Introduction
Triple-negative breast cancer (TNBC) accounts for
approximately 10–20% of all breast cancers, and is negative for
estrogen receptor (ER), progesterone receptor (PR) and human
epidermal growth factor receptor-2 (HER-2) expression (1,2).
Compared with other types of breast cancer, TNBC is characterized
by an increased likelihood of development in younger individuals,
frequently higher grades, a more aggressive subtype and an
increased likelihood to metastasize to distant organs (3,4). For
the treatment of TNBC, due to high tumor heterogeneity and the lack
of specific therapeutic molecular targets, chemotherapy continues
to be the main method of systemic treatment at present (5). However, the majority of patients with
TNBC are not sensitive to chemotherapy, such that most relapse
following chemotherapy and have a poor prognosis (6,7).
Therefore, it remains urgent to reveal the underlying mechanisms of
TNBC progression and develop novel treatment strategies for this
disease.
Transforming growth factor-β (TGF-β) is a cytokine
that is a biologically active polypeptide and regulates the
proliferation, apoptosis, migration and differentiation of various
cancer cell types (8,9). There are currently three known
subtypes of TGF-β, namely TGF-β1, TGF-β2 and TGF-β3, detected in
humans (10). In terms of
physiological functions, TGF-β1 regulates the development of
mammary ducts and acini (11). In
breast cancer, TGF-β1 signaling has a dual effect on tumor growth
and metastasis (12,13). For instance, during the early
stages of tumor progression, TGF-β1 may inhibit this process by
inducing premature breast cancer stem cell aging (14). However, in the later stages, TGF-β1
can promote stem cell-like properties in breast cancer cells
(15). In addition, high levels of
TGF-β1 can enhance the invasive ability of cells and facilitate
tumor growth by inducing epithelial-mesenchymal transition (EMT),
interstitial blood vessel formation or by promoting evasion from
immune surveillance (16–19). Kim et al previously
demonstrated that downregulation of TGF-β1 significantly increased
the migratory ability of TNBC HCC1806 cells, and that patients with
high levels of TGF-β1 expression exhibited a poor prognosis
(20). In a TNBC mouse model,
treatment with TGF-β1-neutralizing antibodies or receptor
serine/threonine kinase inhibitors significantly inhibited the
development of lung and bone metastasis (21). These previous data suggest that
TGF-β1 may be involved in the progression of TNBC; thus, further
research should focus on uncovering the potential value of TGF-β1
in TNBC prognosis.
Survivin, also known as baculoviral IAP
repeat-containing protein 5, is a member of the
apoptosis-inhibiting protein family (22). Survivin can promote cell
proliferation by inhibiting caspase activation and stabilizing
microtubules during cell mitosis to protect cells from apoptosis
(23). Previous studies have found
that survivin is upregulated in a variety of human malignancies
(24–26) and is associated with poor prognosis
(27–30), leading to proposals of survivin
being used as a potential tumor marker and prognostic indicator.
However, the significance of survivin in breast cancer progression
remain controversial. Kennedy et al reported that survivin
is an independent predictor, whereby patients with high survivin
expression tend to have superior prognoses (31). In addition, Yamashita et al
(32) and Hinnis et al
(33) both demonstrated that
survivin is an independent predictor, such that high expression
levels of survivin are associated with poor prognosis. In contrast,
Chu et al (34) documented
that survivin is not an independent predictor or associated with
the recurrence of breast cancer. Notably, molecular characteristics
were not assigned based on ER/PR and HER-2 in the aforementioned
studies. In terms of TNBC, Yamanaka et al found that
inhibition of survivin expression can significantly suppress the
metastasis of TNBC MDA-MB-231 cells, while also significantly
prolonging the survival time of tumor-bearing mice (35). Furthermore, Shi et al
(36) found that survivin
expression was associated with the histological grade and TNM stage
of patients with TNBC, where the survival rate of patients with
survivin-positive TNBC was lower. Observations from these
aforementioned studies suggest that survivin may be involved in the
pathophysiological process of TNBC and therefore attention should
be paid to the potential value of applying survivin as a prognostic
indicator.
In the present study, tissue samples obtained from
patients with TNBC and non-TNBC were used, and the expression of
TGF-β1 and survivin was analyzed and compared between both types of
breast cancer by immunohistochemistry (IHC). The correlation among
the expression levels of these two proteins and the various
clinicopathological parameters was recorded and analyzed. In
addition, factors that can potentially affect prognosis were also
investigated. The aim of the present study was to clarify the
effects and significance of TGF-β1 and survivin protein expression,
either alone or in combination, in regards to the prognosis of TNBC
or non-TNBC.
Materials and methods
Patients and tissue specimens
A total of 142 breast cancer paraffin-embedded
specimens with complete clinicopathological and regular follow-up
data were collected between January 1, 2010 and January 1, 2013
from the Affiliated Hospital of Beihua University (Jilin, China).
The present study was approved by the Ethics Committee of the
Affiliated Hospital of Beihua University and informed consent was
obtained from each patient once the purpose and nature of the study
had been fully explained.
The inclusion criteria for patients in the present
study was as follows: i) all patients were female with TNBC or
non-TNBC, and were diagnosed for the first time; ii) complete
clinicopathological and routine follow-up data were available, and
iii) no chemotherapy, radiotherapy or other antitumor treatments
was performed prior to surgery. All patients underwent radical
mastectomy, and there was no distant metastasis at the time of
initial diagnosis. Post-operative chemotherapy was based on
anthracyclines and paclitaxel drugs for 6–8 cycles. Patients with
>3 axillary lymph node metastases received local radiotherapy.
Patients with non-TNBC were selected as a control group. Notably,
all patients in the control group exhibited no signs of distant
metastasis, and received parallel surgical resection, chemotherapy
and radiotherapy for axillary lymph node metastasis (as previously
described). Among the patients, 90 were diagnosed with TNBC and 52
with non-TNBC using IHC. The main characteristics of patients with
TNBC and non-TNBC are summarized in Table I. All patients were followed up
until December 2016. The median age of the patients with TNBC was
48 years, ranging from 28 to 84 years. In total, 59 cases were
defined as well and moderately differentiated, while 31 cases were
defined as poorly differentiated. Breast tumors were
histopathologically classified according to the WHO classification
(37). The median age of the
non-TNBC patients was 51.5 years, ranging from 38 to 70 years.
Among them, 35 cases were defined as well and moderately
differentiated, whereas 17 cases were categorized as poorly
differentiated. Overall survival (OS) was defined as the period of
time from surgical removal of the primary tumors to death or to the
last follow-up. The range of OS time in patients with TNBC and
non-TNBC in the present study was 10–70 and 15–65 months,
respectively. The median survival time was 36.75 and 39.50 months,
respectively. Progression-free survival (PFS) was defined as the
time from primary tumor resection to deterioration (recurrence or
metastasis) or death. TNBC or non-TNBC relapse and metastasis were
diagnosed by clinical examination, breast ultrasonography, axillary
and cervical lymph ultrasonography, chest computed tomography (CT),
epigastric magnetic resonance imaging (MRI) or MRI scans. The range
of PFS time of patients with TNBC and non-TNBC was 5–70 and 10–65
months, respectively. The median progression time was 25.25 and
35.50 months, respectively.
 | Table I.Clinicopathological characteristics
in patients with TNBC (n=90) and non-TNBC (n=52). |
Table I.
Clinicopathological characteristics
in patients with TNBC (n=90) and non-TNBC (n=52).
|
Characteristics | TNBC patients | Non-TNBC
patients |
|---|
| Age, years |
|
|
|
Median | 48.0 | 51.5 |
|
Range | 28-84 | 38-70 |
| Tumor size, n
(%) |
|
|
|
T2 | 27 (30.0) | 12 (23.1) |
|
T3 | 63 (70.0) | 40 (76.9) |
| Lymph node
metastasis, n (%) |
|
|
|
N0 | 40 (44.4) | 26 (50.0) |
|
NI | 11 (12.2) | 5 (9.6) |
|
N2 | 34 (37.8) | 20 (38.5) |
|
N3 | 5 (5.6) | 1 (1.9) |
| TNM classification,
n (%) |
|
|
| Stage
II | 44 (48.9) | 25 (48.1) |
| Stage
III | 46 (51.1) | 27 (51.9) |
| Histological grade,
n (%) |
|
|
|
Well | 1 (1.1) | 2 (3.8) |
|
Moderate | 58 (64.5) | 33 (63.5) |
|
Poor | 31 (34.4) | 17 (32.7) |
| ER status, n
(%) |
|
|
|
Positive | 0 (0.0) | 39 (75.0) |
|
Negative | 90 (100.0) | 13 (25.0) |
| PR status, n
(%) |
|
|
|
Positive | 0 (0.0) | 21 (40.4) |
|
Negative | 90 (100.0) | 31 (59.6) |
| HER-2 status, n
(%) |
|
|
|
Positive | 0 (0.0) | 17 (32.7) |
|
Negative | 90 (100.0) | 35 (67.3) |
| Ki67 index, n
(%) |
|
|
|
<14% | 0 (0.0) | 7 (13.5) |
|
≥14% | 90 (100.0) | 45 (86.5) |
| Treatment
modality-Curative resection, n (%) |
|
|
|
Yes | 90 (100.0) | 52 (100.0) |
| No | 0 (0.0) | 0 (0.0) |
| Treatment
modality-Postoperative chemotherapy, n (%) |
|
|
|
Yes | 90 (100.0) | 3 (5.8) |
| No | 0 (0.0) | 49 (94.2) |
| Treatment
modality-Postoperative radiotherapy, n (%) |
|
|
|
Yes | 47 (52.2) | 25 (48.1) |
| No | 43 (47.8) | 27 (51.9) |
IHC assay
The paraffin blocks of TNBC and non-TNBC tissues
were selected and sliced continuously to a thickness of 5 µm.
