Introduction
Bladder cancer (BCa) is one of the most prevalent
types of malignant tumor of the urinary system (1). BCas are classified as superficial
papillary or non-papillary carcinoma according to their
constitution (2). Papillary
carcinoma is usually non-invasive but may develop into
non-papillary invasive carcinoma with a high histological grade,
with repeated recurrence after treatment. Conversely, non-papillary
invasive carcinoma usually has a high histological grade and poor
clinical course. Various chromosomal aberrations are involved in
the development and progression of these types of cancer.
Generally, there are two types of chromosomal aberration: Primary,
which is associated with tumor oncogenesis, and secondary, which is
associated with tumor progression (3). Using comparative genomic
hybridization and loss of heterozygosity (LOH) analysis,
aberrations in urothelial carcinoma have been observed on human
chromosomes 1p, 9 and 11p, which contain numerous onco- and tumor
suppressor genes, such as runt-related transcription factor 3,
cyclin-dependent kinase inhibitor 2A and cyclin D1 (4). Advanced BCa is accompanied by
aberrations in human chromosomes 2q, 5q, and 8p (5–7).
Furthermore, insights in the molecular pathology of BCa suggest two
pathways for the development of BCa: Luminal and basal subtype
(8). The q arm of chromosome 9 is
deleted in both molecular subtypes, suggesting that it may be a
primary event in the pathogenesis of BCa (9,10).
Inactivating mutations of tuberous sclerosis 1, which is a key
tumor suppressor gene on 9q, is found in 11-16% of BCa cases,
regardless of stage (11,12). Mutations in Notch homolog 1,
located on 9q, have also been identified in 18% of BCa (13). The aberration rates of these tumor
suppressor genes are inconsistent with the degree of LOH in BCa,
suggesting the presence of novel tumor suppressor genes that
contribute to cancer development.
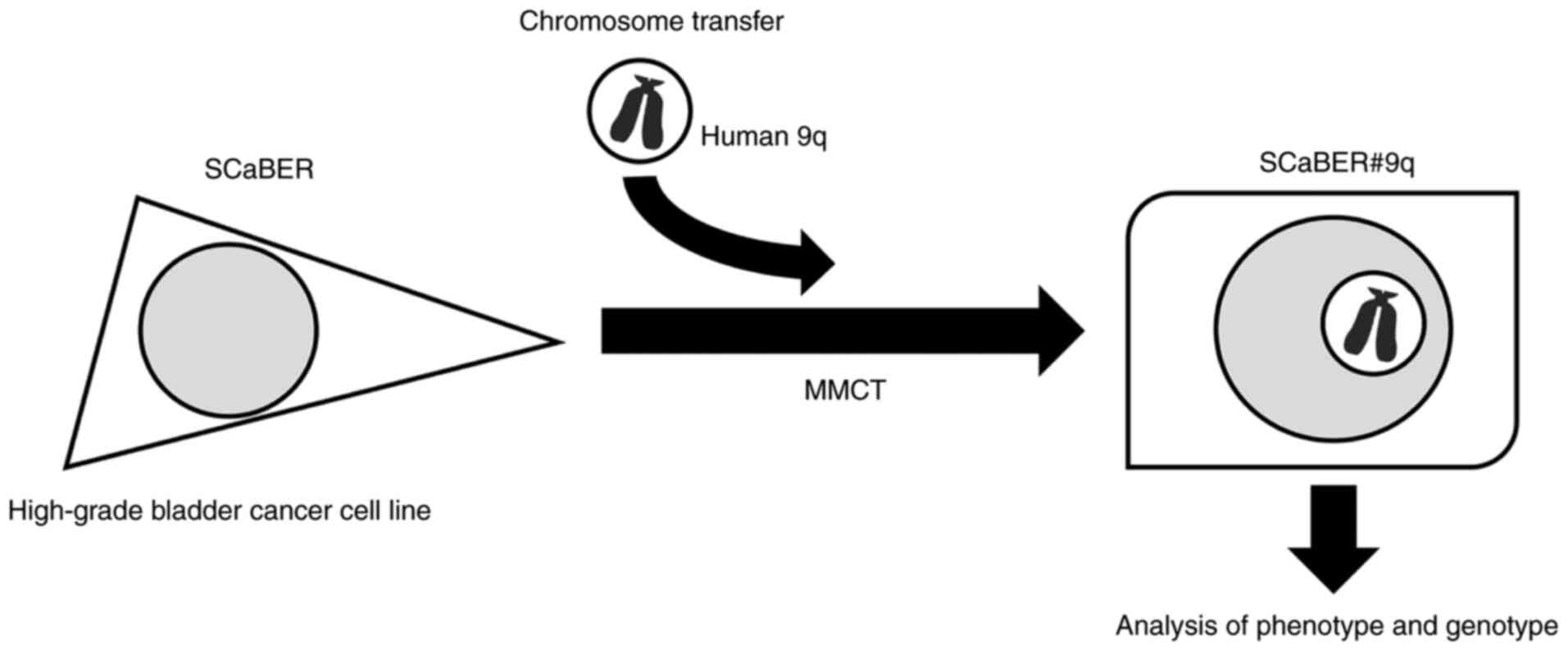
To understand the functional significance of the LOH
at the 9q region in BCa development, human chromosome 9q was
transferred to a BCa cell line using microcell-mediated chromosome
transfer (MMCT) and examined for effects on phenotype.
Materials and methods
Cell lines and culture
SCaBER and RT4 cells were purchased from the
American Type Culture Collection. T24 and 5637 cells were purchased
from the RIKEN Cell Bank. Cells were cultured in DMEM
(Sigma-Aldrich; Merck KGaA) supplemented with 10% fetal bovine
serum (FBS; HyClone; Cytiva), 100 U/ml penicillin and 100 µg/ml
streptomycin (FUJIFILM Wako Pure Chemical Corporation). Mouse A9
cells containing human chromosome 9q or 4, respectively, tagged
with neomycin resistance gene (neo) were maintained in DMEM
supplemented with 10% FBS and 800 µg/ml G418 antibiotic
(Calbiochem; Merck KGaA) and used as chromosome donors. All cell
lines were maintained at 37°C in a humidified incubator with 5%
CO2.
MMCT
Chromosome transfer via chromosome engineering was
performed as previously described (14). A9(neo9q) or A9(neo4) cells were
treated with 0.05 µg/ml colcemid at 37°C for 48 h to induce
formation of micronuclei, which were then purified by cytochalasin
B (10 µg/ml) digestion and centrifugation at 11,900 × g for 60 min
at 34°C. The isolated microcells were then resuspended in
serum-free DMEM and filtered sequentially through 8, 5 and 3 µm
polycarbonate filters (Whatman plc; Cytiva). The purified
microcells were collected by centrifugation at 400 × g for 15 min
at room temperature (RT) and resuspended in serum-free DMEM
containing 50 µg/ml phytohemagglutinin-P (FUJIFILM Wako Pure
Chemical Corporation). The microcells were attached to the cell
monolayer at 37°C for 15 min, fused with recipient cells in 47%
polyethylene glycol solution for 1 min at RT, followed by washing
with serum-free DMEM. Cells then maintained in non-selective medium
(DMEM) for 24 h at 37°C, trypsinized and divided into six 100 mm
dishes containing selection medium (containing 800 µg/ml G418).
Acquisition of microcellular hybrid
clones
The day after microcell fusion, culture medium was
changed to selection medium (containing 800 µg/ml G418). One week
after fusion, no viable cells were observed. Two weeks after
fusion, rapidly proliferating clones were isolated and maintained
by serial passaging, as previously described (14).
