Head and neck cancers (HNCs) constitute a
heterogeneous group of malignant tumors characterized by a high
mortality rate (1). Aggressive
biologic behavior and strong resistance to therapy lead to
>650,000 cases and 330,000 deaths worldwide in 2020 (2).
Genetic mutations and environmental factors are
associated with the genesis and progression of these malignancies.
Tobacco use, Epstein-Barr virus (EBV), human papillomavirus (HPV),
alcohol consumption and exposure to environmental carcinogenic
factors are associated with DNA mutations and lead to dysplastic
lesions in the head and neck area (3,4).
Over the last decades, specific genes, which are responsible for
hereditary types of HNC, have been identified (5). Currently, there are several treatment
options available; however, due to the high rate of relapses, the
search for systemic and local alternative treatments is becoming
more and more important in modern days (6,7).
Entities such as familial oral squamous cell carcinoma (OSCC),
nasopharyngeal cancer and familial malignant melanoma are strongly
related to hereditary DNA mutations (8).
Furthermore, immune dysfunction serves a vital role
in the development and progression of head and neck squamous cell
carcinoma (HNSCC) (9). Previous
data have revealed that immunosuppression signaling (via myeloid
cells or M2 macrophages) prompts the development and progression of
the disease, which is characterized by intra-tumoral heterogeneity,
high mortality and poor prognosis for patients with HNSCC (10).
In the past few years, several epigenetics
mechanisms have been revealed to influence the carcinogenesis of
the head and neck region (11).
Gene silencing by epigenetic mechanisms and hypermethylation of CpG
islands are active areas of research. Deep knowledge of epigenetic
pathways and mechanisms is essential for establishing innovative
approaches to diagnosis and treatment of HNC (12). Thus, epigenetic editing refers to
all epigenetic mechanisms that regulate genome output and the
potential future application of these epigenetic editing mechanisms
for the diagnosis and treatment of HNC disease.
Cancer cells employ a variety of mechanisms to avoid
the immune system and secure malignant progression. Specifically,
distinct ligands on the cancer cell surface interact with immune
cells and activate inhibitor pathways to stop the immune response
(13). Epigenetic drugs
(Vorinostat, Romidepsin) are able to prevent inhibition by
enhancing the expression of tumor antigens, a fundamental function
that restores normal immunogenicity (14). Thus, epigenetic agents can enhance
the host immune system by boosting the immune responses triggered
via immunotherapy (15). This
innovative and promising finding will allow a combined approach
(epigenetic therapy and immunotherapy) for the treatment of several
types of HNC. Overall, the present review discusses the epigenetic
editing mechanisms and the various tumor-related immunosuppressive
signaling routes that trigger drug resistance (chemoresistance) and
immunosuppression. Additionally, the present review discusses the
immune modifications and examines the role of cancer-associated
fibroblasts (CAFs), tumor macrophages and myeloid cells in
tumor-related immunosuppression.
During the last few years, novel promising therapies
for HNC, including immune checkpoint inhibitors, have demonstrated
encouraging results in several clinical trials (16–21).
For instance, nivolumab and pembrolizumab are the main humanized
antibodies specifically designed to block programmed death-ligand 1
(PD-L1) from binding to programmed cell death-1 (PD-1), allowing T
cell activation (Table I). These
drugs have also been used for the treatment of patients with
metastatic HNSCC (22–24). In general, immune checkpoint
signaling is triggered by ligand and receptor interactions
(17), such as cytotoxic
T-lymphocyte-associated protein 4 (CTLA-4) and its ligands CD80 and
CD84, and PD-1 and its ligands PD-L1 and programmed cell death
ligand 2 (25). Specifically,
ipilimumab [Food and Drug Administration (FDA)-approved for
metastatic melanoma] a monoclonal antibody that can re-activate the
immune system by targeting CTLA-4, has been demonstrated to be
associated with overall survival improvements among patients with
squamous cell carcinoma of the head and neck (SCCHN) (26). Similarly, tremelimumab is a fully
human monoclonal antibody against CTLA-4, which blocks the binding
of the antigen-presenting cell ligands B7.1 and B7.2 to CTLA-4
(27). Furthermore, EGFR
inhibitors are also used in HNC treatment. Cetuximab (Erbitux), a
monoclonal antibody given by intravenous infusion, was approved by
the FDA in 2006 as a combination with radiation therapy for the
treatment of SCCHN (28). Blocking
EGFR activity is vital, since EGFR expression is upregulated in 85%
of all HNSCC cases and leads to tumor cell proliferation,
angiogenesis, invasion, tumor recurrence and metastasis. Similarly,
TNF receptor-targeted monoclonal antibodies, such as urelumab
(29), target the extracellular
domain of CD137, triggering activation of CD137-expressing immune
cells and especially cytotoxic T cells. Furthermore, bevacizumab is
a monoclonal antibody that acts as an angiogenesis inhibitor. It
prevents new blood vessel formation by inhibiting vascular
endothelial growth factor A (30).
Similarly, ficlatuzumab is a potent hepatocyte growth factor (HGF)
IgG1 antibody, which inhibits signaling through the HGF/c-Met
signaling pathway and is already in phase II clinical trials
(31,32). Finally, necrosis-targeted IL-12
immunocytokine is a novel immunocytokine designed for delivery of
IL-12 to the tumor microenvironment, which has been demonstrated to
be associated with increased overall antitumor efficacy and
substantial reduction in tumor growth compared to standard
chemotherapy (33).
