Introduction
Tongue squamous cell carcinoma (TSCC) is one of the
most common (90%) oral squamous cell carcinomas and it is estimated
to be responsible for 300,000 newly diagnosed cases and 130,000
cancer-associated deaths worldwide every year (1,2). Its
prevalence is 0.12 per 1,000 individuals in Asia (1,2).
TSCC generally occurs in the middle-aged male population and the
major etiological factors for this disease include tobacco use,
alcohol consumption and betel nut chewing (3,4). The
majority of TSCC cases are noted on the lateral border of the
tongue and the clinical manifestations of this condition consist of
pain, burning sensation and swallowing difficulties (5,6).
Despite the improvements made in TSCC therapy, such as surgical
resection, radiation and chemotherapy, the 5-year survival rate of
patients with TSCC was reported to be ~50% worldwide in 2016 due to
delayed diagnosis, locoregional recurrence and distant metastasis
of the tumors (4,6). Therefore, it is essential to identify
novel and promising biomarkers for the early screening and
follow-up of patients with TSCC.
Circular RNAs (circRNAs/circs) are a specific type
of endogenous non-coding RNAs with covalently closed loop
structure, which are ubiquitously expressed in various cell types
including cancer cells, immune cells, hematopoietic stem cells etc.
(7,8). Accumulating evidence has revealed
that circRNAs exhibit multiple biological functions, including as
microRNA (miRNA/miR) sponges and regulators of transcription,
splicing and translation (7,8).
They are involved in diverse biological processes, including
induction of epithelial-mesenchymal transition (EMT), regulation of
angiogenesis, activation of oncogenic signaling and TSCC
progression (7–10). For example, a previous study by Yao
et al (11) demonstrated that
circ_0001742 expression is increased in TSCC tumor tissues compared
with the corresponding levels noted in adjacent (Ctrl) tissues.
Furthermore, high expression levels of circ_0001742 in tumors are
associated with higher TNM stage and decreased survival rate in
patients with TSCC (11).
Additionally, it has been demonstrated that circ_081069 expression
is upregulated in TSCC tissues compared with the corresponding
expression levels noted in Ctrl normal tissues (11). Furthermore, its upregulation
increased the proliferative and migratory abilities of TSCC cells
(12). These studies indicated the
potential roles of specific circRNAs in the pathological evaluation
of TSCC (11,12). However, to the best of our
knowledge, the comprehensive circRNA expression profile in TSCC has
not been fully investigated. Therefore, the present study aimed to
identify differentially expressed (DE) circRNAs in TSCC tissues
compared with paired Ctrl tissues using microarray analysis, to
further validate the expression levels of 10 candidate circRNAs,
and to examine their association with tumor features and survival
in patients with TSCC.
Materials and methods
Patients
The present study included 60 patients with TSCC who
were recruited between January 2017 and December 2019 and underwent
surgery at the Oral Medicine Department of Cangzhou People's
Hospital (Cangzhou Medical College, Cangzhou, China). The patients
were eligible for analysis in the present study if they met the
following criteria: i) Diagnosis of TSCC confirmed by pathological
examination; ii) aged >18 years old; iii) surgical resection as
primary treatment; iv) availability of fresh-frozen tumor and
paired Ctrl tissues; and v) availability of main clinical data for
study analysis. The patients were excluded according to the
following criteria: i) Neoadjuvant therapy prior to surgery; ii)
complications due to other malignant tumors; iii) known acquired
immune deficiency syndrome; and iv) missing survival follow-up
records. written informed consent was provided by each eligible
patient or his/her guardian if the patients were dead when the
clinical data was collected. The present study was approved by the
Ethics Committee of Cangzhou Medical College (approval no.
20190611-3; Cangzhou, China).
Specimen collection
The tumor tissues and paired Ctrl tissues from each
enrolled patient were collected from the specimen room of Cangzhou
People's Hospital (Cangzhou, China). The tissues were snap-frozen
in liquid nitrogen immediately following resection and stored at
−80°C. In addition, information regarding the clinical features
(Table I) of the patients was
collected from the medical records for study analysis and the
pathological grade/tumor node metastasis (TNM) stage was assessed
based on the 7th edition of the AJCC Cancer Staging Manual
(13). The classification of
diseases was performed according to the International
Classification of Diseases 10th revision (14).
 | Table I.Characteristics of patients with
TSCC. |
Table I.
