Introduction
Gastric cancer (GC) ranks the 5th malignancy and the
3rd cause of cancer-related death according to the global cancer
statistics in 2018 (1). GC
progresses rapidly and numerous of the cancer cases are at a late
stage at the time of diagnosis. The 5-year survival rate of
patients with GC is less than 20% worldwide (2). The poor prognosis in patients with GC
is mainly associated with the recurrence and metastasis of GC
(3). Surgery and chemotherapy are
conventional treatment approaches for patients with GC (4). Target therapies, such as anti-EGFR
signaling agent, have been proven to improve treatment efficacy
(5). However, the molecular
mechanisms underlying GC progression and treatments remain largely
unclear.
Neural precursor cell expressed, developmentally
downregulated 9 (NEDD9) is a well-characterized focal adhesion
scaffold protein (6). High
expression of NEDD9 is observed in the lung, kidney and fetal brain
tissues (7). In cancer cells,
overexpression of NEDD9 is also detected and related to elevated
cell motility (8). Analysis of
NEDD9 expression in gastric tumors suggested that NEDD9
overexpression was associated with metastasis and poor prognosis
(9).
The translation or degradation of messenger RNAs
(mRNAs) can be suppressed by microRNAs (miRNAs) (10), which are endogenous small
non-coding RNA molecules of ~18-25 nucleotides in animals and
plants, comprising one of the most abundant groups of gene
regulatory molecules in multicellular organisms and affecting the
output of numerous protein-coding genes (11). miRNAs are validated as oncogenes or
tumor suppressors, and may predict patient outcome (12). Moreover, NEDD9 can be targeted by
multiple miRNAs in various cancer types: For instance, miR-5195-3p
inhibits cell proliferation by targeting NEDD9 in osteosarcoma
(13), miR-145 suppresses cell
growth and metastasis by targeting NEDD9 in renal cell carcinoma
(14) and miR-1252-5p reduces cell
invasion by targeting NEDD9 in pancreatic cancer (15). However, at present, there are no
studies regarding the miRNAs which can target NEDD9 in GC. In the
present study, it was investigated whether there are other novel
miRNAs that can target NEDD9 and function in GC.
Materials and methods
Collection of specimens
In total, 25 GC tissues and the matched normal
tissues were obtained from patients with GC from the General
Hospital of Southern Theatre Command (Guangzhou, China) from June
2018 until September 2019. The clinical data from patients with GC
are presented in Table I. Patients
who received any chemotherapy or radiotherapy treatments were
excluded from the study. Written informed consent was provided by
all participants before the study. All experiments were approved by
the Ethics Committee of General Hospital of Southern Theater
Command (approval no. GHSTC-2018-06; Guangzhou, China). The tissues
were immediately stored at −80°C before subjected to the future
experiments.
 | Table I.The basic characteristics of patients
with gastric cancer. |
Table I.
The basic characteristics of patients
with gastric cancer.
|
|
| Expression level of
microRNA-4735-3p |
|
|---|
|
|
|
|
|
|---|
| Parameters | Number of
patients | Low, n | High, n | P-value |
|---|
| Sex |
|
|
| 0.69 |
|
Female | 10 | 4 | 6 |
|
| Male | 15 | 8 | 7 |
|
| Age, years |
|
|
| >0.99 |
| ≥55 | 20 | 10 | 10 |
|
| <
55 | 5 | 2 | 3 |
|
| Histology grade |
|
|
| 0.57 |
|
Well-moderately | 11 | 5 | 6 |
|
|
Poorly-signet | 14 | 7 | 7 |
|
| Stage |
|
|
| 0.23 |
|
I–II | 10 | 3 | 7 |
|
|
III–IV | 15 | 9 | 6 |
|
| Size, cm |
|
|
| 0.01 |
|
<3 | 11 | 2 | 9 |
|
| ≥3 | 14 | 10 | 4 |
|
Cell culture
The GC cell line AGS was purchased from American
Type Culture Collection. Cells were maintained in RPMI-1640
supplemented with 10% FBS (both from Gibco; Thermo Fisher
Scientific, Inc.) in a humidified incubator at 37°C with 5%
CO2.
miR-4735-3p overexpression
miR-4735-3p mimic (50 nM;
5′-AAAGGUGCUCAAAUUAGACAU-3′) and miR-NC (50 nM; negative control,
5′-UAGUCUCGGGAGACUCACUACC-3′) mimic were purchased from Guangzhou
RiboBio Co., Ltd. For overexpression of miR-4735-3p, miR-4735-3p
mimic was transfected into AGS cells (2×105 cells/well)
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. Then,
cells were incubated in DMEM for 48 h at 37°C. The protein and mRNA
were extracted from cells at 48 h after transfection.
Wound healing assay
To assess cell migration ability, 1×106
AGS cells were seeded in six-well plates and cultured in serum-free
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.). When the cell
density reached 100% confluence, a wound was made at the centre of
cell monolayer in each well with a 10-µl pipette tip. The culture
medium was then replaced with serum free medium. At 0 and 24 h
after transfection, images of wound area were captured with a light
microscope. The relative area of wound was calculated using
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.) to
reflect cell migration ability.
Cell invasion assay
AGS cells resuspended in 200 µl serum-free RPMI-1640
(both from Gibco; Thermo Fisher Scientific, Inc.) were placed in
the upper chamber of 24-well Transwell plates (8 µm-pore; Corning,
Inc.), while RPMI-1640 containing 20% FBS was added to the lower
chamber and acted as a chemoattractant. The Transwell membranes
were precoated with Matrigel at 4°C for 12 h. At 48 h, the
non-invasive cells in the upper chamber were removed, while the
invasive cells were fixed by 70% ethanol for 15 min at room
temperature and stained by 0.1% crystal violet (Solarbio Science
& Technology Co., Ltd.) for 20 min at room temperature. Images
were captured by an optical microscope.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was isolated from cells and tissues with
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Reverse transcription of
RNA into cDNA was achieved using MMLV-reverse transcriptase
(Promega Corporation) with M-MLV Reaction Buffer and dNTPs (dATP,
dCTP, dGTP and dTTP), at 70°C for 5 min and 37°C for 1 h. qPCR was
performed using the PrimeScript™ RT reagent kit
according to the manufacturer's protocol (Takara, Bio, Inc.) on a
CFX-96 system (Bio-Rad Laboratories, Inc.). The thermocycling
conditions for qPCR were as follows: 95°C for 30 sec followed by 40
cycles of 95°C for 5 sec and 60°C for 30 sec. The relative
expression of genes was normalized to internal controls using the
2−ΔΔCq method (16).