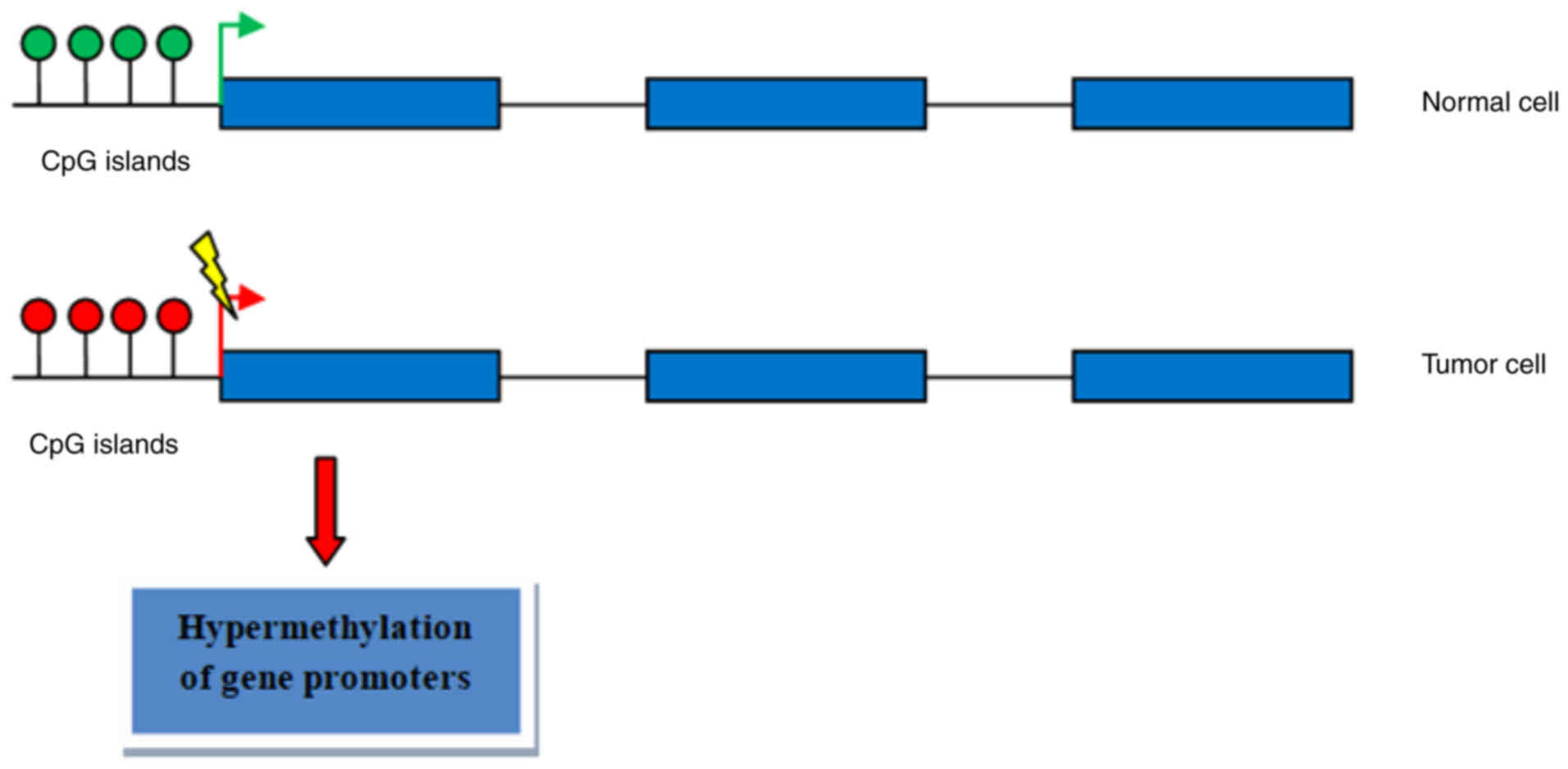
DNA methylation is an epigenetic mechanism involving
the transfer of a methyl group onto the C5 position of the cytosine
to form 5-methylcytosine (34,35).
Methylation usually occurs in CpG islands (Fig. 1) and characteristically acts to
repress gene transcription in HNSCC (36–40).
Aberrant methylation in the promoter region of tumor suppressor
genes could induce their abnormal expression in order to influence
carcinogenesis. Methylation of genes involved in DNA damage repair,
detoxification, cell cycle regulation and apoptosis plays a key
role in tumor development and progression (Table II). Previous studies have revealed
that abnormal O-6-methylguanine-DNA methyltransferase (MGMT)
promoter methylation can markedly increase the risk of
tumorigenesis in HNC (41,42). Furthermore, promoter methylation of
the MGMT, MutL homolog 1 and Ras association domain family 1
isoform A tumor suppressor genes contributes to HNSCC development
(43). Similarly, somatic
mutations and promoter methylation of the ryanodine receptor 2 are
responsible for the invasive pathogenesis of HNSCC (44). Long non-coding RNAs (lncRNAs) may
also affect methylation levels in HNC. For instance, LNCAROD, an
oncogenic lncRNA, is stabilized by m6A methylation and promotes
cancer progression by forming a ternary complex with heat shock
protein family A (Hsp70) member 1A (HSPA1A) and Y-box binding
protein 1 (YBX1) in HNSCC (45).
LNCAROD acts as a scaffold for the interaction between YBX1 and
HSPA1A, preventing proteasomal degradation of YBX1 in HNSCC cells.
Systematic analysis of differentially methylated expressed genes
and site-specific methylation may also be used as potential
prognostic markers in HNC. Specifically, abnormally differentially
methylated genes, including paired box 9, serine/threonine kinase
33, G protein-coupled receptor 150, INSM transcriptional repressor
1 and epoxide hydrolase 3, have been recognized to be associated
with overall survival, and this may aid the identification of
diagnostic biomarkers for early-stage HNC (46). In addition, site-specific
methylation patterns of galanin and galanin receptor 1/2 may serve
as an important site-specific biomarker for the prediction of the
clinical outcome in patients with HNSCC (47). Of note, circulating tumor DNA
(ctDNA) analysis may be important in early diagnosis and eventually
improve the outcomes of patients with HNSCC. Specifically, ctDNA
analysis in HPV-associated oropharyngeal cancer indicated that
three genes, including calmodulin like 5, Dna heat shock protein
family (Hsp40) member C5γ and lymphocyte antigen 6 family member D,
had a high predictive ability as emerging biomarkers and could
determine patient prognosis and real-time surveillance for disease
recurrences (48).
Chromatin architecture is mainly regulated by
histone proteins. DNA is wound around histones to form nucleosomes.
Histone modifications are mostly post translational modifications
to histone proteins and include methylation, phosphorylation,
acetylation, ubiquitylation and sumoylation (49). In HNC, histone modifications can
affect the molecular pathogenesis of the disease. Specifically,
histones may be post-translationally modified at the amino-terminal
ends by acetylation, methylation, phosphorylation, sumoylation,
ubiquitination, and ADP-ribosylation. Recently, the cancer driver
genes isocitrate dehydrogenase [NADP(+)] (IDH)1/2, lysine
demethylase 5C (KDM5C) and KDM6A were revealed to prompt histone
demethylation and hypoxic reprogramming in cancer metabolism
(50). Similarly, a novel
IFNα-induced lncRNA was observed to negatively regulate
immunosuppression by interrupting H3K27 acetylation in HNSCC.
Ectopic expression of lncMX1-215 markedly inhibits the expression
of IFNα-induced, immunosuppression-related molecules, including
PD-L1 and galectin-9 (51). In
addition, enhancer of zeste 2 polycomb repressive complex 2 subunit
(EZH2), a histone-lysine N-methyltransferase enzyme that
participates in histone methylation and transcriptional repression,
serves a key role in HNSCC progression. Specifically, targeting
EZH2 enhances antigen presentation and antitumor immunity, and
circumvents anti-PD-1 resistance in HNC. This is associated with
increased antigen-specific CD8+ T-cell proliferation,
IFNγ production and tumor cell cytotoxicity (52). Of note, EZH2 inhibition reduces the
histone H3K27me3 modification on the β2-microglobulin promoter and
suppresses tumor growth following combination therapy in an
anti-PD-1-resistant model of HNSCC (52). Recently, the histone demethylases
JARID1C/KDM5C and UTX/KDM6A and the gene-encoding metabolic enzymes
IDH1/2 were identified as cancer driver genes that serve a key role
in histone demethylation and hypoxic reprogramming in cancer
metabolism in HNC (50). LncRNAs
may also affect the histone modification mechanism. The lncRNA PVT1
can regulate nasopharyngeal carcinoma cell proliferation via
activation of the lysine acetyltransferase 2A (KAT2A)
acetyltransferase and stabilization of hypoxia-inducible factor 1-α
(HIF-1α) (53). PVT1 serves as a
scaffold for the chromatin modification factor KAT2A, which
mediates histone 3 lysine 9 acetylation, recruiting the nuclear
receptor binding protein transcriptional intermediary factor 1β to
activate nuclear factor 90 transcription, thereby increasing HIF-1α
stability and promoting a malignant phenotype in neural progenitor
cells (NPCs) (53). Another
lncRNA, CCAT1, can promote H3K27-acetylation and modulate the
histone methylation of SPRY4 (sprouty RTK signaling antagonist 4)
and homeobox B13 in the nucleus and cytoplasm, which affects cell
proliferation and migration in esophageal squamous cell carcinoma
(ESCC) (54).