Pathological diagnosis was performed using light microscopy after
hematoxylin and eosin (H&E) staining (hematoxylin staining for
5 min, eosin staining for 20–30 sec, at room temperature). IHC
staining (conventional streptavidin peroxidase method) was then
used to detect the protein expression of TGF-β1 and survivin.
The staining protocol was performed as follows.
First, the paraffin-embedded tissues were dewaxed with xylene and
rehydrated with a descending series of ethanol (the concentration
of gradient ethanol was 100, 95 and 80% respectively). Then, the
slices were placed in a citric acid tissue antigen retrieval
solution (100X; cat. no. mvs-0101; Fuzhou Maixin Biotechnology Co.,
Ltd.), boiled at 95°C for 20 min and then cooled to room
temperature. An endogenous peroxidase blocker (cat. no. kit-9707;
Fuzhou Maixin Biotechnology Co., Ltd.) was added to block
endogenous peroxidase, which required incubation at room
temperature for 10 min to eliminate non-specific staining. The
slides were incubated with non-immunized goat serum (ready-to-use;
cat. no. kit-9707; Fuzhou Maixin Biotechnology Co., Ltd.) for 10
min at room temperature and the serum was removed. Subsequently,
rabbit monoclonal anti-TGF-β1 antibody (1:100; cat. no. ab215715;
Abcam) and rabbit monoclonal anti-survivin antibody (1:100; cat.
no. ab76424; Abcam) were added to the slices and incubated at 4°C
overnight. After washing the slides three times with PBS,
biotin-labeled goat anti-rabbit immunoglobulin (ready-to-use; cat.
no. kit-9707; Fuzhou Maixin Biotechnology Co., Ltd.) was added and
incubated for 10 min at room temperature. Then, in a wet box,
streptavidin-biotin protein peroxidase (cat. no. kit-9707; Fuzhou
Maixin Biotechnology Co., Ltd.) was added to the slices and
incubated at room temperature for 10 min. After washing the sample
with PBS, it was treated with 3,3′-diaminobenzidine (DAB; cat. no.
DAB-0031; Fuzhou Maixin Biotechnology Co., Ltd.) for 5 min at room
temperature and counterstained with hematoxylin at room temperature
for 1 min.
Normal rabbit serum (ready-to-use; cat. no. AR0010;
Wuhan Boster Biological Technology Co., Ltd.) was used as the
negative control instead of rabbit monoclonal anti-TGF-β1 or rabbit
monoclonal anti-survivin antibody. Breast cancer tissues previously
confirmed to express TGF-β1 or survivin protein were used as
positive controls and controls were used in each experiment. The
stained specimens were observed under an optical microscope at ×200
and ×400 magnifications.
IHC evaluation
Each slice was evaluated by using a blind reading
method. All IHC-stained sections were scored by at least three of
four independent and experienced pathologists (NL, CY, DQ and JZ)
who participated in the present study. They had no prior knowledge
of the clinicopathological parameters or of clinical outcomes of
the patients. In total, images taken from five high-power visual
fields (magnification, ×400) were randomly selected per section and
evaluated. The staining results were scored according to the
proportion of positive cells and staining intensity in each
section, where a semi-quantitative analysis was performed by
multiplying the staining intensity score by the positive cell rate
score, generating the immunoreactive score (IRS) (38). The percentage of positive cells was
assessed as follows: i) <10% was defined as 0 points; ii) 10–25%
was scored as 1 point; iii) 26–50% was defined as 2 points; iv)
51–75% was scored as 3 points; v) >75% was scored as 4 points
(39). Staining intensity was
evaluated as follows: i) 0 points for no staining or light yellow;
ii) 1 point for light brown; iii) 2 points for brown; iv) 3 points
for dark brown. In view of the fact that the expression of survivin
in both the cytoplasm and the nucleus is associated with cell
proliferation or survival (40),
Taubert et al also suggested that survivin expression in
both the cytoplasm and the nucleus should be considered together to
evaluate its impact on prognosis (41); therefore, we chose average
fractions of the cytoplasm and nucleus for the evaluation of
survivin expression.
If differential intensities were detected between
the cytoplasm and nucleus, we used the average fraction of the
cytoplasm and nucleus. The higher the IRS, the higher the protein
expression level, which was rated as follows: i) 0, no staining;
ii) 1–4, weak staining; iii) 5–8, moderate staining; iv) 9–12,
strong staining. An IRS of <1 was considered to indicate
negative staining for TGF-β1 or survivin.
Statistical analysis
SPSS 25.0 statistical software (IBM Corp.) was used
for statistical analysis and processing. Mann-Whitney U test was
used to compare the expression levels of TGF-β1 or survivin in TNBC
or non-TNBC tissues. Pearson correlation analysis was used to test
the correlation between TGF-β1 and survivin, while the association
between TGF-β1 or survivin expression and clinicopathological
parameters was tested by using a χ2 test or continuous
correction χ2 test. Kaplan-Meier (K-M) method with
log-rank test was used to construct the OS and PFS curves. Cox
regression was used for the univariate and multivariate analyses of
OS and PFS. P<0.05 was considered to indicate a statistically
significant difference.