Genomic PCR analysis
The presence of the 9p24.2-9q34.3 region on human
chromosome 9q contained in A9(neo9) was verified by PCR using 21
specific sequence-tagged site (STS) markers (D9S54, 9p24.2; D9S268,
9p23; D9S285, 9p22.3; D9S165, 9p21.1; D9S200, 9p13.1; SHGC-103793,
9p12; UT801, 9p11.2; SHGC-141463, 9q12; SHGC-146514, 9q13; D9S15,
9q21.12; D9S1122, 9q21.2; D9S153, 9q21.31; D9S777, 9q22.1; D9S318,
9q22.2; D9S287, 9q22.32; D9S277, 9q31.1; D9S177, 9q33.1; D9S290,
9q34.11; D9S66, 9q34.2; and D9S158, 9q34.3). Primer sequences were
obtained from the National Center for Biotechnology Information
(https://www.ncbi.nlm.nih.gov). PCR was
performed with 35 cycles of 30 sec at 94°C, 30 sec at 58-62°C and
30 sec at 72°C.
RNA isolation and reverse
transcription-quantitative PCR
Total RNA was extracted using RNeasy Mini kit
(Qiagen GmbH) from each cell line and treated with DNase I
(FUJIFILM Wako Pure Chemical Corporation). First-strand cDNA was
synthesized using M-MLV reverse transcriptase (Invitrogen; Thermo
Fisher Scientific, Inc.) with random primers (Invitrogen; Thermo
Fisher Scientific, Inc.), 5× First Strand Buffer (Invitrogen;
Thermo Fisher Scientific, Inc.) and dNTPs (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The temperature protocol was 23°C
for 12 min for primer annealing, followed by 42°C for 50 min for
reverse transcription and then 95°C for 5 min for enzyme
inactivation.
The mRNA expression of PPARG (encoding PPARγ),
forkhead box A1 (FOXA1) and GATA3 was analyzed using specific
primers as follows: PPARG forward, 5′-GACAGGAAAGACAACAGACAAATC −3′
and reverse, 5′-GGGGTGATGTGTTTGAACTTG −3′; FOXA1 forward,
5′-AGGGCTGGATGGTTGTATTG-3′ and reverse, 5′-ACCGGGACGGAGGAGTAG −3′
and GATA3 forward, 5′-GCTTCGGATGCAAGTCCA −3′ and reverse,
5′-GCCCCACAGTTCACACACT-3′. cDNA was amplified using an Applied
Biosystems StepOne thermal cycler system and SYBR Green PCR kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). mRNA levels
were normalized against GAPDH mRNA (PCR primers: Forward,
5′-AGCCACATCGCTCAGACAC-3′ and reverse, 5′-GCCCAATACGACCAAATCC-3′)
using the 2−ΔΔCq method (15). Thermocycling conditions were 10 min
at 95°C for denaturation, followed by 15 sec at 95°C and 60 sec at
60°C for denaturation and annealing/extension for 40 cycles. The
experiments were performed in triplicate.
Heatmap of gene expression
The expression levels of PPARG, FOXA1, and GATA3 of
RT4, T24, SCaBER, SCaBER#4 and SCaBER9q were visualized using a
freely available web server, Heatmapper (http://www.heatmapper.ca.).
Fluorescence in situ hybridization
(FISH)
To identify successful transference of chromosome 9q
in metaphase spreads, chromosomal FISH was performed using plasmid
pSV2neo as a probe. The transferred chromosome 4 was identified
using RP11-84C13 BAC DNA as a probe.
The preparation of chromosome slides, probe
labeling, hybridization, washing and detection of signals were
performed as previously described (16).
Cell proliferation assay
Cell proliferation assay was performed to evaluate
the proliferation capability of cells. SCaBER, SCaBER#4, and
SCaBER#9q cells were seeded at 1.0×105 cells in a 6 cm
dish. All cells were cultured in DMEM supplemented with 10% FBS.
Measurements were made using a hemocytometer on days 1, 2, 3, 4,
and 5 after seeding. Cell counting was performed three times.
Migration assay
Wound healing assay was performed to evaluate the
migration capability of cells. SCaBER, SCaBER#4 and SCaBER#9q cells
were grown to 100% confluence in a 6-cm dish and a scratch was made
with 200-µl pipette tips. The cells were washed with PBS and placed
in serum-free medium (DMEM). The wound width at 0 and 24 h was
measured and evaluated using a digital camera system (NIS-Elements
Documentation, Ver.5.30; Nikon Corporation).
Western blotting
Western blotting was performed as previously
described (17). Membranes were
blotted with rabbit polyclonal antibodies against human PPARγ (cat.
no. #2430; 1:1,000; Cell Signaling Technology, Inc.), PTEN (cat.
no. #9188; 1:1,000; Cell Signaling Technology, Inc.) or α-tubulin
(cat. no. PM054-7; 1:5,000; Medical and Biological Laboratories
Co., Ltd.) and anti-rabbit IgG horseradish peroxidase-linked
antibody (cat. no. #7074; 1:2,000; Cell Signaling Technology,
Inc.), according to the manufacturer's instructions. Immunoreactive
bands were visualized using an enhanced chemiluminescence detection
system (cat. no. 32106; Pierce™ ECL Western Blotting
Substrate; Thermo Fisher Scientific, Inc.). Protein bands on
western blot films were quantified using ImageJ (ver.1.8.0;
National Institutes of Health).
Survival analysis
The Cancer Genome Atlas (TCGA) (18) and National Cancer Institute Genomic
Data Commons (19) public cancer
genome bladder urothelial carcinoma (BLCA) dataset (accessed 20
June, 2021) was visualized and analyzed using UCSC Xena
(xena.ucsc.edu). Overall survival curve was obtained using the
Kaplan-Meier method with the median expression of each gene as the
cutoff (PPARG: <19.31 vs. ≥19.31; FOXA1: <18.80 vs. ≥18.80′
GATA3: <20.52 vs. ≥20.52), and differences in survival were
evaluated with the log-rank test.
Statistical analysis
Data from triplicate experiments are presented as
the mean ± standard error of the mean. Data were analyzed using
one-way ANOVA with post-hoc Tukey's honestly significant difference
test. All statistical analysis was performed using SPSS Statistics
software (version 24.0; IBM Corp.) P<0.05 was considered to
indicate a statistically significant difference.