Chromatin remodeling is vital for the rearrangement
of chromatin from a condensed state to a transcriptionally active
state. This allows access of genomic DNA to the regulatory
transcription machinery proteins and regulates gene expression.
Previous studies have demonstrated that chromatin hypoacetylation,
histone 3 in HNSCC cells is hypoacetylated and that endothelial
cells induce tumor acetylation (55). For instance, TP63 triggers
chromatin remodeling and enhances reprogramming to epidermal
differentiation and thus prompting squamous cell carcinoma
development (56). In addition,
zinc finger and SCAN domain containing 4 (ZSCAN4) prompts
functional histone 3 hyperacetylation at the promoters of OCT3/4
and NANOG, leading to an upregulation of cancer stem cell (CSC)
factors. Consistently, ZSCAN4 depletion leads to downregulation of
CSC markers, a decrease in the formation of tumor spheres and
inhibition of tumor growth (57).
Chromatin remodeling is also initiated by lin-28 homolog B, which
promotes HNC progression via regulation of the insulin-like growth
factor genes (high mobility group AT-hook 2, cyclin D2,
insulin-like growth factor 1 receptor and insulin like growth
factor 2 mRNA binding protein 2) (58). Similarly, lymphoid-specific
helicase (LSH) promotes cancer progression by regulating the
expression of fumarate hydratase (FH), a core component of the
tricarboxylic acid cycle (59).
LSH binds to the FH promoter, recruiting the epigenetic silencer
factor histone H3 Lys 9-specific histone methyltransferase, known
as G9a, to repress FH transcription. Loss-of-function mutations in
SWI/SNF chromatin-remodeling subunit genes are also detected in
HNSCC (60). Actin-like 6A
(ACTL6A), encoding an SWI/SNF subunit linked to stem cell function,
drives Yes1-associated transcriptional regulator (YAP) activation
and is highly expressed together with the p53 family member p63 in
HNSCC This molecular mechanism reveals that ACTL6A and p63
collaborate as oncogenic drivers in HNSCC.
LncRNAs are RNA isoforms larger than 200 nucleotides
that act as regulators of gene expression (61). They are involved in numerous
cellular processes, including cell proliferation, apoptosis and
cellular metabolism, as well as differentiation and development
(62). LncRNA MIR31HG has been
reported to target HIF-1α and P21 to facilitate HNC cancer cell
proliferation and tumorigenesis by promoting cell-cycle progression
(63). Furthermore, lncRNA
LINC00460 enhances HNSCC cell proliferation and
epithelial-mesenchymal transition (EMT)-related metastasis by
facilitating peroxiredoxin-1transfer into the nucleus and
transcription of EMT-related genes, including zinc finger E-box
binding homeobox 1, zinc finger E-box binding homeobox 2 and
vimentin (64). On the other hand,
lncRNA SLC26A4-AS1 can act as a tumor suppressor by inhibiting the
migration, invasion and metastasis of tumor cells in vitro
and in vivo. Of note, SLC26A4-AS1 is able to interact with
DEAD-box helicase 5 (DDX5) and the E3 ligase tripartite motif
containing 25, and promote DDX5 degradation through the
ubiquitin-proteasome pathway (65). In particular, SLC26A4-AS1 inhibits
the expression of multiple DNA double-strand break repair genes and
thyroid cancer metastasis by destabilizing DDX5. In addition, other
lncRNAs have been reported to function as promoters or inhibitors
of cancer metastasis. Specifically, MYOSLID promotes invasion and
metastasis of HNSCC by modulating partial EMT via regulation of
metastasis-related genes, including Slug, podoplanin and laminin
subunit β3 (66).
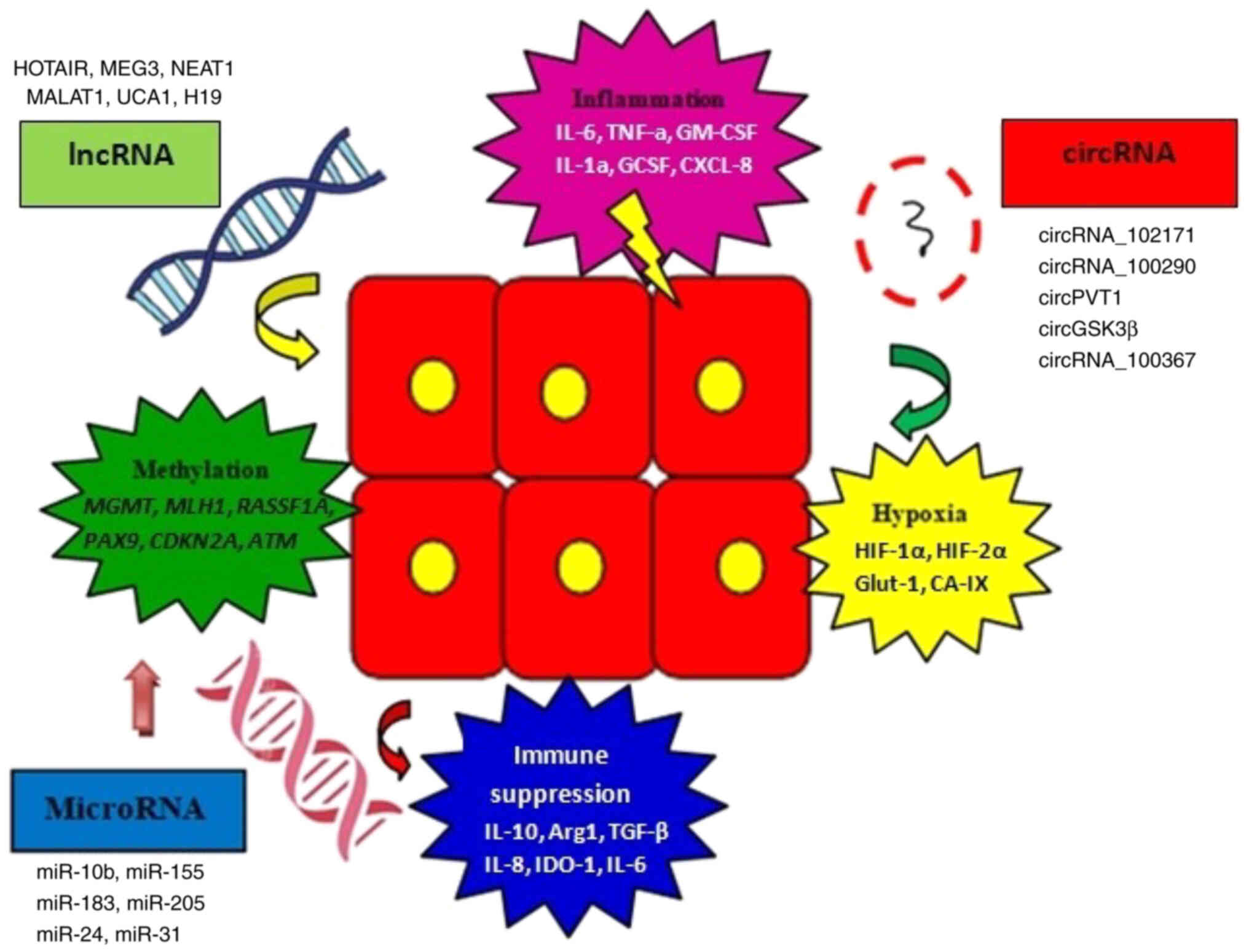
HNC tumor development may also be controlled by
circRNAs present in the tumor microenvironment (67). CircRNAs are single-stranded,
covalently closed RNA molecules that can act as microRNA (miR)
sponges (68). Furthermore,
specific non-coding RNA members (circRNA, lncRNA and miRNA) are
also involved in various tumor-promoting mechanisms like
methylation, hypoxia signaling, immune suppression and
tumor-related inflammation (Fig.
