Introduction
Cutaneous squamous cell carcinoma (CSCC) is common
among elderly individuals and severely impacts the life quality, as
well as poses a threat to their lives (1,2).
CSCC is characterized by small nodules on the surface of the skin,
which gradually become larger and harder to form cauliflower-like
hyperplasia (3). A number of
patients experience ulceration in the lesions, which gradually
increases, accompanied by symptoms such as pus, bleeding and foul
odor (4). In China, CSCC is the
most common skin malignant tumor, and its incidence accounts for
78–90.9% of dermatological malignancies (5). However, if the early diagnosis and
radical resection can be performed, the surgical effects are
satisfactory and the cure rate is high, with potential complete
remission. Therefore, it is necessary to explore early clinical
biomarkers of CSCC and determine the associated molecular
mechanisms.
Previous studies have demonstrated that abnormal
gene expression can affect cell proliferation, invasion and
migration processes, and thus affect the occurrence of tumors
(6). Gene therapy introduces
exogenous genes with certain functions into cells to treat squamous
cell carcinoma (SCC). The metastatic ability of tumor cells is
associated with their ability to induce proteases, as well as
degrade the extracellular matrix and the basement membrane
(7). Matrix metalloproteinases
(MMPs) are a group of proteases that degrade almost all
extracellular matrix and basement membrane components (8). Therefore, gene therapy by antisense
MMP transfection and inhibition of MMP secretion in malignant tumor
cells may be an effective treatment option. In addition, various
genes have been identified to serve crucial roles in CSCC. For
example, Zaravinos et al (9) demonstrated that downregulated Raf-1
kinase inhibitor protein mRNA levels and abnormal BRAF signaling
pathway were involved in the pathogenesis of CSCC. Another study in
cancer cells has also reported that microRNA-21 targets
grainyhead-like transcription factor 3 to upregulate the expression
of PTEN and prevent SCC (10).
Although various genes have been researched, further research is
needed in the clinical application of gene therapy, especially for
patients with grade III and IV CSCC who present with a high degree
of invasion, high recurrence and metastasis rates.
The role of fatty acid-binding protein 7 (FABP7) in
tumor tissues is a controversial issue (11). In a previous study, overexpression
of FABP7 was demonstrated to promote cell proliferation and predict
poor prognosis of clear cell renal cell carcinoma (12). FABP7 is associated melanoma cell
proliferation via modulation of Wnt/β-Catenin signaling (13). Currently, only a few studies have
reported the exact function of FABP7 in cutaneous malignant
melanoma (12,14,15).
Thus, the present study hypothesized that FABP7 may be a diagnostic
biomarker of CSCC, and aimed to determine the detailed mechanism
underlying the potential effects of FABP7 in CSCC.
Notch signaling pathway is involved in regulating
the development of cutaneous tissues (16). Functional genes in Notch pathway
including NCOR2, NCSTN, and MAML2 predict survival of patients with
cutaneous melanoma (17). Thereby,
the mechanism of Notch signaling pathway was also researched in
this study. In this study, we speculated that FABP7 might be a
biomarker for CSCC diagnosis. The present study aims to determine
the molecular mechanism underlying the effects of FABP7 in CSCC,
which may provide a new diagnostic biomarker or treatment target
for CSCC.
Materials and methods
Patient samples and cell lines
A total of 34 punch biopsies and 34 non-lesion
epithelial skin tissue samples were obtained from patients with
CSCC who underwent surgery between January 2018 and January 2019 at
Shanghai Skin Disease Hospital, Tongji University School of
Medicine (Shanghai, China). All patients and their families were
informed about the study; written informed consent was obtained
from all patients, and the study was performed according to the
Declaration of Helsinki. Ethics approval for the study was provided
by the Ethics Committee of the Shanghai Skin Disease Hospital,
Tongji University School of Medicine (Shanghai, China; approval no.
SSDH10561). All tissue samples were snap-frozen in liquid nitrogen
and stored at −70°C until further use for mRNA isolation and
immunohistochemistry (IHC).
Human CSCC cell lines A431, colo-16 and SCL-1 and
the immortalized human normal keratinocyte cell line HaCaT were
purchased from Shanghai Cell Bank of the Chinese Academy of
Sciences. The cells were cultured in DMEM (Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.). When the cells reached 70–80%
confluency, they were trypsinized at room temperature for 3 min and
passaged.
Transfection and grouping
The pLV-puro lentiviral vector (Thermo Fisher
Scientific, Inc.) was digested with the AgeI enzyme, and the
digested product was identified by 1% agarose gel electrophoresis.
According to the FABP7 sequence in GenBank (Gene ID: 2173), the
amplification primers of FABP7 were designed and transferred via
PCR using the PCR amplification kit (mlbio Co., Ltd.). The
following primer sequences were used:
5′-ATCGGATCCATGFTFFAFFCTTTCTGT-3′ (upstream) and
5′-ATAGGATCCATGAGGACTCTCAGCAC-3′ (downstream). The PCR product
contained an AgeI cleavage site sticky end (ACCGGT) at both
ends, which was directly ligated into the downstream of the
digested lentiviral expression vector CMV promoter by ligase
reaction. The reaction conditions were as follows: Pre-denaturation
at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C
for 30 sec, annealing at 55°C for 30 sec and extension at 68°C for
3 min, and a final extension at 68°C for 10 min. The PCR product
was detected by 1% agarose gel electrophoresis. The purified
ligation product was transferred to fresh bacterial competent cells
(JMl09), and the cells were cultured in LB liquid medium at 37°C
for 45 min. The transformed competent cells were transferred to LB
agar medium containing 20 mmol/l MgSO4 and ampicillin
solution (100 mg/ml), and cultured at 37°C for 16 h. Positive
clones were identified by PCR using the CloneJET PCR Cloning Kit
(Thermo Fisher Scientific, Inc.) and sent to the Synbio
technologies Co. ltd. or sequencing, and the vector with the
correct expression sequence of recombinant FABP7 gene was selected.
The following primer sequences were used:
5′-GTCAAGCTTCTAAGTTTGTCTCCATCC-3′ (upstream) and
5′-ATCAAGCTTCCCGACCAGGAACATTTT-3′ (downstream). The FABP7
overexpression plasmid and the two lentiviral helper plasmids in
the lentiviral packaging system were extracted using the plasmid
extraction kit (Qiagen, Inc.). 293T cells were seeded at a density
of 6×106 cells/ml in a 15-cm culture dish, cultured at
37°C with 5% CO2 to 70–80% confluence, and the
lentivirus plasmids were co-transfected into 293T cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), at 37°C for 6 h. The supernatant of 293T cells
transfected for 72 h was collected, centrifuged at 4,000 × g for 10
min at 4°C to remove cell debris, filtered, centrifuged at 7,000 ×
g for 5 min at 4°C, resuspended in ice-cold PBS to detect the
titer, and stored in at −80°C. Based on the transfection, the cells
were divided into FABP7 overexpression (oe-FABP7) and negative
control (NC) groups. The CSCC cell lines (A431 and colo-16) at a
density of 6×105 cells/well in a 6-well culture plate
were infected with the FABP7 with a multiplicity of infection of
20, and the FABP7 gene expression sequence carried by the
lentivirus was integrated into the cell to obtain stable
overexpression. A431 and colo-16 cells were transfected with
lentivirus/medium at a ratio of 1:50. Stable cell lines were
selected by puromycin (Sigma-Aldrich, Merck KGaA) at 5 µg/ml for 2
weeks. The lentivirus vectors contained enhanced green fluorescent
protein (eGFP).