Results
Analysis of mouse A9/human
mono-chromosomal hybrids
The strategy for investigation of tumor suppressor
effect of human chromosome 9 in BCa is shown in Fig. 1. We previously generated a library
of A9 hybrid cells, each containing one of the human chromosomes
(except Y) (20). To confirm the
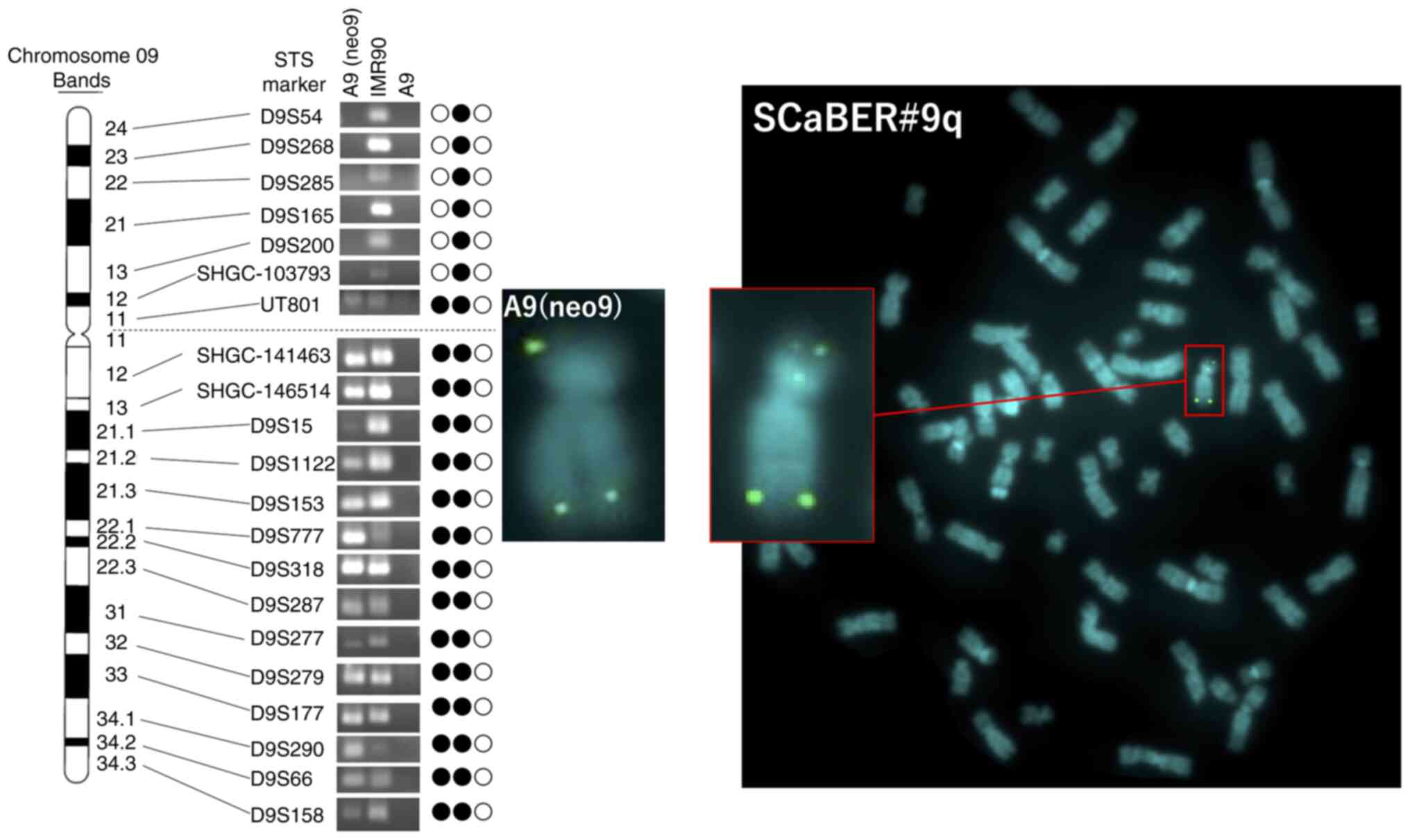
status of human chromosome 9 in A9(neo9) cells, PCR analysis using
21 STS markers located on human chromosome 9 was performed
(Fig. 2). PCR analysis showed
deletion of the short arm loci of human chromosome 9
(#9delp12-pter) in A9(neo9) cells (Fig. 2). FISH analysis was performed in
A9(neo9) cells using neo-plasmid probe. The pSV2neo probe for
#9delp12-pter was randomly integrated into two regions, #9p12 and
#9q34.3 (Fig. 3A). Additionally,
the long arm of human chromosome 9, was independently retained in
mouse A9 cells.
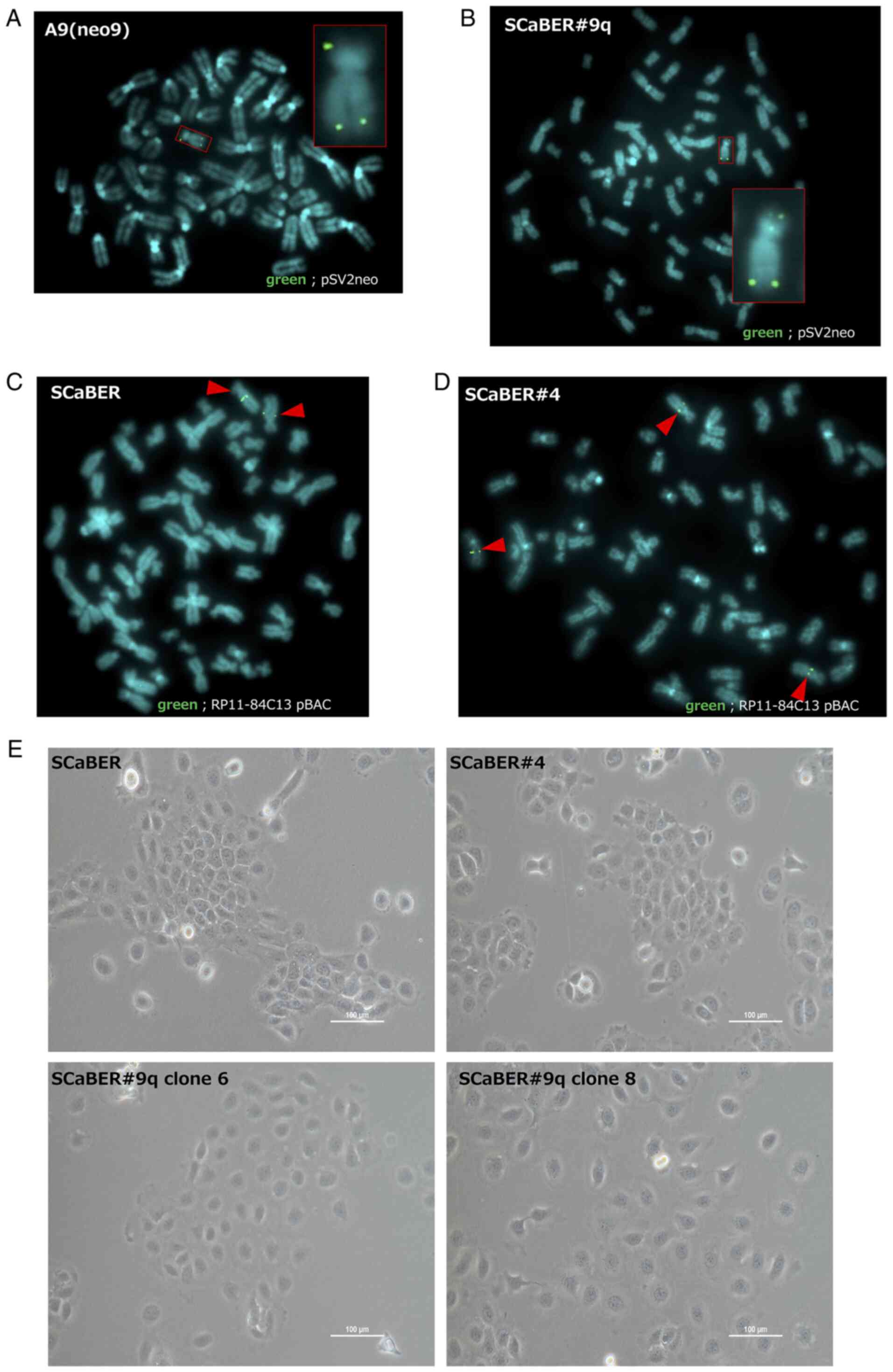
Introduction of human chromosome 9q
into SCaBER cells
Human chromosome 9q was transferred into SCaBER
cells using MMCT. Microcell hybrid cells were isolated via three
successive chromosome transfer experiments and analyzed to confirm
the presence of transferred #9delp12-pter tagged with pSV2neo by
FISH. Transferred #9delp12-pter was stably retained in the
microcell hybrid clone (SCaBER#9q; Fig. 3B). Microcell hybrids with
introduced chromosome 4 (SCaBER#4) were used as a control. The
presence of transferred chromosome 4 in SCaBER#4 was confirmed by
FISH analysis using a PR11-q4C13 BAC probe containing the 4q22.1
genomic DNA region. The parental SCaBER cells had two copies of
chromosome 4, whereas SCaBER#4 microcell hybrid clones had three
copies of chromosome 4; this confirmed the presence of the
transferred chromosome (Fig. 3C and
D).