β-actin and U6 were internal controls for mRNA and miRNA,
respectively. The primers were designed using Primer Premier
software (version 6; Primer Premier) and synthesized by GenScript.
The primer sequences were as follows: miR-4735-3p forward,
5′-AAAGGTGCTCAAATTAGACAT-3′ and reverse,
5′-ATGTCTAATTTGAGCACCTTT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; NEDD9 forward,
5′-ACCATGAATTACGAAGCACCTTA-3′ and reverse,
5′-TAAGGTGCTTCGTAATTCATGGT-3′; β-actin forward,
5′-AAATCTGGCACCACACCTTC-3′; and reverse,
5′-GGGGTGTTGAAGGTCTCAAA-3′.
Protein extraction and western blot
analysis
Lysates were prepared in RIPA lysis buffer
(Sigma-Aldrich; Merck KGaA) following the manufacturer's protocol.
β-actin antibody (1:10,000; cat. no. A1978) was purchased from
Sigma-Aldrich; Merck KGaA. NEDD9 (1:1,000; cat. no. 4044),
E-cadherin (1:1,000; cat. no. ab231303), matrix metalloproteinase
(MMP)2 (1:1,000; cat. no. 40994) and MMP9 (1:1,000; cat. no. 13667)
antibodies were obtained from Cell Signaling Technology, Inc.
Secondary HRP-conjugated antibodies [rabbit anti-mouse IgG H&L
(1:10,000; cat. no. ab6728) and goat anti-rabbit IgG H&L
(1:10,000; cat. no. ab6721)] were purchased from Abcam. Protein
concentration was determined by bicinchoninic acid assay. Briefly,
20 µg protein lysate was added into an SDS-PAGE gel (10%) and
separated by electrophoresis. The proteins were transferred onto a
PVDF membrane, then blocked in 5% non-fat milk at room temperature
for 2 h. Subsequently, the membrane was incubated with the
aforementioned primary antibodies at 4°C overnight. Following the
primary incubation, membranes were incubated with secondary
antibodies at room temperature for 1 h sequentially. The membrane
was washed between the incubation with the primary antibodies and
secondary antibody by 0.2% TBST (2 ml Tween-20 in the 1 l TBST
solution). The membrane was detected with ECL Western Blotting
Substrate (Pierce; Thermo Fisher Scientific, Inc.). ImageJ software
1.48u (National Institutes of Health) was used for densitometric
analysis.
Plasmid construction
Full length of NEDD9 was amplified from AGS cDNA
using PrimeSTAR® GXL DNA Polymerase (Takara Bio, Inc.).
The fragment was annealed into pcDNA3.1 plasmid (1 µg) with T4 DNA
ligase (Takara Bio, Inc.). For overexpression of NEDD9,
pcDNA3.1-NEDD9 was transfected into AGS cells (5×106)
with Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol for 48 h at
37°C. At 48 h after transfection, cells were collected for the
subsequent experiments.
Bioinformatic analysis and dual
luciferase reporter assay
TargetScan 7.1 database (https://www.targetscan.org/vert_71/) was used to
predict the potential miRNAs for NEDD9. The 3′-untranslated region
(UTR) of NEDD9 was amplified from AGS cDNA using
PrimeSTAR® GXL DNA Polymerase (Takara Bio, Inc.) and
ligated into pGL3 basic with T4 DNA ligase (Takara Bio, Inc.). Two
site mutations were introduced into pGL3-NEDD9 wild-type (WT) to
construct pGL3-NEDD9 mutant (Mut) with QuickChange Site-Directed
Mutagenesis kit (Stratagene; Agilent). For dual luciferase reporter
assay, pGL3-NEDD9 WT or pGL3-NEDD9 Mut was co-transfected with
miR-4735-3p mimic or miR-NC mimic into AGS cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, the firefly and Renilla
luciferase activity of each group was detected by a Dual Luciferase
Reporter Assay System (Promega Corporation) according to the
manufacturer's protocol.
RNA immunoprecipitation (RIP)
For RIP assay, anti-Ago2 (catalog no. ab186733,
1:50) and anti-IgG (catalog no. ab205718, 1:50) (both from Abcam)
primary antibodies were used. Nuclei were isolated from cells by
centrifugation for 10 min at 15,000 × g and 4°C, then lysed with
RNA lysis buffer (Promega Corporation) supplemented with protease
and RNase inhibitors, and incubated with primary antibodies at 4°C
overnight. Next, RNA immunoprecipitated with RNA-binding proteins
was isolated after addition of protein A agarose (catalog no.
ab193255; 25 µl) and protein G agarose (catalog no. ab193258; 25
µl) (both from Abcam). Following washes, RNA was purified and
reverse transcribed into cDNA. The expression of targets was
determined by RT-qPCR.