2). CircRNA_102171 has been revealed to interact with catenin β
interacting protein 1to block its interaction with the
β-catenin/transcription factor 3/transcription factor 4/lymphoid
enhancer binding factor 1 complex, leading to activation of the
Wnt/β-catenin signaling pathway in papillary thyroid cancer
(69). Similarly, circRNA_100290
can function as a competing endogenous RNA to regulate CDK6
expression in oral cancer by sponging up miR-29b family members
(70). CircPVT1 may act as an
oncogene in HNSCC, where it is transcriptionally enhanced by the
mut-p53/YAP/TEA domain transcription factor 1 complex and alters
the expression of miR-497-5p and genes involved in regulation of
cell proliferation (71). Another
circRNA member, circGSK3β, may trigger ESCC cell migration and
invasion via direct interaction with GSK3β and inhibition of GSK3β
activity (72). Certain circRNAs
also serve important roles in regulating the radioresistance of
ESCC (73). In particular,
circRNA_100367 enhances the radioresistance of ESCC cells
(KYSE-150R) via the miR-217/Wnt3 signaling pathway.
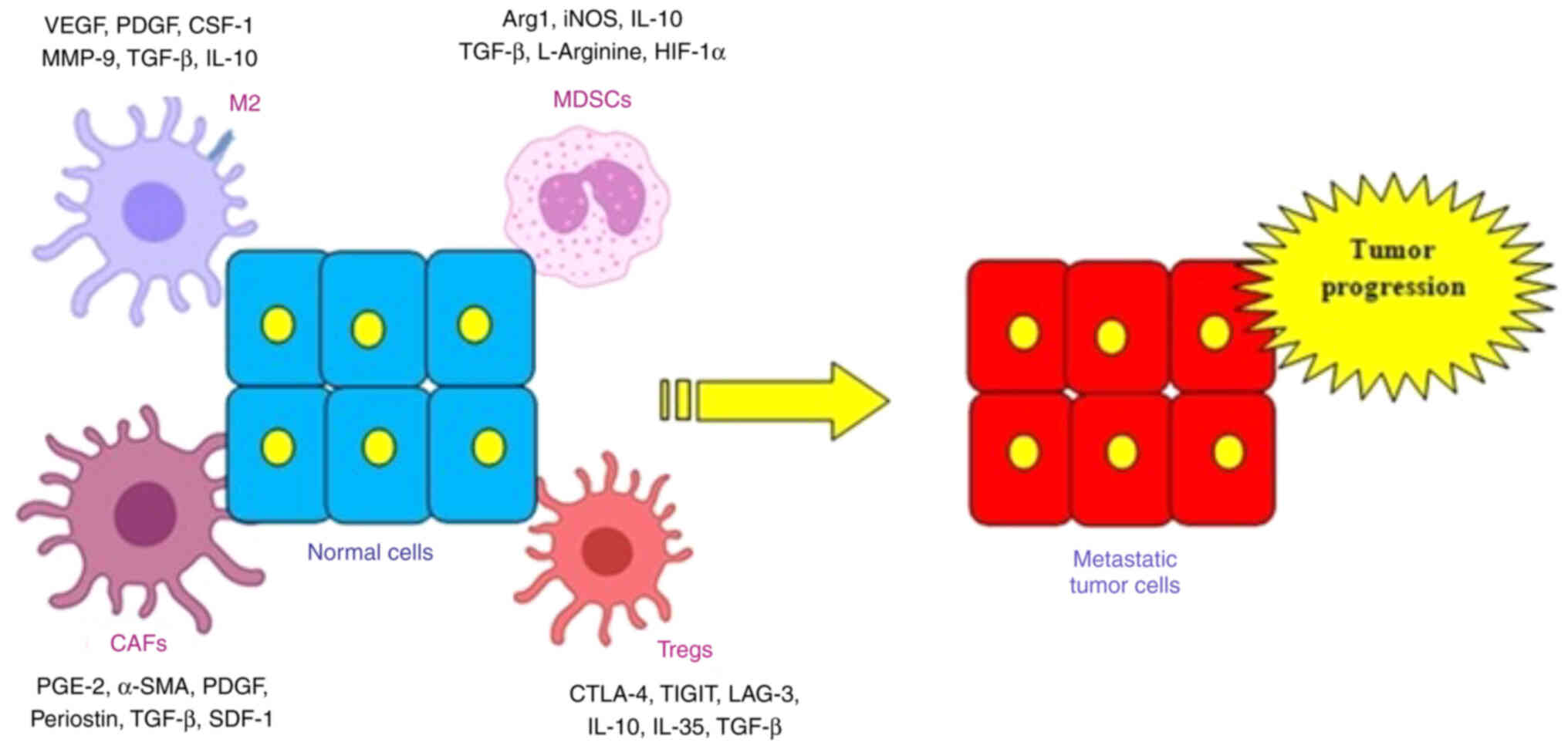
HNC development and progression are also regulated
by the CAFs signaling cascade (Fig.
3). CAFs are a group of activated fibroblasts with marked
heterogeneity and plasticity in the tumor microenvironment. They
secrete a variety of active factors [inside the TME, that promote
extensive angiogenesis signaling (via TGFb), increased matrix
deposition (via PDGF, SDF-1), and tissue remodeling (a-SMA, PGE-2,
periostin)]. CAF-derived exosomal miR-196a can confer cisplatin
resistance in HNC by targeting cyclin dependent kinase inhibitor 1B
and inhibitor of growth family member 5, indicating that miR-196a
may serve as a promising predictor of and potential therapeutic
target for cisplatin resistance in HNC (74). CAFs may also affect HNC stemness
inside the tumor microenvironment. For instance, periostin secreted
by CAFs promotes cancer stemness in HNC via activation of the
protein tyrosine kinase 7 (inactive)-Wnt/β-catenin signaling
pathway (75). Similarly, when
HNSCC cells are cocultured with normal fibroblasts, this
upregulates autophagy via IL6, IL8 and basic fibroblast growth
factor, promoting HNC progression (76). CAF-derived IL-6 may also prompt HNC
progression via osteopontin (OPN)-NF-κB signaling, which is
associated with poor patient prognosis (77). CAFs may also affect the functional
polarization of tumor-associated macrophages (TAMs) in OSCC.
Specifically, CAFs promote an immunosuppressive microenvironment
through the induction and accumulation of protumoral macrophages,
which is also associated with lymphatic invasion, vascular
invasion, lymph node involvement and TNM stage (78).