Results
TGF-β1 and survivin expression in TNBC
and non-TNBC samples
Among the 90 cases of TNBC, 24 (26.7%) were tested
negative for TGF-β1 expression, whereas 66 (73.3%) had positive
TGF-β1 expression. By contrast, 19 (21.1%) tested negative for
survivin expression and 71 (78.9%) had positive expression
(Table II). Among the 52 cases of
non-TNBC, 23 (44.2%) were negative for TGF-β1 and 29 (55.8%) were
positive for TGF-β1. In addition, 14 (26.9%) were negative for
survivin and 38 (73.1%) were positive for surviving (Table III). Positive TGF-β1 protein
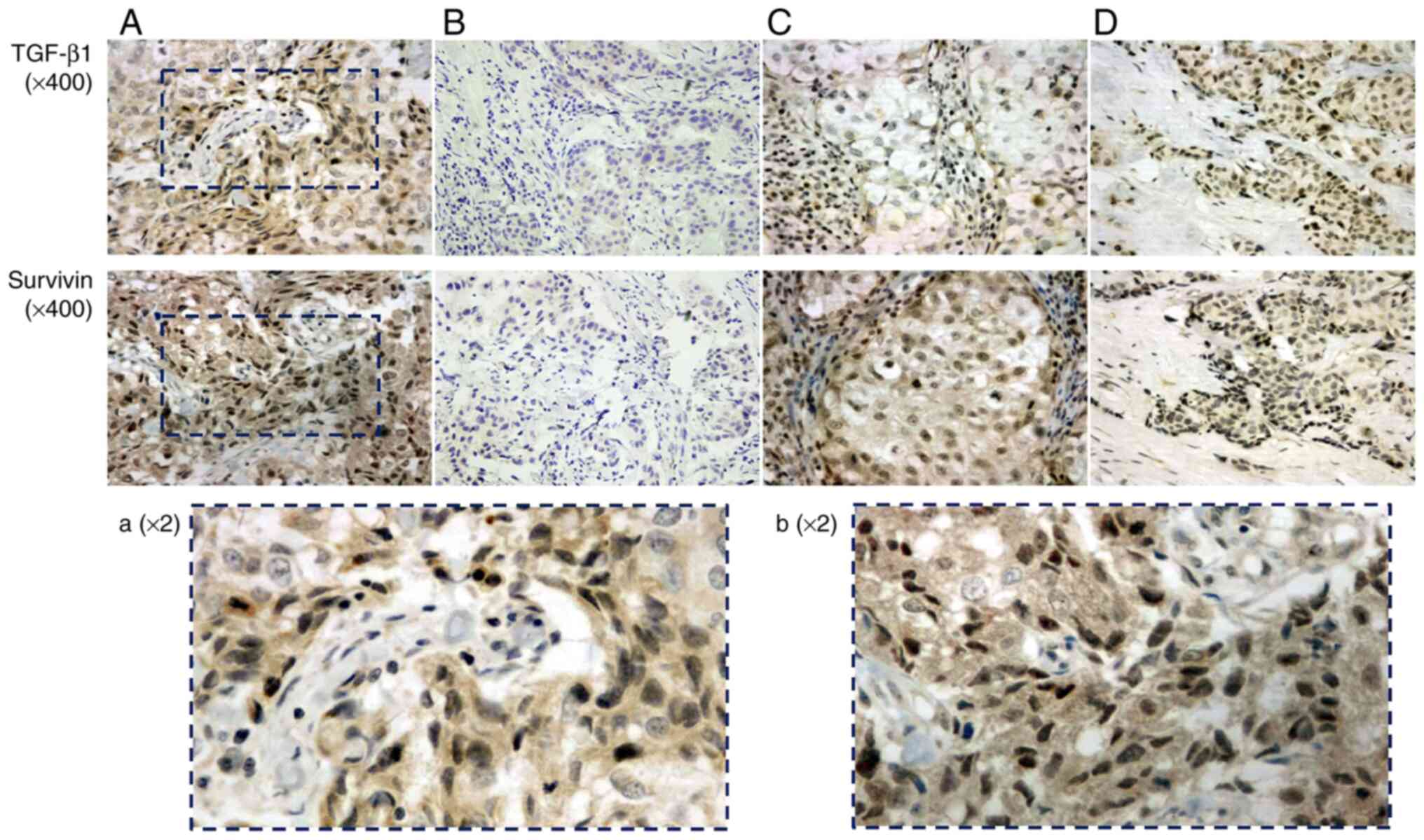
staining was mainly distributed in the cytoplasm, while positive
survivin protein staining was detected both in the nucleus and
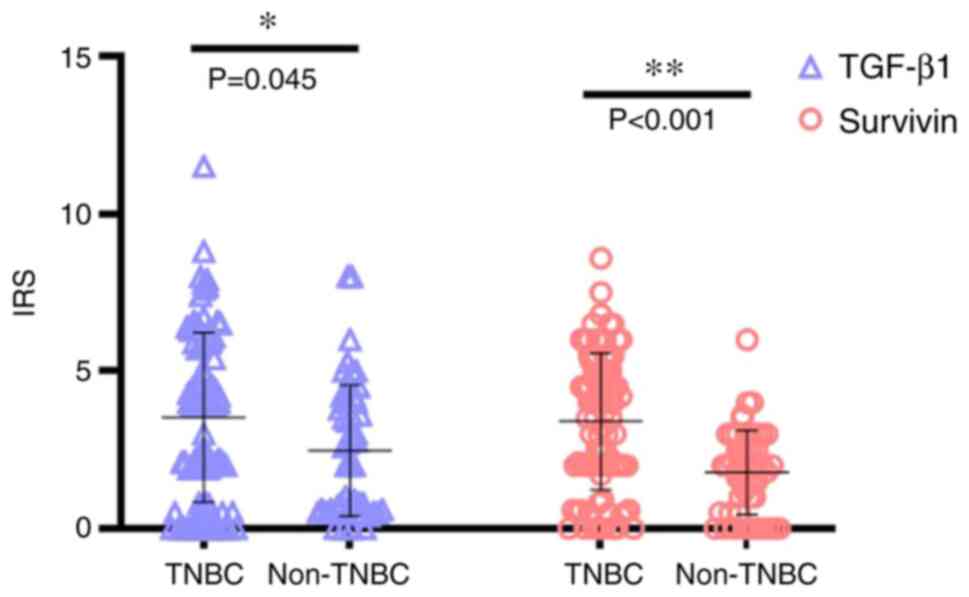
cytoplasm (Fig. 1). Comparing the
expression levels of these two proteins in TNBC and non-TNBC,
TGF-β1 expression levels were significantly higher in TNBC compared
with those in non-TNBC (z=−2.009; P=0.045). Similarly, the levels
of survivin protein expression in TNBC were also significantly
higher compared with those in non-TNBC (z=−4.417; P<0.001;
Fig. 2).
 | Table II.Association analysis of
clinicopathological parameters with TGF-β1 and survivin expression
in patients with TNBC (n=90). |
Table II.
Association analysis of
clinicopathological parameters with TGF-β1 and survivin expression
in patients with TNBC (n=90).
|
|
| TGF-β1 |
|
| Survivin |
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Variable | n | (−) | (+) | χ2 | P-value | (−) | (+) | χ2 | P-value |
|---|
| Total (TNBC) | 90 | 24 | 66 |
|
| 19 | 71 |
|
|
| Age, years |
|
|
|
|
|
|
|
|
|
|
≤48 | 51 | 14 | 37 | 0.037a | 0.847 | 10 | 41 | 0.160a | 0.689 |
|
>48 | 39 | 10 | 29 |
|
| 9 | 30 |
|
|
| Tumor size |
|
|
|
|
|
|
|
|
|
| T2 | 27 | 10 | 17 | 2.121a | 0.145 | 10 | 17 | 5.874a | 0.015b |
| T3 | 63 | 14 | 49 |
|
| 9 | 54 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
|
|
Positive | 50 | 10 | 40 | 2.557a | 0.110 | 8 | 42 | 1.765a | 0.184 |
|
Negative | 40 | 14 | 26 |
|
| 11 | 29 |
|
|
| TNM
classification |
|
|
|
|
|
|
|
|
|
| Stage
II | 44 | 15 | 29 | 2.426a | 0.119 | 12 | 32 | 1.962a | 0.161 |
| Stage
III | 46 | 9 | 37 |
|
| 7 | 39 |
|
|
| Tumor
differentiation |
|
|
|
|
|
|
|
|
|
|
Well/moderate | 59 | 16 | 43 | 0.018a | 0.894 | 12 | 47 | 0.061a | 0.804 |
|
Poor | 31 | 8 | 23 |
|
| 7 | 24 |
|
|
 | Table III.Association analysis of
clinicopathological parameters with TGF-β1 and survivin expression
in patients with non-TNBC (n=52). |
Table III.
Association analysis of
clinicopathological parameters with TGF-β1 and survivin expression
in patients with non-TNBC (n=52).