Statistical analysis
All data were analyzed with GraphPad Prism 6.0
(GraphPad Software, Inc.) and presented as the mean ± SD. The
differences of two groups were compared with Students' t test
(unpaired Students' t-test for cells, paired Students' t-test for
tissues). Pearson's correlation analysis was used to detect the
correlation between miR-4735-3p and NEDD9. For three groups, the
comparisons were conducted using one-way ANOVA followed by Newman
Keuls analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-4735-3p is inversely related with
NEDD9 in GC tissues
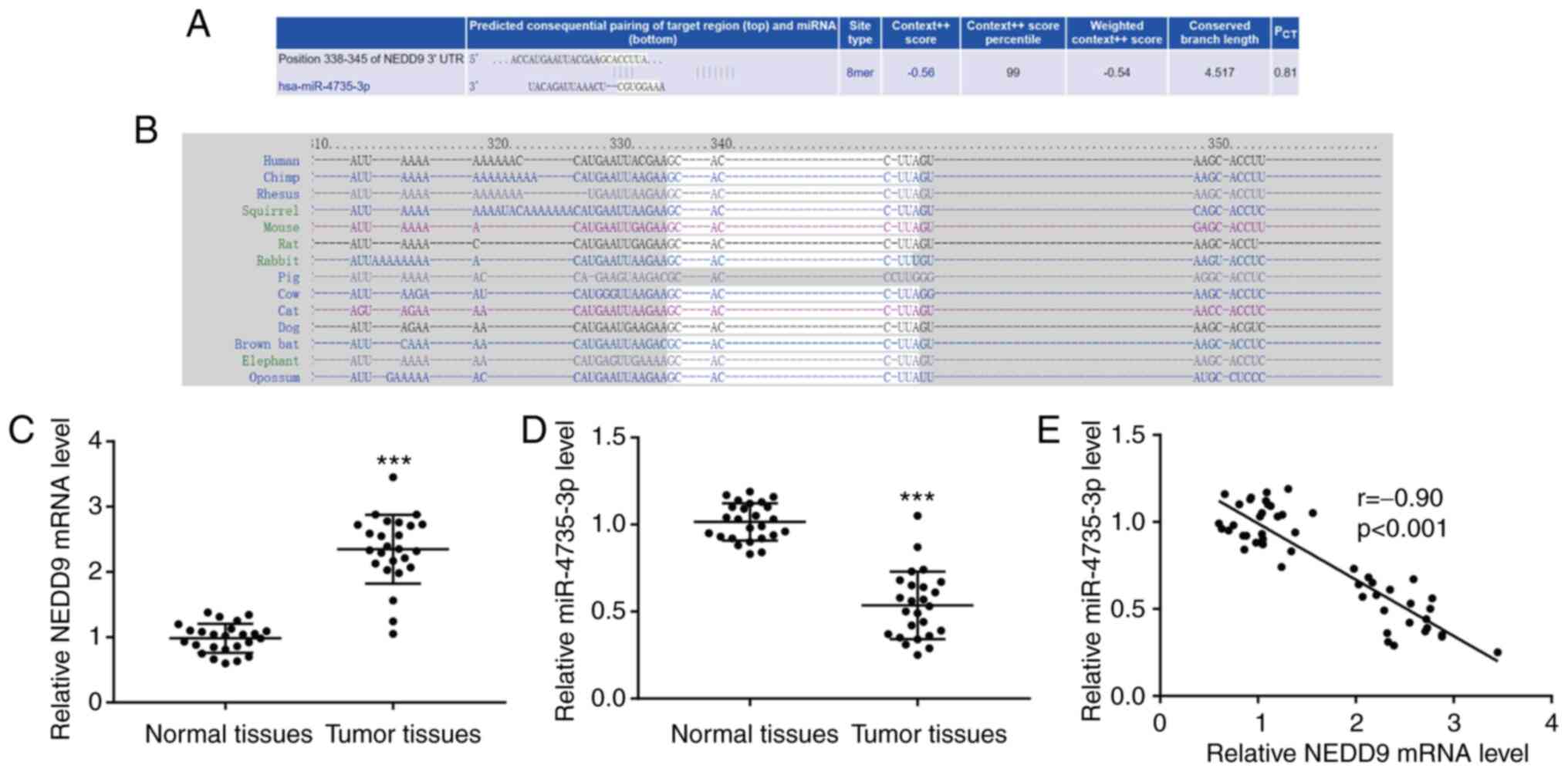
TargetScan 7.1 exhibited that there was a putative
binding site between miR-4735-3p and NEDD9 3′UTR, and the context++
score percentile between miR-4735-3p and NEDD9 was as high as 99
(Fig. 1A). The putative binding
site between miR-4735-3p and NEDD9 3′UTR was conserved in numerous
species, including chimp, rhesus and squirrel (Fig. 1B). In GC tissues, an increase of
NEDD9 mRNA level was observed (Fig.
1C) and a decrease of miR-4735-3p level (Fig. 1D) compared with normal tissues. It
was observed that miR-4735-3p expression was negatively correlated
with GC tumor size, but not sex, age, histology grade or stage
(Table I). Notably, using