The morphological features of microcell hybrid
clones generated via introduction of a human chromosome were
microscopically examined. SCaBER#9q cells were flatter and larger
than parental SCaBER and SCaBER#4 cells (Fig. 3E). Thus, human chromosome 9q may
have carried genes that regulated this transformed phenotype in
SCaBER cells.
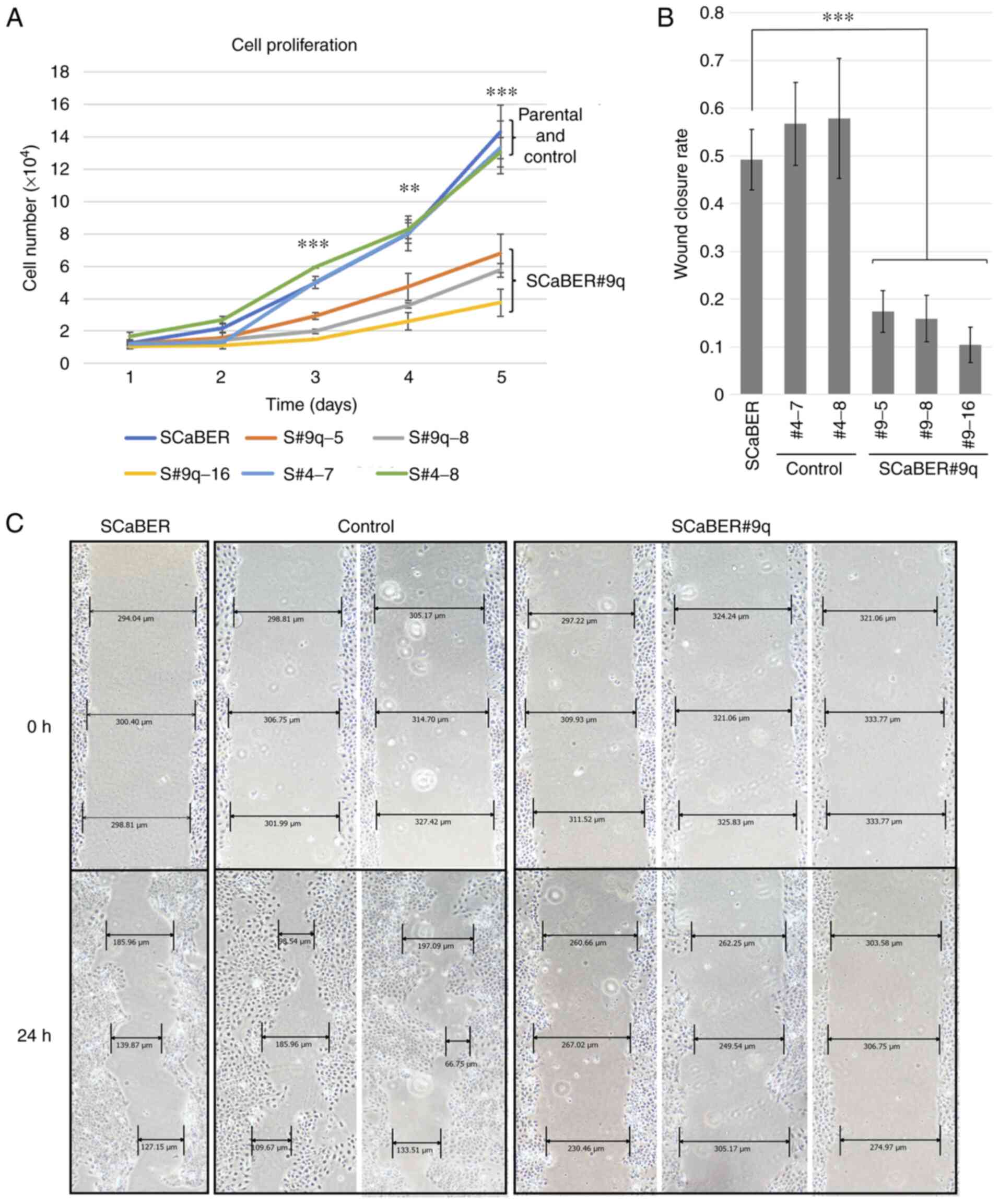
Effect of chromosome 9q introduction
on cell proliferation and migration ability
The proliferation rate of microcell hybrid clones
was examined to determine the effects of chromosome transfer on
cell proliferation. SCaBER#9q cells exhibited significantly
decreased proliferation compared with control SCaBER#4 and parental
cells at days 3-5 (Fig. 4A).
To evaluate the role of chromosome 9q in regulation
of malignant potential of basal BCa, wound healing assay was
performed using SCaBER microcell hybrid clones. Compared with
parental SCaBER and SCaBER#4 cells, SCaBER#9q cells showed
decreased mobility (Fig. 4B and
C). This indicated that tumor suppressor genes involved in cell
proliferation and migration in SCaBER cells were present on 9q.
Expression profile analysis of luminal
markers
The luminal subtype of BCa has a lower proliferative
and migratory potential than the basal subtype and is less
malignant (8). GATA3, FOXA1, and
PPARγ are specifically upregulated in the luminal subtype (as
previously determined by RNA sequencing analysis) (21). Furthermore, overexpression of GATA3
and FOXA1, which are tumor suppressive, is accompanied by
activation of PPARγ in transformation of basal into luminal BCa
cells (22–26). Additionally, LOH on the long arm of
chromosome 9 is commonly observed in premalignant lesions of
bladder carcinogenesis, suggesting that chromosome 9q encodes tumor
suppressor genes that serve a key role in development and
progression of BCa. The expression levels of luminal markers
(FOXA1, GATA3 and PPARG) was examined; compared with SCaBER cells,
SCaBER#9q cells exhibited a 2.4-, 1.9- and 5.2-fold increase in the
expression of the aforementioned luminal markers (Fig. 5A-C).