Characteristics of patients with
TSCC.
| Characteristics | Patients with TSCC
(n=60) | Female patients
(n=17) | Male patients
(n=43) |
|---|
| Age, years (mean ±
SD) | 56.3±11.1 | 51.7±11.4 | 58.1±10.6 |
| Age, n (%) |
|
|
|
| 18–39
years | 4 (6.7) | 2 (11.8) | 2 (4.7) |
| 40–49
years | 9 (15.0) | 5 (29.4) | 4 (9.3) |
| 50–59
years | 24 (40.0) | 6 (35.3) | 18 (41.9) |
| 60–69
years | 16 (26.7) | 3 (17.6) | 13 (30.2) |
| 70–79
years | 7 (11.7) | 1 (5.9) | 6 (14.0) |
| Sex, n (%) |
|
|
|
|
Male | 43 (71.7) | 0 (0.0) | 43 (100.0) |
|
Female | 17 (28.3) | 17 (100.0) | 0 (0.0) |
| Han nationality, n
(%) | 60 (100.0) | 17 (100.0) | 43 (100.0) |
| Origin, n (%) |
|
|
|
| Eastern
region of Hebei province, China | 37 (61.7) | 11 (64.7) | 26 (60.5) |
| Other
regions of Hebei province, China | 18 (30.0) | 4 (23.5) | 14 (32.6) |
| Other
provinces, China | 5 (8.3) | 2 (11.8) | 3 (7.0) |
| Pathological grade,
n (%) |
|
|
|
| G1 | 14 (23.3) | 5 (29.4) | 9 (20.9) |
| G2 | 33 (55.0) | 9 (52.9) | 24 (55.8) |
| G3 | 13 (21.7) | 3 (17.6) | 10 (23.3) |
| ICD-10
classification code, n (%) |
|
|
|
| C02.1,
M8070/31 | 3 (5.0) | 2 (11.8) | 1 (2.3) |
| C02.1,
M8070/32 | 8 (13.3) | 3 (17.6) | 5 (11.6) |
| C02.1,
M8070/33 | 3 (5.0) | 0 (0.0) | 3 (7.0) |
| C02.8,
M8070/31 | 2 (3.3) | 1 (5.9) | 1 (2.3) |
| C02.8,
M8070/32 | 5 (8.3) | 1 (5.9) | 4 (9.3) |
| C02.8,
M8070/33 | 1 (1.7) | 1 (5.9) | 0 (0.0) |
| C02.9,
M8070/31 | 9 (15.0) | 2 (11.8) | 7 (16.3) |
| C02.9,
M8070/32 | 20 (33.3) | 5 (29.4) | 15 (34.9) |
| C02.9,
M8070/33 | 9 (15.0) | 2 (11.8) | 7 (16.3) |
| T stage, n (%) |
|
|
|
| T1 | 11 (18.3) | 2 (11.8) | 9 (20.9) |
| T2 | 37 (61.7) | 15 (88.2) | 22 (51.2) |
| T3 | 12 (20.0) | 0 (0.0) | 12 (27.9) |
| N stage, n (%) |
|
|
|
| N0 | 43 (71.7) | 17 (100.0) | 26 (60.5) |
| N1 | 13 (21.7) | 0 (0.0) | 13 (30.2) |
| N2 | 4 (6.6) | 0 (0.0) | 4 (9.3) |
| TNM stage, n
(%) |
|
|
|
| I | 11 (18.3) | 2 (11.8) | 9 (20.9) |
| II | 31 (51.7) | 15 (88.2) | 16 (37.2) |
|
III | 14 (23.3) | 0 (0.0) | 14 (32.6) |
| IV | 4 (6.7) | 0 (0.0) | 4 (9.3) |
| Adjuvant
radiotherapy, n (%) |
|
|
|
|
Yes | 44 (73.3) | 10 (58.8) | 34 (79.1) |
| No | 16 (26.7) | 7 (41.2) | 9 (20.9) |
Microarray analysis
Microarray analysis was performed in five pairs of
TSCC and Ctrl tissues, which were randomly selected from the total
number of specimens of the enrolled patients (including 3 males and
2 females; mean age, 47.2±8.8 years; age range, 36–58 years). The
separation of total RNA in the specimens was performed using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The integrity and quantification analysis of total RNA was
performed using an Agilent 2100 Bioanalyzer (Agilent Technologies,
Inc.) and NanoDrop ND-1000 spectrophotometer (Thermo Fisher
Scientific, Inc.), respectively. The linear RNAs were removed from
the total RNA molecules using RNase R (Epicentre; Illumina, Inc.).
Microarray analysis was conducted on the Platform GPL19978
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL19978)
with the use of Agilent-069978 Arraystar Human CircRNA microarray
V1 (Agilent Technologies, Inc.) and the Arraystar Human Circular
RNA Microarray V1.0 (6×7K; Arraystar, Inc.), which contains 5,396
probes specific for the human circRNA backsplice junction region.
The detailed procedure was performed as described in a previous
study (15).