Pearson's correlation analysis, a significant negative correlation
was detected between miR-4735-3p and NEDD9 (Fig. 1E), suggesting that there may be a
regulatory association between miR-4735-3p and NEDD9 in GC.
NEDD9 is negatively regulated by
miR-4735-3p in GC cells
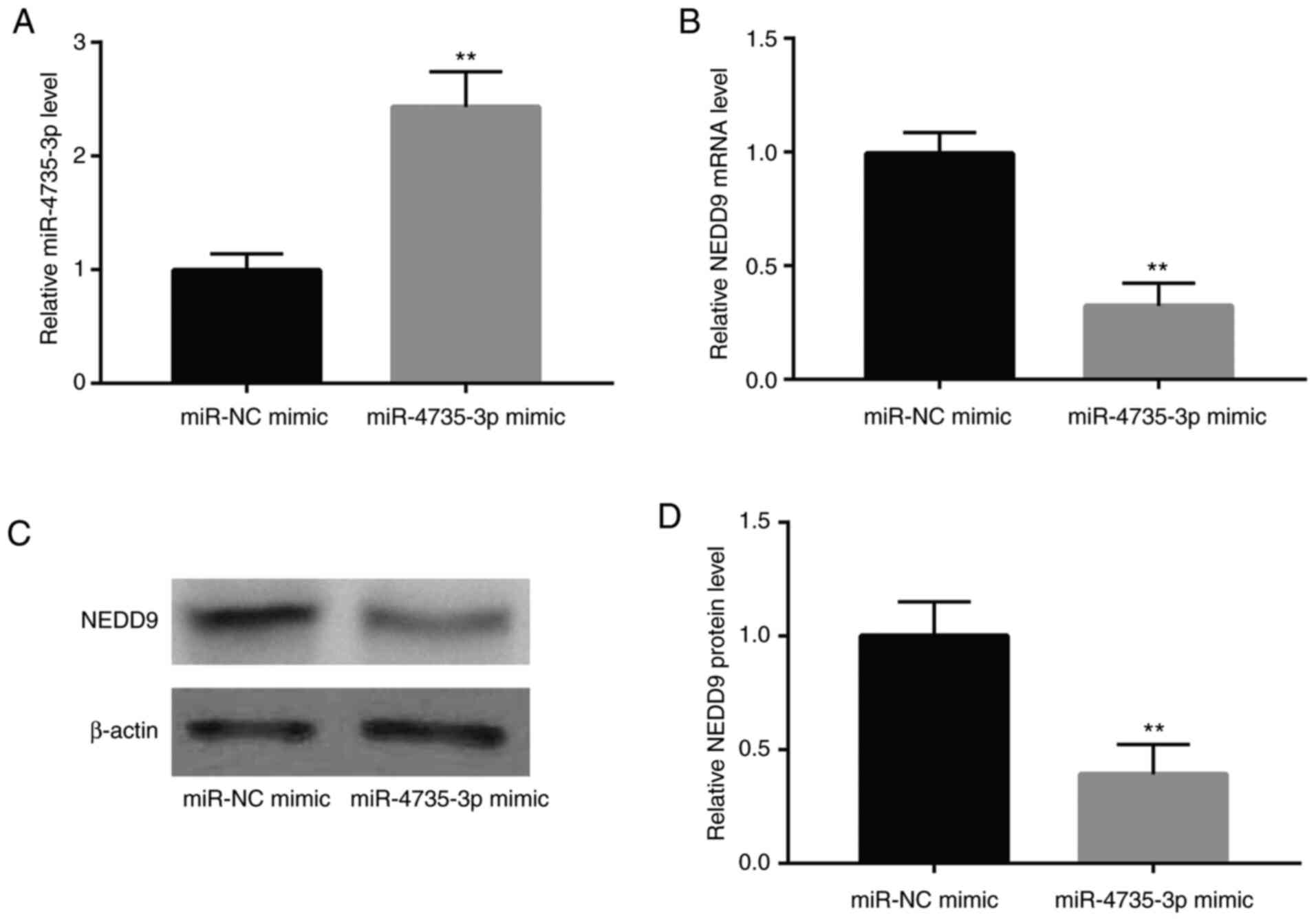
To investigate whether miR-4735-3p regulated NEDD9
expression in GC, miR-4735-3p mimic was used to overexpress
miR-4735-3p in GC cell line AGS. As revealed in Fig. 2A, transfection of miR-4735-3p mimic
elevated miR-4735-3p expression in AGS cells. RT-qPCR and western
blotting showed that miR-4735-3p mimic decreased NEDD9 mRNA and
protein levels (Fig. 2B-D). These
data indicated that NEDD9 was negatively regulated by miR-4735-3p
in GC cells.
miR-4735-3p directly binds to NEDD9 in
GC cells
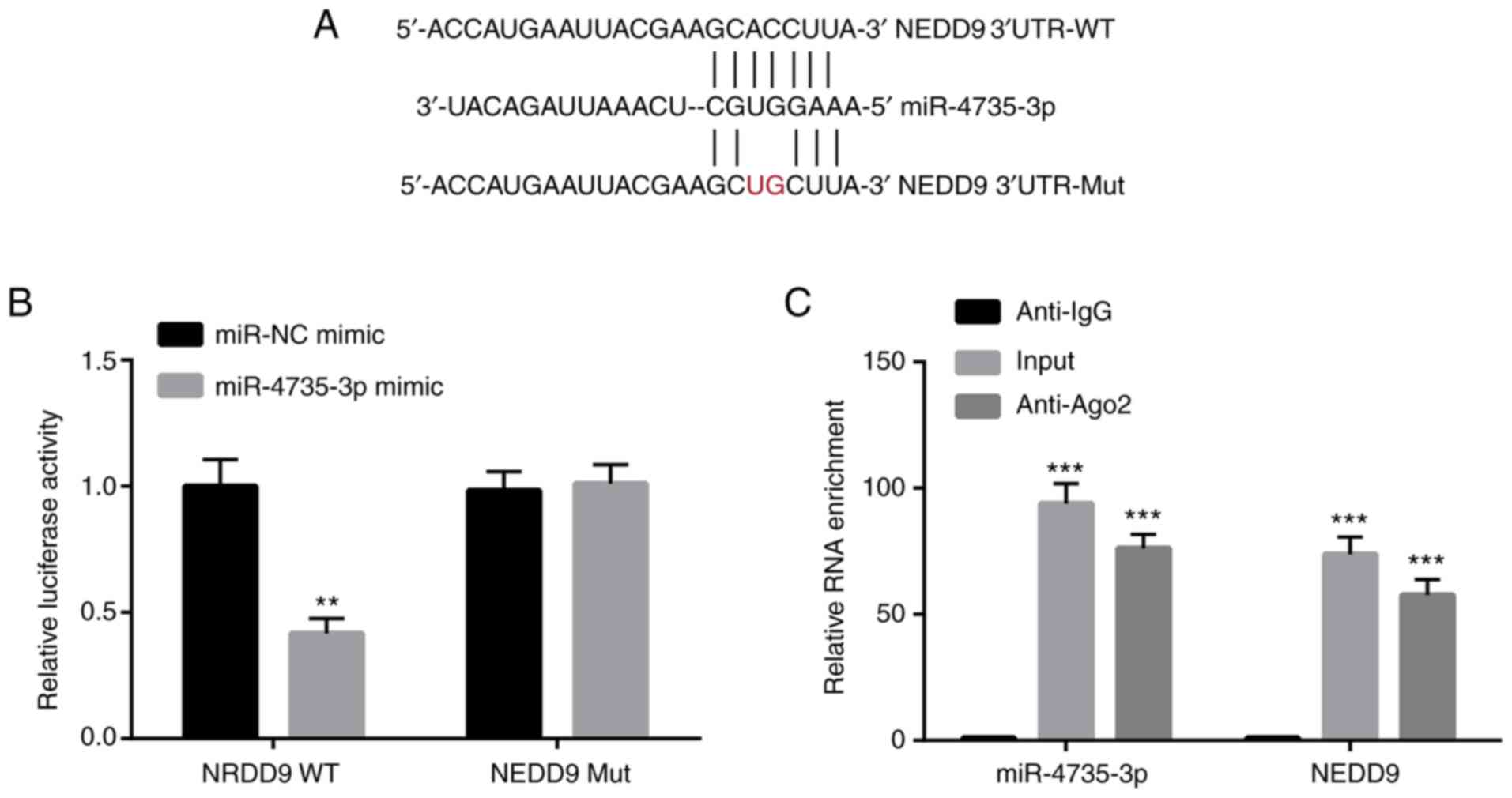
To further explore whether miR-4735-3p directly
regulated NEDD9, dual luciferase assay was used. Mutations at two
nucleotides were introduced into NEDD9 3′UTR (from CC to UG)
(Fig. 3A). miR-4735-3p mimic
significantly decreased the relative luciferase activity of AGS
cells transfected with NEDD9 3′UTR-WT; while no effect was observed
in the relative luciferase activity of AGS cells transfected with
NEDD9 3′UTR-Mut (Fig. 3B).
