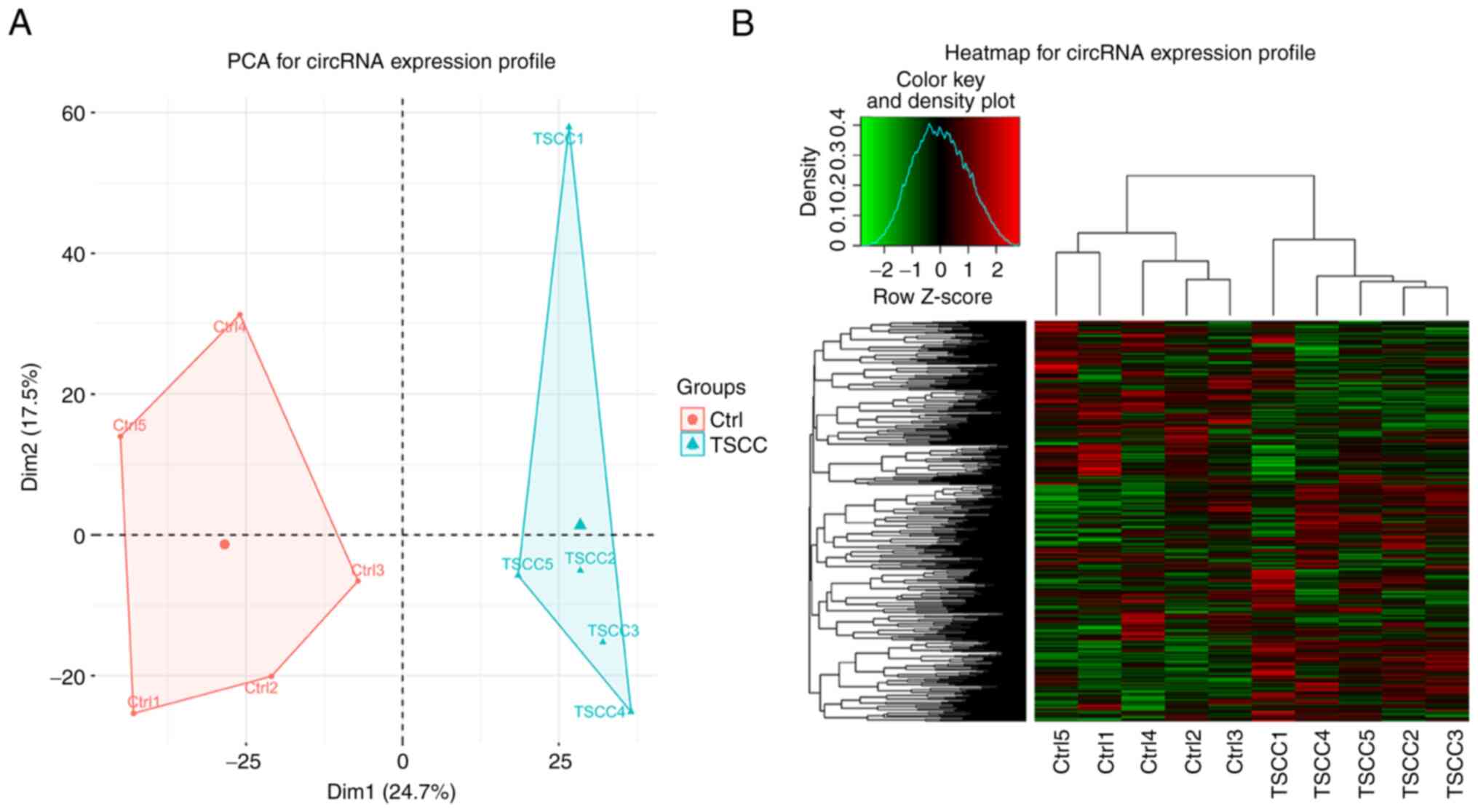
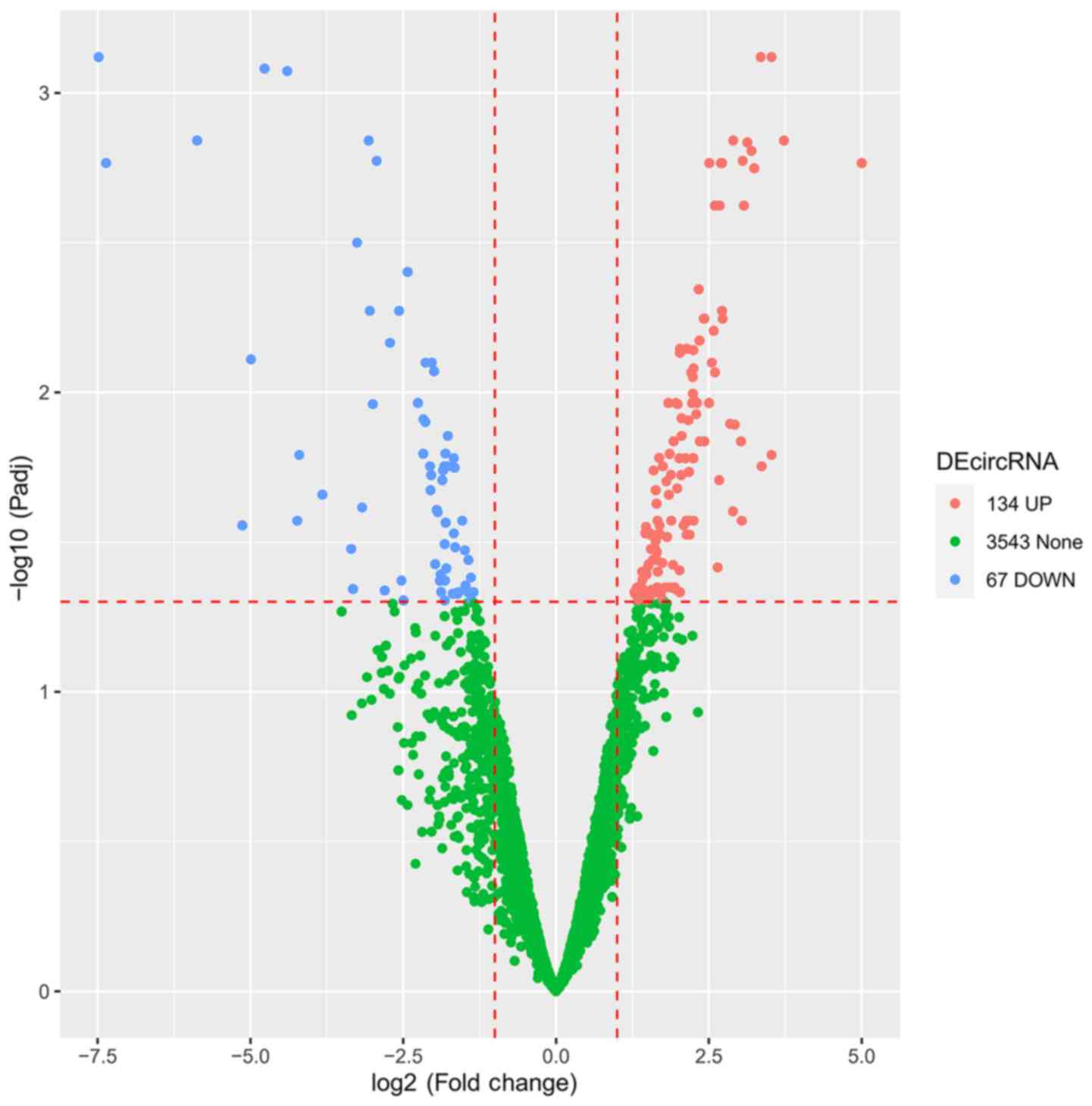
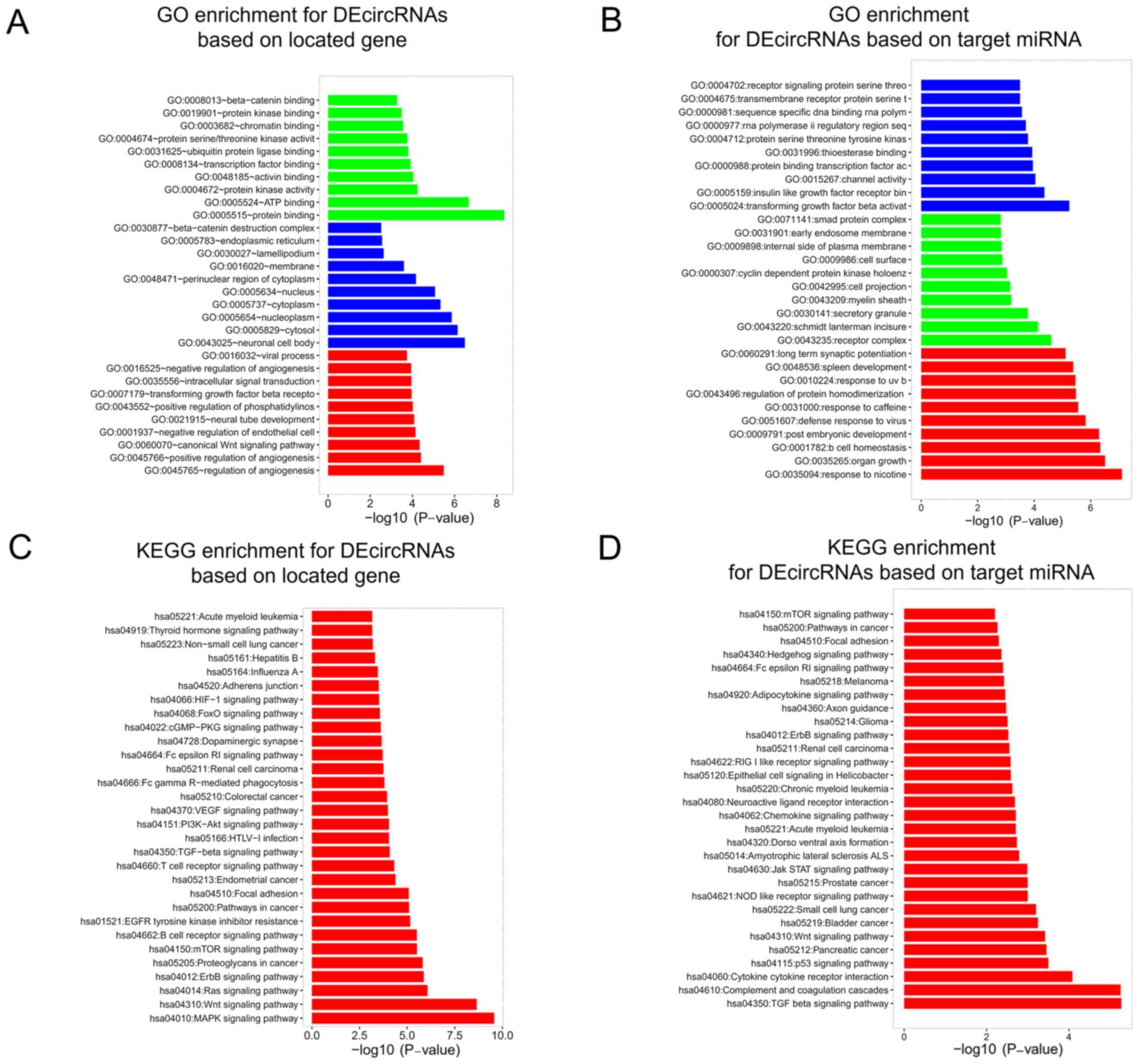
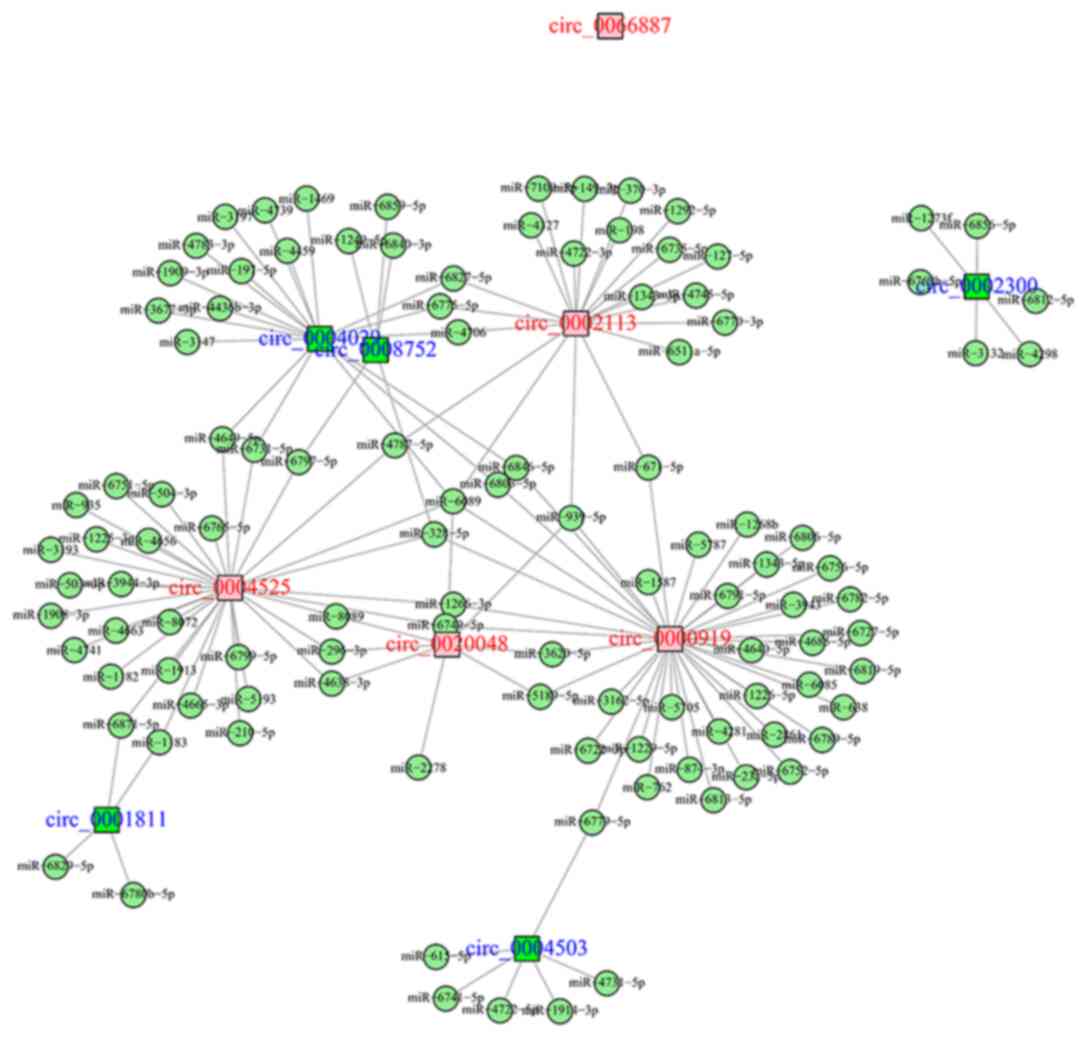
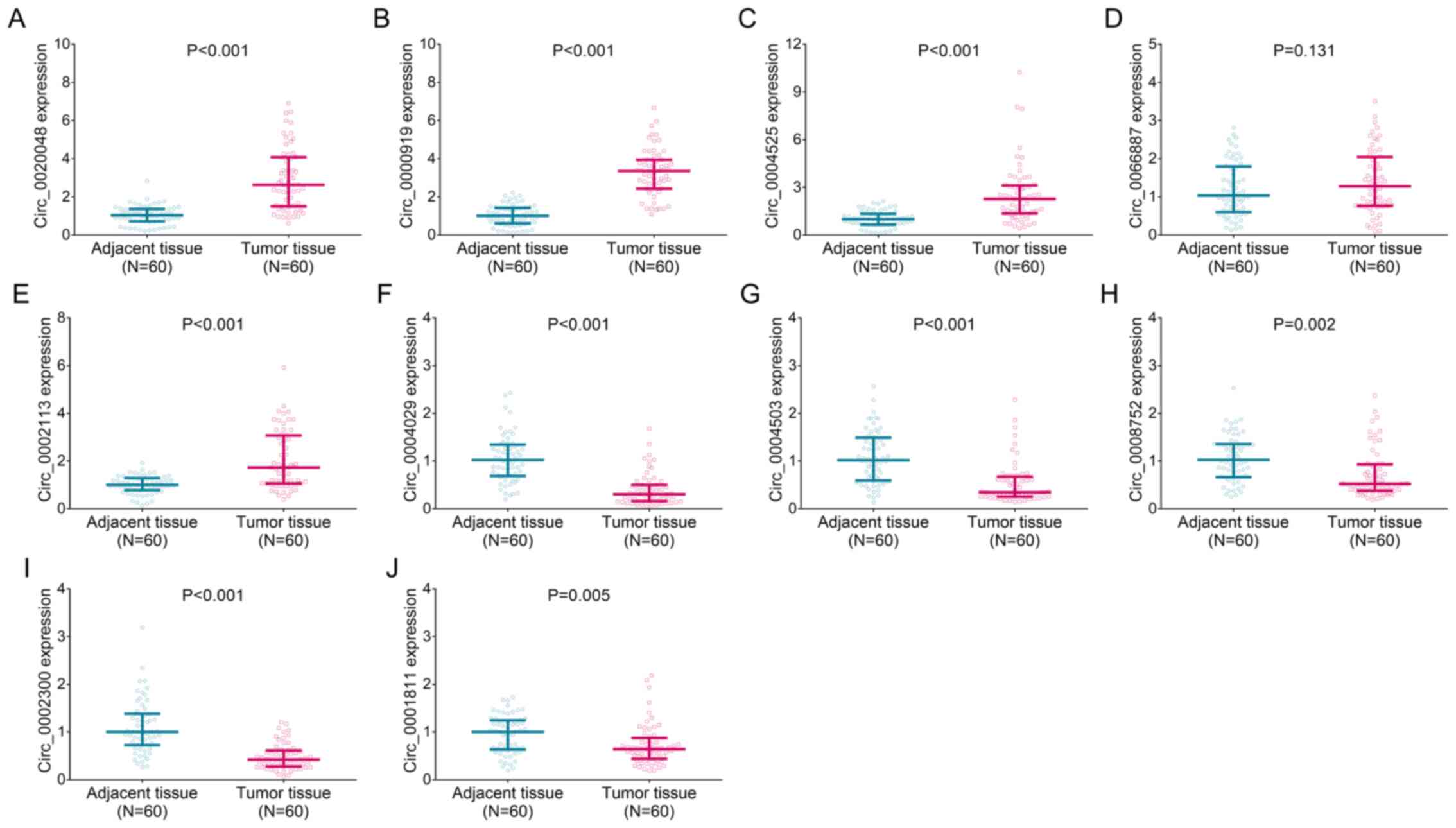
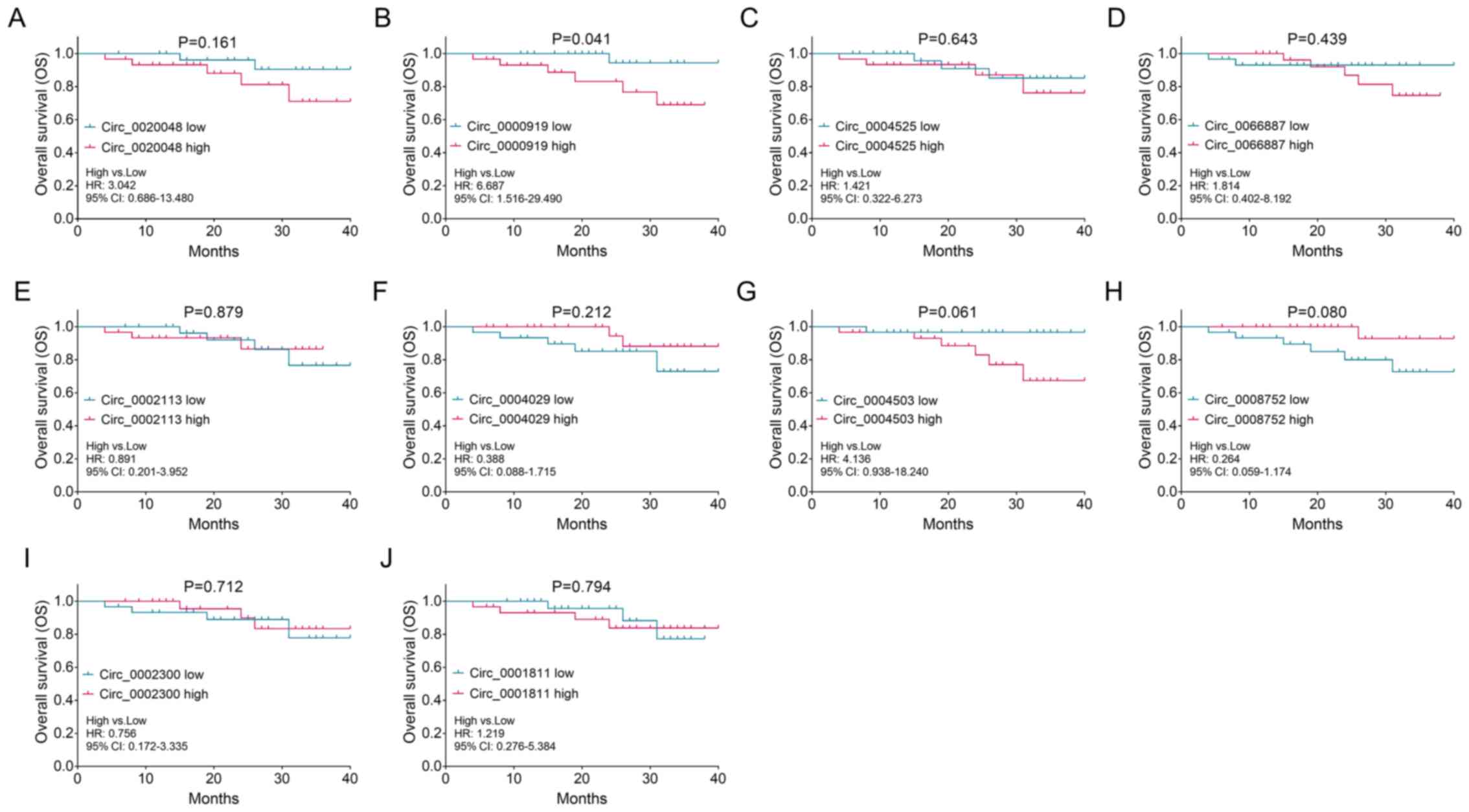
|
1
|
Kim YJ and Kim JH: Increasing incidence
and improving survival of oral tongue squamous cell carcinoma. Sci
Rep. 10:78772020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shrestha AD, Vedsted P, Kallestrup P and
Neupane D: Prevalence and incidence of oral cancer in low- and
middle-income countries: A scoping review. Eur J Cancer Care
(Engl). 29:e132072020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohta K and Yoshimura H: Squamous cell
carcinoma of the dorsal tongue. CMAJ. 191:E13102019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Machiels JP, René Leemans C, Golusinski W,
Grau C, Licitra L and Gregoire V: EHNS Executive Board. Electronic
address. simplesecretariat@ehns.orgESMO
Guidelines Committee. Electronic address. simpleclinicalguidelines@esmo.orgESTRO
Executive Board. Electronic address. simpleinfo@estro.orgSquamous cell
carcinoma of the oral cavity, larynx, oropharynx and hypopharynx:
EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 31:1462–1475. 2020. View Article : Google Scholar
|
|
5
|
Okubo M, Iwai T, Nakashima H, Koizumi T,
Oguri S, Hirota M, Mitsudo K and Tohnai I: Squamous Cell Carcinoma
of the Tongue Dorsum: Incidence and Treatment Considerations.