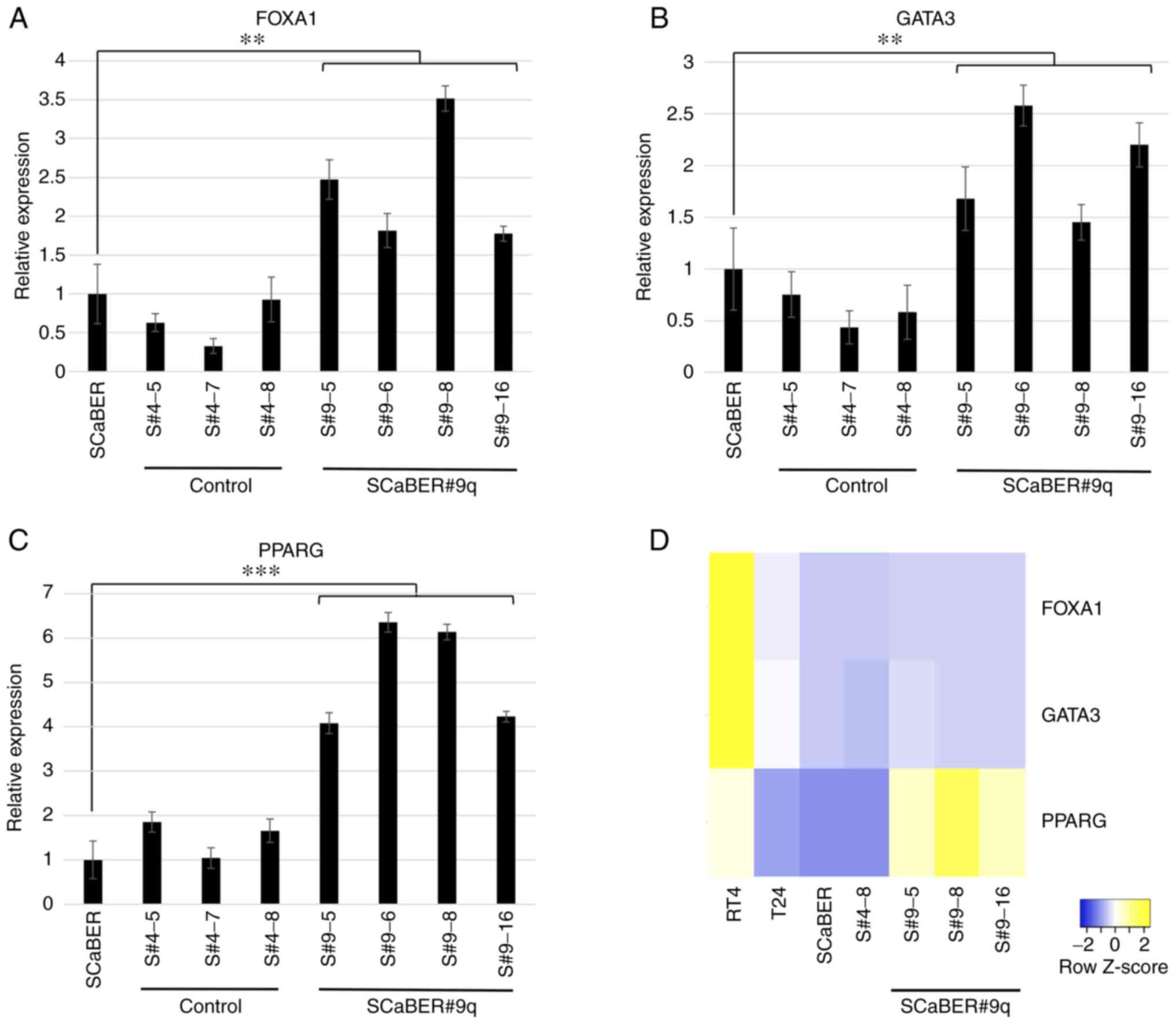 | Figure 5.Reverse transcription-quantitative
PCR of the luminal markers FOXA1, GATA3 and PPARG from cells
transferred with 9q and control cells. Human bladder cancer cells
transferred with long arm of chromosome 9 (SCaBER#9q) showed 2.4-,
1.9-, and 5.2-fold increase in (A) FOXA1, (B) GATA3 and (C) PPARG
expression, respectively, compared with SCaBER and SCaBER#4 cells.
Data are presented as the mean ± SEM of triplicate experiments.
**P<0.01, ***P<0.001. (D) Heatmap of expression levels of
FOXA1, GATA3 and PPARG compared with other cell lines (RT4,
luminal; T24, non-type; 5637, basal). Gene expression levels were
normalized against GAPDH mRNA using the 2−ΔΔCq method.
FOXA1, forkhead box A1. |
A heatmap of differential molecular subtypes was
used to evaluate the relative expression of luminal markers
(Fig. 5D). The relative expression
level of luminal markers was high in RT4 cells (typical luminal
subtype), whereas SCaBER (basal) and T24 cells (non-type) exhibited
low expression levels (27). Only
expression of PPARG was higher in SCaBER#9q cells than in parental,
SCaBER#4 and RT4 cells (Fig. 5D).
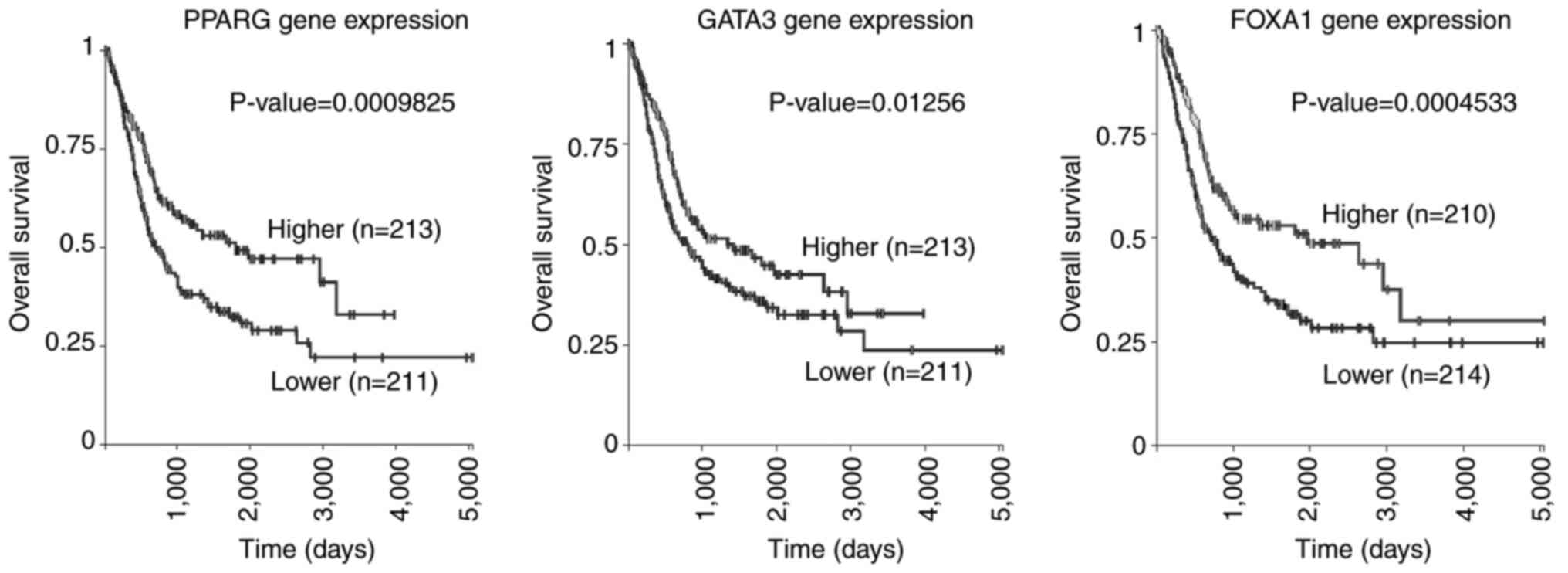
The expression profiles of three luminal marker genes in BCa was
further investigated using BLCA dataset. Higher expression of all
luminal marker genes was associated with improved survival in
patients with BCa (Fig. 6). These
results suggested that tumor suppressor genes on 9q may serve an
important role in determining the molecular subtype of BCa, which
is associated with development of malignancy.