MDSCs are a heterogeneous group of immature myeloid
cells implicated in the regulation of immune responses. MDSCs are
directly implicated in the pathogenesis of HNC. Therefore, studies
target these myeloid cell subsets in order to reverse
immunosuppression (79,80). Recently, STAT3 inhibition in
combination with radiation, improved tumor growth delay in HNC via
downregulation of MDSCs, decrease in Tregs and upregulation of
effector T cells and M1 macrophage levels (81). In addition, distinct populations of
immune-suppressive macrophages differentiate from monocytic MDSCs
via increased expression of S100A9 protein and transcription factor
CCAAT/enhancer binding proteinβ (82). Furthermore, stimulator of
interferon response cGAMP interactor (STING) serves a vital role in
both differentiation and regulation of MDSC expression levels.
STING represses NPC-derived MDSC induction by enhancing suppressor
of cytokine signaling 1 (SOCS1) expression in both tumor cells and
MDSCs (83). SOCS1 physically
interacts with STAT3 through its SH2 domain to prevent STAT3
phosphorylation and dimerization, resulting in reduced MDSC
induction via inhibition of granulocyte-macrophage
colony-stimulating factor (GM-CSF) and IL-6 production (83). The expansion of MDSCs is also
critical for tumor propagation. EBV latent membrane protein 1
(LMP1) may promote MDSC expansion in the tumor microenvironment by
inducing extra-mitochondrial glycolysis in malignant cells, which
triggers immune escape initially. Specifically, LMP1 promotes the
expression of multiple glycolytic genes, including glucose
transporter 1 (84). This
metabolic reprogramming results in increased levels of the Nod-like
receptor family protein 3 inflammasome, cyclooxygenase 2 and
phosphorylated-p65 and, consequently, increased production of
IL-1β, IL-6 and GM-CSF (84).
T-cell immunoglobulin mucin 3 (TIM3) may also affect MDSC
differentiation. TIM3 expression is increased in patients with
recurrent HNSCC (85).
CD8+ T cells and CD11b+ CD33+
MDSCs are associated with increased TIM3 expression and effector
T-cell dysfunction in human HNSCC (85).
Another significant factor contributing to HNC
development are TAMs, which exhibit important functions in
facilitating the metastatic cascade of tumor cells (86). Specifically, Snail-overexpressing
cancer cells may promote M2 polarization of TAMs by delivering
miR-21-abundant exosomes in HNC (87). Furthermore, high miR-21 expression
is associated with release of miR-21-abundant tumor-derived
exosomes and a higher level of snail family transcriptional
repressor 1and the M2 marker mannose receptor C-type 1b (87). In addition, CAFs may affect the
functional polarization of TAMs. The expression of M2 biomarkers
CD68, CD14, CD163, CD200R, CD206, major histocompatibility complex,
class I, G, CD80 and CD86 is higher in CD14-positive cells
co-cultured with the culture supernatants of CAFs in OSCC than in
control cells (78). The gene
expression levels of arginase-I, IL10 and TGF-β are increased in
CAF-educated cells, and T cell proliferation is strongly
suppressed, and the neutralization of TGF-β, IL-10 or arginase-I
markedly restores T cell proliferation (78). Furthermore, interplay between
cancer cells and M2 macrophages is necessary for miR-550a-3-5p
downregulation and HPV-positive OSCC progression. MiR-550a-3-5p,
downregulated by E6 oncoprotein, inhibits M2 macrophage
polarization via YAP/C-C motif chemokine ligand 2 signaling, which
in turn abrogates the EMT program in HPV-positive OSCC cells
(88). Previous studies have
revealed a novel paracrine loop between cancer cells and
macrophages (89,90). M1-like TAMs activated by
exosome-transferred thrombospondin 1 promote malignant migration in
OSCC (91).
Understanding the epigenetic modifications and the
molecular crosstalk between tumor and immune cells is vital for
future therapeutic modalities against head and neck carcinogenesis.
Additionally, targeting the tumor microenvironment to reverse drug
resistance and inhibit immunosuppressive tumor networks emerges as
a useful tool for current immunotherapeutic approaches. Targeting
these aberrant patterns of signaling could provide biomarkers for
early detection and diagnosis for HNSCC and will improve the
therapy and extend overall survival. Furthermore, the deregulation
of tumor suppressor genes and oncogenes by epigenetic mechanism is
related to HNSCC tumorigenesis. Therefore, current trials are
ongoing to evaluate the role of novel immune checkpoint inhibitors
in order to retain effective tumor control and improve the quality
of life for patients with HNSCC. Deciphering these signaling
mechanisms and their crosstalk with immune and tumor cells may
bring novel perspectives for HNSCC therapeutic approaches, and
thus, a number of them may be used as potential therapeutic targets
or in combination with current immunotherapy regimens.
Not applicable.
Funding: No funding was received.
Not applicable.
SG, SP, AP, NT, PV, IS, NA, TK and KD contributed to
manuscript drafting and to the critical revisions of the
intellectual content. SG, SP and KD performed the literature search
and selection and were responsible for the general study
supervision. All authors read and approved the final version of the
manuscript to be published. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Auperin A: Epidemiology of head and neck
cancers: An update. Curr Opin Oncol. 32:178–186. 2020. View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer Statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dos Santos ES, Wagner VP, Cabral Ramos J,
Lambert DW, Castilho RM and Paes Leme AF: Epigenetic modulation of
the tumor microenvironment in head and neck cancer: Challenges and
opportunities. Crit Rev Oncol Hematol. 164:1033972021. View Article : Google Scholar
|
|
5
|
Prime SS, Thakker NS, Pring M, Guest PG
and Paterson IC: A review of inherited cancer syndromes and their
relevance to oral squamous cell carcinoma. Oral Oncol. 37:1–16.
2001. View Article : Google Scholar
|
|
6
|
Bennardo L, Bennardo F, Giudice A,
Passante M, Dastoli S, Morrone P, Provenzano E, Patruno C and
Nisticò SP: Local chemotherapy as an adjuvant treatment in
unresectable squamous cell carcinoma: What do we know so far? Curr
Oncol. 28:2317–2325. 2021. View Article : Google Scholar
|
|
7
|
Pentangelo G, Nisticò SP, Provenzano E,
Cisale GY and Bennardo L: Topical 5% imiquimod sequential to
surgery for HPV-related squamous cell carcinoma of the lip.
Medicina (Kaunas). 57:5632021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Venugopal R, Bavle RM, Konda P,
Muniswamappa S and Makarla S: Familial cancers of head and neck
region. J Clin Diagn Res. 11:ZE01–ZE06. 2017.PubMed/NCBI
|
|
9
|
Plavc G, Jesenko T, Oražem M and Strojan
P: Challenges in combining immunotherapy with radiotherapy in
recurrent/metastatic head and neck cancer. Cancers (Basel).
12:31972020. View Article : Google Scholar
|
|
10
|
Caudell JJ, Torres-Roca JF, Gillies RJ,
Enderling H, Kim S, Rishi A, Moros EG and Harrison LB: The future
of personalised radiotherapy for head and neck cancer. Lancet
Oncol. 18:e266–e273. 2017. View Article : Google Scholar
|
|
11
|
Bakhtiar SM, Ali A and Barh D: Epigenetics
in head and neck cancer. Methods Mol Biol. 1238:751–769. 2015.