Furthermore, RIP assay demonstrated that miR-4735-3p and NEDD9 were
highly enriched in the Ago2 group compared with the anti-IgG group
(Fig. 3C). The aforementioned
results confirmed that miR-4735-3p directly regulated NEDD9 in GC
cells.
miR-4735-3p overexpression suppresses
GC cell migration and invasion by regulating NEDD9
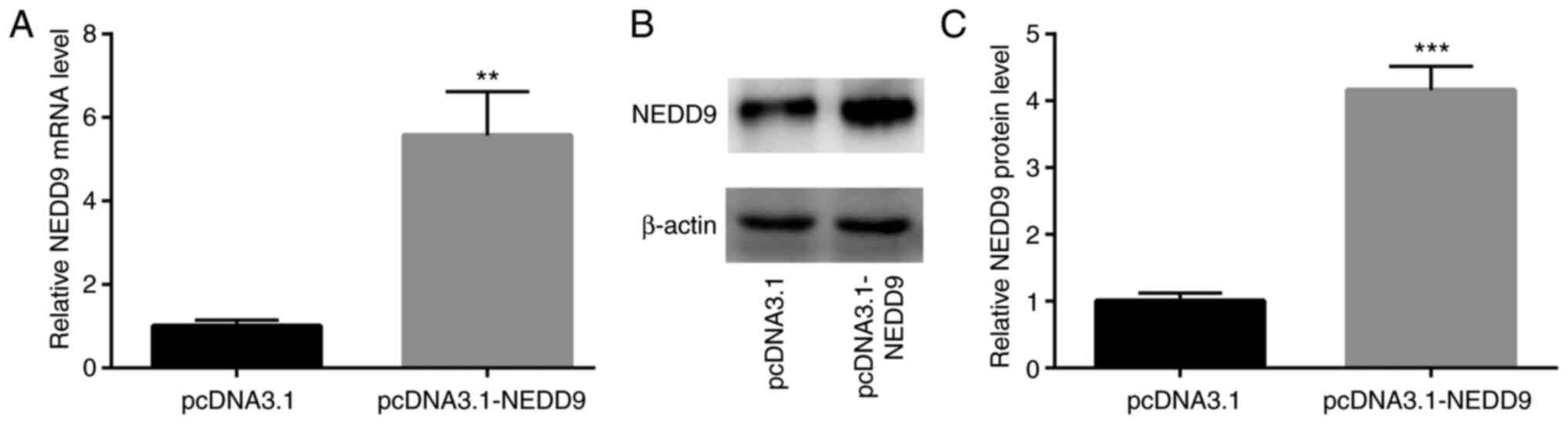
Next, transfection of recombinant NEDD9 was used to
study the function of NEDD9 in GC cells. As revealed in Fig. 4A-C, transfection of NEDD9 elevated
NEDD9 mRNA and protein expression in AGS cells.
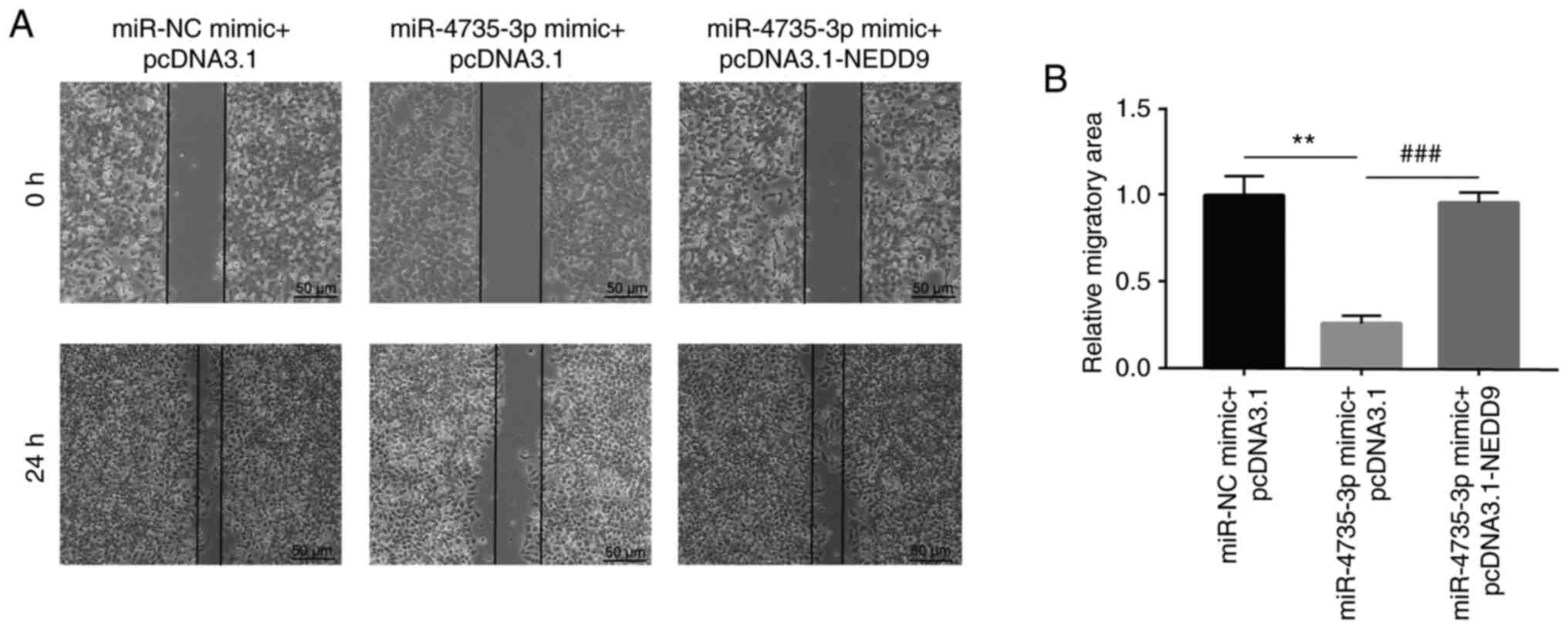
In a wound healing assay, miR-4735-3p mimic
significantly inhibited migration of AGS cells; in addition,
transfection of recombinant NEDD9 reversed miR-4735-3p-induced
inhibition of AGS cell migration (Fig.