Reverse transcription-quantitative
(RT-q)PCR assay
RT-qPCR was used to detect the expression levels of
FABP7 in tissues and cells before and after transfection. The
tissue samples were ground, and total RNA was extracted with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and chloroform, precipitated with isopropanol, washed with
75% ethanol, dried, dissolved in a diethyl pyrocarbonate-containing
solution and stored at −80°C until use. PCR amplification was
performed for total RNA using the PCR amplification kit (Takara
Bio, Inc.). For cells, total RNA was extracted using the Tripure
Isolation reagent (Sangon Biotech Co., Ltd.) according to the
manufacturer's instructions. The A260/A280 ratio was determined
using a DU-800 UV spectrophotometer (Beckman Coulter, Inc.) to
determine RNA purity. A total of 1 µg RNA was collected from each
sample to synthesize the first strand of cDNA using the M-MuLV
First-strand cDNA Synthesis kit (Shanghai Shenggong Bioengineering
Technology Service Co., Ltd.) according to the manufacturer's
instructions in a 20-µl reaction system. The reaction was
terminated at 70°C for 10 min, and the system was placed on ice for
subsequent experiments. mRNA expression levels were detected by
qPCR. qPCR was subsequently performed using the SYBR GREEN RT-PCR
kit (Qiagen GmbH). The following thermocycling conditions were used
for qPCR: 95°C for 5 min, denaturation at 95°C for 10 sec,
annealing at 60°C for 15 sec, extension at 72°C for 20 sec, a total
of 40 cycles. Quantitative molecular detection Real-Time PCR
(BioRad-CFX system; Bio-Rad Laboratories, Inc.) was used. GAPDH was
used as internal reference, and the gene-specific primers were as
follows: FABP7 forward, 5′-TCAGGAAGGCGGCAAAGTGGT-3′ and reverse,
5′-CATAACAGCGAACAGCAACGACATC-3′; and GAPDH forward,
5′-TCCCTGAGCTGAACGGGAAG-3′ and reverse, 5′-GGAGGAGTGGGTGTCGCTGT-3′.
Data were analyzed using the 2−ΔΔCq method (18).
IHC assay
The specimens were fixed in 4% neutral formalin
solution at 4°C overnight, embedded in paraffin and cut into 4 µm
slices, and then placed in the center of a slide and heated in an
oven at 69°C overnight. The Pv-9000 polymer detection system
(Guangzhou Dingguo biology Co. ltd.) was used to detect the protein
expression of FABP7, Ki67, Notch 1 and Notch 3 according to the
manufacturer's instructions. The tissue sections were treated with
xylene at room temperature twice for 5 min and washed with 95%
ethanol four times. The samples were subsequently rinsed with tap
water, placed in a hydrogen peroxide solution at room temperature
for 10 min. After cooling, the sample was washed in PBS. The
primary antibody (FABP7; cat. no. ab32423; 5 µg/ml; 1:800; Abcam)
was added to the samples, incubated at 37°C for 1 h and rinsed with
PBS. Subsequently, the secondary antibody (cat. no. ab150077;
1:1,000; Abcam) was added to the sample and incubated for 20 min at
37°C. The 3,3′-diaminobenzidine (DAB) coloring solution was added,
and the sample was stained with hematoxylin at room temperature for
3 min. Hydrochloric acid alcohol was used to remove the excess
hematoxylin, and 1% ammonia water solution was used for bluing. The
sample was dehydrated, made transparent by xylene and sealed.
Protein expression was calculated with the positive staining cells
under a light microscope (Olympus Corporation, Tokyo Japan,
magnification, ×40-400) in five randomly selected visual fields.
The images were analyzed using Photoshop (Adobe Systems, Inc.).
Database prediction
The STRING database (cn.string-db.org/) was used to
predict FABP7 associated signal pathways according to the previous
study (19).
Western blotting
Western blotting was used to determine the protein
expression levels of FABP7 in CSCC tissues and cells before and
after transfection, as well as to detect the expression of
proliferation-, invasion- and Notch pathway-associated proteins.
The primary antibodies used were as follows: Anti-FABP7 (cat. no.
ab253552); Rabbit monoclonal (EPR3821) anti-proliferating cell
nuclear antigen (PCNA; cat. no. ab92552); rabbit monoclonal
(EPR21043) anti-SNAIL (cat. no. ab216347); rabbit monoclonal
(EPR1791-4) anti-N-cadherin (cat. no. ab202030); mouse monoclonal
(10E4E6) anti-Twist (cat. no. ab175430); rabbit monoclonal
(EPR1184) anti-MMP2 (cat. no. ab92526; rabbit monoclonal
(EPR17888-71) anti-MMP7 (cat. no. ab207299); rabbit anti-activated
Notch1 (cat. no. ab8925); and rabbit polyclonal anti-NOTCH3 (cat.
no. ab23426) and anti-GAPDH (cat. no. ab8245). All primary
antibodies were purchased from Abcam and used at 1:1,000 dilution.
Extraction of total cellular proteins and western blotting were
performed as previously described (20). A total 40 µg protein/lane was added
to a 10% polyacrylamide gel, separated by electrophoresis and
electroporated to a PVDF membrane. The transferred PVDF membrane
was rinsed 3 times with 0.1% TBS-Tween-20 (TBST) and blocked with a
0.5% skim milk powder blocking solution at 37°C for 1 h. The
membrane was placed into a working solution containing the primary
antibodies and gently shake overnight at 4°C. The next day, the
samples were washed three times with TBST and incubated with
horseradish peroxidase-conjugated goat anti-rabbit IgG (1:1,000;
cat. no. ab6721; Abcam) at room temperature for 1 h. Color
development was performed at 4°C, avoiding light using a DAB
Chromogenic kit (mlbio Co., Ltd.), according to the manufacturer's
instructions. The ImageQuant LAS 4000 mini (General Electric
Company) was used to calculate the relative protein expression
levels. Western Blotting quantified using Quantity One Software
(ChemiDoc XRS, Bio-Rad Laboratories, Inc.).
Cell counting kit-8 (CCK-8) assay
A431 and colo-16 cells were seeded in 6-well plates
at a density of 2×104. Following 24-h culture, the cells
were infected with the empty vector control or FABP7 overexpression
virus. The cells were harvested at 96 h post-infection and
inoculated in 96-well plates at a density of 2,000 cells/well.
Subsequently, at 12, 24, 48 and 72 h, 10 µl CCK-8 solution (Dojindo
Molecular Technologies, Inc.) was added and incubated at 37°C for 2
h. A microplate reader was used to measure the absorbance at 450
nm, and cell proliferation curves were plotted.
