Introduction
Lung cancer, including non-small cell lung cancer
(NSCLC) and SCLC, is one of the major causes of cancer-related
death worldwide (1). NSCLC
accounts for 80-85% of lung cancers and lung adenocarcinoma (LUAD)
is the most prevalent sub-type of NSCLC (2). Difficulties in early diagnosis, high
metastatic potential and the occurrence of treatment resistance in
advanced disease are the reasons for the poor survival of patients
with LUAD. Numerous targeted treatment drugs, such as gefitinib,
erlotinib, and crizotinib, have been widely used in clinical
treatment (3,4). Although molecularly targeted
therapies have produced promising clinical outcomes, only a
minority of patients with LUAD are ideal candidates for targeted
therapies (5). Furthermore, curing
patients with LUAD remains challenging due to target genetic
resistance mutations or off-target mechanisms of resistance
(6–8). Therefore, it is necessary to
investigate more effective molecular pathological diagnosis and
markers for predicting the prognosis of patients with LUAD.
The 14-3-3 protein family consists of at least seven
isoforms, namely β, γ, ε, η, ξ, σ and τ/θ, which are present in
mammalian cells (9). 14-3-3
proteins are involved in a variety of cell signal transduction
processes, such as cell proliferation, cell cycle regulation,
apoptosis and malignant transformation (10,11).
In the 14-3-3 protein family, 14-3-3σ is most associated with tumor
occurrence and development (12).
14-3-3ơ was originally identified as a human mammary epithelium
marker 1 and as a tumor suppressor gene (13,14),
which was determined to be reduced or lost in numerous types of
solid tumor (15). Loss or
reduction of 14-3-3σ by CpG methylation or p53 mutation contributes
to the progression of different types of carcinomas, including
early stages of tumor development (16–21).
This suggests that the function of 14-3-3σ may help prevent the
malignant transformation of epithelial cells. However, a growing
body of evidence suggested that 14-3-3σ does not act as a tumor
suppressor in cancers. Enhanced 14-3-3σ has been observed in
pancreatic cancer (22), gastric
carcinomas (23) and prostate
cancer (24), and which is
positively associated with aggressive tumor behavior and poor
prognosis (15,25,26).
Therefore, the biological role of 14-3-3ơ in tumorigenesis and
progression of human cancers varies according to the specific tumor
type.
In lung cancer, 14-3-3 protein was elevated in tumor
tissues compared to normal tissues (9,27,28).
Upregulation of 14-3-3σ was also observed in LUAD (27) and NSCLC tissues (29). Increased expression of 14-3-3 may
be due to the decreased DNA methylation of 14-3-3σ (30) and has been associated with
cisplatin resistance in NSCLC cells (29). These results suggest that 14-3-3σ
may be a promising biomarker for the molecular pathology, diagnosis
and prognosis of patients with LUAD. However, only a small number
of studies have reported on this possibility (29,31).
Furthermore, the biological function of 14-3-3ơ in tumorigenesis
and progression of LUAD has remained to be fully elucidated. In the
present study, the prognostic significance of 14-3-3σ expression in
LUAD was assessed by a bioinformatics analysis of the clinical
features and survival information of patients from a public
database and in clinical patients from our hospital. Furthermore,
in vitro and in vivo experiments were performed to
investigate the effect of 14-3-3σ expression on cell proliferation,
colony formation and anchorage-independent growth, as well as tumor
growth, of LUAD. Overall, 14-3-3ơ was indicated to be involved in
tumor progression and high expression of 14-3-3σ was associated
with poor prognosis of patients with LUAD.
Materials and methods
Cell lines and culture
The human bronchial epithelial immortalized cell
line Beas-2B was purchased from the American Type Culture
Collection and cultured in Bronchial Epithelial Basal Medium (BEBM;
Lonza) with 10% fetal bovine serum (FBS; Corning, Inc.) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
The lung cancer cell lines NCI-H1299, NCI-H358, A-549 and NCI-H23
were purchased from Zhong Qiao Xin Zhou Biotechnology Co., Ltd.
NCI-H1299, NCI-H358 and NCI-H23 were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
and 1% penicillin/streptomycin; A-549 was grown in F-12K medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
and 1% penicillin/streptomycin. All cells were maintained in a
humidified incubator with 5% CO2 at 37°C.
Clinical specimens
To detect the protein level of 14-3-3ơ in LUAD
tissue, a total of four LUAD tissue samples and non-tumor adjacent
tissues samples were derived from biopsy samples and pathologically
confirmed as LUAD from December 2020 to August 2021. Specific
information on the patients is provided in Table I. The present study was performed
according to the Declaration of Helsinki and approved by Ethics
Committee of the People's Hospital of Beilun District [Ningbo,
China; no. 2021-23(YS)]. Each patient provided written informed
consent prior to surgery.
 | Table I.Clinicopathological data of the
patients whose samples were used for western blot analysis. |
Table I.
Clinicopathological data of the
patients whose samples were used for western blot analysis.
| ID | Sex | Age, years | Pathologic
type | TNM stage |
|---|
| 3397749 | Male | 51 | Infiltrating
adenocarcinoma of the left upper lung | T1cN0M0 |
| 2296185 | Female | 41 | Infiltrating
adenocarcinoma of the left upper lung | T1bN0M0 |
| 2295463 | Male | 43 | Infiltrating
adenocarcinoma of the left upper lung | T1bN0M0 |
| 2287639 | Male | 41 | Infiltrating
adenocarcinoma of the right upper lung | T1cN0M0 |
To detect the relationship between 14-3-3ơ and
prognosis of patients with LUAD, a total of 106 paraffin-embedded
LUAD specimens from the archives of the Department of Pathology at
People's Hospital of Beilun (Ningbo, China) were included in the
present study. All patients had undergone surgery from February
2002 to October 2003. All patients were pathologically confirmed as
LUAD. Furthermore, they had no chemotherapy, radiation therapy or
history of surgery. The patients, 66 males (62.3%) and 40 females
(37.7%), ranged in age from 39 to 77 years (mean, 59 years). All
patients received radical surgery for primary tumor and lymph
nodes, and postoperative cisplatin-based adjuvant chemotherapy. The
patients did not receive any radiotherapy or chemotherapy prior to
surgery. The histological type and stage were determined according
to the classification for NSCLC by the World Health Organization
and the International Union against Cancer Tumor-Nodes-Metastasis
staging system (32). All patients
provided written informed consent for tissue use prior to surgery
and this study was approved by the Institute Research Ethics
Committee of People's Hospital of Beilun District (Ningbo, China).
All patients had follow-up records for over 6 years. After the
completion of therapy, patients were observed every 3 months for
the first 3 years and every 6 months thereafter. Overall survival
(OS) was defined as the time from diagnosis to the date of death,
or at the latest date if patients were still alive.
Progression-free survival (PFS) was determined from the first day
of treatment to the earliest signs of disease progression as
identified by CT or MRI, or death from any cause.
Plasmids, siRNA and transfection
Plasmids of HA-14-3-3σ were obtained from Professor
Jie Xu from The Second Affiliated Hospital of the Third Military
Medical University (Army Medical University, Chongqing, China). The
targeting plasmids were delivered into NCI-H1299 LUAD cells using
the Lipofectamine® 2000 transfection reagent (Cat No:
11668019; Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. siRNA designed to target 14-3-3σ
(si-14-3-3σ) was purchased from Sangon Biotech Co., Ltd., including
si-14-3-3σ-1# (5′-GGAUCCCACUCUUCUUGCA-3′) and si-14-3-3σ-2#
(5′-GACCAUGUUUCCUCUCAAU-3′). The sequence for the scrambled control
siRNA was 5′-AUUGUAUGCGAUCGCAGACUU-3′. For transient transfection,
control and si-14-3-3σ siRNA (#1 and #2) were mixed with
Lipofectamine® 2000 and then added to the cell culture
medium of A-549 LUAD cells according to the manufacturer's
protocol. Finally, western blot analysis was used to assess the
transfection efficiency.