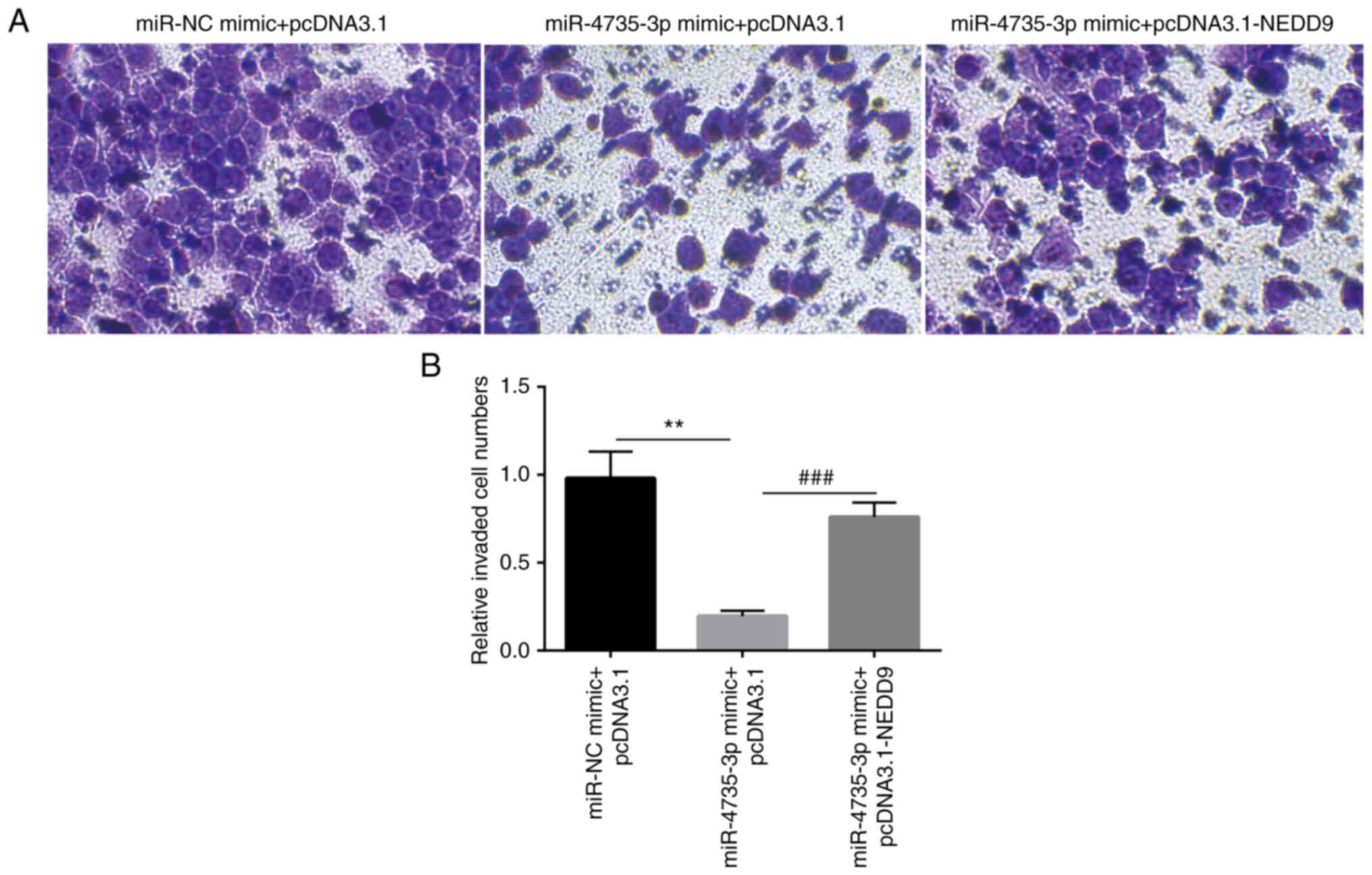
5A and B). In a Matrigel invasion assay, miR-4735-3p
overexpression significantly inhibited invasion of AGS cells; in
addition, transfection of recombinant NEDD9 reversed
miR-4735-3p-induced inhibition of AGS cell invasion (Fig. 6A and B).
These data collectively showed that miR-4735-3p was
a tumor suppressor in GC cells by regulation of NEDD9.