|
|
| TGF-β1 |
|
| Survivin |
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Variable | n | (−) | (+) | χ2 | P-value | (−) | (+) | χ2 | P-value |
|---|
| Total
(non-TNBC) | 52 | 23 | 29 |
|
| 14 | 38 |
|
|
| Age, years |
|
|
|
|
|
|
|
|
|
|
≤51.5 | 26 | 13 | 13 | 0.702a | 0.402 | 9 | 17 | 1.564a | 0.211 |
|
>51.5 | 26 | 10 | 16 |
|
| 5 | 21 |
|
|
| Tumor size |
|
|
|
|
|
|
|
|
|
| T2 | 12 | 5 | 7 | 0.042a | 0.838 | 4 | 8 | 0.040b | 0.842 |
| T3 | 40 | 18 | 22 |
|
| 10 | 30 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
|
|
|
Negative | 26 | 10 | 16 | 0.702a | 0.402 | 6 | 20 | 0.391a | 0.532 |
|
Positive | 26 | 13 | 13 |
|
| 8 | 18 |
|
|
| TNM
classification |
|
|
|
|
|
|
|
|
|
| Stage
II | 25 | 9 | 16 | 1.322a | 0.250 | 6 | 19 | 0.209a | 0.647 |
| Stage
III | 27 | 14 | 13 |
|
| 8 | 19 |
|
|
| Tumor
differentiation |
|
|
|
|
|
|
|
|
|
|
Well/moderate | 35 | 12 | 23 | 4.293a | 0.038c | 8 | 27 | 0.378b | 0.538 |
|
Poor | 17 | 11 | 6 |
|
| 6 | 11 |
|
|
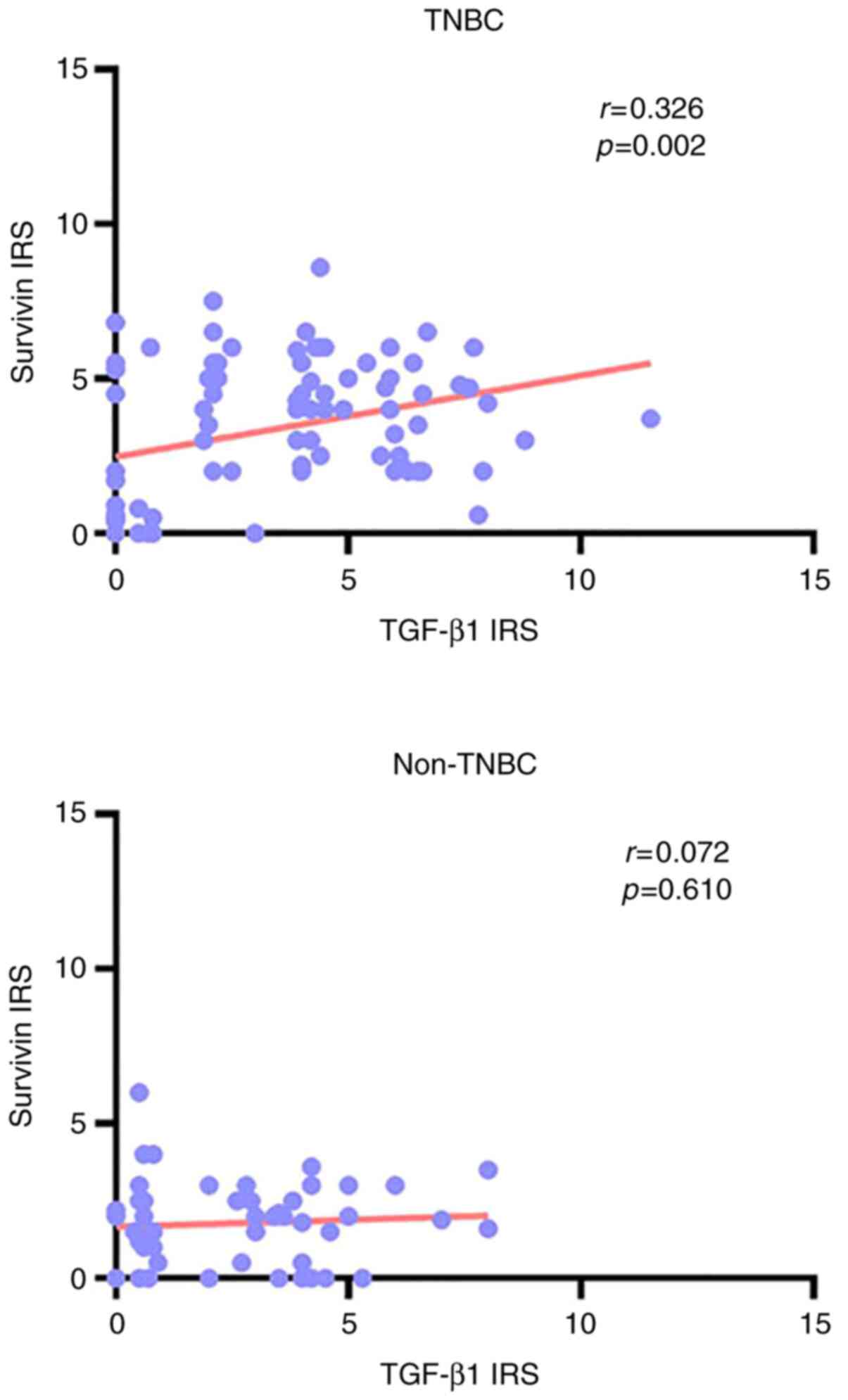
According to the expression levels of TGF-β1 and
survivin that were detected in each patient in this study (that is,
the IRS value of these two proteins in each patient), the
correlation between TGF-β1 and survivin in TNBC and non-TNBC
samples was determined using Pearson correlation analysis.
According to the Pearson correlation coefficient and the P-value,
the expression of TGF-β1 was found to be positively correlated with
that of survivin in TNBC (r=0.326; P=0.002), but no correlation was
identified between TGF-β1 or survivin expression in non-TNBC
(r=0.072; P=0.610; Fig. 3).
Association between TGF-β1 or survivin
expression and clinicopathological parameters
To study the effect of TGF-β1 or survivin on patient
prognosis, the potential association between TGF-β1 or survivin
expression and the traditional prognostic markers, including age,
tumor diameter, tumor histological grade and lymph node metastasis,
was analyzed. In TNBC, TGF-β1 protein expression was not found to
be associated with any of the clinicopathological parameters
(P>0.05; Table II), whereas
survivin protein expression was not associated with any of the
clinicopathological parameters (P>0.05; Table II) except for tumor diameter
(P=0.015; Table II). In the
non-TNBC cases, TGF-β1 protein expression was not associated with
any of the clinicopathological parameters (P>0.05; Table III), except for histological
tumor grade (P=0.038; Table
III), while survivin protein expression was not associated with
any of the clinicopathological parameters (P>0.05; Table III).
Association between TGF-β1 or survivin
expression levels and the survival of patients with TNBC and
non-TNBC
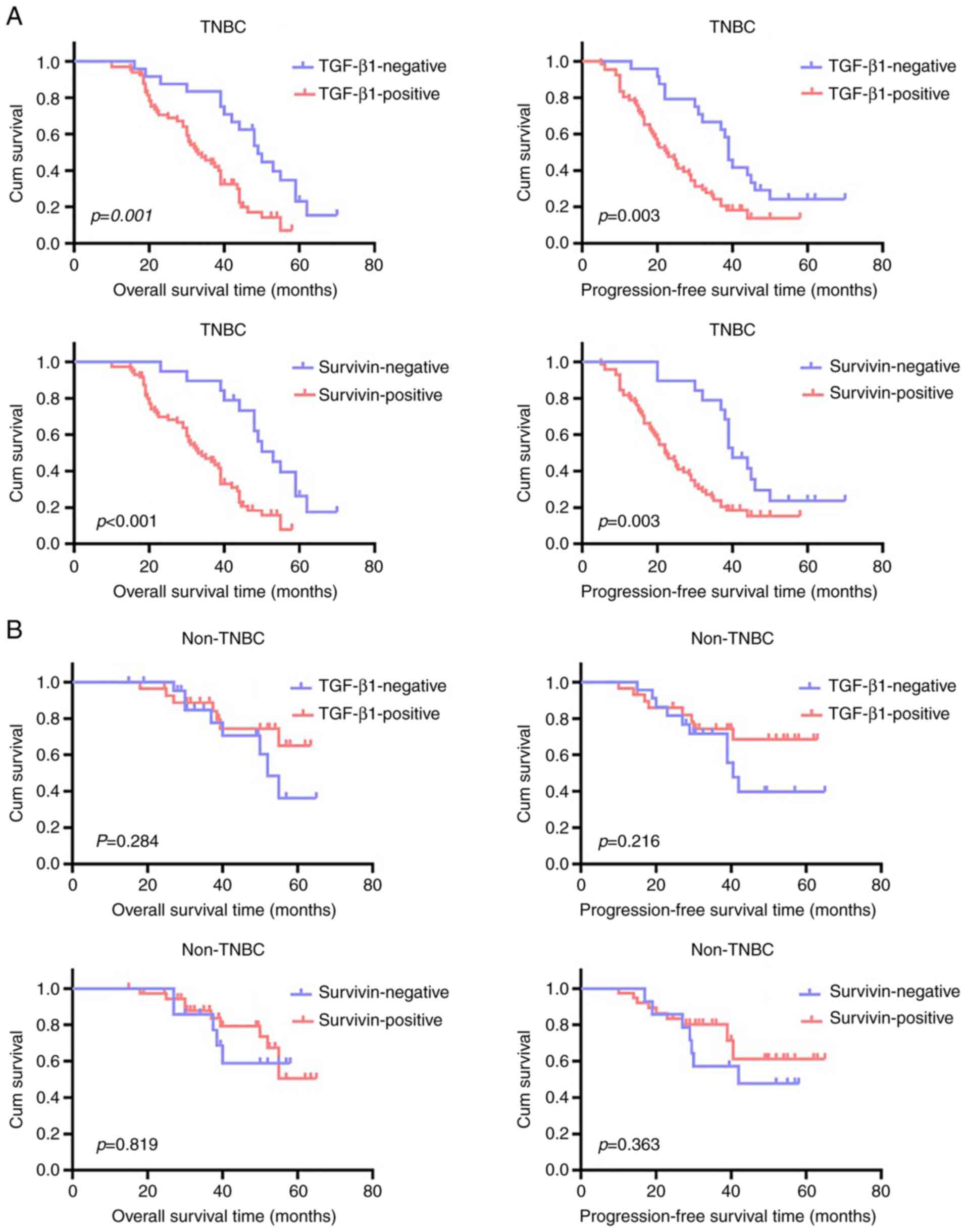
K-M analysis showed that the OS time (P=0.001) and
PFS time (P=0.003) of patients with TGF-β1-positive TNBC was
significantly shortened compared with that in patients with
TGF-β1-negative TNBC (Fig. 4A). In
addition, both the OS (P<0.001) and PFS (P=0.003) times of
patients with survivin-negative TNBC were significantly longer
compared with those of patients with survivin-positive TNBC
(Fig. 4A). By contrast, in
patients with non-TNBC, neither TGF-β1 nor survivin protein
expression levels conferred significant differences in OS time
(P=0.284 for TGF-β1 and P=0.819 for survivin; Fig. 4B) and PFS time (P=0.216 for TGF-β1
and P=0.363 for survivin; Fig.