ATP-lite cell proliferation assay
Cells were seeded into 96-well plates in triplicate
at an initial density of 0.2×104 cells/well. Cells were
processed using the ATP-lite assay (cat. no. 6016739; Perkin-Elmer,
Inc.) to assess cell proliferation at various time-points (24, 48,
72, 96 and 120 h) according to the manufacturer's protocol
(33). Luminescence was read using
a LD 400 plate reader (Beckman Coulter, Inc.). Each experiment was
performed in triplicate.
Colony-formation assay
After transduction with targeting plasmids or small
interfering (si)RNA, the stably transfected A-549 or NCI-H1299
cells were seeded into 6-cm dishes in triplicate at a density of
0.5×103 cells/well and incubated at 37°C for 14 days.
The colonies were fixed with 4% paraformaldehyde, stained with
crystal violet and then counted (33).
Soft agar assay
A standard soft agar assay was done as detailed
previously (34). In brief,
5×103 of parental cells or their transfectants
(14-3-3σ-overexpressing or 14-3-3σ silenced, along with the vector
controls), were suspended in 0.33% agar (cat. no. A5431;
MilliporeSigma) containing 10% FCS in 6-cm dishes. After culture at
37°C for 14 days, the cells were stained with
p-iodonitrotetrazolium (cat. no. V900870; 1 mg/ml;
MilliporeSigma) overnight at 37°C and the colonies with cell
numbers of more than eight were counted in five randomly selected
areas from each dish.
Western blot analysis
Total protein was extracted from cells using RIPA
lysis buffer (50 mM Tris-pH 7.4, 150 mM NaCl, 1% Triton X-100, 1%
sodium deoxycholate and 0.1% SDS; cat. no. P0013B; Beyotime
Institute of Biotechnology) supplemented with phosphatase (cat. no.
B15001; Bimake) and protease inhibitors (cat. no. B14001; Bimake).
BCA assay kit (cat. no. P0011; Beyotime Institute of Biotechnology)
was used to determine the protein concentration and 40 µg protein
was loaded onto the gel per lane. The equal amounts of total
protein were separated by SDS-PAGE (8–15%) and then transferred
onto nitrocellulose (NC) membranes (EMD Millipore). The NC
membranes were blocked with 5% nonfat milk (cat. no. P0216;
Beyotime Institute of Biotechnology) for 1 h at room temperature,
followed by incubation with primary antibodies overnight at 4°C.
The following primary antibodies were used: 14-3-3σ (cat. no.
sc-100638; 1:1,000 dilution; Santa Cruz Biotechnology, Inc.), poly
(ADP-ribose) polymerase (PARP; cat. no. 13371-1-AP; 1:1,000
dilution; ProteinTech Group, Inc.), Caspase-3 (cat. no. 66470-2-Ig;
1:1,000 dilution; ProteinTech Group, Inc.), GAPDH (cat. no.
60004-1-Ig; 1:1,000 dilution; ProteinTech Group, Inc.) and
hemagglutinin (HA)-tag (cat. no. 51064-2-AP; 1:1,000 dilution;
ProteinTech Group, Inc.). The NC membranes were then incubated with
the following corresponding secondary antibodies: Goat anti-rabbit
IgG (cat. no. SA00001-2; 1:2,000 dilution; ProteinTech Group,
Inc.); goat anti-mouse IgG (cat. no. SA00001-1; 1:2,000 dilution;
ProteinTech Group, Inc.); goat anti-rat IgG (cat. no. SA00001-15;
1:2,000 dilution; ProteinTech Group, Inc.) for 1 h at room
temperature. ECL reagent (cat. no. 32106; Thermo Fisher Scientific,
Inc.) was used to detect the signals.
Immunohistochemical (IHC) analysis and
scoring
IHC analysis was used to study 14-3-3σ protein
expression in human LUAD tissues and normal adjacent tissues as
controls. The slides were dewaxed in xylene, rehydrated with an
alcohol gradient, immersed in 3% hydrogen peroxide for 10 min to
block endogenous peroxidase activity and subjected to antigen
retrieval by pressure cooking in Tris/EDTA (pH=8.0) for 10 min. The
slides were then incubated with 14-3-3σ primary antibody overnight
at 4°C. After incubation with the secondary antibody for 30 min at
37°C, the specimens were stained with a DAB staining kit (cat. no.
ab64238; Abcam). Finally, the sections were counterstained with
hematoxylin, dehydrated and fixed. The brown granules in the
cytoplasm were considered as positive staining for 14-3-3σ. For the
evaluation of nuclear and cytoplasmic staining, the staining
intensity was scored as follows (35): Negative (score 0); weak (score 1);
moderate (score 2) and strong (score 3). The degree of staining was
divided into four categories according to the percentage of stained
cells in the field: Negative (score 0%), 0-25% (score 1), 26-75%
(score 2), 56-100% (score 3) and 76-100% (score 4). The results
reported as the expression score were the product of the above two
scores. Protein expression was then evaluated by jointly assessing
the intensity and extent of staining. IHC staining was assessed and
scored by two independent researchers (JL and CDZ), who were
blinded to the clinicopathological data.
Selection of cutoff score for 14-3-3σ
expression
A receiver operating characteristic (ROC) curve
analysis was performed for the selection of the cutoff value of the
14-3-3σ IHC score for OS and PFS, as described previously (36). In brief, the sensitivity and
specificity of the outcome being studied for each score were
plotted to generate an ROC curve. The score localized closest to
the point of maximum sensitivity and specificity, the point on the
curve (0.0, 1.0), was selected as the cutoff score leading to the
highest number of tumors correctly classified as with or without
the respective outcome. To facilitate the ROC curve analysis, the
patients' outcome characteristics were dichotomized by survival
[death vs. other outcome (censored, alive or death from other
causes)].
Animal experiment
All experimental procedures using mice were
performed in accordance with protocols approved by the Laboratory
Animal Welfare and Ethics Committee of The Third People's Hospital
of Yunnan Province (Kunming, China). A total of eight BALB/C nude
mice (nu/nu; male; aged, 4–6 weeks; body weight, 18–20 g) were
purchased from Beijing Vital River Laboratory Animal Technology
Co., Ltd. and each experimental group consisted of four mice. In
brief, 5×105 NCI-H1299 cells stably transfected with
Vector or HA-14-3-3σ were mixed 1:1 with Matrigel® in a
total volume of 0.2 ml and injected subcutaneously into the right
flank of each mouse (37). Tumor
size and mouse body weight were measured twice a week with a
vernier caliper and precise analytical balance, respectively. On
day 28 after tumor cell injection, mice were euthanized by carbon
dioxide asphyxiation (30% vol/min) when certain tumors reached the
size limits set by the guidelines ‘Tumor Induction in Mice and
Rats-UCSF Animal Care and Use’ (38). The largest sizes of tumor in the
study were no bigger than 16 mm. At 10 min after the animals were
euthanized by carbon dioxide asphyxiation, the absence of vital
signs was checked to confirm that the animals had died. In this
study, the nude mice did not suddenly lose 20-25% weight
(cachexia); therefore, no other humane endpoint was used in this
experiment except for tumor size limitation. Tumors were excised,
weighed and frozen or fixed in 4% paraformaldehyde and
paraffin-embedded for IHC analysis. The mean tumor volume (TV) was
calculated according to the following equation: TV=(L ×
W2)/2, where L is the length and W is the width of the
tumor.
IHC staining of mouse tumors was performed as
described above. In brief, after deparaffinization, rehydration,
antigen retrieval and blocking, the tissue slides were incubated
overnight at 4°C with the indicated antibodies. The following
primary antibodies were used: Ki-67 (1:1,000 dilution; cat. no.
9449; Cell Signaling Technology, Inc.), p21 (1:200 dilution; cat.
no. 10355-1-AP; ProteinTech Group, Inc.), p27 (1:200 dilution; cat.
no. 25614-1-AP; ProteinTech Group, Inc.) and cleaved-Caspase-3
(Asp175) (1:200 dilution; cat. no. 9664; Cell Signaling Technology,
Inc.).