miR-4735-3p downregulates MMP2 and
MMP9 in GC cells
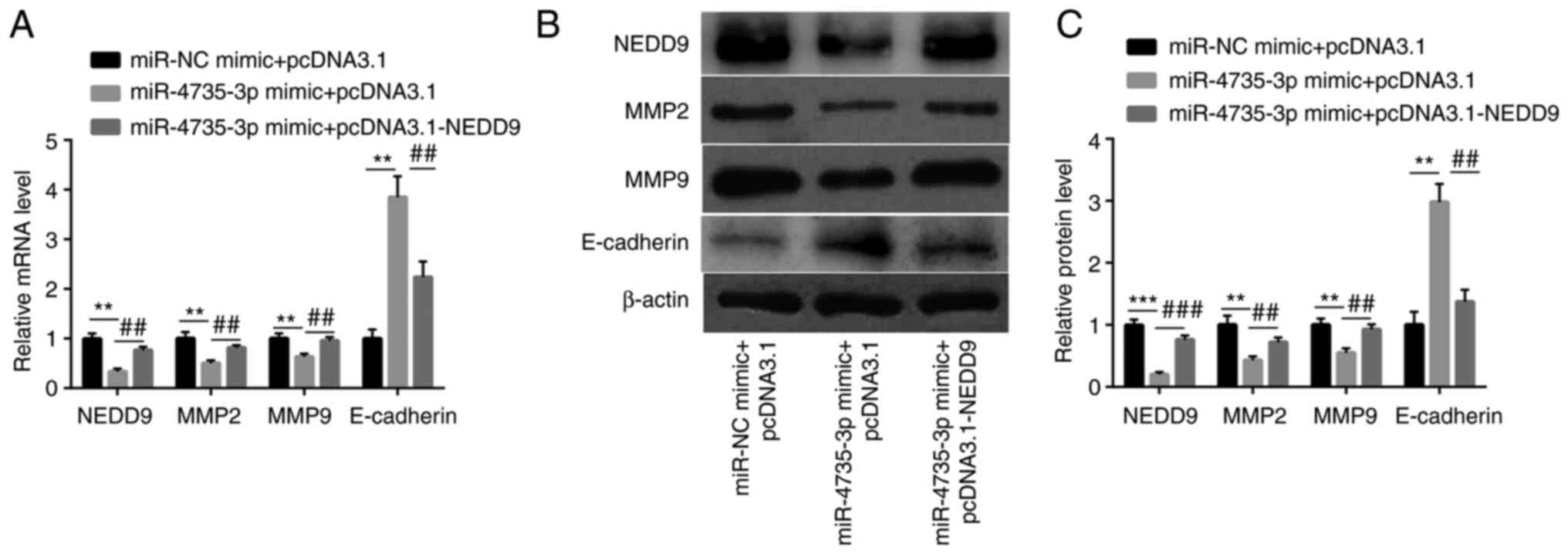
In AGS cells, miR-4735-3p mimic decreased NEDD9 mRNA
and protein levels accompanied with downregulation of MMP2 and
MMP9, while increased E-cadherin mRNA and protein levels.
Furthermore, transfection of recombinant NEDD9 led to an elevation
of NEDD9, MMP2 and MMP9 proteins and a decrease of E-cadherin
(Fig. 7A-C). These data suggested
that miR-4735-3p could downregulate MMP2 and MMP9 and upregulate
E-cadherin by suppression of NEDD9.
Discussion
Recently, the investigations on NEDD9 in GC have
suggested that high NEDD9 expression was associated with poor
differentiation, venous invasion, invasive depth, preset lymph node
metastasis, distant metastasis and high clinical stage in patients
with GC (17). Later, an in
vivo study revealed that silencing of NEDD9 reduced the tumor
volume in nude mice bearing gastric tumor (18); the in vitro assays indicated
that decreased level of NEDD9 inhibited GC cell proliferation and
migration (19,20). In the present study, it was also
found that NEDD9 level was upregulated in tumor tissues of patients
with GC, suggesting its oncogenic role in GC. Moreover, during the
preparation of our article, several papers published to exhibit the
oncogenic role of NEDD9 in GC; for instance, higher NEDD9 level was
discovered in intestinal type GC compared with the adjacent
non-neoplastic mucosa (21) and
higher epithelial NEDD9 expression was related to higher mortality
(22), which further proved the
present findings.
miR-4735-3p acts as an oncogene or tumor suppressor
in different cancer types. For example, miR-4735-3p acted as an
oncogene in non-small cell lung cancer (NSCLC), as it was increased
in NSCLC tissues and cell lines (23); however, it also served as a tumor
suppressor in other types of cancer, as patients with low
miR-4735-3p expression exerted an improved overall survival than
those with high expression in prostate cancer (24). Long non-coding (lnc)RNA ARAP1-AS1
aggravated the development of bladder cancer by sponging
miR-4735-3p/NOTCH2 axis (25).
LncRNA ARAP1-AS1 promoted the progression of ovarian cancer by
sponging miR-4735-3p/PLAGL2 axis (26). In consistency with previous studies
(24–26), a decreased expression of
miR-4735-3p was observed in GC tissues compared with matched normal
tissues. In addition, using RT-qPCR, western blotting, dual
luciferase reporter and RIP assay, it was found that NEDD9 was
inversely associated with and targeted by miR-4735-3p in GC
tissues.
Moreover, function assays indicated that miR-4735-3p
regulated GC cell migration and invasion by suppression of NEDD9.
However, the potential molecules related to metastasis of GC and
NEDD9 are left to be explored.
As acknowledged, MMPs are endopeptidases involved in
metastasis of cancer cells (27).
Among 26 MMP family members, MMP2 and MMP9 were extensively studied
due to their pivotal role in mediating cell invasion and migration
in various cell types (28,29).
E-cadherin acted as an invasion-suppressor molecule (30). It was reported that NEDD9 promoted
metastasis of oral squamous cell carcinoma cells via upregulation
of MMP9 (31), suggesting that
NEDD9 regulated cell motility via MMPs. Furthermore, it was
reported that downregulation of NEDD9 increased the expression
levels of E-cadherin in colon cancer (32). Consistently, in the present study,
it was identified that miR-4735-3p could regulate E-cadherin, MMP2
and MMP9 by suppression of NEDD9.
In conclusion, the present study demonstrated a
tumor suppressor role of miR-4735-3p in GC by directly targeting
NEDD9.
However, there remain 2 questions to be addressed:
i) in the present study, the relationship between miRNA-4735-3p and
NEDD9 was mainly investigated by examining the mRNA level. The
protein level of NEDD9 in normal tissues and tumor tissues was not
tested, which will be evaluated in future work; ii) in the present
study, the function regulation of miRNA-4735-3p/NEDD9 was mainly
evaluated by overexpression of miRNA-4735-3p and NEDD9; Cell lines
with knock-out of NEDD9 will be examined in future studies.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YM conceptualized the study, performed the
experiments and wrote the manuscript. YM confirms the authenticity
of all the raw data. YM read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of General Hospital of Southern Theater Command
(Guangzhou, China). Written informed consent was provided by all
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray JA, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Matsuoka T and Yashiro M: Rho/ROCK
signaling in motility and metastasis of gastric cancer. World J
Gastroenterol. 20:13756–13766. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kang YK, Kang WK, Shin DB, Chen J, Xiong
J, Wang J, Lichinitser M, Guan Z, Khasanov R, Zheng L, et al:
Capecitabine/cisplatin versus 5-fluorouracil/cisplatin as
first-line therapy in patients with advanced gastric cancer: A
randomised phase III noninferiority trial. Ann Oncol. 20:666–673.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yuan DD, Zhu ZX, Zhang X and Liu J:
Targeted therapy for gastric cancer: Current status and future
directions (Review). Oncol Rep. 35:1245–1254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
O'Neill GM, Fashena SJ and Golemis EA:
Integrin signalling: A new Cas(t) of characters enters the stage.