View Article : Google Scholar
|
|
12
|
Gazdzicka J, Golabek K, Strzelczyk JK and
Ostrowska Z: Epigenetic modifications in head and neck cancer.
Biochem Genet. 58:213–244. 2020. View Article : Google Scholar
|
|
13
|
Armeanu S, Bitzer M, Lauer UM, Venturelli
S, Pathil A, Krusch M, Kaiser S, Jobst J, Smirnow I, Wagner A, et
al: Natural killer cell-mediated lysis of hepatoma cells via
specific induction of NKG2D ligands by the histone deacetylase
inhibitor sodium valproate. Cancer Res. 65:6321–6329. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chiappinelli KB, Zahnow CA, Ahuja N and
Baylin SB: Combining epigenetic and immunotherapy to combat cancer.
Cancer Res. 76:1683–1689. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dunn J and Rao S: Epigenetics and
immunotherapy: The current state of play. Mol Immunol. 87:227–239.
2017. View Article : Google Scholar
|
|
16
|
Leemans CR, Snijders PJF and Brakenhoff
RH: The molecular landscape of head and neck cancer. Nat Rev
Cancer. 18:269–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ferris RL: Immunology and immunotherapy of
head and neck cancer. J Clin Oncol. 33:3293–3304. 2015. View Article : Google Scholar
|
|
18
|
Motzer RJ, Penkov K, Haanen J, Rini B,
Albiges L, Campbell MT, Venugopal B, Kollmannsberger C, Negrier S,
Uemura M, et al: Avelumab plus Axitinib versus sunitinib for
advanced Renal-Cell carcinoma. N Engl J Med. 380:1103–1115. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chu L, Chen Y, Liu Q, Liang F, Wang S, Liu
Q, Yu H, Wu X, Zhang J, Deng J, et al: A phase II study of apatinib
in patients with chemotherapy-refractory esophageal squamous cell
carcinoma (ESO-Shanghai 11). Oncologist. 26:e925–e935. 2021.
View Article : Google Scholar
|
|
20
|
Mollica Poeta V, Massara M, Capucetti A
and Bonecchi R: Chemokines and Chemokine receptors: New targets for
cancer immunotherapy. Front Immunol. 10:3792019. View Article : Google Scholar
|
|
21
|
Pooler DB, Ness DB, Sarantopoulos J,
Squittieri N, Ravichandran S, Britten CD, Amaravadi RK,
Vaishampayan U, LoRusso P, Shapiro GI, et al: The effect of
sonidegib (LDE225) on the pharmacokinetics of bupropion and
warfarin in patients with advanced solid tumours. Br J Clin
Pharmacol. 87:1291–1302. 2021. View Article : Google Scholar
|
|
22
|
Karam SD and Raben D: Radioimmunotherapy
for the treatment of head and neck cancer. Lancet Oncol.
20:e404–e416. 2019. View Article : Google Scholar
|
|
23
|
Ferris RL, Blumenschein G Jr, Fayette J,
Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE,
Even C, et al: Nivolumab for Recurrent Squamous-Cell Carcinoma of
the Head and Neck. N Engl J Med. 375:1856–1867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burtness B, Harrington KJ, Greil R,
Soulières D, Tahara M, de Castro G Jr, Psyrri A, Basté N, Neupane
P, Bratland Å, et al: Pembrolizumab alone or with chemotherapy
versus cetuximab with chemotherapy for recurrent or metastatic
squamous cell carcinoma of the head and neck (KEYNOTE-048): A
randomised, open-label, phase 3 study. Lancet. 394:1915–1928. 2019.
View Article : Google Scholar
|
|
25
|
Billan S, Kaidar-Person O and Gil Z:
Treatment after progression in the era of immunotherapy. Lancet
Oncol. 21:e463–e476. 2020. View Article : Google Scholar
|
|
26
|
Janjigian YY, Shitara K, Moehler M,
Garrido M, Salman P, Shen L, Wyrwicz L, Yamaguchi K, Skoczylas T,
Campos Bragagnoli A, et al: First-line nivolumab plus chemotherapy
versus chemotherapy alone for advanced gastric, gastro-oesophageal
junction, and oesophageal adenocarcinoma (CheckMate 649): A
randomised, open-label, phase 3 trial. Lancet. 398:27–40. 2021.
View Article : Google Scholar
|
|
27
|
Tarhini AA: Tremelimumab: A review of
development to date in solid tumors. Immunotherapy. 5:215–229.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu Y, Han L, Sheng Y and Wu S: Cetuximab
monotherapy for relapsing high-grade mucoepidermoid carcinoma: A
case report and review of the literature. Oral Oncol.
107:1048242020. View Article : Google Scholar
|
|
29
|
Chester C, Sanmamed MF, Wang J and Melero
I: Immunotherapy targeting 4-1BB: Mechanistic rationale, clinical
results, and future strategies. Blood. 131:49–57. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Garcia J, Hurwitz HI, Sandler AB, Miles D,
Coleman RL, Deurloo R and Chinot OL: Bevacizumab
(Avastin®) in cancer treatment: A review of 15 years of
clinical experience and future outlook. Cancer Treat Rev.
86:1020172020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Scagliotti GV, Novello S and von Pawel J:
The emerging role of MET/HGF inhibitors in oncology. Cancer Treat
Rev. 39:793–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shih YH, Chen PC and Chu CY: Severe
refractory scarring alopecia associated with combinational use of
ficlatuzumab (AV-299) and gefitinib. J Clin Oncol. 31:e335–e337.
2013. View Article : Google Scholar
|
|
33
|
Greiner JW, Morillon YM II and Schlom J:
NHS-IL12, a tumor-targeting immunocytokine. Immunotargets Ther.
10:155–169. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View Article : Google Scholar
|
|
35
|
Koch A, Joosten SC, Feng Z, de Ruijter TC,
Draht MX, Melotte V, Smits KM, Veeck J, Herman JG, Van Neste L, et
al: Analysis of DNA methylation in cancer: Location revisited. Nat
Rev Clin Oncol. 15:459–466. 2018. View Article : Google Scholar
|
|
36
|
Klutstein M, Nejman D, Greenfield R and
Cedar H: DNA methylation in cancer and aging. Cancer Res.
76:3446–3450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eads CA, Lord RV, Wickramasinghe K, Long
TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester
TR, Skinner KA and Laird PW: Epigenetic patterns in the progression
of esophageal adenocarcinoma. Cancer Res. 61:3410–3418.
2001.PubMed/NCBI
|
|
38
|
Chen Y, Jiang X, Li X, Yan D, Liu J, Yang
J and Yan S: The methylation modification of m6A regulators
contributes to the prognosis of head and neck squamous cell
carcinoma. Ann Transl Med. 9:13462021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou C, Ye M, Ni S, Li Q, Ye D, Li J, Shen
Z and Deng H: DNA methylation biomarkers for head and neck squamous
cell carcinoma. Epigenetics. 13:398–409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hier J, Vachon O, Bernstein A, Ibrahim I,
Mlynarek A, Hier M, Alaoui-Jamali MA, Maschietto M and da Silva SD:
Portrait of DNA methylated genes predictive of poor prognosis in
head and neck cancer and the implication for targeted therapy. Sci
Rep. 11:100122021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhao JJ, Li HY, Wang D, Yao H and Sun DW:
Abnormal MGMT promoter methylation may contribute to the risk of
esophageal cancer: A meta-analysis of cohort studies. Tumour Biol.