4B).
Univariate and multivariate analysis
of the prognostic variables in patients with TNBC or non-TNBC
Univariate analysis revealed that in patients with
TNBC, poorly differentiated tumors, TGF-β1-positive and
survivin-positive expression significantly predicted a shorter OS
times (P=0.008, P=0.001 and P=0.001, respectively; Table IV) and increased risks of disease
progression (P=0.001, P=0.004 and P=0.004, respectively; Table IV). However, the status of tumor
differentiation, levels of TGF-β1 and survivin expression in
patients with non-TNBC were not found to predict OS time (P=0.195,
P=0.295 and P=0.821, respectively; Table IV) or PFS time (P=0.105, P=0.225
and P=0.370; respectively; Table
IV). Subsequently, since the levels of TGF-β1 protein
expression were found to be positively correlated with those of
survivin protein expression in patients with TNBC, the patients
were divided into four subgroups according to their expression
profiles of TGF-β1 and survivin: TGF-β1/survivin co-negative group,
TGF-β1/survivin co-positive group,
TGF-β1-negative/survivin-positive group and
TGF-β1-positive/survivin-negative group. Univariate analysis showed
that TGF-β1/survivin co-expression (co-negative vs. co-positive)
was significantly associated with OS time (P=0.001, Table IV) and PFS time (P=0.003, Table IV), while neither
TGF-β1-negative/survivin-positive expression (vs. TGF-β1/survivin
co-negative expression) nor TGF-β1-positive/survivin-negative
expression (vs. TGF-β1/survivin co-negative expression) were
associated with OS time (P=0.077 and P=0.417 Table IV) and PFS time (P=0.412 and
P=0.960 p>0.05; Table IV) in
patients with TNBC. In patients with non-TNBC, none of the four
subgroups were associated with OS time (P>0.05; Table IV) or PFS time (P>0.05;
Table IV). Other
clinicopathological parameters, including age, tumor size and lymph
node metastasis, were not found to be significantly associated with
the OS time (P>0.05; Table IV)
or PFS time (P>0.05; Table IV)
in both patients with TNBC and patients with non-TNBC. Multivariate
Cox regression analysis showed that in patients with TNBC,
expression of either TGF-β1 or survivin alone was not an
independent predictor of prognosis (P=0.572 and P=0.059,
respectively; Table V) in OS time
or progression of deterioration (P=0.365 and 0.126, respectively;
Table V) in PFS time. However, the
status of tumor differentiation remained to be a significant
independent predictor (P=0.003 and P<0.001, respectively;
Table V), whether in OS or PFS
time. After the comprehensive consideration of the TGF-β1/survivin
co-expression profile and tumor differentiation, both were found to
be independent predictors of OS (P=0.004 and P=0.002 respectively;
Table V) and PFS (P=0.006 and
P<0.001 respectively; Table V)
in patients with TNBC. In patients with non-TNBC, neither the tumor
differentiation status nor the expression profile of TGF-β1 and
survivin, either alone or in combination, were significant
predictors for OS (P>0.05; Table
V) and PFS (P>0.05; Table
V).
 | Table IV.Univariate analysis of
clinicopathological parameters for overall (OS) and
progression-free survival (PFS) in patients with TNBC and
non-TNBC. |
Table IV.
Univariate analysis of
clinicopathological parameters for overall (OS) and
progression-free survival (PFS) in patients with TNBC and
non-TNBC.
| A, OS |
|---|
|
|---|
| Variable | TNBC univariate Cox
analysis HR (95% CI) | P-value | Non-TNBC univariate
Cox analysis HR (95% CI) | P-value |
|---|
| Agea | 1.118
(0.689-1.814) | 0.652 | 1.370
(0.495-3.788) | 0.544 |
| Tumor
sizeb | 1.317
(0.784-2.211) | 0.298 | 1.509
(0.472-4.820) | 0.487 |
| Tumor
differentiationc | 1.950
(1.189-3.198) | 0.008d | 1.977
(0.705-5.544) | 0.195 |
| Lymph node
metastasise | 0.946
(0.583-1.537) | 0.824 | 1.641
(0.583-4.623) | 0.349 |
| TGF-β1e | 2.685
(1.467-4.913) | 0.001d | 0.579
(0.209-1.608) | 0.295 |
|
Survivine | 3.221
(1.635-6.348) | 0.001d | 0.883
(0.302-2.586) | 0.821 |
| TGF-β1/survivin
co-expression |
| 0.007d |
| 0.741 |
|
TGF-β1(−)/survivin(−) | 1 |
| 1 |
|
|
TGF-β1(−)/survivin(+) | 2.688
(0.899-8.035) | 0.077 | 0.986
(0.199-4.890) | 0.986 |
|
TGF-β1(+)/survivin(−) | 2.149
(0.269-17.182) | 0.417 | 0.711
(0.118-4.272) | 0.709 |
|
TGF-β1(+)/survivin(+) | 3.512
(1.720-7.169) | 0.001d | 0.500
(0.091-2.743) | 0.425 |
|
| B, PFS |
|
|
Variable | TNBC univariate
Cox analysis HR (95% CI) | P-value | Non-TNBC
univariate Cox analysis HR (95% CI) | P-value |
|
| Agea | 1.139
(0.711-1.827) | 0.588 | 1.104
(0.437-2.788) | 0.834 |
| Tumor
sizeb | 1.271
(0.761-2.122) | 0.360 | 1.054
(0.372-2.982) | 0.922 |
| Tumor
differentiationc | 2.256
(1.386-3.673) | 0.001d | 2.151
(0.851-5.435) | 0.105 |
| Lymph node
metastasise | 0.869
(0.543-1.391) | 0.558 | 1.780
(0.690-4.595) | 0.233 |
| TGF-β1e | 2.284
(1.309-3.983) | 0.004d | 0.561
(0.221-1.428) | 0.225 |
|
Survivine | 2.429
(1.332-4.431) | 0.004d | 0.648
(0.251-1.673) | 0.370 |
| TGF-β1/survivin
co-expression |
| 0.020d |
| 0.489 |
|
TGF-β1(−)/survivin (−) | 1 |
| 1 |
|
|
TGF-β1(−)/survivin (+) | 1.542
(0.548-4.344) | 0.412 | 0.736
(0.190-2.851) | 0.657 |
|
TGF-β1(+)/survivin (−) | 1.053
(0.137-8.099) | 0.960 | 0.682
(0.152-3.049) | 0.616 |
|
TGF-β1(+)/survivin (+) | 2.615
(1.393-4.910) | 0.003d | 0.334
(0.075-1.496) | 0.152 |
 | Table V.Multivariate analysis of
clinicopathological parameters for overall (OS) and
progression-free survival (PFS) in patients with TNBC and
non-TNBC. |
Table V.