Colony formation assay
A431 and colo-16 cells infected with the empty
vector control or FABP7 overexpression virus for 96 h were
inoculated into 6-well plates at a density of 200 cells/well and
cultured in the 37°C with 5% CO2 for 14 days.
Subsequently, the cells were washed twice with PBS, fixed with 4%
paraformaldehyde at room temperature for 15 min, washed twice with
PBS, stained with Giemsa at room temperature for 20 min, washed
three times with deionized water and cloned, photographed and
counted. Colonies were then counted under an invert microscope
(Leica, Germy). Only colonies >32 cells were scored.
Cell scratch assay
A431 and colo-16 cells were incubated and treated by
FABP7 overexpression transfection. A total of 5×105
cells/well (A431 and colo-16) were inoculated and cultured
overnight. The next day, a 200-µl pipette tip for was used to
create a wound. The cells were washed three times with PBS, the
cells in the wound were removed, and 2 ml cell culture medium
containing 2% fetal bovine serum was added. The cells were placed
in a 37°C incubator with 5% CO2 and cultured for 24 h.
Images were captured at 0 and 24 h, and the width of wound was
measured.
Transwell invasion assay
A431 and colo-16 cells were diluted in serum-free
DMEM medium and inoculated into the upper chambers of Transwell
inserts at ~2×105 cells/well. The lower chamber was
filled with DMEM containing 10% serum, and the cells were incubated
at 37°C for 24 h. The upper chamber was removed and washed with
PBS, fixed with 10% methanol for 20 min and stained with 0.1%
crystal violet at room temperature for 20 min. Following three
rinses with PBS, the cells on the upper surface of the membrane
were scraped with a cotton swab. The number of cells that migrated
to the lower surface of the membrane was counted under an inverted
microscope (Leica Microsystems GmbH; ×200 magnification), and the
experiment was repeated three times.
Tumor formation experiment
All animal experiments were performed in accordance
with the principles and procedures approved by the Animal
Experimentation Ethics Committee of the Shanghai Skin Disease
Hospital (Approval number: AN12047J), Tongji University School of
Medicine. A total of 12 specific pathogen-free-Balb/c nude male
mice (age, 6–8 weeks; weight, 18–22 g) were purchased from Beijing
Weitong Lihua Experimental Animal Co., Ltd. and housed in a
periodically UV-sterilized environment at 22°C and 50% humidity.
The mice had free access to water and food. Animal health and
behavior were monitored daily. The mice were divided into NC (n=6)
and oe-FABP7 (n=6) groups. A431 cells at the logarithmic phase were
transfected, the concentration of the cell suspension was adjusted
using serum-free DMEM medium, and the cells (2×106) were
subcutaneously inoculated into the left flanks of the mice for
modeling. Notable tumor growth at the injection site was used to
verify successful modeling. The tumor diameters were measured every
7 days by a vernier caliper, and the volume was calculated as
follows: V=0.52 × length × width2. The mice were
sacrificed at 35 days post-modeling by anesthesia with sodium
pentobarbital (50 mg/kg) and rapid cervical dislocation. The
maximum tumour volume was 1.5 mm3. No animals were found
dead during the experiment.
Statistical analysis
SPSS 10.0 statistical software (SPSS, Inc.) was used
for all statistical analyses. All experiments were repeated three
times for statistical analysis. The data are presented as the mean
± SD of ≥3 replicates. Unpaired two-tailed Student's t-test was
performed to compare the data between the two groups. The
χ2 test was used to compare the differences in
categorical variables between two groups. One-way ANOVA was used to
determine the difference between >2 groups, while a t-test was
used to analyze differences between two groups. Kaplan-Meier curves
and Cox regression analysis were used for survival and multivariate
analysis respectively, which were used to determine the
associations between the biomarker expression and patient survival.
P<0.05 was considered to indicate a statistically significant
difference.
Results
FABP7 is expressed at low levels in
CSCC tissues and cell lines
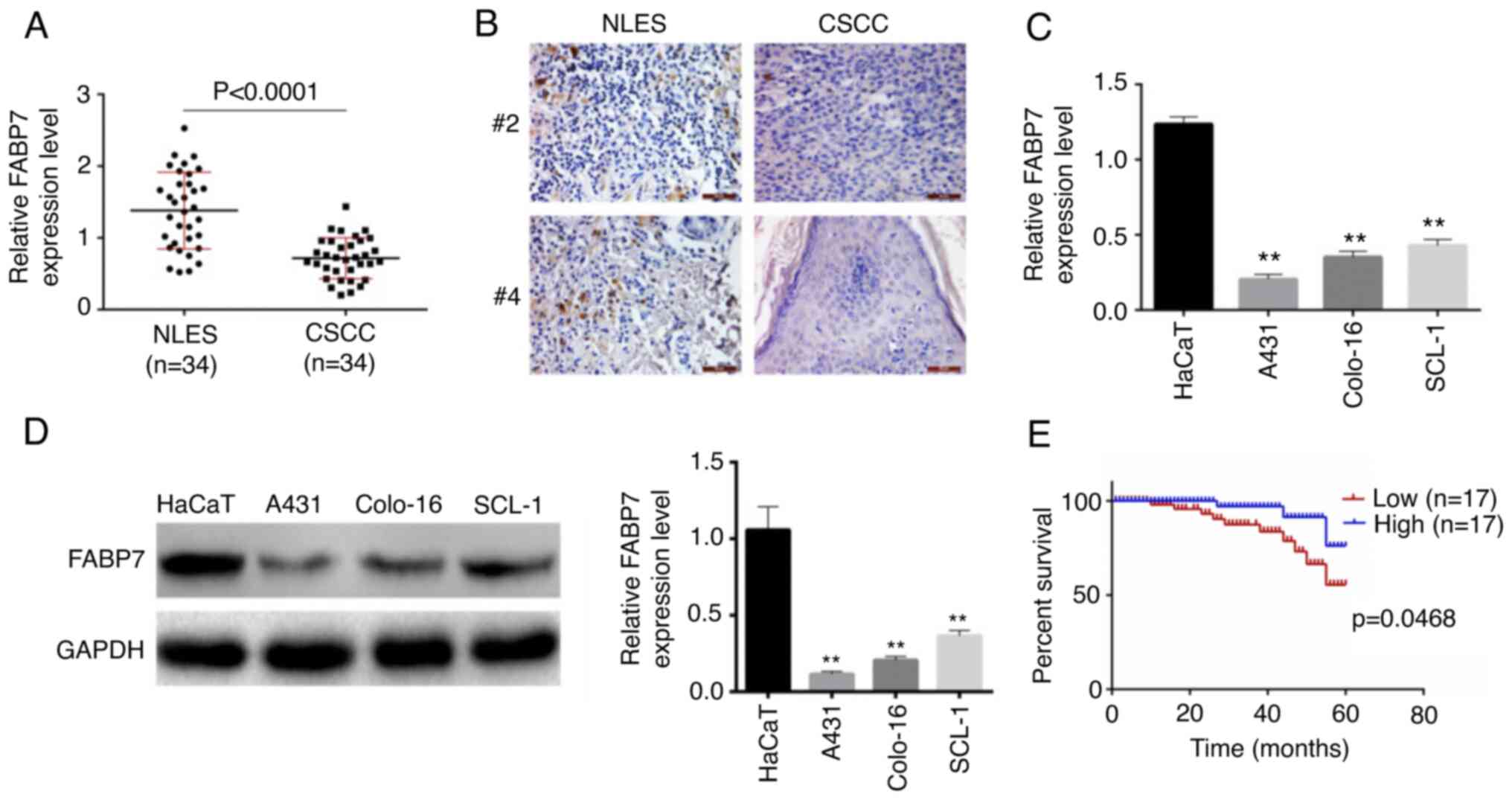
RT-qPCR was used to determine the expression levels
of FABP7 in the punch biopsies from CSCC (n=34) and non-lesion
epithelial skin (n=34) samples. As presented in Fig. 1A, the expression levels of FABP7
were significantly lower in CSCC tissues compared with those in the
normal epithelial samples (P<0.0001). Similar results were
obtained by immunohistochemistry (Fig.