Statistical analysis
All data were expressed as the mean ± standard
deviation and statistical charts were prepared using GraphPad Prism
7.0 software (GraphPad Software, Inc.) and SPSS 20.0 (IBM
Corporation). Transcripts per million (TPM) values were used and
survival analysis was applied for assessing differential expression
of 14-3-3σ mRNA levels in UALCAN. For survival analysis, the
χ2 test or Fisher's exact test was used to assess the
association between 14-3-3σ protein expression and
clinicopathological variables. The multivariate Cox proportional
hazards model was used to estimate the hazard ratios and 95%
confidence intervals for patient outcomes. The associations between
14-3-3σ protein expression and OS or PFS were determined by
Kaplan-Meier analysis. The log-rank test was performed to assess
differences in survival probability between patient subsets.
Differences between two groups were analyzed using an unpaired
Student's t-test, while the expression of 14-3-3σ in tumor tissues
and matched normal tissues was compared using a paired Student's
t-test. Comparisons of multiple groups were performed using one-way
ANOVA followed by Dunnett's or Tukey's test. P<0.05 was
considered to indicate a statistically significant difference.
Results
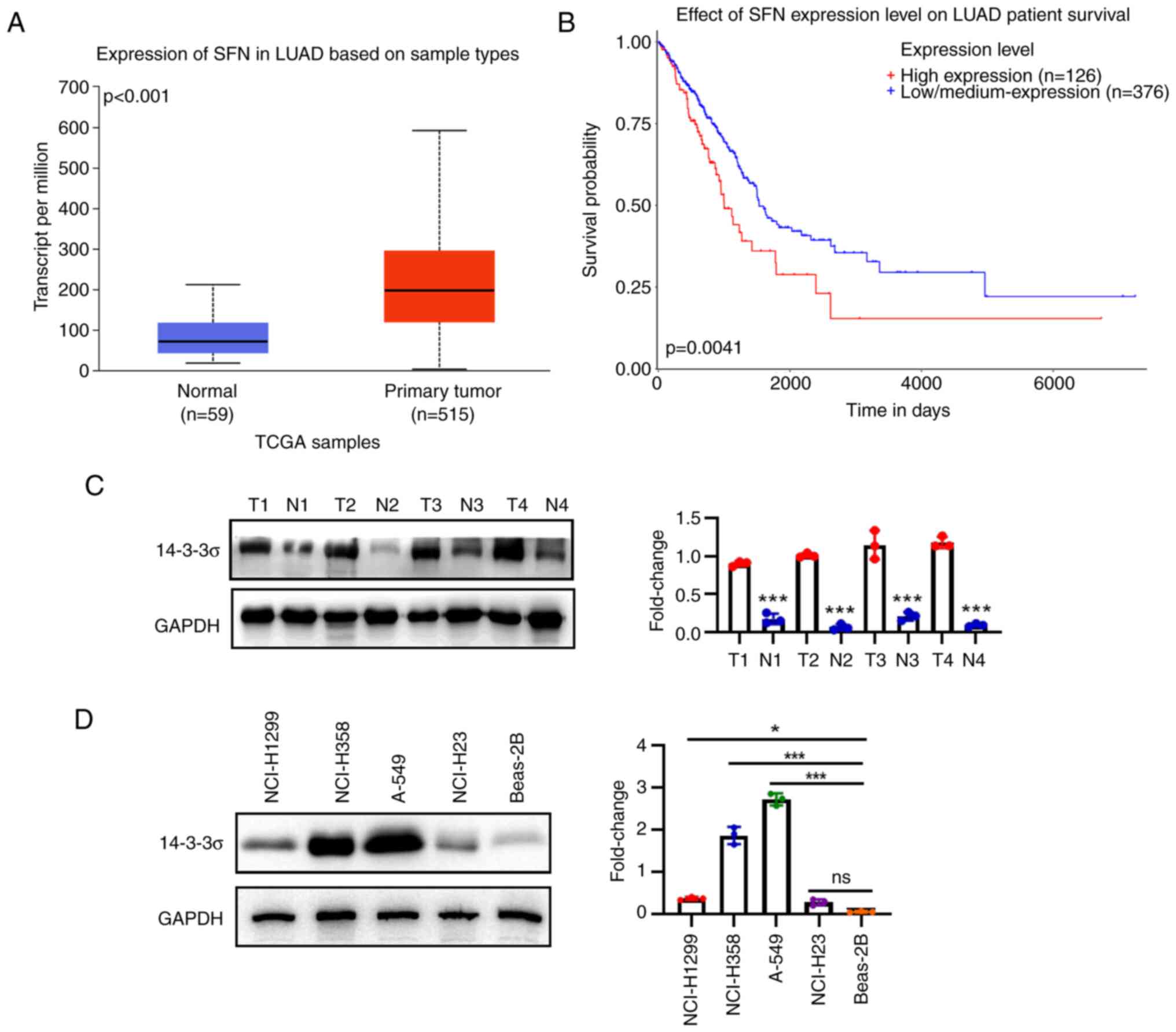
Integrated analysis indicates that
14-3-3σ is an oncogene in LUAD
It was previously reported that 14-3-3σ expression
is present in LUAD (27), but the
biofunction role of 14-3-3ơ and its clinicopathological/prognostic
significance in LUAD have remained elusive. In the present study,
14-3-3σ mRNA expression was analyzed in LUAD using the data from
UALCAN (http://ualcan.path.uab.edu.).
Significant differences were identified between LUAD tissues and
their normal counterparts (Fig.
1A). For the dataset from UALCAN, Kaplan-Meier survival
analysis also indicated that patients with higher 14-3-3σ mRNA
expression had poor overall survival (Fig. 1B). Next, the protein expression of
14-3-3σ was examined in LUAD and adjacent normal tissues by western
blot analysis (Fig. 1C), and it
was observed that 14-3-3σ was upregulated in LUAD tissues, whereas
its expression was low in the normal adjacent tissue. Consistent
with this result, the expression of 14-3-3σ in four established
LUAD cell lines (A-549, NCI-H1299, NCI-H358 and NCI-H23) and a
human bronchial epithelial cell line (Beas-2B) was analyzed by
western blot analysis, indicating that 14-3-3σ was highly expressed
in LUAD cell lines (Fig. 1D).
Taken together, 14-3-3σ may function as an oncogene for LUAD.
14-3-3σ expression and survival:
Univariate analysis
Consistent with the western blot results, IHC
analysis revealed that 14-3-3σ was present in the cytoplasm and
highly expressed in LUAD tissues (Fig.
2Aa1 and a2), while it was weakly
expressed in normal paired lung tissues (Fig. 2Ac1 and c2).
Next, ROC curve analysis was employed to determine a cutoff score
for 14-3-3σ expression to predict survival (Fig. 2B). The cutoff score for 14-3-3σ to
predict OS and PFS in patients with LUAD (n=106) was 3.8
(P<0.001) and 4.1 (P=0.017), respectively. Therefore, a 14-3-3σ
expression score of 4.0 (>4.0 vs. ≤4.0) was selected as the
unified cutoff point for survival analysis in patients with LUAD.
The ROC-derived 14-3-3σ cutoff score for patients with LUAD was 4
and the cohort (n=106) was divided into a high (52/106, 49.1%) and
a low (54/106, 50.9%) expression group. Kaplan-Meier analysis
further indicated that high expression of 14-3-3σ predicted an
inferior OS and PFS in patients with LUAD (P<0.001, Fig. 2C).