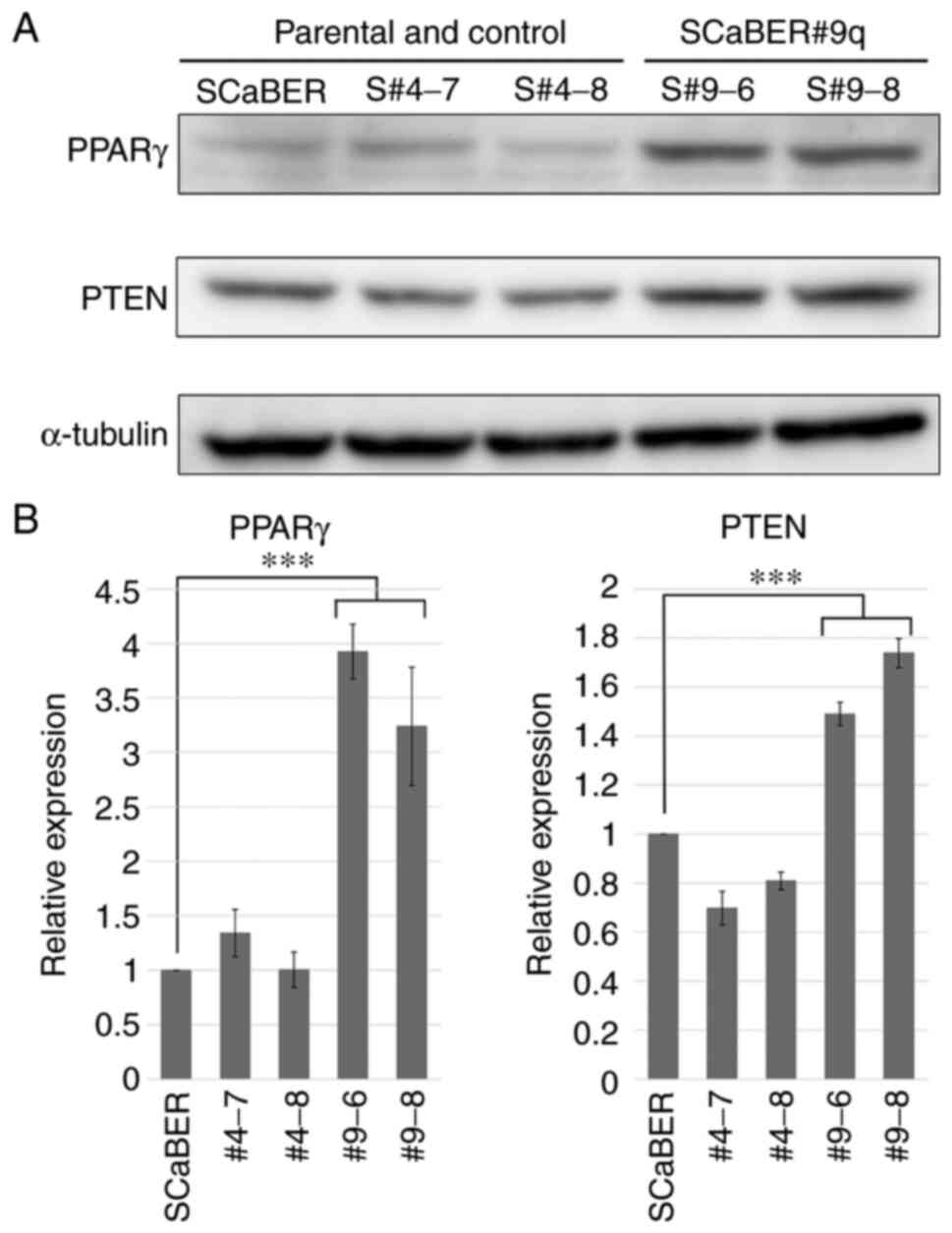
Analysis of protein expression of
luminal markers
Western blotting was performed to confirm that
expression of luminal markers (GATA3, FOXA1. and PPARG) was also
increased at the protein level. Only PPARγ was significantly
increased in SCaBER#9q cells (3.0-4.4-fold) compared with parental
cells (Figs. 7A and B and S1). PPARγ inhibits proliferation,
metastasis and invasion of cancer by induction of PTEN expression
(28). The expression levels of
PTEN were also increased in SCaBER#9q cells (Fig. 7A and B). This suggested that
chromosome 9q carried genes that regulate luminal marker of
BCa.
Discussion
BCa drug treatment has been based on cisplatin-based
chemotherapy for 30 years but the development of immunotherapy
(pembrolizumab) using immune checkpoint inhibitors has presented a
change for BCa treatment. However, pembrolizumab only has a 21.1%
response and 7.0% complete response rate (29). Therefore, there is a need for
precision medicine, identification of biomarkers that can be used
as predictors of therapeutic efficacy and development of novel
therapies. Greater understanding of the molecular mechanisms
underlying the development and progression of BCa is therefore key.
Identification of novel tumor suppressor genes involved in
progression of BCa may clarify the mechanism of its development and
lead to identification of new therapeutic targets. The present
study showed that malignant phenotypes, such as cell proliferation
and migration, are suppressed in the SCaBER human high-grade basal
BCa cell line following introduction of the long arm of human
chromosome 9, resulting in enhanced expression of the luminal
marker PPARγ. This suggested that chromosome 9q carried genes that
directly or indirectly regulated the PPARG luminal gene in SCaBER
cells.
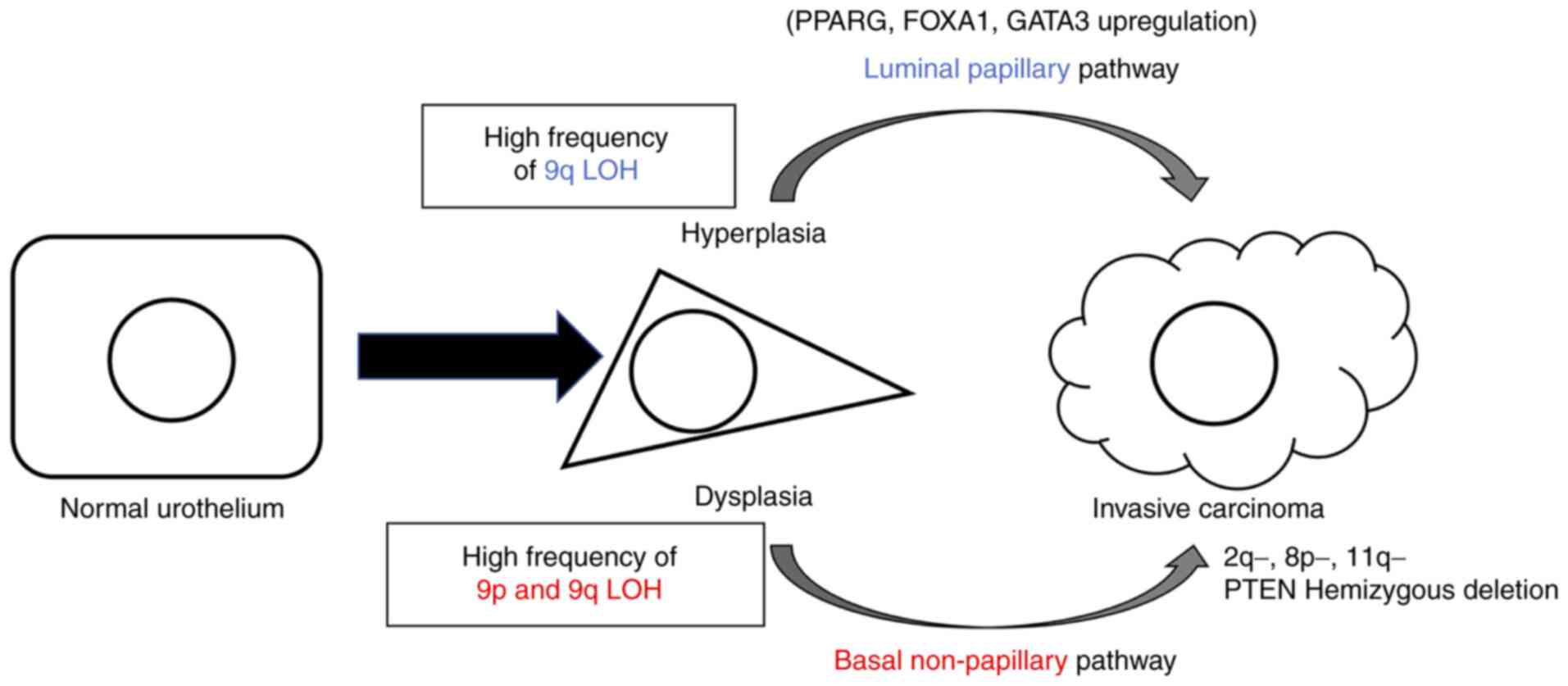
TCGA, MD Anderson Cancer Center and other research
groups have reported that urothelial carcinoma can be classified
into basal and luminal subtypes by gene expression profiling
(30,31). Based on this classification and
pathological characteristics, two pathways of carcinogenesis have
been proposed: Papillary/luminal pathway, which leads to
hyperplasia, papillary non-invasive and invasive carcinoma, and the
non-papillary/basal pathway, which leads to flat dysplasia,
carcinoma in situ and non-papillary invasive carcinoma
(Fig. 8) (8). GATA3, FOXA1, and PPARγ are
upregulated in the luminal pathway and overexpression of GATA3 and
FOXA1 and PPARγ activation drive transdifferentiation from the
basal to luminal phenotype (23).