1B). In addition, the expression levels of FABP7 in human CSCC
cell lines A431, colo-16 and SCL-1, and the immortalized human
normal keratinocyte cell line HaCaT, were detected by RT-qPCR
(Fig. 1C) and western blotting
(Fig. 1D). The results
demonstrated that FABP7 levels were significantly lower in A431,
colo-16 and SCL-1 cell lines compared with those in HaCaT cells
(P<0.01). Based on RT-qPCR results, the median value (0.78) of
FABP7 expression level of all patients was chosen as threshold, 34
patients with CSCC were divided into FABP7 high- and low-expression
groups. Patients in the FABP7 low-expression group exhibited a
lower survival rate compared with those in the FABP7 high
expression group (Fig. 1E). In
addition, the clinicopathological characteristics of the patients
were recorded and presented in Table
I. The age and sex were not different between the two groups.
However, the patients in the FABP7 low-expression group presented
with a higher Tumor-Node-Metastasis stage based on TNM stage
(21), histological grade and
degree of differentiation, compared with those in the FABP7
high-expression group. Based on the Cox multivariate analysis, the
age and sex between two groups were exponent of B [exp (B)] >1,
although no significant differences were identified. Notably, the
expression levels of FABP7 were exp (B) <1 and P<0.05,
indicating that it may be a suitable prognostic factor for
cutaneous squamous cell carcinoma (Table II).
 | Table I.Association between FABP7 expression
levels and patient clinicopathological characteristics in cutaneous
squamous cell carcinoma. |
Table I.
Association between FABP7 expression
levels and patient clinicopathological characteristics in cutaneous
squamous cell carcinoma.
|
|
| FABP7 expression,
n |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Patients, n | High | Low | P-value |
|---|
| Total | 34 | 17 | 17 |
|
| Age, years |
|
|
| 0.241 |
|
<45 | 13 | 8 | 5 |
|
|
≥45 | 21 | 9 | 12 |
|
| Sex |
|
|
| 0.500 |
|
Female | 15 | 8 | 7 |
|
|
Male | 19 | 9 | 10 |
|
| TNM Stage |
|
|
| 0.042 |
|
I–II | 16 | 11 | 5 |
|
|
III–IV | 18 | 6 | 12 |
|
| Histological
grade |
|
|
| 0.015 |
|
I–II | 16 | 12 | 4 |
|
|
III–IV | 18 | 5 | 13 |
|
|
Differentiation |
|
|
| 0.040 |
|
High | 14 | 10 | 4 |
|
|
Low | 20 | 7 | 13 |
|
 | Table II.Cox regression analysis for
multivariate analysis. |
Table II.
Cox regression analysis for
multivariate analysis.
| Variable | B | SE | Wald | df | P-value | Exp (B) | 95% CI for Exp
(B) |
|---|
| Age | 0.120 | 0.675 | 0.031 | 1 | 0.859 | 1.127 | 0.300-4.234 |
| Sex | 0.968 | 0.588 | 2.706 | 1 | 0.100 | 2.633 | 0.831-8.342 |
| FABP7
expression | −1.802 | 0.754 | 5.710 | 1 | 0.017 | 0.165 | 0.038-0.723 |
FABP7 overexpression inhibits A431 and
colo-16 cell proliferation, invasion and migration abilities
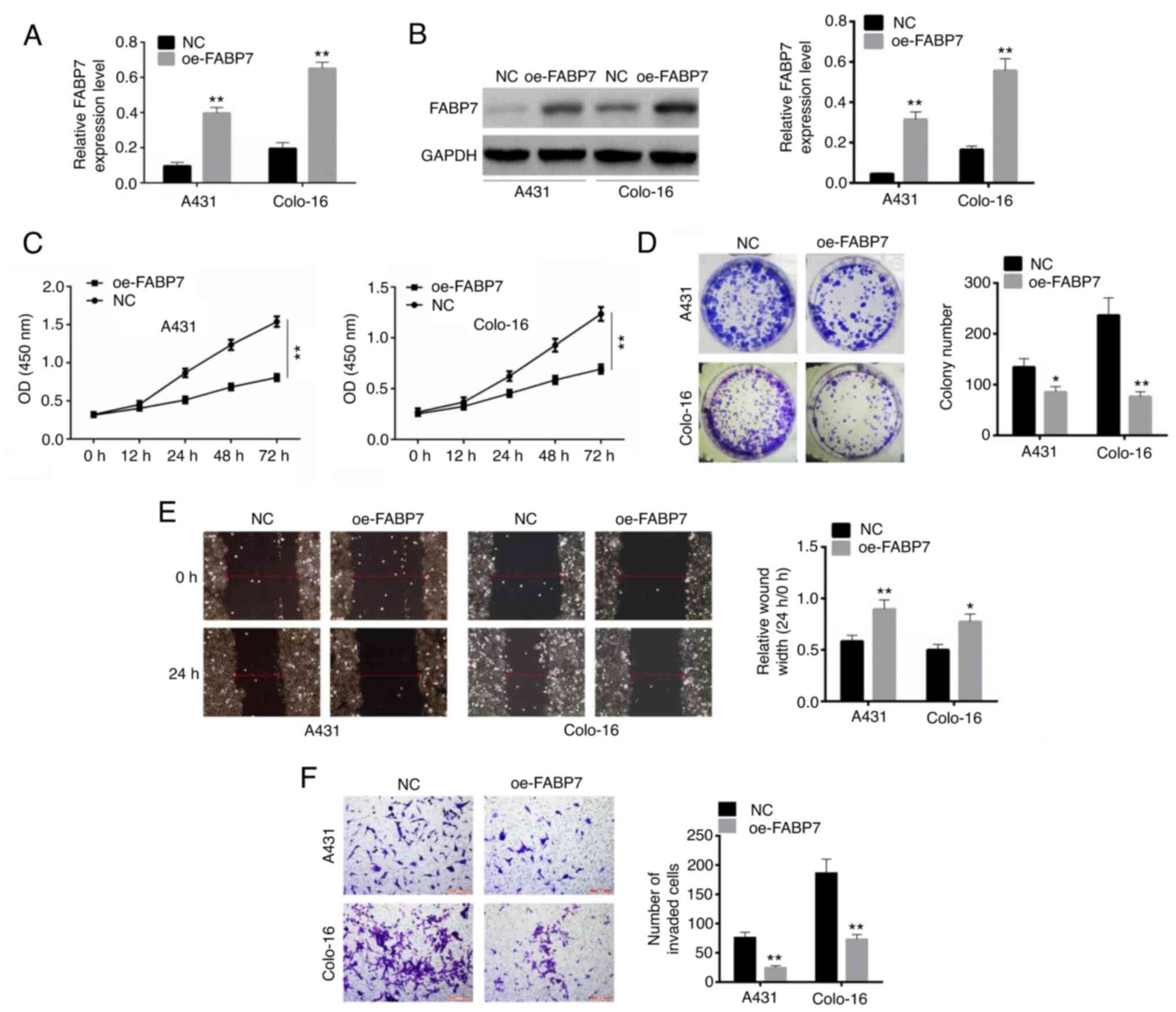
RT-qPCR and western blot assays were used to detect
the transfection efficiency of oe-FABP7 in A431 and colo-16 cells.