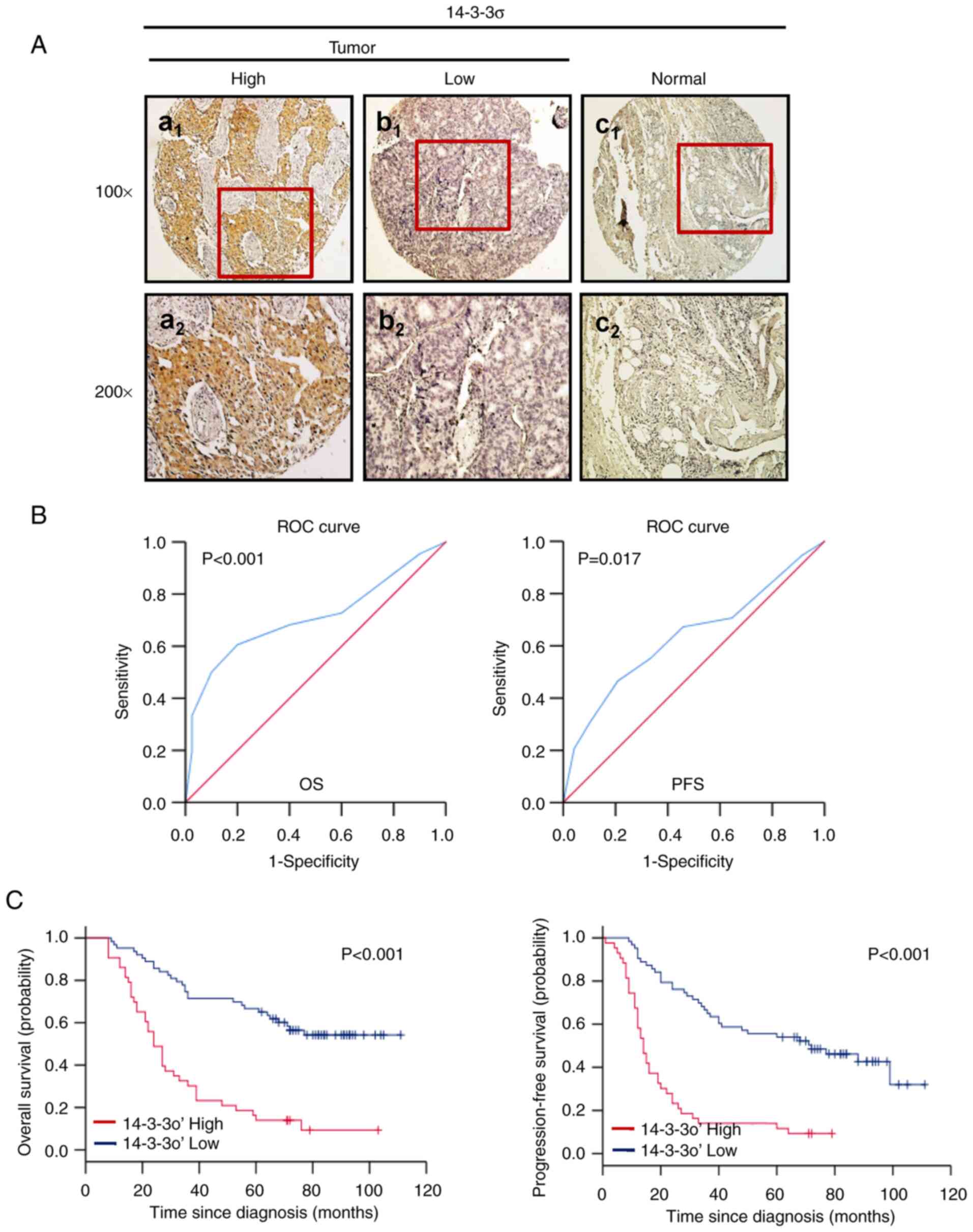 | Figure 2.Protein levels of 14-3-3ơ are
significantly upregulated in LUAD and predict poor patient
survival. (A) Representative IHC staining of 14-3-3ơ in LUAD and
normal adjacent tissues: (a1) 14-3-3ơ was strongly
expressed in the cytoplasm of LUAD tissue; (b1) 14-3-3ơ
was weakly expressed in the cytoplasm of LUAD tissue;
(c1) 14-3-3ơ was negatively expressed in paired normal
adjacent tissues from the same case (×100). (a2,
b2 and c2) provide magnified windows (×200)
from a1, b1 and c1, respectively.
The images of the IHC staining are representative of all samples.
(B) ROC curve analyses of the predictive value of 14-3-3ơ in
patients with LUAD. OS (left) or PFS (right) in the overall
patients. For each IHC score, the sensitivity and specificity for
the outcome being studied were plotted, thus generating an ROC
curve. The optimal cutoff score for 14-3-3ơ for OS and PFS was 4.1
and 3.8, respectively. (C) Kaplan-Meier survival analysis of the
impact of 14-3-3ơ expression on survival of patients with LUAD.
High expression of 14-3-3ơ was significantly associated with poor
OS (left) and PFS (right) in the dataset. LUAD, lung
adenocarcinoma; OS, overall survival; PFS, progression-free
survival; ROC, receiver operating characteristic; IHC,
immunohistochemistry. |
14-3-3σ expression in LUAD tissues and
clinical features of patients
The association between 14-3-3σ expression and the
clinical characteristics of patients with LUAD was then examined.
As presented in Table II, high
expression of 14-3-3σ in LUAD tissues was significantly positively
associated with the tumor stage (P=0.001). In addition, high
expression of 14-3-3σ was associated with tumor size (P=0.036) and
recurrence (P<0.001, Table
II). However, no association between 14-3-3σ and other patient
characteristics was observed, including patient age, sex or node
stage.
 | Table II.Association of 14-3-3ơ expression
with characteristics of patients with lung adenocarcinoma. |
Table II.
Association of 14-3-3ơ expression
with characteristics of patients with lung adenocarcinoma.
|
|
| 14-3-3ơ |
|
|---|
|
|
|
|
|
|---|
| Variable | Total n | High | Low | P-value |
|---|
| Age, years |
|
|
| 0.078 |
|
≥59.00 | 56 | 32 | 24 |
|
|
<59.00 | 50 | 20 | 30 |
|
| Sex |
|
|
| 0.803 |
|
Male | 66 | 33 | 33 |
|
|
Female | 40 | 19 | 21 |
|
| Tumor size
(mm) |
|
|
| 0.036 |
|
<20 | 7 | 5 | 6 |
|
|
20-50 | 53 | 18 | 31 |
|
|
>50 | 46 | 29 | 17 |
|
| Tumor stage |
|
|
| 0.001 |
|
T1+T2 | 52 | 17 | 35 |
|
|
T3+T4 | 54 | 35 | 19 |
|
| Node stage |
|
|
| 0.227 |
|
N0+N1 | 45 | 19 | 26 |
|
|
N2+N3 | 61 | 33 | 28 |
|
| Recurrence |
|
|
| <0.001 |
|
Positive | 61 | 46 | 15 |
|
|
Negative | 45 | 6 | 39 |
|
Multivariate Cox regression
analysis
To avoid the influence of various factors in the
univariate analysis, the expression of 14-3-3σ as well as other
parameters were examined in a multivariate Cox regression analysis
(Table III). Among the patients
with LUAD, 14-3-3σ was indeed determined to be a significant
independent prognostic factor for OS (hazard ratio, 4.878; 95% CI,
2.895-8.219; P<0.001; Table
III) and PFS (hazard ratio, 3.041; 95% CI, 1.878-4.923;
P<0.001; Table III).
Furthermore, the node stage was also identified as an independent
prognostic parameter for OS (hazard ratio, 2.396; 95% CI,
1.115-5.148; P=0.025; Table III)
and PFS (hazard ratio, 2.471; 95% CI, 1.168-5.228; P=0.018;
Table III) in the patients with
LUAD. However, other important prognostic factors, including age,
sex, smoking, tumor stage and metastasis, were no significant
independent prognostic factors for LUAD according to this analysis,
implying that a larger cohort may be required in future
studies.
 | Table III.Results of multivariate Cox
proportional hazards analysis in lung adenocarcinoma. |
Table III.
Results of multivariate Cox
proportional hazards analysis in lung adenocarcinoma.
|
| For death | For survival with
relapse |
|---|
|
|
|
|
|---|
| Variable | Hazard ratio | 95% confidence
interval | P-value | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Age (≥59.00 vs.
