CR and Goel A: MicroRNAs as potential liquid biopsy biomarkers in
colorectal cancer: A systematic review. Biochim Biophys Acta Rev
Cancer. 1870:274–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takeuchi K, Naito M, Kawai S, et al: Study
profile of the Japan multi-institutional collaborative cohort
(J-MICC) study. J Epidemiol. 31:660–668. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:5116–5121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Asai Y, Naito M, Suzuki M, Tomoda A,
Kuwabara M, Fukada Y, Okamoto A, Oishi S, Ikeda K, Nakamura T, et
al: Baseline data of Shizuoka area in the Japan multi-institutional
collaborative cohort study (J-MICC Study). Nagoya J Med Sci.
71:137–144. 2009.PubMed/NCBI
|
|
15
|
Morita E, Hamajima N, Hishida A, Aoyama K,
Okada R, Kawai S, Tomita K, Kuriki S, Tamura T, Naito M, et al:
Study profile on baseline survey of Daiko Study in the Japan
multi-institutional collaborative cohort study (J-MICC Study).
Nagoya J Med Sci. 73:187–195. 2011.PubMed/NCBI
|
|
16
|
Schwarzenbach H, da Silva AM, Calin G and
Pantel K: Data normalization strategies for MicroRNA
quantification. Clin Chem. 61:1333–1342. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie B, Ding Q, Han H and Wu D: miRCancer:
A microRNA-cancer association database constructed by text mining
on literature. Bioinformatics. 29:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Storey JD: The positive false discovery
rate: A Bayesian interpretation and the q-value. Ann. Statist.
31:2013–2035. 2003. View Article : Google Scholar
|
|
19
|
Coronel-Hernández J, López-Urrutia E,
Contreras-Romero C, Delgado-Waldo I, Figueroa-González G,
Campos-Parra AD, Salgado-García R, Martínez-Gutierrez A,
Rodríguez-Morales M, Jacobo-Herrera N, et al: Cell migration and
proliferation are regulated by miR-26a-5p in colorectal cancer via
the PTEN-AKT axis. Cancer Cell Int. 19:802019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
López-Urrutia E, Coronel-Hernández J,
García-Castillo V, Contreras-Romero C, Martínez-Gutierrez A,
Estrada-Galicia D, Terrazas LI, López-Camarillo C,
Maldonado-Martínez H, Jacobo-Herrera N and Pérez-Plasencia C:
MiR-26a downregulates retinoblastoma in colorectal cancer. Tumour
Biol. Apr 26–2017.(Epub ahead of print). doi:
10.1177/1010428317695945. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chai B, Guo Y, Cui X, Liu J, Suo Y, Dou Z
and Li N: MiR-223-3p promotes the proliferation, invasion and
migration of colon cancer cells by negative regulating PRDM1. Am J
Transl Res. 11:4516–4523. 2019.PubMed/NCBI
|
|
22
|
Okugawa Y, Yao L, Toiyama Y, Yamamoto A,
Shigemori T, Yin C, Omura Y, Ide S, Kitajima T, Shimura T, et al:
Prognostic impact of sarcopenia and its correlation with
circulating miR-21 in colorectal cancer patients. Oncol Rep.
39:1555–1564. 2018.PubMed/NCBI
|
|
23
|
Suzuki HI, Young RA and Sharp PA:
Super-enhancer-mediated RNA processing revealed by integrative
MicroRNA network analysis. Cell. 168:1000–1014.e15. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Asakura K, Kadota T, Matsuzaki J, Yoshida
Y, Yamamoto Y, Nakagawa K, Takizawa S, Aoki Y, Nakamura E, Miura J,
et al: A miRNA-based diagnostic model predicts resectable lung
cancer in humans with high accuracy. Commun Biol. 3:1342020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamada H, Suzuki K, Fujii R, Kawado M,
Hashimoto S, Watanabe Y, Iso H, Fujino Y, Wakai K and Tamakoshi A:
Circulating miR-21-5p, miR-29a, and miR-126 are associated with
premature death risk due to cancer and cardiovascular disease: The
JACC study. Sci Rep. 11:52982021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Balzano F, Deiana M, Giudici SD, Oggiano
A, Baralla A, Pasella S, Mannu A, Pescatori M, Porcu B, Fanciulli
G, et al: miRNA stability in frozen plasma samples. Molecules.
20:19030–19040. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang K, Yuan Y, Cho JH, McClarty S, Baxter
D and Galas DJ: Comparing the MicroRNA spectrum between serum and
plasma. PLoS One. 7:e415612012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barrera-Ramirez J, Lavoie JR, Maganti HB,
Stanford WL, Ito C, Sabloff M, Brand M, Rosu-Myles M, Le Y, Allan
DS, et al: Micro-RNA profiling of exosomes from marrow-derived
mesenchymal stromal cells in patients with acute myeloid leukemia:
Implications in leukemogenesis. Stem Cell Rev Rep. 13:817–825.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang H, Hu Z and Chen L: Enhanced plasma
miR-26a-5p promotes the progression of bladder cancer via targeting
PTEN. Oncol Lett. 16:4223–4228. 2018.PubMed/NCBI
|
|
30
|
Miyamoto K, Seki N, Matsushita R, Yonemori
M, Yoshino H, Nakagawa M and Enokida H: Tumour-suppressive
miRNA-26a-5p and miR-26b-5p inhibit cell aggressiveness by
regulating PLOD2 in bladder cancer. Br J Cancer. 115:354–363. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ye MF, Lin D, Li WJ, Xu HP and Zhang J:
MiR-26a-5p serves as an oncogenic MicroRNA in non-small cell lung
cancer by targeting FAF1. Cancer Manag Res. 12:7131–7142. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang ZM, Ge HF, Yang CC, Cai Y, Chen Z,
Tian WZ and Tao JL: MicroRNA-26a-5p inhibits breast cancer cell
growth by suppressing RNF6 expression. Kaohsiung J Med Sci.
35:467–473. 2019.PubMed/NCBI
|