Following transfection, the levels of FABP7 were significantly
overexpressed in both A431 and colo-16 cell lines compared with
those in the NC-transfected cells (P<0.01; Fig. 2A and B). Following a 72-h
transfection, the optical density in the oe-FABP7 groups was
significantly lower compared with that in the control groups, which
confirmed that cell viability was inhibited (P<0.01; Fig. 2C). In addition, the numbers of
colonies formed by cells transfected with the oe-FABP7 vector were
significantly lower compared with those in the corresponding
control groups (P<0.01; Fig.
2D). Similarly, the relative wound width in oe-FABP7 group was
larger than control (P<0.01; Fig.
2E). The results of the Transwell invasion assay confirmed that
invasion was inhibited in oe-FABP7 groups compared with the
NC-transfected cells (P<0.01; Fig.
2F).
FABP7 overexpression inhibits cell
proliferation, invasion and the Notch signaling pathway
PCNA, Snail, N-cadherin, Twist, MMP-2 and MMP-7 are
crucial proteins associated with cell proliferation and invasion
(22). Following transfection,
these proteins were detected by western blotting. The expression
levels of all these proliferation- and invasion-associated proteins
(PCNA, Snall, n-Cad, Twist, MMP-2 and MMP-7) were lower in the
oe-FABP7 groups compared with those in the NC groups (P<0.01;
Fig. 3A). Based on the STRING
database, FABP7 is associated with the Kyoto Encyclopedia of Genes
and Genomes Notch pathway. Notably, overexpression of FABP7
inhibited the expression levels of Notch 1 and Notch 3 compared
with those in the NC groups (P<0.01; Fig. 3B). Therefore, FABP7 overexpression
may inhibit the proliferation, invasion and migration of A431 and
colo-16 cells.
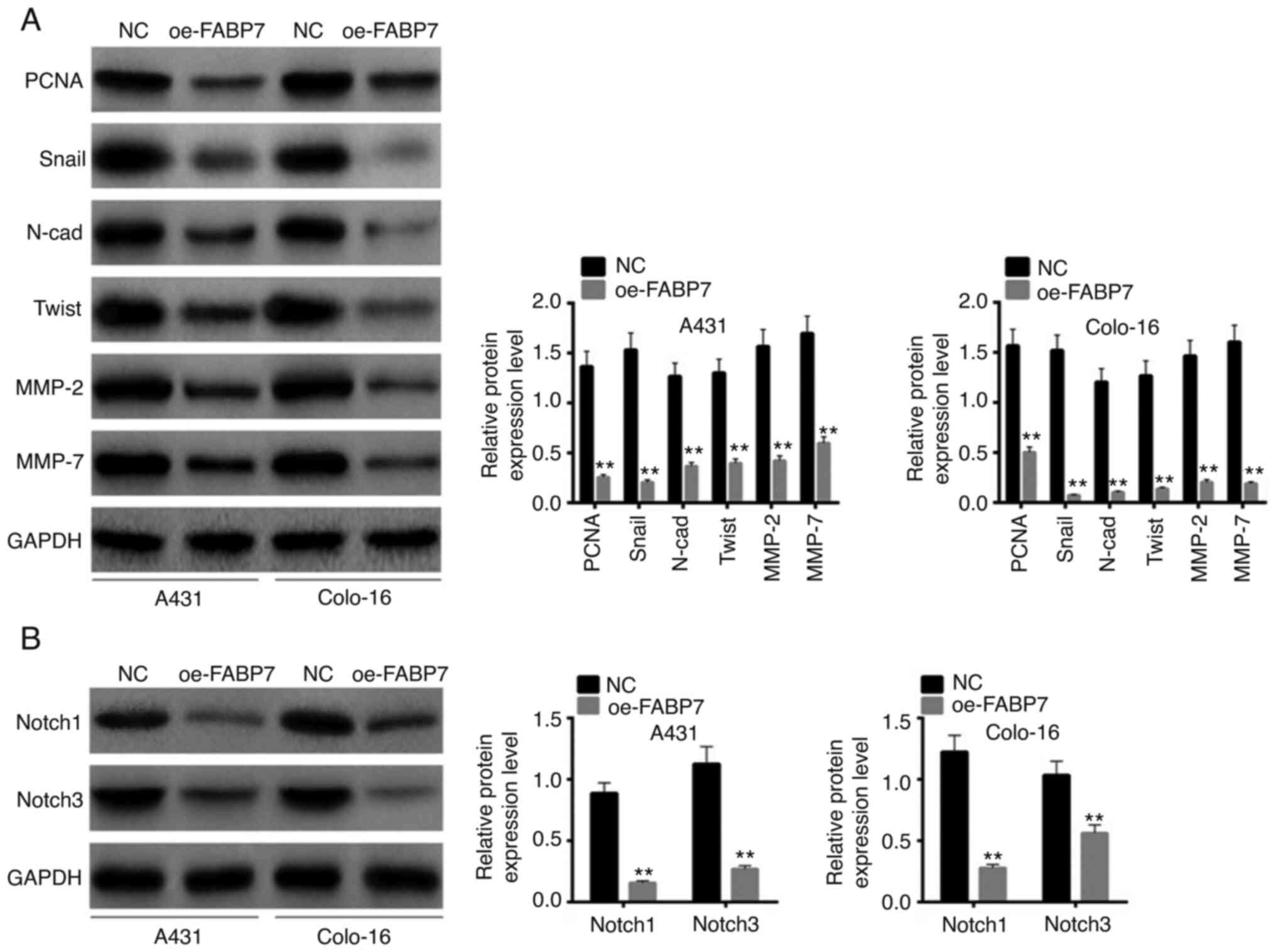 | Figure 3.Effects of FABP7 overexpression on
proliferation-, invasion- and Notch pathway-associated proteins.