<59 years) | 1.390 | (0.888-2.176) | 0.150 | 1.168 | (0.749-1.822) | 0.492 |
| Sex (male vs.
female) | 0.624 | (0.301-1.292) | 0.204 | 0.869 | (0.407-1.858) | 0.718 |
| Smoking (yes vs.
no) | 0.939 | (0.468-1.883) | 0.859 | 1.052 | (0.510-2.168) | 0.892 |
| Tumor size, mm |
|
|
|
|
|
|
|
<20 | 1.024 | (0.357-2.937) | 0.965 | 0.781 | (0.275-2.218) | 0.642 |
|
20-50 | 1.287 | (0.758-2.185) | 0.351 | 1.153 | (0.679-1.958) | 0.598 |
|
>50 | 1 (Reference) | 1 |
| 1 (Reference) | 1 |
|
| Tumor stage
(T4 + T3 vs. T2 +
T1) | 1.130 | (0.667-1.915) | 0.649 | 1.307 | (0.777-2.201) | 0.313 |
| Nodal stage
(N3 + N2 vs. N1 +
N0) | 2.396 | (1.115-5.148) | 0.025 | 2.471 | (1.168-5.228) | 0.018 |
| 14-3-3ơ (high vs.
low) | 4.878 | (2.895-8.219) | <0.001 | 3.041 | (1.878-4.923) | <0.001 |
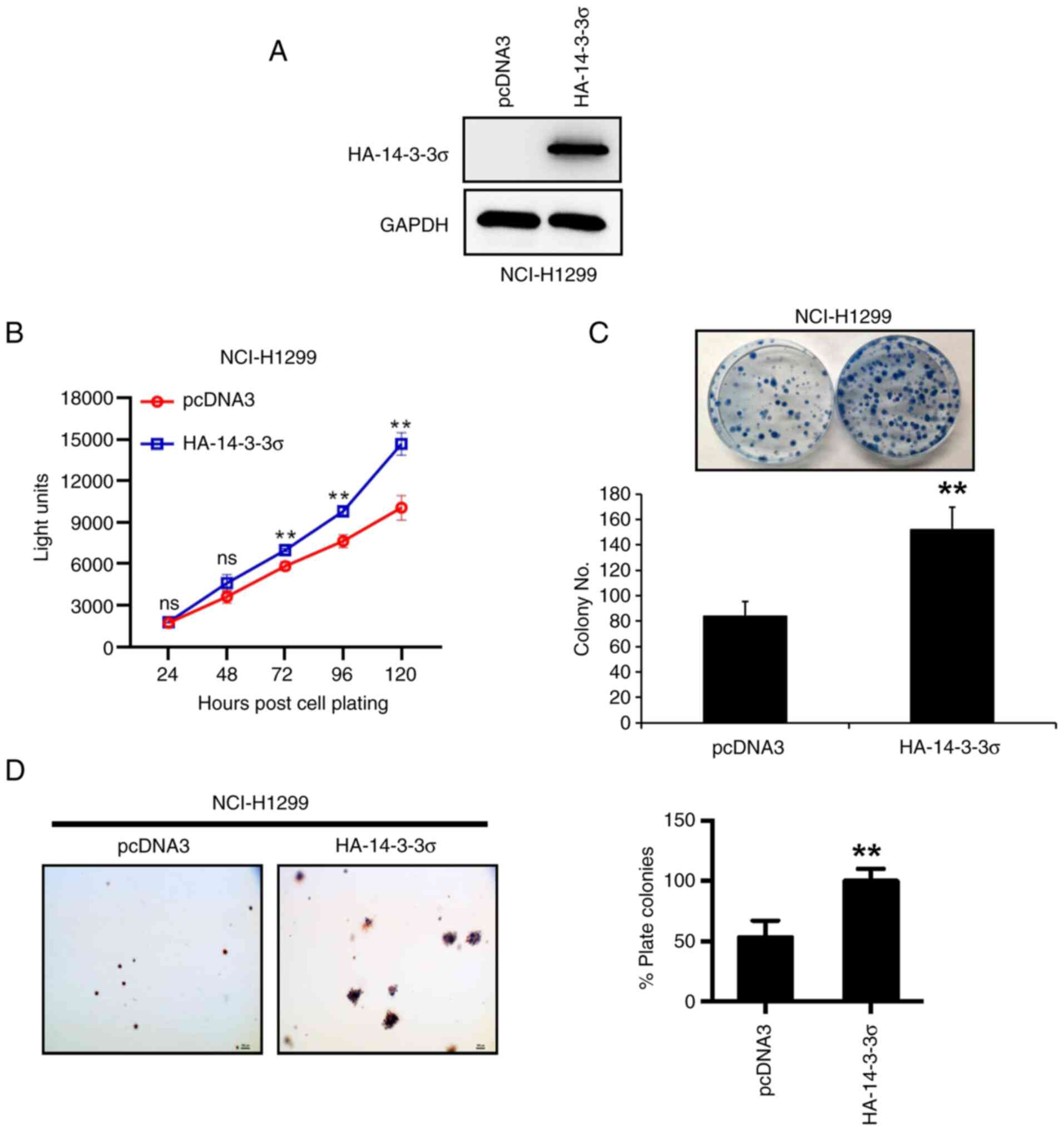
14-3-3σ overexpression promotes growth
of LUAD cells in vitro
The above results suggested that high expression of
14-3-3σ was associated with progression and poor prognosis of LUAD.
Thus, the biological function of 14-3-3σ in LUAD was further
investigated in vitro. First, 14-3-3σ plasmid was
transfected into NCI-H1299 LUAD cells (Fig. 3A), as the native cell line
expressed 14-3-3σ at a low level (Fig.
1D), and it was indicated that overexpression of 14-3-3σ
promoted cell proliferation and survival, as measured by ATP-lite
(Fig. 3B) and colony formation
assays (Fig. 3C), respectively.
Furthermore, the anchorage-independent growth of NCI-H1299 cells,
measured by the soft agar assay, was increased upon 14-3-3σ
overexpression (Fig. 3D). Thus,
14-3-3σ appeared to have growth-promoting activity in LUAD
cells.
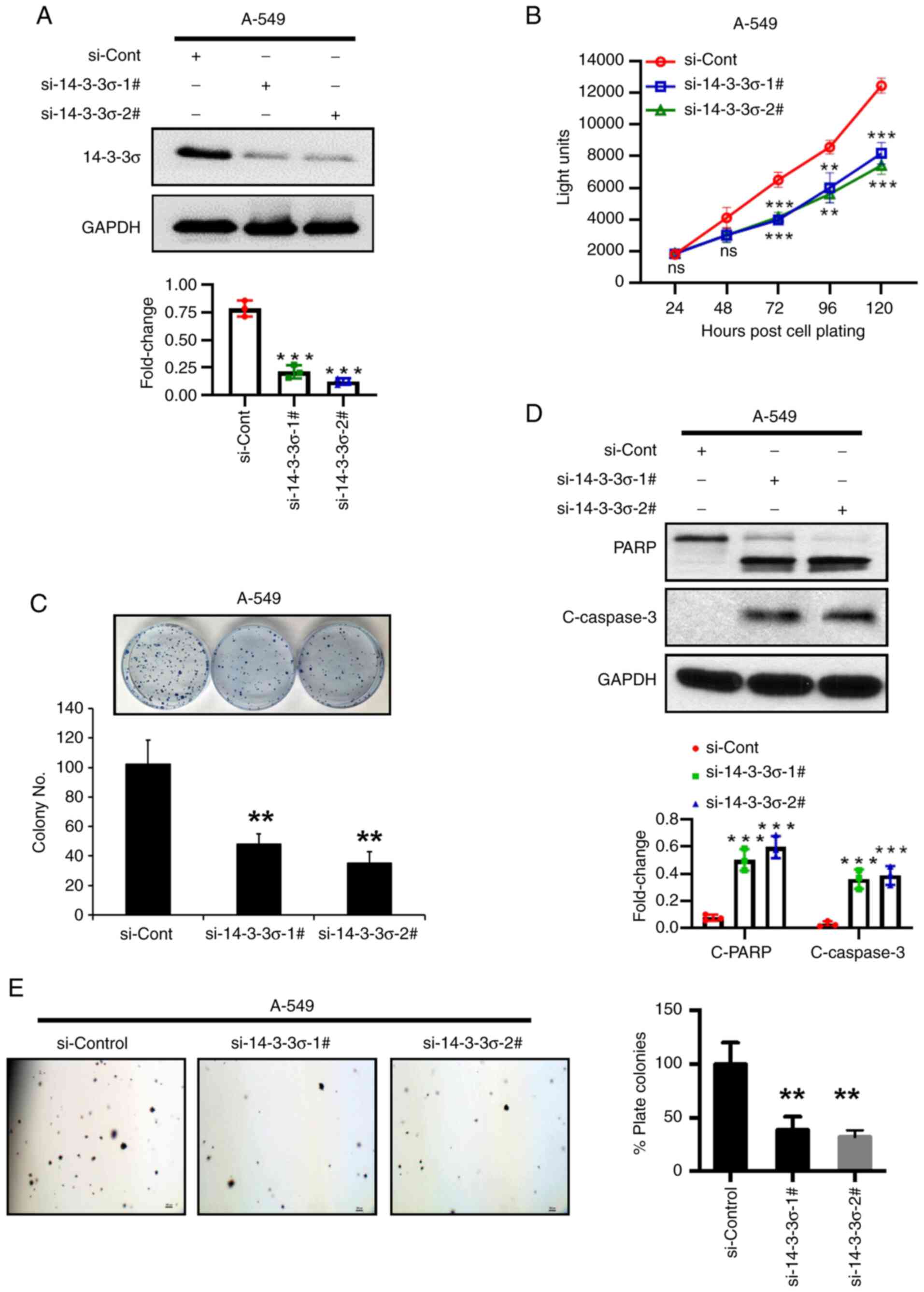
14-3-3σ silencing inhibits LUAD cell
growth by inducing apoptosis
To further validate 14-3-3σ as an oncogene for LUAD,
siRNA-based knockdown experiments were performed in A-549 LUAD
cells, as the native cell line has high expression of 14-3-3σ
(Fig. 1D). The results indicated
that 14-3-3σ depletion suppressed cell proliferation and the
colony-forming ability of the cells (Fig. 4A-C). The nature of survival
inhibition upon 14-3-3σ depletion was demonstrated to be induction
of apoptosis, as evidenced by an increase in PARP cleavage and
Caspase-3 cleavage (Fig. 4D).