Multivariate analysis of
clinicopathological parameters for overall (OS) and
progression-free survival (PFS) in patients with TNBC and
non-TNBC.
| A, OS-Comprehensive
consideration of tumor differentiation, TGF-β1 and survivin |
|---|
|
|---|
| Variable | TNBC Multivariate
Cox analysis HR (95% CI) | P-value | non-TNBC
Multivariate Cox analysis HR (95% CI) | P-value |
|---|
| Tumor
differentiation | 2.127
(1.289-3.510) | 0.003a | 1.708
(0.534-5.464) | 0.367 |
| TGF-β1 | 1.308
(0.516-3.321) | 0.572 | 0.726
(0.229-2.299) | 0.586 |
| Survivin | 2.782
(0.961-8.051) | 0.059 | 0.850
(0.289-2.503) | 0.769 |
|
| B,
OS-comprehensive consideration of tumor differentiation and
TGF-β1/survivin co-expression |
|
|
Variable | TNBC
Multivariate Cox analysis HR (95% CI) | P-value | non-TNBC
Multivariate Cox analysis HR (95% CI) | P-value |
|
| Tumor
differentiation | 2.197
(1.322-3.652) | 0.002a | 1.740
(0.543-5.570) | 0.351 |
| TGF-β/survivin
co-expression |
| 0.004a |
| 0.924 |
|
TGF-β1(−)/survivin(−) | 1 |
| 1 |
|
|
TGF-β1(−)/survivin(+) | 3.388
(1.116-10.290) | 0.031a | 1.060
(0.212-5.287) | 0.944 |
|
TGF-β1(+)/survivin(−) | 3.319
(0.404-27.297) | 0.264 | 0.971
(0.144-6.532) | 0.976 |
|
TGF-β1(+)/survivin(+) | 3.855
(1.876-7.922) |
<0.001a | 0.673
(0.111-4.093) | 0.667 |
|
| C,
PFS-Comprehensive consideration of tumor differentiation, TGF-β1
and survivin |
|
|
Variable | TNBC
Multivariate Cox analysis HR (95% CI) | P-value | non-TNBC
Multivariate Cox analysis HR (95% CI) | P-value |
|
| Tumor
differentiation | 2.656
(1.608-4.388) |
<0.001a | 1.768
(0.621-5.037) | 0.286 |
| TGF-β1 | 1.493
(0.627-3.556) | 0.365 | 0.690
(0.242-1.967) | 0.488 |
| Survivin | 2.099
(0.812-5.423) | 0.126 | 0.662
(0.253-1.733) | 0.401 |
|
| D,
PFS-comprehensive consideration of tumor differentiation and
TGF-β1/survivin co-expression |
|
|
Variable | TNBC
Multivariate Cox analysis HR (95% CI) | P-value | non-TNBC
Multivariate Cox analysis HR (95% CI) | P-value |
|
| Tumor
differentiation | 2.675
(1.606-4.455) |
<0.001a | 1.857
(0.651-5.293) | 0.247 |
| TGF-β/survivin
co-expression |
| 0.006a |
| 0.727 |
|
TGF-β1(−)/survivin(−) | 1 |
| 1 |
|
|
TGF-β1(−)/survivin(+) | 2.180
(0.756-6.291) | 0.149 | 0.885
(0.222-3.525) | 0.862 |
|
TGF-β1(+)/survivin(−) | 1.733
(0.22-13.644) | 0.602 | 1.010
(0.197-5.161) | 0.991 |
|
TGF-β1(+)/survivin(+) | 3.160
(1.663-6.007) |
<0.001a | 0.491
(0.097-2.498) | 0.392 |
Discussion
The present study found that the expression levels
of transforming growth factor-β1 (TGF-β1) protein in
triple-negative breast cancer (TNBC) tissues were significantly
higher compared with those in their non-TNBC counterparts. A
previous study performed in 1,881 breast cancer tissue samples by
Hachim et al also found that the expression level of TGF-β1
mRNA in patients with TNBC was significantly higher compared with
that in other types of breast cancer (42). In addition, Kim et al found
that TGF-β1 mRNA expression and the invasive abilities of TNBC
cells (MDA-MB-231, Hs578T and HCC1806) were significantly higher
compared with those of non-TNBC cells (BT474, ZR75-1 and SKBR3)
(43). The authors also revealed
that treatment with the dual selective TGF-β1 receptor (RI/RII)
inhibitor LY2109761 completely inhibited the invasiveness of the
TNBC cells (43). However, the
molecular mechanism underlying the differential expression profile
of TGF-β1 in TNBC and non-TNBC remains unclear. It has been found
that expression of the circular RNA ankyrin repeat and sterile α
motif domain containing 1b (circANKS1B) was closely correlated with
the invasion, metastasis and poor prognosis of breast cancer
(including all subtypes of breast cancer) (44); in addition, circANKS1B expression
was found to be low in non-TNBC tissues or cells (MCF-7), but was
significantly higher in TNBC tissues or cells (MDA-MB-231)
(44). Increased circANKS1B
expression was found to increase the expression of the
transcription factor upstream transcription factor 1, to upregulate
the expression of TGF-β1 by directly binding to the promoter. Thus,
TGF-β1/Smad signaling is activated to enhance
epithelial-mesenchymal transition (EMT) and metastasis (44). This may partially explain the
differential expression of TGF-β1 in TNBC and non-TNBC tissues
found in the present study. Compared with those in patients with
TGF-β1-positive TNBC, patients with TGF-β1-negative expression
exhibited longer overall survival (OS) and progression-free
survival (PFS) times, in addition to a better prognosis, although
multivariate Cox analysis revealed that TGF-β1 expression was not
an independent predictor in patients with TNBC, whether in OS or
PFS time. In addition, no significant difference was found in OS or
PFS between patients with TGF-β1-positive non-TNBC and patients
with TGF-β1-negative non-TNBC. This suggest that TGF-β1 expression
may be more beneficial in predicting the prognosis of patients with
TNBC, in which TGF-β1 may serve as a key signaling component.
Similarly, although survivin was also not observed to be an
independent predictor of OS and PFS in patients with TNBC, the
survival rate of patients with survivin-negative expression was
significantly higher compared with that with survivin-positive
expression. However, in patients with non-TNBC, survivin protein
expression was not associated with OS or PFS. Therefore, the
expression of survivin appeared to be of higher importance for the
malignant progression of TNBC instead of non-TNBC, where patients
with TNBC showing higher expression levels of survivin tended to
exhibit more severe malignancy, resulting in poorer prognoses.
Shi et al also drew a similar conclusion in a
previous study with TNBC, whereby survivin-positive patients tended
to have shorter OS and disease-free survival times (36), but Dogu et al and Jha et
al did not observe any effects exerted by survivin on the
prognosis of patients with TNBC (45,46).
These discrepancies may be attributed to the different antibodies
used, distinct sample groups or the cut-off criteria set prior to
evaluation. Although some studies found that survivin was a
prognostic marker of breast cancer (47,48),
further research is required with regards to its significance in
the progression of TNBC.
Another finding in the present study was that there
was a positive correlation between the expression levels of TGF-β1
and survivin in TNBC, but not in non-TNBC. Therefore, further study
on the significance of both TGF-β1 and survivin expression combined
with the prognosis of patients with TNBC and patients with non-TNBC
was performed. In this subsequent study, it was found that the
survival time of patients with TGF-β1/survivin co-positivity was
significantly shorter compared with that of patients who were
negative for both TGF-β1 and survivin expression in TNBC. This
suggests that the positive expression of TGF-β1 and survivin
combined can be used as an independent predictor of prognosis for
patients with TNBC. However, in non-TNBC, combined TGF-β1 and
survivin expression was not associated with the OS or PFS of the
patients, which may mean that the combined expression profile of
TGF-β1 and survivin can mediate different effects in TNBC and
non-TNBC, such that the combined positivity of TGF-β1 and survivin
can predict the prognosis in patients with TNBC, but not in
patients with non-TNBC.
Results of a previous study demonstrated that TGF-β1
was a negative regulator of survivin in a healthy prostatic
epithelial cell line and in malignant tumors during the early
stages of development (49).
TGF-β1 activates retinoblastoma (Rb) by inducing the low
phosphorylation of Rb, causing Rb to bind to E2F transcription
factor 4 to form a repressor complex on the survivin promoter.
Simultaneously, the TGF-β1/survivin regulatory axis remains intact
to ensure sensitivity to apoptotic signals (50). Thus, TGF-β1 inhibits the expression
of survivin in the non-TNBC MCF-7 cell line (51). Results of a further previous study
demonstrated that TGF-β1 could not inhibit survivin expression;
however, TGF-β1 may promote survivin expression in glioblastoma
(52). TGF-β can induce survivin
gene expression by activating the NF-κB subunit p65/RelA in mouse
4T1 TNBC cells (53), while in
human MDA-MB-231 TNBC cells, TGF-β1 can upregulate survivin
expression by activating the Wnt/β-catenin pathway (54,55).
Indeed, upregulation of Wnt/β-catenin signaling in TNBC compared
with that in non-TNBC and normal healthy tissues has been
frequently observed (56–58). This may be the reason underlying
the observation that the expression levels of survivin in TNBC were
higher compared with those in non-TNBC in the present study. The
regulatory relationship between TGF-β1 and survivin may be
bidirectional. Although the specific mechanism underlying this
phenomenon was not found, it may facilitate the understanding of
the reason behind the prognosis of patients positive for both
TGF-β1 and survivin expression being worse than that of patients
negative for both TGF-β1 and survivin expression in the present
study. In this patient subgroup, TGF-β1 may serve as the initiator
of survivin expression, which may then inhibit cell apoptosis by
promoting survivin expression and the proliferation of tumor cells.