(A) The expression levels of PCNA, Snail, N-cad, Twist, MMP-2 and
MMP-7 were detected by western blotting. (B) Notch pathway-related
proteins were detected by western blotting. **P<0.01 vs. NC.
FABP7, fatty acid-binding protein 7; oe, overexpression vector; NC,
negative control; PCNA, proliferating cell nuclear antigen; N-cad,
N-cadherinl; MMP, matrix metalloproteinase. |
FABP7 overexpression inhibits CSCC
growth in vivo
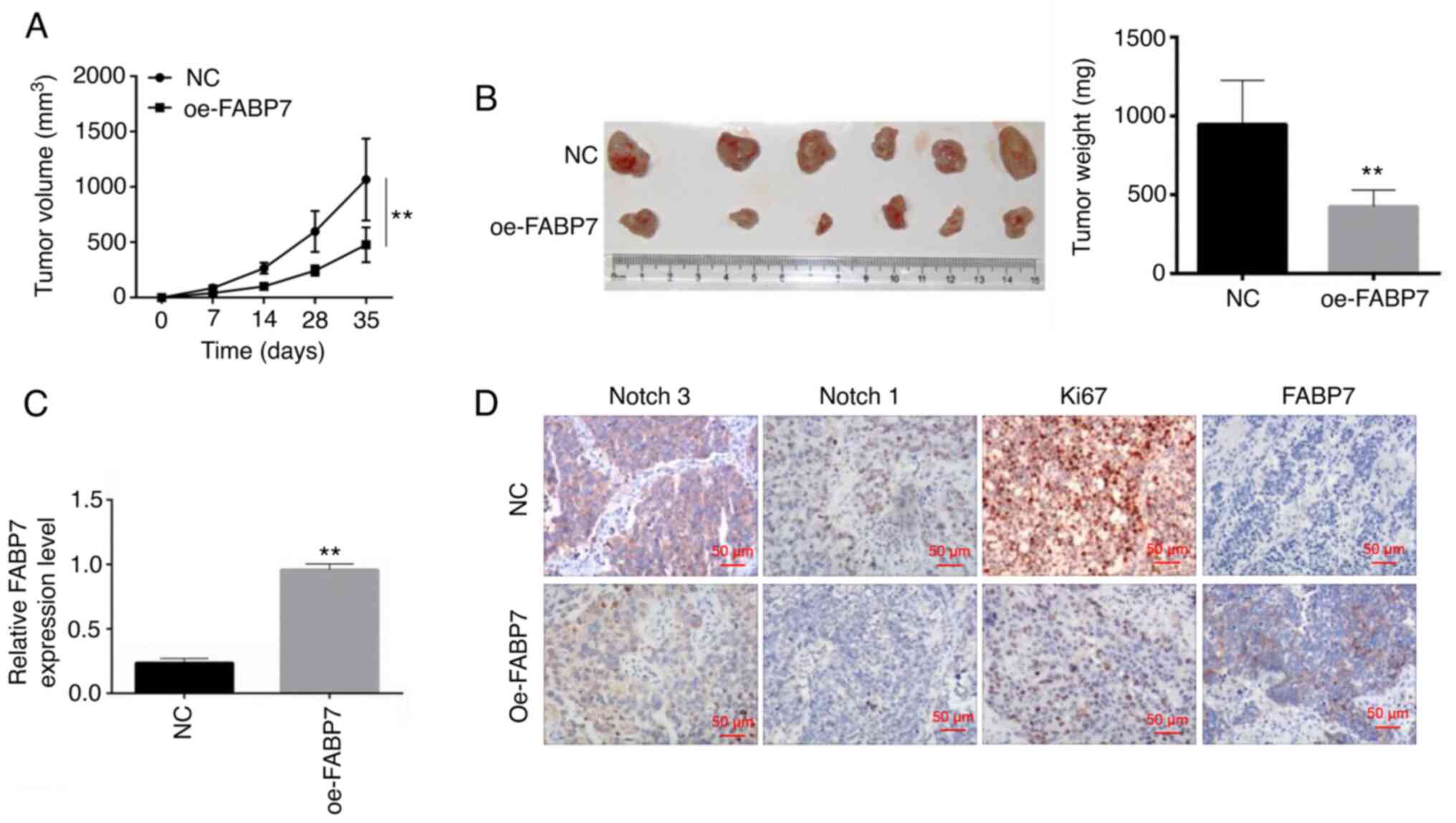
At 35 days post-inoculation, the volume and weight
of mouse xenograft tumors were significantly lower in the oe-FABP7
group compared with those in the control group (P<0.01; Fig. 4A and B), suggesting that tumor
growth was inhibited by oe-FABP7 in vivo. RT-qPCR was used
to detect FABP7 expression in the tumor tissues; the results
demonstrated that the levels of FABP7 were decreased in vivo
in the FABP7 overexpression group compared with those in the NC
group (P<0.01; Fig. 4C).
Immunohistochemistry was used to detect the expression of FABP7,
Ki67, Notch 1 and Notch 3. The results demonstrated that
overexpression of FABP7 inhibited the protein expression levels of
Notch 1 and Notch 3 compared with those in the control group
(Fig. 4D).
Discussion
In order to determine the molecular mechanism
underlying the effects of FABP7 in CSCC, FABP7 overexpression
models in vivo and in vitro were constructed, and
cell viability, invasion and the Notch pathway signaling pathway
were analyzed in the present study. The results demonstrated that
FABP7 expression levels were significantly lower in human CSCC
tissues and cells compared with those in non-tumor tissues and
cells. Transfection with oe-FABP7 inhibited the viability, invasion
and migration of A431 and colo-16 cells compared with those
observed in the control cells. In addition, overexpression of FABP7
reduced the expression levels of proteins associated with cell
proliferation, invasion and the Notch signaling pathway compared
with those in the NC groups. FABP7 overexpression reduced the
growth of CSCC tumors in vivo, and inhibited the expression
levels of Ki67, Notch 1 and Notch 3 compared with those in the
control group.
FABP7 is a protein-coding gene involved in various
diseases and cell functions. For example, FABP7 has been reported
to be associated with optic nerve glioma and neoplasm (23). The peroxisome
proliferator-activated receptor (PPAR) signaling pathway serves an
important role in rat brain astrocytes (24); Nijsten et al (25) have reported that PPARα, PPARβ and
PPARγ are abnormally expressed in CSCC samples and associated with
immunoreactivity. PPARβ/δ KO mice displayed increased inflammation
in response to 12-O-tetradecanoylphorbol-13-acetate (TPA) treatment
(26). Notably, Slipicevic et
al (11) demonstrated that
FABP7 enhances the invasion and proliferation of melanoma cells via
the ERK and MAPK pathways, and this pathway is crucial for the
pathogenesis of cutaneous disease (27). For instance, microRNA-148a inhibits
the MAPK pathway and suppresses the development of CSCC (28). In addition, high mobility group box
1 has also been demonstrated to participate in the MAPK signaling
pathway and regulate the metastasis of CSCC (29). Various long non-coding RNAs or
microRNAs (miRs) induce the expression of ERK and regulate the
proliferation of CSCC cells including long non-coding RNA PICSAR,
miR-124 and miR-214 (30,31). Therefore, we hypothesized that
FABP7 may inhibit the viability, invasion and migration of CSCC
cells via the PPAR and MAPK/ERK1/2 pathways.