Furthermore, the anchorage-independent growth of A-549 LUAD cells
was inhibited by up to 60% on 14-3-3σ depletion (Fig. 4E). Thus, growth suppression upon
14-3-3σ depletion may be mediated via apoptosis induction.
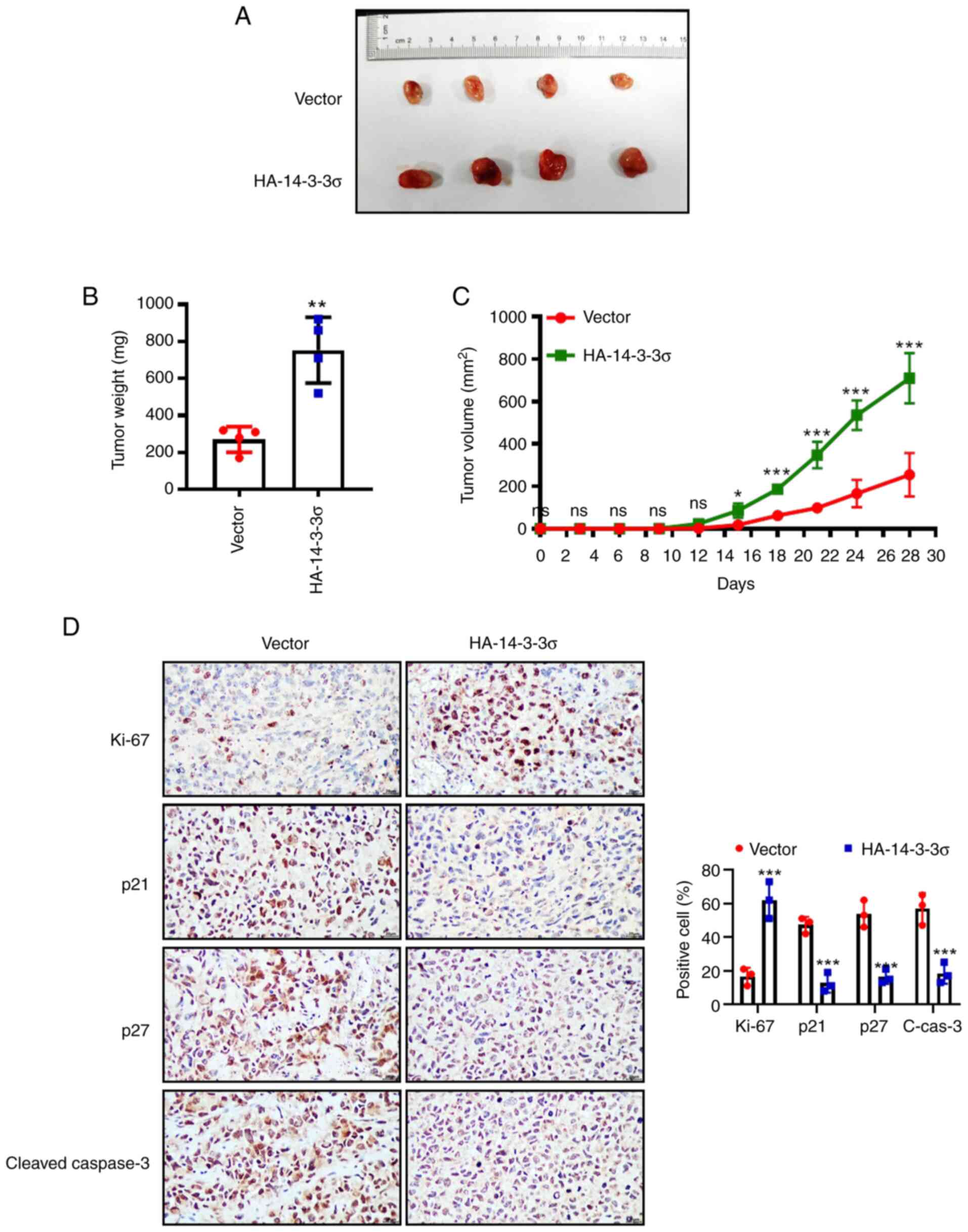
Overexpression of 14-3-3σ promotes
LUAD tumor growth in vivo
Next, the above in vitro findings were
validated in a xenograft model. NCI-H1299 cells with stable
overexpression of 14-3-3σ were inoculated into the right flank of
nude mice and, as expected, overexpression of 14-3-3σ profoundly
promoted tumor growth in vivo, increasing the tumor size and
weight compared to the vector control (Fig. 5A-C). Likewise, IHC staining of
tumor tissues also revealed that overexpression of 14-3-3σ markedly
promoted tumor growth (increase of Ki-67 and decrease of p21 and
p27) and inhibited apoptosis (decrease of cleaved-Caspase-3)
(Fig. 5D). Collectively, the
results of the in vitro cell experiments and in vivo
xenograft models consistently demonstrated that overexpression of
14-3-3σ significantly promoted the growth of LUAD. At the same
time, clinical data analysis also confirmed that 14-3-3σ high
expression was positively correlated with poor prognosis of
patients with LUAD.
Discussion
LUAD is one of the most common causes of global
cancer-related mortalities worldwide (39). Although previous studies have
indicated that numerous abnormally expressed genes in LUAD may help
classify prognostic risks (40–42),
there is still an urgent requirement for novel molecular markers to
identify tumor progression and predict prognosis. The 14-3-3 family
of proteins have received considerable attention in the past few
years due to their involvement in cancers by regulating a variety
of cellular processes (10,11).
14-3-3ơ was originally identified as a tumor suppressor gene
(13–15), which was upregulated by p53 upon
DNA damage, and reported to sequester the essential mitosis
initiation complex cdc2-cyclin B1 into the nucleus, thus preventing
the initiation of mitosis (42–45).
As a result, 14-3-3σ induces G2 arrest, allowing DNA damage repair
to take place (18–20). However, there is increasing
evidence that 14-3-3σ is an oncogene in cancers (22–24).
Although 14-3-3σ expression was also observed in LUAD (27), its significance in the prognosis
and function of patients with LUAD has remained largely
elusive.