35:10085–10093. 2014. View Article : Google Scholar
|
|
42
|
Ji X, Guan C, Jiang X and Li H: Diagnostic
accuracy of DNA methylation for head and neck cancer varies by
sample type and number of markers tested. Oncotarget.
7:80019–80032. 2016. View Article : Google Scholar
|
|
43
|
Koutsimpelas D, Pongsapich W, Heinrich U,
Mann S, Mann WJ and Brieger J: Promoter methylation of MGMT, MLH1
and RASSF1A tumor suppressor genes in head and neck squamous cell
carcinoma: Pharmacological genome demethylation reduces
proliferation of head and neck squamous carcinoma cells. Oncol Rep.
27:1135–1141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schmitt K, Molfenter B, Laureano NK, Tawk
B, Bieg M, Hostench XP, Weichenhan D, Ullrich ND, Shang V, Richter
D, et al: Somatic mutations and promotor methylation of the
ryanodine receptor 2 is a common event in the pathogenesis of head
and neck cancer. Int J Cancer. 145:3299–3310. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y,
Mei Y, Tan Y, Li X, Zeng Z, et al: LNCAROD is stabilized by m6A
methylation and promotes cancer progression via forming a ternary
complex with HSPA1A and YBX1 in head and neck squamous cell
carcinoma. Mol Oncol. 14:1282–1296. 2020. View Article : Google Scholar
|
|
46
|
Bai G, Song J, Yuan Y, Chen Z, Tian Y, Yin
X, Niu Y and Liu J: Systematic analysis of differentially
methylated expressed genes and site-specific methylation as
potential prognostic markers in head and neck cancer. J Cell
Physiol. 234:22687–22702. 2019. View Article : Google Scholar
|
|
47
|
Misawa K, Mochizuki D, Endo S, Mima M,
Misawa Y, Imai A, Shinmura K, Kanazawa T, Carey TE and Mineta H:
Site-specific methylation patterns of the GAL and GALR1/2 genes in
head and neck cancer: Potential utility as biomarkers for
prognosis. Mol Carcinog. 56:1107–1116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Misawa K, Imai A, Matsui H, Kanai A,
Misawa Y, Mochizuki D, Mima M, Yamada S, Kurokawa T, Nakagawa T and
Mineta H: Identification of novel methylation markers in
HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue
verification and validation testing in ctDNA. Oncogene.
39:4741–4755. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Castilho RM, Squarize CH and Almeida LO:
Epigenetic modifications and head and neck cancer: Implications for
tumor progression and resistance to therapy. Int J Mol Sci.
18:15062017. View Article : Google Scholar
|
|
50
|
Chang S, Yim S and Park H: The cancer
driver genes IDH1/2, JARID1C/KDM5C, and UTX/KDM6A: Crosstalk
between histone demethylation and hypoxic reprogramming in cancer
metabolism. Exp Mol Med. 51:1–17. 2017. View Article : Google Scholar
|
|
51
|
Ma H, Chang H, Yang W, Lu Y, Hu J and Jin
SA: Novel IFNα-induced long noncoding RNA negatively regulates
immunosuppression by interrupting H3K27 acetylation in head and
neck squamous cell carcinoma. Mol Cancer. 19:42020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhou L, Mudianto T, Ma X, Riley R and
Uppaluri R: Targeting EZH2 enhances antigen presentation, antitumor
immunity, and circumvents anti-PD-1 resistance in head and neck
cancer. Clin Cancer Res. 26:290–300. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang Y, Chen W, Lian J, Zhang H, Yu B,
Zhang M, Wei F, Wu J, Jiang J, Jia Y, et al: The lncRNA PVT1
regulates nasopharyngeal carcinoma cell proliferation via
activating the KAT2A acetyltransferase and stabilizing HIF-1α. Cell
Death Differ. 27:695–710. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang E, Han L, Yin D, He X, Hong L, Si X,
Qiu M, Xu T, De W, Xu L, et al: H3K27 acetylation activated-long
non-coding RNA CCAT1 affects cell proliferation and migration by
regulating SPRY4 and HOXB13 expression in esophageal squamous cell
carcinoma. Nucleic Acids Res. 45:3086–3101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kadoch C: Diverse compositions and
functions of chromatin remodeling machines in cancer. Sci Transl
Med. 11:eaay10182019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yi M, Tan Y, Wang L, Cai J, Li X, Zeng Z,
Xiong W, Li G, Li X, Tan P and Xiang B: TP63 links chromatin
remodeling and enhancer reprogramming to epidermal differentiation
and squamous cell carcinoma development. Cell Mol Life Sci.
77:4325–4346. 2020. View Article : Google Scholar
|
|
57
|
Portney BA, Arad M, Gupta A, Brown RA,
Khatri R, Lin PN, Hebert AM, Angster KH, Silipino LE, Meltzer WA,
et al: ZSCAN4 facilitates chromatin remodeling and promotes the
cancer stem cell phenotype. Oncogene. 39:4970–4982. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Alajez NM, Shi W, Wong D, Lenarduzzi M,
Waldron J, Weinreb I and Liu FF: Lin28b promotes head and neck
cancer progression via modulation of the insulin-like growth factor
survival pathway. Oncotarget. 3:1641–1652. 2012. View Article : Google Scholar
|
|
59
|
He X, Yan B, Liu S, Jia J, Lai W, Xin X,
Tang CE, Luo D, Tan T, Jiang Y, et al: Chromatin remodeling factor
LSH drives cancer progression by suppressing the activity of
fumarate hydratase. Cancer Res. 76:5743–5755. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Saladi SV, Ross K, Karaayvaz M, Tata PR,
Mou H, Rajagopal J, Ramaswamy S and Ellisen LW: ACTL6A Is
Co-Amplified with p63 in squamous cell carcinoma to drive YAP
activation, regenerative proliferation, and poor prognosis. Cancer
Cell. 31:35–49. 2017. View Article : Google Scholar
|
|
61
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar
|
|
62
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar
|
|
63
|
Wang R, Ma Z, Feng L, Yang Y, Tan C, Shi
Q, Lian M, He S, Ma H and Fang J: LncRNA MIR31HG targets HIF1A and
P21 to facilitate head and neck cancer cell proliferation and
tumorigenesis by promoting cell-cycle progression. Mol Cancer.
17:1622017. View Article : Google Scholar
|
|
64
|
Jiangm Y, Cao W, Wu K, Qin X, Wang X, Li
Y, Yu B, Zhang Z, Wang X, Yan M, et al: LncRNA LINC00460 promotes
EMT in head and neck squamous cell carcinoma by facilitating
peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res.