Overexpression of FABP7 was demonstrated to inhibit
the expression of proliferation- and invasion-associated proteins
(PCNA, Snail, N-cadherin, Twist, MMP-2 and MMP-7) as well as Notch
pathway-related proteins (Notch 1 and Notch 3) in the present
study. Invasion and migration of tumor cells are the main features
of malignant tumors and the primary cause of malignant
tumor-associated death (32). The
occurrence of invasion and migration involves complex signaling
pathways, such as MAPK and PI3K signaling pathways in tumor cells
and the microenvironment in which they were located. Activation and
interaction of these signaling pathways are involved in tumor
survival and growth (33). A
previous study reported that PCNA is expressed at high levels in
CSCC, which is associated with grade (34). In addition, Gong et al
(35) reported that The positivity
rates of Snail expression were 0% in normal cervical tissues, 32.0%
in CIN tissues, and 66.2% in CSCC tissues. Furthermore,
Co-expression of HIF-1alpha, TWIST and Snail in primary tumors of
patients with head and neck cancers correlated with worst prognosis
(36,37). Similarly, Ahmed Haji Omar et
al (38) demonstrated that
MMPs participate in the initiation, metastasis, invasion and
evasiveness of CSCC. In addition, the Notch pathway-associated
proteins are important genes with high mutation rate in primary
CSCC (39). Thus, overexpression
of FABP7 may inhibit the proliferation and invasion of CSCC cells
by regulating the Notch signaling pathway.
Although the molecular mechanism of FABP7 was
studied in cells and nude mice, the present study had certain
limitations. First, the number of cases included in the current
study was small, and the statistical analysis of experimental
results was limited. In addition, due to the experimental
conditions, only human CSCC cell lines (A431, colo-16, and SCL-1)
and immortalized human normal keratinocyte cell line (HaCaT) were
used in the present study. The reliability of these data needs to
be further verified. Second, the experiments were not blinded.
Blind experiments will be performed in future studies. Third, the
target gene may be regulated by various genes, creating a
complicated interaction network. The present study only focused on
the Notch pathway, proliferation and invasion. Finally, the
functions of proliferation and invasion-associated proteins and
Notch pathway related proteins were obtained from previous studies,
but were not verified in the present study. The aforementioned
limitations will be addressed in our future studies. For instance,
a multi-center, double-blind, large-sample study will be designed
and conducted. Additional cell lines and animal models will be used
to verify the results, and other genes and associated microRNAs and
pathways will be studied to construct a regulatory network and
identify potential targets for CSCC diagnosis and treatment.
In conclusion, the results of the present study
demonstrated that FABP7 inhibited the proliferation and invasion of
CSCC cells by participating in the Notch signaling pathway, and may
be a diagnostic biomarker or treatment target for CSCC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZS, YZ and XW designed the study. ZS, YG and DZ
performed the experiments. GZ and YZ analyzed the data. ZS, YZ and
XW wrote the manuscript. All authors read and approved the final
manuscript, and confirmed the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Shanghai Skin Disease Hospital, Tongji University
School of Medicine (approval no. SSDH-IEC-SG-029-3.1; Shanghai,
China). Written informed consent was obtained from all patients.
All animal experiments were performed in accordance with the
principles and procedures approved by the Animal Experimentation
Ethics Committee of the Shanghai Skin Disease Hospital, Tongji
University School of Medicine (Approval number:
SSDH-AEEC-SG-331-2.1).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Llewellyn CD, Linklater K, Bell J, Johnson
NW and Warnakulasuriya KA: Squamous cell carcinoma of the oral
cavity in patients aged 45 years and under: A descriptive analysis
of 116 cases diagnosed in the South East of England from 1990 to
1997. Oral Oncol. 39:106–114. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vauterin TJ, Veness MJ, Morgan GJ, Poulsen
MG and O'Brien CJ: Patterns of lymph node spread of cutaneous
squamous cell carcinoma of the head and neck. Head Neck.
28:785–791. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haytoglu NS, Gurel MS, Erdemir VA,
Leblebici C and Haytoglu TG: An unusual case of sporotrichoid
nodules: Metastatic cutaneous squamous cell carcinoma. Dermatol
Online J. 19:181742013.PubMed/NCBI
|
|
4
|
Hahn SB, Kim DJ and Jeon CH: Clinical
study of Marjolin's ulcer. Yonsei Med J. 31:234–241. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao B: Clinical dermatology. Jiangsu
Science and Technology Press; Nanjing: pp. p7072001
|
|
6
|
Aupperle KR, Boyle DL, Hendrix M, Seftor
EA, Zvaifler NJ, Barbosa M and Firestein GS: Regulation of
synoviocyte proliferation, apoptosis, and invasion by the p53 tumor
suppressor gene. Am J Pathol. 152:1091–1098. 1998.PubMed/NCBI
|
|
7
|
Sloane BF and Honn KV: Cysteine
proteinases and metastasis. Cancer Metastasis Rev. 3:249–263. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miwa S, Miyagawa S, Soeda J and Kawasaki
S: Matrix metalloproteinase-7 expression and biologic
aggressiveness of cholangiocellular carcinoma. Cancer. 94:428–434.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zaravinos A, Kanellou P, Baritaki S,
Bonavida B and Spandidos DA: BRAF and RKIP are significantly
decreased in cutaneous squamous cell carcinoma. Cell Cycle.
8:1402–1408. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Darido C, Georgy SR, Wilanowski T, Dworkin
S, Auden A, Zhao Q, Rank G, Srivastava S, Finlay MJ, Papenfuss AT,
et al: Targeting of the tumor suppressor GRHL3 by a
miR-21-dependent proto-oncogenic network results in PTEN loss and
tumorigenesis. Cancer Cell. 20:635–648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Slipicevic A, Jørgensen K, Skrede M,
Rosnes AK, Trøen G, Davidson B and Flørenes VA: The fatty acid
binding protein 7 (FABP7) is involved in proliferation and invasion
of melanoma cells. BMC Cancer. 8:2762008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou J, Deng Z, Chen Y, Gao Y, Wu D, Zhu
G, Li L, Song W, Wang X, Wu K and He D: Overexpression of FABP7
promotes cell growth and predicts poor prognosis of clear cell
renal cell carcinoma. Urol Oncol. 33:113.e9–e17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Umaru BA, Kagawa Y, Shil SK, Arakawa N,
Pan Y, Miyazaki H, Kobayashi S, Yang S, Cheng A, Wang Y, et al:
Ligand Bound fatty acid binding protein 7 (FABP7) drives melanoma
cell proliferation via modulation of Wnt/β-catenin signaling. Pharm
Res. 38:479–490. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tani G, Tomuschat C, O'Donnell AM, Coyle D
and Puri P: Increased population of immature enteric glial cells in
the resected proximal ganglionic bowel of Hirschsprung's disease
patients. J Surg Res. 218:150–155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goto Y, Koyanagi K, Narita N, Kawakami Y,
Takata M, Uchiyama A, Nguyen L, Nguyen T, Ye X, Morton DL and Hoon
DS: Aberrant fatty acid-binding protein-7 gene expression in
cutaneous malignant melanoma. J Invest Dermatol. 130:221–229. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu YY, Zheng MH, Zhang R, Liang YM and Han
H: Notch signaling pathway and cancer metastasis. Adv Exp Med Biol.