In the present study, transcription levels of PPARG, FOXA1 and
GATA3 were upregulated in SCaBER#9q cells compared with parental
and SCaBER#4 cells. However at the protein level, only PPARγ was
upregulated in SCaBER#9q cells. Translation is affected by various
regulatory factors, such as the cap-binding protein eIF4E and
microRNAs (32,33). Therefore, the discrepancy between
mRNA and protein expression of luminal genes in SCaBER#9q cells may
be because of specific regulatory factors of translation.
PPARs, which are members of the nuclear receptor
superfamily, can be divided into three subtypes: PPARα, β and γ
(34). Previous studies have shown
that PPARγ serves a key role in occurrence and progression of BCa
via regulation of proliferation, apoptosis, metastasis, reactive
oxygen species and lipid metabolism (26,35–38).
High expression of PPARγ indicates better prognosis for patients
with more differentiated, non-invasive tumors with low
proliferative potential (39).
Aberration of chromosome 9, including deletions and
LOH, is frequently observed in both non-muscle and muscle invasive
BCa (>50%). In particular, p16, a tumor suppressor gene located
on chromosome 9p, plays a key role in the progression of non-muscle
invasive BCa (40–44) but the functional role of novel
suppressor genes on 9q in BCa remains unclear. In the present
study, expression levels of PPARγ and PTEN were significantly
increased by introduction of 9q to basal BCa cells (SCaBER),
suggesting a transformation from poorly differentiated to more
highly differentiated, low-grade BCa. The proliferative and
migratory ability of SCaBER#9q cells also decreased. These results
support a previous study showing that PPARγ inhibits proliferation,
metastasis and invasion of cancer by inducing PTEN expression
(28). However, there are
conflicting reports on the association between PPARγ and PTEN in
BCa (17,45). The present findings may provide
insight into PTEN regulatory pathways.
In conclusion, the present study provided evidence
that the long arm of human chromosome 9 contained genes that
regulate PPARγ. We previously identified paired-like homeodomain 1
as a novel tumor suppressor gene on human chromosome 5 that
regulates telomerase activity chromosome transfer and gene
expression profiling analysis (46). Future identification and
characterization of putative PPARγ regulatory genes on 9q should
facilitate understanding of the molecular mechanisms involved in
the development of BCa.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by Japan Society for the
Promotion of Science KAKENHI (grant no. 21K09422).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
TYa, TO and HK designed the experiments and analyzed
the data. TYa, TO and RS performed experiments. TO and HK wrote the
manuscript. TYu, NY, HI, SM, KH and MH analyzed data. TO and HK
confirm the authenticity of all the raw data. HK and AT conceived
and supervised the project. All authors revised and edited the
manuscript. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kakizoe T, Tobisu K, Takai K, Tanaka Y,
Kishi K and Teshima S: Relationship between papillary and nodular
transitional cell carcinoma in the human urinary bladder. Cancer
Res. 48:2299–2303. 1988.PubMed/NCBI
|
|
3
|
Luis NM, López-Knowles E and Real FX:
Molecular biology of bladder cancer. Clin Transl Oncol. 9:5–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fadl-Elmula I: Chromosomal changes in
uroepithelial carcinomas. Cell Chromosome. 4:12005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lindgren D, Sjödahl G, Lauss M, Staaf J,
Chebil G, Lövgren K, Gudjonsson S, Liedberg F, Patschan O, Månsson
W, et al: Integrated genomic and gene expression profiling
identifies two major genomic circuits in urothelial carcinoma. PLoS
One. 7:e388632012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nishiyama N, Arai E, Nagashio R, Fujimoto
H, Hosoda F, Shibata T, Tsukamoto T, Yokoi S, Imoto I, Inazawa J
and Kanai Y: Copy number alterations in urothelial carcinomas:
Their clinicopathological significance and correlation with DNA
methylation alterations. Carcinogenesis. 32:462–469. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hurst CD, Platt FM, Taylor CF and Knowles
MA: Novel tumor subgroups of urothelial carcinoma of the bladder
defined by integrated genomic analysis. Clin Cancer Res.
18:5865–5877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Czerniak B, Dinney C and McConkey D:
Origins of bladder cancer. Annu Rev Pathol. 11:149–174. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hartmann A, Schlake G, Zaak D, Hungerhuber
E, Hofstetter A, Hofstaedter F and Knuechel R: Occurrence of
chromosome 9 and p53 alterations in multifocal dysplasia and
carcinoma in situ of human urinary bladder. Cancer Res. 62:809–818.
2002.PubMed/NCBI
|
|
10
|
Hopman AH, Moesker O, Smeets AW, Pauwels
RP, Vooijs GP and Ramaekers FC: Numerical chromosome 1, 7, 9, and
11 aberrations in bladder cancer detected by in situ hybridization.
Cancer Res. 51:644–651. 1991.PubMed/NCBI
|
|
11
|
Platt FM, Hurst CD, Taylor CF, Gregory WM,
Harnden P and Knowles MA: Spectrum of phosphatidylinositol 3-kinase
pathway gene alterations in bladder cancer. Clin Cancer Res.
15:6008–6017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sjödahl G, Lauss M, Gudjonsson S, Liedberg
F, Halldén C, Chebil G, Månsson W, Höglund M and Lindgren D: A
systematic study of gene mutations in urothelial carcinoma;
inactivating mutations in TSC2 and PIK3R1. PLoS One. 6:e185832011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rampias T, Vgenopoulou P, Avgeris M,
Polyzos A, Stravodimos K, Valavanis C, Scorilas A and Klinakis A: A
new tumor suppressor role for the Notch pathway in bladder cancer.
Nat Med. 20:1199–1205. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kugoh H, Ohira T and Oshimura M: Studies
of tumor suppressor genes via chromosome engineering. Cancers
(Basel). 8:42015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Uejima H, Mitsuya K, Kugoh H, Horikawa I
and Oshimura M: Normal human chromosome 2 induces cellular
senescence in the human cervical carcinoma cell line SiHa. Genes
Chromosomes Cancer. 14:120–127. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Zhang Y, Yu C, Zhang P, Gu S, Wang
G, Xiao H and Li S: Bergenin inhibits bladder cancer progression
via activating the PPARγ/PTEN/Akt signal pathway. Drug Dev Res.