38:3652019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yuan J, Song Y, Pan W, Li Y, Xu Y, Xie M,
Shen Y, Zhang N, Liu J, Hua H, et al: LncRNA SLC26A4-AS1 suppresses
the MRN complex-mediated DNA repair signaling and thyroid cancer
metastasis by destabilizing DDX5. Oncogene. 39:6664–6676. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Xiong HG, Li H, Xiao Y, Yang QC, Yang LL,
Chen L, Bu LL, Zhang WF, Zhang JL and Sun ZJ: Long noncoding RNA
MYOSLID promotes invasion and metastasis by modulating the partial
epithelial-mesenchymal transition program in head and neck squamous
cell carcinoma. J Exp Clin Cancer Res. 38:2782019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bi W, Huang J, Nie C, Liu B, He G, Han J,
Pang R, Ding Z, Xu J and Zhang J: CircRNA circRNA_102171 promotes
papillary thyroid cancer progression through modulating
CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin
Cancer Res. 37:2752018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen L, Zhang S, Wu J, Cui J, Zhong L,
Zeng L and Ge S: CircRNA_100290 plays a role in oral cancer by
functioning as a sponge of the miR-29 family. Oncogene.
36:4551–4561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Verduci L, Ferraiuolo M, Sacconi A, Ganci
F, Vitale J, Colombo T, Paci P, Strano S, Macino G, Rajewsky N and
Blandino G: The oncogenic role of circPVT1 in head and neck
squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD
transcription-competent complex. Genome Biol. 18:2372017.
View Article : Google Scholar
|
|
72
|
Hu X, Wu D, He X, Zhao H, He Z, Lin J,
Wang K, Wang W, Pan Z, Lin H and Wang M: circGSK3β promotes
metastasis in esophageal squamous cell carcinoma by augmenting
β-catenin signaling. Mol Cancer. 18:1602019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang
S, Gu H, Wang X, Zhao D and Fan R: CircRNA_100367 regulated the
radiation sensitivity of esophageal squamous cell carcinomas
through miR-217/Wnt3 pathway. Aging (Albany NY). 11:12412–12427.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Qin X, Guo H, Wang X, Zhu X, Yan M, Wang
X, Xu Q, Shi J, Lu E, Chen W and Zhang J: Exosomal miR-196a derived
from cancer-associated fibroblasts confers cisplatin resistance in
head and neck cancer through targeting CDKN1B and ING5. Genome
Biol. 20:122019. View Article : Google Scholar
|
|
75
|
Yu B, Wu K, Wang X, Zhang J, Wang L, Jiang
Y, Zhu X, Chen W and Yan M: Periostin secreted by cancer-associated
fibroblasts promotes cancer stemness in head and neck cancer by
activating protein tyrosine kinase 7. Cell Death Dis. 9:10822018.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
New J, Arnold L, Ananth M, Alvi S,
Thornton M, Werner L, Tawfik O, Dai H, Shnayder Y, Kakarala K, et
al: Secretory autophagy in cancer-associated fibroblasts promotes
head and neck cancer progression and offers a novel therapeutic
target. Cancer Res. 77:6679–6691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Qin X, Yan M, Wang X, Xu Q, Wang X, Zhu X,
Shi J, Li Z, Zhang J and Chen W: Cancer-associated
fibroblast-derived IL-6 promotes head and neck cancer progression
via the osteopontin-NF-kappa B signaling pathway. Theranostics.
8:921–940. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Takahashi H, Sakakura K, Kudo T, Toyoda M,
Kaira K, Oyama T and Chikamatsu K: Cancer-associated fibroblasts
promote an immunosuppressive microenvironment through the induction
and accumulation of protumoral macrophages. Oncotarget.
8:8633–8647. 2017. View Article : Google Scholar
|
|
79
|
Davis RJ, Van Waes C and Allen CT:
Overcoming barriers to effective immunotherapy: MDSCs, TAMs, and
Tregs as mediators of the immunosuppressive microenvironment in
head and neck cancer. Oral Oncol. 58:59–70. 2016. View Article : Google Scholar
|
|
80
|
Greene S, Robbins Y, Mydlarz WK, Huynh AP,
Schmitt NC, Friedman J, Horn LA, Palena C, Schlom J, Maeda DY, et
al: Inhibition of MDSC trafficking with SX-682, a CXCR1/2
inhibitor, enhances NK-cell immunotherapy in head and neck cancer
models. Clin Cancer Res. 26:1420–1431. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Oweida AJ, Darragh L, Phan A, Binder D,
Bhatia S, Mueller A, Court BV, Milner D, Raben D, Woessner R, et
al: STAT3 modulation of regulatory T cells in response to radiation
therapy in head and neck cancer. J Natl Cancer Inst. 111:1339–1349.
2019. View Article : Google Scholar
|
|
82
|
Kwak T, Wang F, Deng H, Condamine T, Kumar
V, Perego M, Kossenkov A, Montaner LJ, Xu X, Xu W, et al: Distinct
populations of immune-suppressive macrophages differentiate from
monocytic myeloid-derived suppressor cells in cancer. Cell Rep.
33:1085712020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang CX, Ye SB, Ni JJ, Cai TT, Liu YN,
Huang DJ, Mai HQ, Chen QY, He J, Zhang XS, et al: STING signaling
remodels the tumor microenvironment by antagonizing myeloid-derived
suppressor cell expansion. Cell Death Differ. 26:2314–2328. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Cai TT, Ye SB, Liu YN, He J, Chen QY, Mai
HQ, Zhang CX, Cui J, Zhang XS, Busson P, et al: LMP1-mediated
glycolysis induces myeloid-derived suppressor cell expansion in
nasopharyngeal carcinoma. PLoS Pathog. 13:e10065032017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Liu JF, Ma SR, Mao L, Bu LL, Yu GT, Li YC,
Huang CF, Deng WW, Kulkarni AB, Zhang WF and Sun ZJ: T-cell
immunoglobulin mucin 3 blockade drives an antitumor immune response
in head and neck cancer. Mol Oncol. 11:235–247. 2017. View Article : Google Scholar
|
|
86
|
Larionova I, Cherdyntseva N, Liu T,
Patysheva M, Rakina M and Kzhyshkowska J: Interaction of
tumor-associated macrophages and cancer chemotherapy.
Oncoimmunology. 8:15960042019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Hsieh CH, Tai SK and Yang MH:
Snail-overexpressing cancer cells promote M2-Like polarization of
tumor-associated macrophages by delivering MiR-21-Abundant
Exosomes. Neoplasia. 20:775–788. 2018. View Article : Google Scholar
|
|
88
|
Cao MX, Zhang WL, Yu XH, Wu JS, Qiao XW,
Huang MC, Wang K, Wu JB, Tang YJ, Jiang J, et al: Interplay between
cancer cells and M2 macrophages is necessary for miR-550a-3-5p
down-regulation-mediated HPV-positive OSCC progression. J Exp Clin
Cancer Res. 39:1022020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Rigo A, Gottardi M, Zamò A, Mauri P,
Bonifacio M, Krampera M, Damiani E, Pizzolo G and Vinante F:
Macrophages may promote cancer growth via a GM-CSF/HB-EGF paracrine
loop that is enhanced by CXCL12. Mol Cancer. 9:2732010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang F, Li P, Liu S, Yang M, Zeng S, Deng
J, Chen D, Yi Y and Liu H: β-Catenin-CCL2 feedback loop mediates
crosstalk between cancer cells and macrophages that regulates
breast cancer stem cells. Oncogene. 40:5854–5865. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Xiao M, Zhang J and Chen W and Chen W:
M1-like tumor-associated macrophages activated by
exosome-transferred THBS1 promote malignant migration in oral
squamous cell carcinoma. J Exp Clin Cancer Res. 37:1432018.
View Article : Google Scholar : PubMed/NCBI
|