727:186–198. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang W, Liu H, Liu Z, Zhu D, Amos CI,
Fang S, Lee JE and Wei Q: Functional variants in notch pathway
genes NCOR2, NCSTN, and MAML2 predict survival of patients with
cutaneous melanoma. Cancer Epidemiol Biomarkers Prev. 24:1101–1110.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dressler F, Whalen JA, Reinhardt BN and
Steere AC: Western blotting in the serodiagnosis of Lyme disease. J
Infect Dis. 167:392–400. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cañueto J, Cardeñoso E, Garcia J,
Santos-Briz A, Castellanos A, Fernández-López E, Blanco-Gómez A,
Perez-Losada J and Román-Curto C: EGFR expression is associated
with poor outcome in cutaneous squamous cell carcinoma. Br J
Dermatol. 176:279–1287. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu C, Bain X, Zhang L, Hu Y, Wu Y, Pei T
and Han X: Long noncoding RNA LINC00968 inhibits proliferation,
migration and invasion of lung adenocarcinoma through targeting
miR-22-5p/CDC14A axis. 3 Biotech. 11:4332021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang Y, Bollen AW, Aldape KD and Gupta N:
Nuclear FABP7 immunoreactivity is preferentially expressed in
infiltrative glioma and is associated with poor prognosis in
EGFR-overexpressing glioblastoma. BMC Cancer. 6:972006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tripathi S, Kushwaha R, Mishra J, Gupta
MK, Kumar H, Sanyal S, Singh D, Sanyal S, Sahasrabuddhe AA, Kamthan
M, et al: Docosahexaenoic acid up-regulates both PI3K/AKT-dependent
FABP7-PPARγ interaction and MKP3 that enhance GFAP in developing
rat brain astrocytes. J Neurochem. 140:96–113. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nijsten T, Geluyckens E, Colpaert C and
Lambert J: Peroxisome proliferator-activated receptors in squamous
cell carcinoma and its precursors. J Cutan Pathol. 32:340–347.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Man MQ, Barish GD, Schmuth M, Crumrine D,
Barak Y, Chang S, Jiang Y, Evans RM, Elias PM and Feingold KR:
Deficiency of PPARbeta/delta in the epidermis results in defective
cutaneous permeability barrier homeostasis and increased
inflammation. J Invest Dermatol. 128:370–377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agelopoulos K, Rülander F, Dangelmaier J,
Lotts T, Osada N, Metze D, Luger TA, Loser K and Ständer S:
Neurokinin 1 receptor antagonists exhibit peripheral effects in
prurigo nodularis including reduced ERK1/2 activation. J Eur Acad
Dermatol Venereol. 33:2371–2379. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luo Q, Li W, Zhao T, Tian X, Liu Y and
Zhang X: Role of miR-148a in cutaneous squamous cell carcinoma by
repression of MAPK pathway. Arch Biochem Biophys. 583:47–54. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun Y, Tu Y, He LI, Ji C and Cheng BO:
High mobility group box 1 regulates tumor metastasis in cutaneous
squamous cell carcinoma via the PI3K/AKT and MAPK signaling
pathways. Oncol Lett. 11:59–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Piipponen M, Nissinen L, Farshchian M,
Riihilä P, Kivisaari A, Kallajoki M, Peltonen J, Peltonen S and
Kähäri VM: Long noncoding RNA PICSAR promotes growth of cutaneous
squamous cell carcinoma by regulating ERK1/2 activity. J Invest
Dermatol. 136:1701–1710. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamane K, Jinnin M, Etoh T, Kobayashi Y,
Shimozono N, Fukushima S, Masuguchi S, Maruo K, Inoue Y, Ishihara
T, et al: Down-regulation of miR-124/-214 in cutaneous squamous
cell carcinoma mediates abnormal cell proliferation via the
induction of ERK. J Mol Med (Berl). 91:69–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Karbowniczek M, Chosia M and Domagała W:
Nuclear morphometry of MIB-1 positive and negative tumor cells in
primary and metastatic malignant melanoma of the skin. Pol J
Pathol. 50:235–241. 1999.PubMed/NCBI
|
|
33
|
Pourreyron C, Chen M, McGrath JA,
Salas-Alanis JC, South AP and Leigh IM: High levels of type VII
collagen expression in recessive dystrophic epidermolysis bullosa
cutaneous squamous cell carcinoma keratinocytes increases PI3K and
MAPK signalling, cell migration and invasion. Br J Dermatol.
170:1256–1265. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo Y, Wang Q, Tian P and Jia Y: Highly
expressed CHAF1A and PCNA are positively associated with malignancy
of cervical squamous cell carcinoma. Xi Bao Yu Fen Zi Mian Yi Xue
Za Zhi. 33:1696–1701. 2017.(In Chinese). PubMed/NCBI
|
|
35
|
Gong X, Tao Y, Zhou L, Yu L, Wu S, Song W,
Wang D and Cheng Z: Expressions of Snail, Slug and KAI1 proteins in
cervical carcinoma and their clinicopathological significance. Nan
Fang Yi Ke Da Xue Xue Bao. 35:1733–1738. 2015.(In Chinese).
PubMed/NCBI
|
|
36
|
Yao J, Caballero OL, Huang Y, Lin C,
Rimoldi D, Behren A, Cebon JS, Hung MC, Weinstein JN, Strausberg RL
and Zhao Q: Altered expression and splicing of ESRP1 in malignant
melanoma correlates with epithelial-mesenchymal status and
tumor-associated immune cytolytic activity. Cancer Immunol Res.
4:552–561. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang MH, Wu MZ, Chiou SH, Chen PM, Chang
SY, Liu CJ, Teng SC and Wu KJ: Direct regulation of TWIST by
HIF-1alpha promotes metastasis. Nat Cell Biol. 10:295–305. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ahmed Haji Omar A, Haglund C, Virolainen
S, Häyry V, Atula T, Kontio R, Salo T, Sorsa T and Hagström J:
MMP-7, MMP-8, and MMP-9 in oral and cutaneous squamous cell
carcinomas. Oral Surg Oral Med Oral Pathol Oral Radiol.
119:459–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Toland AE: Frequent somatic mutations of
chromatin remodeling genes in metastatic cutaneous squamous cell
carcinoma. Dermatol Online J. 22:2016.
|