Indian J Otolaryngol Head Neck Surg. 69:6–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Montero PH and Patel SG: Cancer of the
oral cavity. Surg Oncol Clin N Am. 24:491–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ge P, Zhang J, Zhou L, Lv MQ, Li YX, Wang
J and Zhou DX: CircRNA expression profile and functional analysis
in testicular tissue of patients with non-obstructive azoospermia.
Reprod Biol Endocrinol. 17:1002019. View Article : Google Scholar
|
|
8
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar
|
|
9
|
Fan HY, Jiang J, Tang YJ, Liang XH and
Tang YL: circRNAs: A New Chapter in Oral Squamous Cell Carcinoma
Biology. OncoTargets Ther. 13:9071–9083. 2020. View Article : Google Scholar
|
|
10
|
Wang YF, Li BW, Sun S, Li X, Su W, Wang
ZH, Wang F, Zhang W and Yang HY: Circular RNA Expression in Oral
Squamous Cell Carcinoma. Front Oncol. 8:3982018. View Article : Google Scholar
|
|
11
|
Yao Y, Bi L and Zhang C: Circular
RNA_0001742 has potential to predict advanced tumor stage and poor
survival profiles in tongue squamous cell carcinoma management. J
Clin Lab Anal. 34:e233302020. View Article : Google Scholar
|
|
12
|
Wei T, Ye P, Yu GY and Zhang ZY: Circular
RNA expression profiling identifies specific circular RNAs in
tongue squamous cell carcinoma. Mol Med Rep. 21:1727–1738.
2020.PubMed/NCBI
|
|
13
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: the 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar
|
|
14
|
World Health Organization, . The ICD-10
classification of mental and behavioural disorders: diagnostic
criteria for research. World Health Organization; Geneva,
Switzerland: 1993, https://apps.who.int/iris/handle/10665/37108October
25–2016
|
|
15
|
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y,
Sun B, Wang Y, Wang L and Huang Q: Circular RNAs expression
profiles in human gastric cancer. Sci Rep. 7:90602017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Prats AC, David F, Diallo LH, Roussel E,
Tatin F, Garmy-Susini B and Lacazette E: Circular RNA, the Key for
Translation. Int J Mol Sci. 21:E85912020. View Article : Google Scholar
|
|
18
|
Tucker D, Zheng W, Zhang DH and Dong X:
Circular RNA and its potential as prostate cancer biomarkers. World
J Clin Oncol. 11:563–572. 2020. View Article : Google Scholar
|
|
19
|
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie
JW, Wang JB, Lin JX, Chen QY, Cao LL, et al: Circular RNA
circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to
facilitate gastric cancer invasion and metastasis. Cancer Lett.
471:38–48. 2020. View Article : Google Scholar
|
|
20
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martin-Orozco E, Sanchez-Fernandez A,
Ortiz-Parra I and Ayala-San Nicolas M: WNT Signaling in Tumors: The
Way to Evade Drugs and Immunity. Front Immunol. 10:28542019.
View Article : Google Scholar
|
|
22
|
Basu S, Cheriyamundath S and Ben-Ze'ev A:
Cell-cell adhesion: Linking Wnt/β-catenin signaling with partial
EMT and stemness traits in tumorigenesis. F1000 Res. 7:72018.
View Article : Google Scholar
|
|
23
|
Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y and
Hu LL: ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med.
19:1997–2007. 2020.PubMed/NCBI
|
|
24
|
Liu R, Shen L, Lin C, He J, Wang Q, Qi Z,
Zhang Q, Zhou M and Wang Z: MiR-1587 Regulates DNA Damage Repair
and the Radiosensitivity of CRC Cells via Targeting LIG4. Dose
Response. 18:15593258209369062020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park EJ, Jung HJ, Choi HJ, Jang HJ, Park
HJ, Nejsum LN and Kwon TH: Exosomes co-expressing AQP5-targeting
miRNAs and IL-4 receptor-binding peptide inhibit the migration of
human breast cancer cells. FASEB J. 34:3379–3398. 2020. View Article : Google Scholar : PubMed/NCBI
|