Trends Cell Biol. 10:111–119. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Law SF, Estojak J, Wang B, Mysliwiec T,
Kruh G and Golemis EA: Human enhancer of filamentation 1, a novel
p130cas-like docking protein, associates with focal adhesion kinase
and induces pseudohyphal growth in saccharomyces cerevisiae. Mol
Cell Biol. 16:3327–3337. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim M, Gans JD, Nogueira C, Wang A, Paik
JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al:
Comparative oncogenomics identifies NEDD9 as a melanoma metastasis
gene. Cell. 125:1269–1281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Q, Wang H, Ma Y, Zhang J, He X, Ma J
and Zhao ZS: Overexpression of Nedd9 is a prognostic marker of
human gastric cancer. Med Oncol. 31:332014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Shi G, Zhu D, Jin Y and Yang X:
miR-5195-3p suppresses cell proliferation and induces apoptosis by
directly targeting NEDD9 in osteosarcoma. Cancer Biother
Radiopharm. 34:405–412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu R, Ji Z, Li X, Zhai Q, Zhao C, Jiang Z,
Zhang S, Nie L and Yu Z: miR-145 functions as tumor suppressor and
targets two oncogenes, ANGPT2 and NEDD9, in renal cell carcinoma. J
Cancer Res Clin Oncol. 140:387–397. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xue Y, Wu T, Sheng Y, Zhong Y, Hu B and
Bao C: MicroRNA-1252-5p, regulated by Myb, inhibits invasion and
epithelial-mesenchymal transition of pancreatic cancer cells by
targeting NEDD9. Aging (Albany NY). 13:18924–18945. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi R, Wang L, Wang T, Xu J, Wang F and Xu
M: NEDD9 overexpression correlates with the progression and
prognosis in gastric carcinoma. Med Oncol. 31:8522014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang S, Wu L, Liu Q, Chen K and Zhang X:
Study on effect of the expression of siRNA in gastric cancer
bearing nude mice transplanted tumor NEDD9 gene. Pak J Pharm Sci.
27 (5 Suppl):S1651–S1656. 2014.
|
|
19
|
Zhang S, Wu L, Liu Q, Chen K and Zhang X:
Impact on growth and invasion of gastric cancer cell lines by
silencing NEDD9. Onco Targets Ther. 8:223–231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao S, Min P, Liu L, Zhang L, Zhang Y,
Wang Y, Zhao X, Ma Y, Xie H, Zhu C, et al: NEDD9 facilitates
hypoxia-induced gastric cancer cell migration via MICAL1 related
Rac1 activation. Front Pharmacol. 10:2912019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Elaidy NF, Harb OA, Mohamed AM, Hemeda R,
Taha HF, Samir A, Elsayed AM, Osman G and Hendawy EIE: Prognostic
significances of NEDD-9 and FOXL-1 expression in intestinal type
gastric carcinoma: An immunohistochemical study. J Gastrointest
Cancer. 52:728–737. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lerotić I, Vuković P, Hrabar D, Misir Z,
Kruljac I, Pavić T, Forgač J, Ćaćić P and Ulamec M: Expression of
NEDD9 and connexin-43 in neoplastic and stromal cells of gastric
adenocarcinoma. Bosn J Basic Med Sci. 21:542–548. 2021.PubMed/NCBI
|
|
23
|
Wang D, Han X, Li C and Bai W: FBXL3 is
regulated by miRNA-4735-3p and suppresses cell proliferation and
migration in non-small cell lung cancer. Pathol Res Pract.
215:358–365. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou W, Huang S, Jiang Q and Yuan T:
Suppression of miR-4735-3p in androgen receptor-expressing prostate
cancer cells increases cell death during chemotherapy. Am J Transl
Res. 9:3714–3722. 2017.PubMed/NCBI
|
|
25
|
Teng J, Ai X, Jia Z, Wang K, Guan Y and
Guo Y: Long non-coding RNA ARAP1-AS1 promotes the progression of
bladder cancer by regulating miR-4735-3p/NOTCH2 axis. Cancer Biol
Ther. 20:552–561. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li C, Dong B, Xu X, Li Y, Wang Y and Li X:
LncRNA ARAP1-AS1 aggravates the malignant phenotypes of ovarian
cancer cells through sponging miR-4735-3p to enhance PLAGL2
expression. Cytotechnology. 73:363–372. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li L and Li H: Role of microRNA-mediated
MMP regulation in the treatment and diagnosis of malignant tumors.
Cancer Biol Ther. 14:796–805. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ren F, Tang R, Zhang X, Madushi WM, Luo D,
Dang Y, Li Z, Wei K and Chen G: Overexpression of MMP family
members functions as prognostic biomarker for breast cancer
patients: A systematic review and meta-analysis. PLoS One.
10:e01355442015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fioruci-Fontanelli BA, Chuffa LG, Mendes
LO, Pinheiro PF, Delella FK, Kurokawa CS, Felisbino SL and Martinez
FE: MMP-2 and MMP-9 activities and TIMP-1 and TIMP-2 expression in
the prostatic tissue of two ethanol-preferring rat models. Anal
Cell Pathol (Amst). 2015:9545482015.PubMed/NCBI
|
|
30
|
Mareel M, Bracke M, Van Roy F and Vakaet
L: Expression of E-cadherin in embryogenetic ingression and cancer
invasion. Int J Dev Biol. 37:227–235. 1993.PubMed/NCBI
|
|
31
|
Grauzam S, Brock AM, Holmes CO, Tiedeken
JA, Boniface SG, Pierson BN, Patterson DG, Coaxum SD, Neskey DM and
Rosenzweig SA: NEDD9 stimulated MMP9 secretion is required for
invadopodia formation in oral squamous cell carcinoma. Oncotarget.
9:25503–25516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng H, Wu J, Huang Q, Yang X, Yang K, Qiu
Y, Ren J, Shen R and Qi H: NEDD9 promotes invasion and migration of
colorectal cancer cell line HCT116 via JNK/EMT. Oncol Lett.
18:4022–4029. 2019.PubMed/NCBI
|