This may also transform TGF-β1 from a tumor suppressor into a tumor
promoter. However, it should be noted that no associations between
combined TGF-β1 and survivin expression and prognosis were found in
patients with non-TNBC, suggesting that the TGF-β1/survivin
signaling pathway serves a greater role in the malignant
progression of TNBC.
In conclusion, the results of the present study
showed that for patients with TNBC, the prognosis of those that
tested negative for TGF-β1 or survivin expression was superior
compared with that in patients positive for TGF-β1 or survivin
expression. Although the expression levels of either TGF-β1 or
survivin alone could not be used as an independent predictor, there
was an interesting finding whereby the expression of TGF-β1 and
survivin was positively correlated in the tissue samples of
patients with TNBC. However, this was not found in the samples of
patients with non-TNBC. According to the results of the present
study, the combined levels of TGF-β1/survivin expression can be
used as an independent prognostic factor for patients with TNBC,
but not in their non-TNBC counterparts. Since TNBC is highly
malignant with no specific treatment options available, analysis of
TNBC and non-TNBC samples is of importance for the optimization of
clinical treatment regimens, which may help to avoid improper or
excessive treatment. Thus, further attention should be paid to the
combined expression levels of TGF-β1 and survivin in patients with
TNBC. Of course, the present study had its limitation as it
detected the expression of proteins in paraffin samples only by
immunohistochemistry, and future research will attempt to confirm
the results of the present study by analyzing TGF-β1 and survivin
mRNA and protein levels in fresh TNBC and non-TNBC samples.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Science and
Technology projects in Jilin Province Department of Education
(grant no. JJKH20210052KJ), and the Department of Science and
Technology of Jilin Province (grant no. YDZJ202101ZYTS090).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
NL and CY contributed to the study design. DQ, CY
and NL contributed to data analysis. NL, JZ and JJ contributed to
the collection of the tissue samples and patient data. NL and CY
wrote the manuscript. NL and CY confirm the authenticity of all the
raw data. All authors read and approved the manuscript and agree to
be accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work (including the data)
are appropriately investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Affiliated Hospital of Beihua University [grant no. (2018)10],
and informed consent was obtained from each patient once the
purpose and nature of the study had been fully explained.
Patient consent for publication
Informed consent was obtained from all patients
regarding the publication of the data and associated images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bianchini G, Balko JM, Mayer IA, Sanders
ME and Gianni L: Triple-negative breast cancer: Challenges and
opportunities of a heterogeneous disease. Nat Rev Clin Oncol.
13:674–690. 2016. View Article : Google Scholar
|
|
2
|
Marra A, Viale G and Curigliano G: Recent
advances in triple negative breast cancer: The immunotherapy era.
BMC Med. 17:902019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garrido-Castro A, Lin N and Polyak K:
Insights into molecular classifications of triple-negative breast
cancer: Improving patient selection for treatment. Cancer Discov.
9:176–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boyle P: Triple-negative breast cancer:
Epidemiological considerations and recommendations. Ann Oncol. 23
(Suppl 6):vi7–v12. 2012. View Article : Google Scholar
|
|
5
|
Zhang C, Han Y, Huang H, Min L, Qu L and
Shou C: Integrated analysis of expression profiling data identifies
three genes in correlation with poor prognosis of triple-negative
breast cancer. Int J Oncol. 44:2025–2033. 2014. View Article : Google Scholar
|
|
6
|
Azim HA Jr, Michiels S, Bedard PL, Singhal
SK, Criscitiello C, Ignatiadis M, Haibe-Kains B, Piccart MJ,
Sotiriou C and Loi S: Elucidating prognosis and biology of breast
cancer arising in young women using gene expression profiling. Clin
Cancer Res. 18:1341–1351. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abramson VG, Lehmann BD, Ballinger TJ and
Pietenpol JA: Subtyping of triple-negative breast cancer:
Implications for therapy. Cancer. 121:8–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jakowlew SB: Transforming growth
factor-beta in cancer and metastasis. Cancer Metastasis Rev.
25:435–457. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Elliott RL and Blobe GC: Role of
transforming growth factor beta in human cancer. J Clin Oncol.
23:2078–2093. 2005. View Article : Google Scholar
|
|
10
|
Kaminska B, Wesolowska A and Danilkiewicz
M: TGF beta signalling and its role in tumour pathogenesis. Acta
Biochim Pol. 52:329–337. 2005. View Article : Google Scholar
|
|
11
|
Moses H and Barcellos-Hoff M: TGF-beta
biology in mammary development and breast cancer. Cold Spring Harb
Perspect Biol. 3:a0032772011. View Article : Google Scholar
|
|
12
|
Suriyamurthy S, Baker D, Ten Dijke P and
Iyengar PV: Epigenetic reprogramming of TGF-β Signaling in breast
cancer. Cancers. 11:7262019. View Article : Google Scholar
|
|
13
|
Li CJ, Chu PY, Yiang GT and Wu MY: The
molecular mechanism of epithelial-mesenchymal transition for breast
carcinogenesis. Biomolecules. 9:4762019. View Article : Google Scholar
|
|
14
|
Siegel PM and Massagué J: Cytostatic and
apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev
Cancer. 3:807–821. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bhola NE, Balko JM, Dugger TC, Kuba MG,
Sánchez V, Sanders M, Stanford J, Cook RS and Arteaga CL: TGF-β
inhibition enhances chemotherapy action against triple-negative
breast cancer. J Clin Invest. 123:1348–1358. 2013. View Article : Google Scholar
|
|
16
|
Lee YJ, Park JH and Oh SM: Activation of
NF-κB by TOPK upregulates Snail/Slug expression in TGF-β1 signaling
to induce epithelial-mesenchymal transition and invasion of breast
cancer cells. Biochem Biophys Res Commun. 530:122–129. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Xu C, Liu X, Yang Y, Cao L, Xiang
G, Liu F, Wang S, Liu J, Meng Q, et al: TGF-β1 stimulates
epithelial-mesenchymal transition and cancer-associated
myoepithelial cell during the progression from in situ to invasive
breast cancer. Cancer Cell Int. 19:3432019. View Article : Google Scholar
|
|
18
|
Syed V: TGF-β signaling in cancer. J Cell
Biochem. 117:1279–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yeo HL, Fan TC, Lin RJ, Yu JC, Liao GS,
Chen ES, Ho MY, Lin WD, Chen K, Chen CH, et al: Sialylation of
vasorin by ST3Gal1 facilitates TGF-β1-mediated tumor angiogenesis
and progression. Int J Cancer. 144:1996–2007. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim S, Lee J, You D, Jeong Y, Jeon M, Yu
J, Kim SW, Nam SJ and Lee JE: Berberine suppresses cell motility
through downregulation of TGF-β1 in triple negative breast cancer
cells. Cell Physiol Biochem. 45:795–807. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tan AR, Alexe G and Reiss M: Transforming
growth factor-beta signaling: Emerging stem cell target in
metastatic breast cancer? Breast Cancer Res Treat. 115:453–495.
2009. View Article : Google Scholar
|
|
22
|
Srinivasula SM and Ashwell JD: IAPs:
What's in a name? Mol Cell. 30:123–135. 2008. View Article : Google Scholar
|
|
23
|
Singh M, Chaudhry P, Fabi F and Asselin E:
Cisplatin-induced caspase activation mediates PTEN cleavage in
ovarian cancer cells: A potential mechanism of chemoresistance. BMC
Cancer. 13:2332013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma J, Tian K, Du J, Wu Z, Wang L and Zhang
J: High expression of survivin independently correlates with tumor
progression and mortality in patients with skull base chordomas. J
Neurosurg. 132:140–149. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khan Z, Khan N, Tiwari RP, Patro IK,
Prasad GB and Bisen PS: Down-regulation of survivin by oxaliplatin
diminishes radioresistance of head and neck squamous carcinoma
cells. Radiother Oncol. 96:267–273. 2010. View Article : Google Scholar
|
|
26
|
Lorenzetti MA, Mosna MJ, De Matteo EN,
García Lombardi M, Colli SL and Preciado MV: Overexpression of
survivin in pediatric Hodgkin lymphoma tumor cells:
Characterization of protein expression and splice-variants
transcription profile. Exp Mol Pathol. 108:24–31. 2019. View Article : Google Scholar
|
|
27
|
Stavropoulos A, Varras M, Vasilakaki T,
Varra VK, Varra FN, Tsavari A, Nonni A, Kavantzas N and Lazaris AC:
Expression of anti-apoptotic protein survivin in human endometrial
carcinoma: Clinical and pathological associations as a separate
factor and in combination with concomitant PTEN and p53 expression.