In the present study, certain findings regarding the
important role of 14-3-3ơ in LUAD were presented and supporting
evidence for the following was provided: i) 14-3-3ơ is strongly
expressed in both LUAD cells and tissues, Similar to the findings
in previous studies (27); ii)
overexpression of 14-3-3ơ significantly promotes cell growth and
survival in both in vitro and in vivo LUAD cancer
models, whereas 14-3-3ơ depletion produces opposite effects; iii)
14-3-3ơ depletion also induces cell apoptosis, as evidenced by the
increase of PARP cleavage and Caspase-3 cleavage; iv) high
expression of 14-3-3ơ positively correlates with tumor size, tumor
stage and recurrence, worse OS and PFS in LUAD; v) multivariate
analyses further revealed that 14-3-3ơ is an independent prognostic
biomarker for patients with LUAD. Taken together, the findings of
the present study provided evidence that overexpression of 14-3-3ơ
may contribute to an increased degree of malignancy and unfavorable
prognostic phenotype in LUAD.
Data reported by certain studies are completely
contradictory regarding the prognostic impact of 14-3-3σ in
different human cancers. Loss of expression of 14-3-3σ has been
documented to be associated with poor prognosis in patients with
breast cancer (46,47), endometrial (48) and ovarian cancers (49). However, consistent with findings in
pancreatic cancer (50) and
colorectal cancer (51), the
results of the present study suggested that high expression of
14-3-3σ was positively associated with poor survival. The
underlying mechanism(s) by which 14-3-3σ affected cancer prognosis
may depend on the intrinsic properties of the tumor type. More
recently, studies have indicated that overexpression of 14-3-3σ is
associated with tumor progression in pancreatic cancer (26,50),
which may further explain the present findings that higher 14-3-3σ
expression was predominantly detected in more advanced tumor
stages.
In conclusion, in the present study, 14-3-3σ was
identified as an independent prognostic biomarker for OS and PFS in
LUAD. The results suggested that 14-3-3σ has clinical value in
predicting the prognosis of LUAD and identifying patients with LUAD
at high risk of progression and recurrence. Although the present
study reported the effects of 14-3-3σ in LUAD, the regulatory
mechanisms remain to be elucidated. In the future, further research
will be performed to investigate how 14-3-3σ regulates cellular
processes in LUAD.
Acknowledgements
The authors would like to thank Professor Jie Xu
(The Second Affiliated Hospital of the Third Military Medical
University, Army Medical University, Chongqing, China) for
providing cell lines, plasmids and research cooperation.
Funding
This work was supported by the National Natural Science
Foundation of China (grant no. 81860017 to HFL) and Yunnan
Provincial Department of Education Science Research Fund Project
(grant no. 2019J0792 to JL).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Author's contributions
HFL and YBC were involved in the conception and
design of the research. HFL edited and revised the manuscript. JFF,
JL and CDZ performed the experiments, interpreted the results of
the experiments and prepared the figures. YBC and JG drafted the
manuscript. JG was also responsible for the collation of clinical
data. HFL and YBC confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The protocol for the study involving human
participants was reviewed and approved by the Ethics Committee of
the People's Hospital of Beilun District of China [Ningbo, China;
no. 2021-23(YS)]. The patients/participants provided their written
informed consent to participate in this study. The animal
experiment was approved by the Institutional Animal Care and Use
Committee of the People's Hospital of Beilun District of China
(Ningbo, China; no. NBU20220098).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Robinson KW and Sandler AB: The role of
MET receptor tyrosine kinase in non-small cell lung cancer and
clinical development of targeted anti-MET agents. Oncologist.
18:115–122. 2013. View Article : Google Scholar
|
|
3
|
Kobayashi Y and Mitsudomi T: Not all
epidermal growth factor receptor mutations in lung cancer are
created equal: Perspectives for individualized treatment strategy.
Cancer Sci. 107:1179–1186. 2016. View Article : Google Scholar
|
|
4
|
Karachaliou N, Santarpia M, Cao MZ,
Teixido C, Sosa AE, Berenguer J, Capote AR, Altavilla G and Rosell
R: Anaplastic lymphoma kinase inhibitors in phase I and phase II
clinical trials for non-small cell lung cancer. Expert Opin
Investig Drugs. 26:713–722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roviello G: The distinctive nature of
adenocarcinoma of the lung. Onco Targets Ther. 8:2399–2406. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sacco JJ, Al-Akhrass H and Wilson CM:
Challenges and strategies in precision medicine for non-small-cell
lung cancer. Curr Pharm Des. 22:4374–4385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gainor JF, Dardaei L, Yoda S, Friboulet L,
Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K,
Singh M, et al: Molecular mechanisms of resistance to first- and
second-generation ALK inhibitors in ALK-rearranged lung cancer.
Cancer Discov. 6:1118–1133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kobayashi S, Boggon TJ, Dayaram T, Janne
PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG and Halmos
B: EGFR mutation and resistance of non-small-cell lung cancer to
gefitinib. N Engl J Med. 352:786–792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Raungrut P, Wongkotsila A,
Lirdprapamongkol K, Svasti J, Geater SL, Phukaoloun M, Suwiwat S
and Thongsuksai P: Prognostic significance of 14-3-3gamma
overexpression in advanced non-small cell lung cancer. Asian Pac J
Cancer Prev. 15:3513–3518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Porter GW, Khuri FR and Fu H: Dynamic
14-3-3/client protein interactions integrate survival and apoptotic
pathways. Semin Cancer Biol. 16:193–202. 2006. View Article : Google Scholar
|
|
11
|
Morrison DK: The 14-3-3 proteins:
Integrators of diverse signaling cues that impact cell fate and
cancer development. Trends Cell Biol. 19:16–23. 2009. View Article : Google Scholar
|
|
12
|
Huang Y, Yang M and Huang W: 14-3-3 σ: A
potential biomolecule for cancer therapy. Clin Chim Acta.
511:50–58. 2020. View Article : Google Scholar
|
|
13
|
Ling C, Zuo D, Xue B, Muthuswamy S and
Muller WJ: A novel role for 14-3-3sigma in regulating epithelial
cell polarity. Genes Dev. 24:947–956. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ling C, Su VM, Zuo D and Muller WJ: Loss
of the 14-3-3sigma tumor suppressor is a critical event in
ErbB2-mediated tumor progression. Cancer Discov. 2:68–81. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boudreau A, Tanner K, Wang D, Geyer FC,
Reis-Filho JS and Bissell MJ: 14-3-3sigma stabilizes a complex of
soluble actin and intermediate filament to enable breast tumor
invasion. Proc Natl Acad Sci USA. 110:E3937–E3944. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lodygin D and Hermeking H: The role of
epigenetic inactivation of 14-3-3sigma in human cancer. Cell Res.
15:237–246. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nacht M, Ferguson AT, Zhang W, Petroziello
JM, Cook BP, Gao YH, Maguire S, Riley D, Coppola G, Landes GM, et
al: Combining serial analysis of gene expression and array
technologies to identify genes differentially expressed in breast
cancer. Cancer Res. 59:5464–5470. 1999.PubMed/NCBI
|
|
18
|
Ferguson AT, Evron E, Umbricht CB, Pandita
TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA,
Stampfer MR and Sukumar S: High frequency of hypermethylation at
the 14-3-3 sigma locus leads to gene silencing in breast cancer.
Proc Natl Acad Sci USA. 97:6049–6054. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lodygin D, Diebold J and Hermeking H:
Prostate cancer is characterized by epigenetic silencing of
14-3-3sigma expression. Oncogene. 23:9034–9041. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iwata N, Yamamoto H, Sasaki S, Itoh F,
Suzuki H, Kikuchi T, Kaneto H, Iku S, Ozeki I, Karino Y, et al:
Frequent hypermethylation of CpG islands and loss of expression of
the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene.
19:5298–5302. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lodygin D, Epanchintsev A, Menssen A,
Diebold J and Hermeking H: Functional epigenomics identifies genes
frequently silenced in prostate cancer. Cancer Res. 65:4218–4227.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okada T, Masuda N, Fukai Y, Shimura T,
Nishida Y, Hosouchi Y, Kashiwabara K, Nakajima T and Kuwano H:
Immunohistochemical expression of 14-3-3 sigma protein in
intraductal papillary-mucinous tumor and invasive ductal carcinoma
of the pancreas. Anticancer Res. 26((4B)): 3105–3110.