82:278–286. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chin L, Hahn WC, Getz G and Meyerson M:
Making sense of cancer genomic data. Genes Dev. 25:534–555. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grossman RL, Heath AP, Ferretti V, Varmus
HE, Lowy DR, Kibbe WA and Staudt LM: Toward a shared vision for
cancer genomic data. N Engl J Med. 375:1109–1112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kugoh H, Mitsuya K, Meguro M, Shigenami K,
Schulz TC and Oshimura M: Mouse A9 cells containing single human
chromosomes for analysis of genomic imprinting. DNA Res. 6:165–172.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eriksson P, Aine M, Veerla S, Liedberg F,
Sjödahl G and Höglund M: Molecular subtypes of urothelial carcinoma
are defined by specific gene regulatory systems. BMC Med Genomics.
8:252015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hustler A, Eardley I, Hinley J, Pearson J,
Wezel F, Radvanyi F, Baker SC and Southgate J: Differential
transcription factor expression by human epithelial cells of buccal
and urothelial derivation. Exp Cell Res. 369:284–294. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Warrick JI, Walter V, Yamashita H, Chung
E, Shuman L, Amponsa VO, Zheng Z, Chan W, Whitcomb TL, Yue F, et
al: FOXA1, GATA3 and PPARγ cooperate to drive luminal subtype in
bladder cancer: A molecular analysis of established human cell
lines. Sci Rep. 6:385312016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Osei-Amponsa V, Buckwalter JM, Shuman L,
Zheng Z, Yamashita H, Walter V, Wildermuth T, Ellis-Mohl J, Liu C,
Warrick JI, et al: Hypermethylation of FOXA1 and allelic loss of
PTEN drive squamous differentiation and promote heterogeneity in
bladder cancer. Oncogene. 39:1302–1317. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Ishiguro H, Kawahara T, Kashiwagi E,
Izumi K and Miyamoto H: Loss of GATA3 in bladder cancer promotes
cell migration and invasion. Cancer Biol Ther. 15:428–435. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng S, Qian K, Wang Y, Wang G, Liu X,
Xiao Y and Wang X: PPARγ inhibition regulates the cell cycle,
proliferation and motility of bladder cancer cells. J Cell Mol Med.
23:3724–3736. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hau AM, Nakasaki M, Nakashima K, Krish G
and Hansel DE: Differential mTOR pathway profiles in bladder cancer
cell line subtypes to predict sensitivity to mTOR inhibition. Urol
Oncol. 35:593–599. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin MS, Huang JX, Chen WC, Zhang BF, Fang
J, Zhou Q, Hu Y and Gao HJ: Expression of PPARγ and PTEN in human
colorectal cancer: An immunohistochemical study using tissue
microarray methodology. Oncol Lett. 2:1219–1224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bellmunt J, de Wit R, Vaughn DJ, Fradet Y,
Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK,
et al: Pembrolizumab as second-line therapy for advanced urothelial
carcinoma. N Engl J Med. 376:1015–1026. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Choi W, Czerniak B, Ochoa A, Su X,
Siefker-Radtke A, Dinney C and McConkey DJ: Intrinsic basal and
luminal subtypes of muscle-invasive bladder cancer. Nat Rev Urol.
11:400–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Robertson AG, Kim J, Al-Ahmadie H,
Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA,
Akbani R, et al: Comprehensive molecular characterization of
muscle-invasive bladder cancer. Cell. 171:540–556.e25. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sonenberg N and Hinnebusch AG: New modes
of translational control in development, behavior, and disease. Mol
Cell. 28:721–729. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou Y, Liang H, Liao Z, Wang Y, Hu X,
Chen X, Xu L and Hu Z: miR-203 enhances let-7 biogenesis by
targeting LIN28B to suppress tumor growth in lung cancer. Sci Rep.
7:426802017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lemberger T, Braissant O, Juge-Aubry C,
Keller H, Saladin R, Staels B, Auwerx J, Burger AG, Meier CA and
Wahli W: PPAR tissue distribution and interactions with other
hormone-signaling pathways. Ann N Y Acad Sci. 804:231–251. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cheng S, Wang G, Wang Y, Cai L, Qian K, Ju
L, Liu X, Xiao Y and Wang X: Fatty acid oxidation inhibitor
etomoxir suppresses tumor progression and induces cell cycle arrest
via PPARγ-mediated pathway in bladder cancer. Clin Sci (Lond).
133:1745–1758. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cao R, Wang G, Qian K, Chen L, Ju L, Qian
G, Wu CL, Dan HC, Jiang W, Wu M, et al: TM4SF1 regulates apoptosis,
cell cycle and ROS metabolism via the PPARγ-SIRT1 feedback loop in
human bladder cancer cells. Cancer Lett. 414:278–293. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cao R, Wang G, Qian K, Chen L, Qian G, Xie
C, Dan HC, Jiang W, Wu M, Wu CL, et al: Silencing of HJURP induces
dysregulation of cell cycle and ROS metabolism in bladder cancer
cells via PPARγ-SIRT1 feedback loop. J Cancer. 8:2282–2295. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang G, Cao R, Wang Y, Qian G, Dan HC,
Jiang W, Ju L, Wu M, Xiao Y and Wang X: Simvastatin induces cell
cycle arrest and inhibits proliferation of bladder cancer cells via
PPARγ signalling pathway. Sci Rep. 6:357832016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mylona E, Giannopoulou I, Diamantopoulou
K, Bakarakos P, Nomikos A, Zervas A and Nakopoulou L: Peroxisome
proliferator-activated receptor gamma expression in urothelial
carcinomas of the bladder: Association with differentiation,
proliferation and clinical outcome. Eur J Surg Oncol. 35:197–201.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cairns P, Mao L, Merlo A, Lee DJ, Schwab
D, Eby Y, Tokino K, van der Riet P, Blaugrund JE and Sidransky D:
Rates of p16 (MTS1) mutations in primary tumors with 9p loss.
Science. 265:415–417. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Williamson MP, Elder PA, Shaw ME, Devlin J
and Knowles MA: p16 (CDKN2) is a major deletion target at 9p21 in
bladder cancer. Hum Mol Genet. 4:1569–1577. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ploussard G, Dubosq F, Soliman H, Verine
J, Desgrandchamps F, De Thé H and Mongiat-Artus P: Prognostic value
of loss of heterozygosity at chromosome 9p in non-muscle-invasive
bladder cancer. Urology. 76:513.e13–e18. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Krüger S, Mahnken A, Kausch I and Feller
AC: P16 immunoreactivity is an independent predictor of tumor
progression in minimally invasive urothelial bladder carcinoma. Eur
Urol. 47:463–467. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bartoletti R, Cai T, Nesi G, Roberta
Girardi L, Baroni G and Dal Canto M: Loss of P16 expression and
chromosome 9p21 LOH in predicting outcome of patients affected by
superficial bladder cancer. J Surg Res. 143:422–427. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang Z, Xu H, Ji J, Shi X, Lyu J, Zhu Y,
Yu H and Wang F: Heterogeneity of PTEN and PPAR-γ in cancer and
their prognostic application to bladder cancer. Exp Ther Med.
18:3177–3183. 2019.PubMed/NCBI
|
|
46
|
Qi DL, Ohhira T, Fujisaki C, Inoue T, Ohta
T, Osaki M, Ohshiro E, Seko T, Aoki S, Oshimura M and Kugoh H:
Identification of PITX1 as a TERT suppressor gene located on human
chromosome 5. Mol Cell Biol. 31:1624–1636. 2011. View Article : Google Scholar : PubMed/NCBI
|