Oncol Lett. 20:1033–1054. 2020. View Article : Google Scholar
|
|
28
|
Hanif A, Lee S, Gupta M, Chander A,
Kannisto ED, Punnanitinont A, Fenstermaker R, Ciesielski M, Attwood
K, Qiu J, et al: Exploring the role of survivin in neuroendocrine
neoplasms. Oncotarget. 11:2246–2258. 2020. View Article : Google Scholar
|
|
29
|
Kapiris I, Nastos K, Karakatsanis A,
Theodosopoulos T, Karandrea D, Kondi Pafiti A and Contis J:
Survivin expression in hepatocellular carcinoma. Correlation with
clinicopathological characteristics and overall survival. J BUON.
24:1934–1942. 2019.PubMed/NCBI
|
|
30
|
Veiga GLD, Silva RDMD, Pereira EC, Azzalis
LA, Alves BDCA, Gehrke FS, Gascón TM and Fonseca FLA: The role of
survivin as a biomarker and potential prognostic factor for breast
cancer. Rev Assoc Med Bras (1992). 65:893–901. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kennedy SM, O'Driscoll L, Purcell R,
Fitz-Simons N, McDermott EW, Hill AD, O'Higgins NJ, Parkinson M,
Linehan R and Clynes M: Prognostic importance of survivin in breast
cancer. Br J Cancer. 88:1077–1083. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamashita S, Masuda Y, Kurizaki T, Haga Y,
Murayama T, Ikei S, Kamei M, Takeno S and Kawahara K: Survivin
expression predicts early recurrence in early-stage breast cancer.
Anticancer Res. 27:2803–2808. 2007.PubMed/NCBI
|
|
33
|
Hinnis AR, Luckett JC and Walker RA:
Survivin is an independent predictor of short-term survival in poor
prognostic breast cancer patients. Br J Cancer. 96:639–645. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chu JS, Shew JY and Huang CS:
Immunohistochemical analysis of survivin expression in primary
breast cancers. J Formos Med Assoc. 103:925–931. 2004.
|
|
35
|
Yamanaka K, Nakata M, Kaneko N, Fushiki H,
Kita A, Nakahara T, Koutoku H and Sasamata M: YM155, a selective
survivin suppressant, inhibits tumor spread and prolongs survival
in a spontaneous metastatic model of human triple negative breast
cancer. Int J Oncol. 39:569–575. 2011.
|
|
36
|
Shi CT, Ma J, Shi QF, Zhang Y and Wang HN:
High survivin and low zinc finger of the cerebellum 1 expression
indicates poor prognosis in triple-negative breast carcinoma. J
Breast Cancer. 22:248–259. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lakhani SR EI, Schnitt SJ, Tan PH and van
de Vijver MJ: WHO classification of tumors of the breast. 4th
edtion. Lyon: IARC Press; 2012
|
|
38
|
Wang Q, Zhao ZB, Wang G, Hui Z, Wang MH,
Pan JF and Zheng H: High expression of KIF26B in breast cancer
associates with poor prognosis. PLoS One. 8:e616402013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tong D, Qu H, Meng X, Jiang Y, Liu D, Ye
S, Chen H, Jin Y, Fu S and Geng J: S-allylmercaptocysteine promotes
MAPK inhibitor-induced apoptosis by activating the TGF-β signaling
pathway in cancer cells. Oncol Rep. 32:1124–1132. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mahotka C, Wenzel M, Springer E, Gabbert
HE and Gerharz CD: Survivin-deltaEx3 and survivin-2B: Two novel
splice variants of the apoptosis inhibitor survivin with different
antiapoptotic properties. Cancer Res. 59:6097–6102. 1999.PubMed/NCBI
|
|
41
|
Taubert H, Heidenreich C, Holzhausen HJ,
Schulz A, Bache M, Kappler M, Eckert AW, Würl P, Melcher I,
Hauptmann K, et al: Expression of survivin detected by
immunohistochemistry in the cytoplasm and in the nucleus is
associated with prognosis of leiomyosarcoma and synovial sarcoma
patients. BMC Cancer. 10:652010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hachim MY, Hachim IY, Dai M, Ali S and
Lebrun JJ: Differential expression of TGFβ isoforms in breast
cancer highlights different roles during breast cancer progression.
Tumour Biol. 40:10104283177482542018. View Article : Google Scholar
|
|
43
|
Kim S, Lee J, Jeon M, Lee JE and Nam SJ:
Zerumbone suppresses the motility and tumorigenecity of triple
negative breast cancer cells via the inhibition of TGF-β1 signaling
pathway. Oncotarget. 7:1544–1558. 2016. View Article : Google Scholar
|
|
44
|
Zeng K, He B, Yang BB, Xu T, Chen X, Xu M,
Liu X, Sun H, Pan Y and Wang S: The pro-metastasis effect of
circANKS1B in breast cancer. Mol Cancer. 17:1602018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dogu GG, Ozkan M, Ozturk F, Dikilitas M,
Er O and Ozturk A: Triple-negative breast cancer:
Immunohistochemical correlation with basaloid markers and
prognostic value of survivin. Med Oncol. 27:34–39. 2010. View Article : Google Scholar
|
|
46
|
Jha K, Shukla M and Pandey M: Survivin
expression and targeting in breast cancer. Surg Oncol. 21:125–131.
2012. View Article : Google Scholar
|
|
47
|
Li Y, Ma X, Wu X, Liu X and Liu L:
Prognostic significance of survivin in breast cancer:
Meta-analysis. Breast J. 20:514–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dai JB, Zhu B, Lin WJ, Gao HY, Dai H,
Zheng L, Shi WH and Chen WX: Identification of prognostic
significance of BIRC5 in breast cancer using integrative
bioinformatics analysis. Biosci Rep. 40:BSR201936782020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Song K, Shankar E, Yang J, Bane K,
Wahdan-Alaswad R and Danielpour D: Critical role of a
survivin/TGF-β/mTORC1 axis in IGF-I-mediated growth of prostate
epithelial cells. PLoS One. 8:e618962013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang J, Song K, Krebs TL, Jackson MW and
Danielpour D: Rb/E2F4 and Smad2/3 link survivin to TGF-beta-induced
apoptosis and tumor progression. Oncogene. 27:5326–5338. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Perera CN, Chin HG, Duru N and Camarillo
IG: Leptin-regulated gene expression in MCF-7 breast cancer cells:
Mechanistic insights into leptin-regulated mammary tumor growth and
progression. J Endocrinol. 199:221–233. 2008. View Article : Google Scholar
|
|
52
|
Chen W, Zhong X, Wei Y, Liu Y, Yi Q, Zhang
G, He L, Chen F, Liu Y and Luo J: TGF-β regulates survivin to
affect cell cycle and the expression of EGFR and MMP9 in
glioblastoma. Mol Neurobiol. 53:1648–1653. 2016. View Article : Google Scholar
|
|
53
|
Neil JR, Tian M and Schiemann WP: X-linked
inhibitor of apoptosis protein and its E3 ligase activity promote
transforming growth factor-{beta}-mediated nuclear factor-{kappa}B
activation during breast cancer progression. J Biol Chem.
284:21209–21217. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gangrade A, Pathak V, Augelli-Szafran CE,
Wei HX, Oliver P, Suto M and Buchsbaum DJ: Preferential inhibition
of Wnt/β-catenin signaling by novel benzimidazole compounds in
triple-negative breast cancer. Int J Mol Sci. 19:15242018.
View Article : Google Scholar
|
|
55
|
Hseu YC, Lin YC, Rajendran P, Thigarajan
V, Mathew DC, Lin KY, Way TD, Liao JW and Yang HL: Antrodia
salmonea suppresses invasion and metastasis in triple-negative
breast cancer cells by reversing EMT through the NF-κB and
Wnt/β-catenin signaling pathway. Food Chem Toxicol. 124:219–230.
2019. View Article : Google Scholar
|
|
56
|
Yang Z, Ji L, Jiang G, Liu R, Liu Z, Yang
Y, Ma Q and Zhao H: FL118, a novel camptothecin analogue,
suppressed migration and invasion of human breast cancer cells by
inhibiting epithelial-mesenchymal transition via the Wnt/β-catenin
signaling pathway. Biosci Trends. 12:40–46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ma F, Li W, Liu C, Li W, Yu H, Lei B, Ren
Y, Li Z, Pang D and Qian C: MiR-23a promotes TGF-β1-induced EMT and
tumor metastasis in breast cancer cells by directly targeting CDH1
and activating Wnt/β-catenin signaling. Oncotarget. 8:69538–69550.
2017. View Article : Google Scholar
|
|
58
|
Chen X, Duan N, Zhang C and Zhang W:
Survivin and tumorigenesis: Molecular mechanisms and therapeutic
strategies. J Cancer. 7:314–323. 2016. View Article : Google Scholar : PubMed/NCBI
|