2006.PubMed/NCBI
|
|
23
|
Kuramitsu Y, Baron B, Yoshino S, Zhang X,
Tanaka T, Yashiro M, Hirakawa K, Oka M and Nakamura K: Proteomic
differential display analysis shows up-regulation of 14-3-3 sigma
protein in human scirrhous-type gastric carcinoma cells. Anticancer
Res. 30:4459–4465. 2010.PubMed/NCBI
|
|
24
|
Ito Y, Miyoshi E, Uda E, Yoshida H, Uruno
T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Matsuura N, et al:
14-3-3 sigma possibly plays a constitutive role in papillary
carcinoma, but not in follicular tumor of the thyroid. Cancer Lett.
200:161–166. 2003. View Article : Google Scholar
|
|
25
|
Neal CL and Yu D: 14-3-3ζ as a prognostic
marker and therapeutic target for cancer. Expert Opin Ther Targets.
14:1343–1354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guweidhi A, Kleeff J, Giese N, El Fitori
J, Ketterer K, Giese T, Büchler MW, Korc M and Friess H: Enhanced
expression of 14-3-3sigma in pancreatic cancer and its role in cell
cycle regulation and apoptosis. Carcinogenesis. 25:1575–1585. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shiba-Ishii A, Kano J, Morishita Y, Sato
Y, Minami Y and Noguchi M: High expression of stratifin is a
universal abnormality during the course of malignant progression of
early-stage lung adenocarcinoma. Int J Cancer. 129:2445–2453. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qi W, Liu X, Qiao D and Martinez JD:
Isoform-specific expression of 14-3-3 proteins in human lung cancer
tissues. Int J Cancer. 113:359–363. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cetintas VB, Tetik A, Cok G, Kucukaslan
AS, Kosova B, Gunduz C, Veral A and Eroglu Z: Role of 14-3-3sigma
in resistance to cisplatin in non-small cell lung cancer cells.
Cell Biol Int. 37:78–86. 2013. View Article : Google Scholar
|
|
30
|
Radhakrishnan VM, Jensen TJ, Cui H,
Futscher BW and Martinez JD: Hypomethylation of the 14-3-3sigma
promoter leads to increased expression in non-small cell lung
cancer. Genes Chromosomes Cancer. 50:830–836. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang R, Yan B, Li Z, Jiang Y, Mao C, Wang
X and Zhou X: Long non-coding RNA HOX transcript antisense RNA
promotes expression of 14-3-3sigma in non-small cell lung cancer.
Exp Ther Med. 14:4503–4508. 2017.PubMed/NCBI
|
|
32
|
Carbone L: Pain management standards in
the eighth edition of the guide for the care and use of laboratory
animals. J Am Assoc Lab Anim Sci. 51:322–328. 2012.
|
|
33
|
Xu J, Zhou W, Yang F, Chen G, Li H, Zhao
Y, Liu P, Li H, Tan M, Xiong X and Sun Y: The β-TrCP-FBXW2-SKP2
axis regulates lung cancer cell growth with FBXW2 acting as a
tumour suppressor. Nat Commun. 8:140022017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gu Q, Tan M and Sun Y: SAG/ROC2/Rbx2 is a
novel activator protein-1 target that promotes c-Jun degradation
and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced
neoplastic transformation. Cancer Res. 67:3616–3625. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shang W, Liang X, Li S, Li T, Zheng L,
Shao W, Wang Y, Liu F, Ma L and Jia J: Orphan nuclear receptor
Nurr1 promotes Helicobacter pylori-associated gastric
carcinogenesis by directly enhancing CDK4 expression. EBioMedicine.
53:1026722020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhou WH, Tang F, Xu J, Wu X, Yang SB, Feng
ZY, Ding YG, Wan XB, Guan Z, Li HG, et al: Low expression of Beclin
1, associated with high Bcl-xL, predicts a malignant phenotype and
poor prognosis of gastric cancer. Autophagy. 8:389–400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou W, Xu J, Tan M, Li H, Li H, Wei W and
Sun Y: UBE2M is a stress-inducible dual E2 for neddylation and
ubiquitylation that promotes targeted degradation of UBE2F. Mol
Cell. 70:1008–1024. 2018. View Article : Google Scholar
|
|
38
|
Travis WD, Brambilla E, Burke AP, Marx A
and Nicholson AG: Introduction to The 2015 world health
organization classification of tumors of the lung, pleura, thymus,
and heart. J Thorac Oncol. 10:1240–1242. 2015. View Article : Google Scholar
|
|
39
|
Zhang Y, Yuan Y, Li Y, Zhang P, Chen P and
Sun S: An inverse interaction between HOXA11 and HOXA11-AS is
associated with cisplatin resistance in lung adenocarcinoma.
Epigenetics. 14:949–960. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jin CY, Du L, Nuerlan AH, Wang XL, Yang YW
and Guo R: High expression of RRM2 as an independent predictive
factor of poor prognosis in patients with lung adenocarcinoma.
Aging (Albany NY). 13:3518–3535. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu Y and Tian X: Analysis of genes
associated with prognosis of lung adenocarcinoma based on GEO and
TCGA databases. Medicine (Baltimore). 99:e201832020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Y, Liu X, Liu L, Li J, Hu Q and Sun
R: Expression and prognostic significance of m6A-related genes in
lung adenocarcinoma. Med Sci Monit. 26:e9196442020.
|
|
43
|
Dellinger RW, Karjian PL and Neuteboom ST:
NB1011 induces Ser15 phosphorylation of p53 and activates the G2/M
checkpoint. Anticancer Drugs. 14:449–455. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Holm R, Ali T, Svendsrud DH, Nesland JM,
Kristensen GB and Lyng H: Expression of 14-3-3sigma in cervical
squamous cell carcinomas: Relationship with clinical outcome. Oncol
Rep. 22:11–15. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Horie-Inoue K and Inoue S: Epigenetic and
proteolytic inactivation of 14-3-3sigma in breast and prostate
cancers. Semin Cancer Biol. 16:235–239. 2006. View Article : Google Scholar
|
|
46
|
Simpson PT, Gale T, Reis-Filho JS, Jones
C, Parry S, Steele D, Cossu A, Budroni M, Palmieri G and Lakhani
SR: Distribution and significance of 14-3-3sigma, a novel
myoepithelial marker, in normal, benign, and malignant breast
tissue. J Pathol. 202:274–285. 2004. View Article : Google Scholar
|
|
47
|
Neal CL, Yao J, Yang W, Zhou X, Nguyen NT,
Lu J, Danes CG, Guo H, Lan KH, Ensor J, et al: 14-3-3zeta
overexpression defines high risk for breast cancer recurrence and
promotes cancer cell survival. Cancer Res. 69:3425–3432. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nakayama H, Sano T, Motegi A, Oyama T and
Nakajima T: Increasing 14-3-3 sigma expression with declining
estrogen receptor alpha and estrogen-responsive finger protein
expression defines malignant progression of endometrial carcinoma.
Pathol Int. 55:707–715. 2005. View Article : Google Scholar
|
|
49
|
Akahira J, Sugihashi Y, Suzuki T, Ito K,
Niikura H, Moriya T, Nitta M, Okamura H, Inoue S, Sasano H, et al:
Decreased expression of 14-3-3 sigma is associated with advanced
disease in human epithelial ovarian cancer: Its correlation with
aberrant DNA methylation. Clin Cancer Res. 10:2687–2693. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li Z, Dong Z, Myer D, Yip-Schneider M, Liu
J, Cui P, Schmidt CM and Zhang JT: Role of 14-3-3sigma in poor
prognosis and in radiation and drug resistance of human pancreatic
cancers. BMC Cancer. 10:5982010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Perathoner A, Pirkebner D, Brandacher G,
Spizzo G, Stadlmann S, Obrist P, Margreiter R and Amberger A:
14-3-3sigma expression is an independent prognostic parameter for
poor survival in colorectal carcinoma patients. Clin Cancer Res.
11:3274–3279. 2005. View Article : Google Scholar : PubMed/NCBI
|