Bioinformatics analysis
Differential expression analysis between five TSCC
and Ctrl tissues was performed using the Limma package (https://cran.r-project.org/bin/windows/base/) in R
software v.4.0.2. Principal component analysis (PCA) and heatmap
analysis for circRNA expression profiling were conducted using the
Factoextra (https://cran.r-project.org/web/packages/factoextra/index.html)
and pheatmap packages v.4.0.2 (https://cran.r-project.org/web/packages/pheatmap/index.html)
in the R software. The DEcircRNAs were identified based on the fold
change (FC) ≥2.0 and adjusted P-value (BH multiple test correction)
<0.05. These indices are shown in the volcano plot. Gene
Ontology (GO) (http://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) (https://david.ncifcrf.gov) enrichment analyses were
performed to identify DEcircRNAs based on the gene location and
target miRNAs (miranda criterion, total score >120 & total
energy <-20). The circRNA-miRNA regulatory network of the top 10
DEcircRNAs (based on rank of log2FC) was predicted by miRanda
v.3.3a (http://www.microrna.org/microrna/getDownloads.do). The
annotation for the target miRNA was obtained from miRwalk database
v.3 (http://mirwalk.umm.uni-heidelberg.de/), which covered
the GO database (molecular function, cellular component and
biological process), the pathway database (KEGG database), the
human phenotype database (https://hpo.jax.org/app/) and the disease database
(http://www.obofoundry.org/ontology/doid.html).
Reverse transcription-quantitative PCR
(RT-qPCR)
According to the Log2FC absolute value, the top five
upregulated and downregulated DEcircRNAs were selected as candidate
DEcircRNAs for further investigation. RT-qPCR was performed to
determine the relative expression levels of 10 candidate circRNAs
in tumor and paired Ctrl tissues derived from 60 patients with
TSCC. Briefly, total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The detection
of circRNAs was performed following digestion of the linear RNA
using RNase R (Epicentre; Illumina, Inc.). Subsequently, the
synthesis of cDNA was carried out (denaturation at 65°C for 5 min,
reverse transcription at 42°C for 18 min and inactivation at 98°C
for 5 min) using the PrimeScript™ RT reagent kit (Takara Bio,
Inc.), followed by its amplification using SYBR-Green Premix
DimerEraser™ (Takara Bio, Inc.). Lastly, the relative expression
levels of the circRNAs were calculated using the 2-ΔΔCq method
(16). The thermocycling
conditions for qPCR were as follows: Pre-denaturation at 95°C for 3
min; followed by 40 cycles of denaturation at 95°C for 15 sec and
annealing/elongation at 61°C for 20 sec. GAPDH was used as an
internal reference for circRNAs. The primers are shown in Table SI.
Survival analysis
The survival follow-up data of the patients were
collected from the records and the last follow-up date was May 31,
2020. Overall survival (OS) was evaluated from the date of surgery
to the date of death.
Statistical analysis
The estimation of the sample size was performed
based on the expression levels of 10 candidate circRNAs in the
microarray analysis. All experimental studies were conducted in
triplicate. The statistically significant differences in the
expression levels of the 10 candidate circRNAs between tumor and
Ctrl tissues were evaluated based on a significance level of 0.05
and a power of 0.80. The required sample size included 57 pairs of
tumor and Ctrl tissues. Considering an attrition rate of 5%, the
final sample size was set to 60 pairs of tumor and Ctrl tissues.
Descriptive analysis was performed for the characteristics of the
patients, using numbers, percentages or mean ± SD. The relative
expression levels of circRNAs were presented as the median with the
interquartile range and differences in the expression levels
between tumor and Ctrl tissues were determined using the Wilcoxon
signed-rank test. The correlation analysis between tumor circRNA
expression and clinical features was carried out using the
Spearman's rank correlation test. A Kaplan-Meier curve was used to
display the OS. According to the median level of circRNA
expression, the latter was divided into circRNA high and low
expression. The association between the expression levels of
circRNAs in the tumor samples and OS was evaluated using the
log-rank test. SPSS software (v22.0; IBM Corp.) and GraphPad Prism
software (v7.01; GraphPad Software, Inc.) were used for data
analysis and graphical presentation. P<0.05 was considered to
indicate a statistically significant difference.
Results
Clinical characteristics of patients
with TSCC
A total of 60 patients with TSCC with a mean age of
56.3±11.1 years were included. The number of male and female
patients was 43 (71.7%) and 17 (28.3%), respectively (Table I). The evaluation of the
pathological grade of the patients yielded the following results: A
total of 14 (23.3%) patients presented with G1 TSCC, whereas 33
(55.0%) and 13 (21.7%) exhibited G2 and G3 TSCC tumors,
respectively. The TNM stage classification included 11 (18.3%)
patients with stage I, 31 (51.7%) patients with stage II, 14
(23.3%) patients with stage III and 4 (6.7%) patients with stage IV
tumors. In addition, 44 (73.3%) patients received adjuvant
radiotherapy. Detailed information of patients with TSCC in the
present study is shown in Table
I.
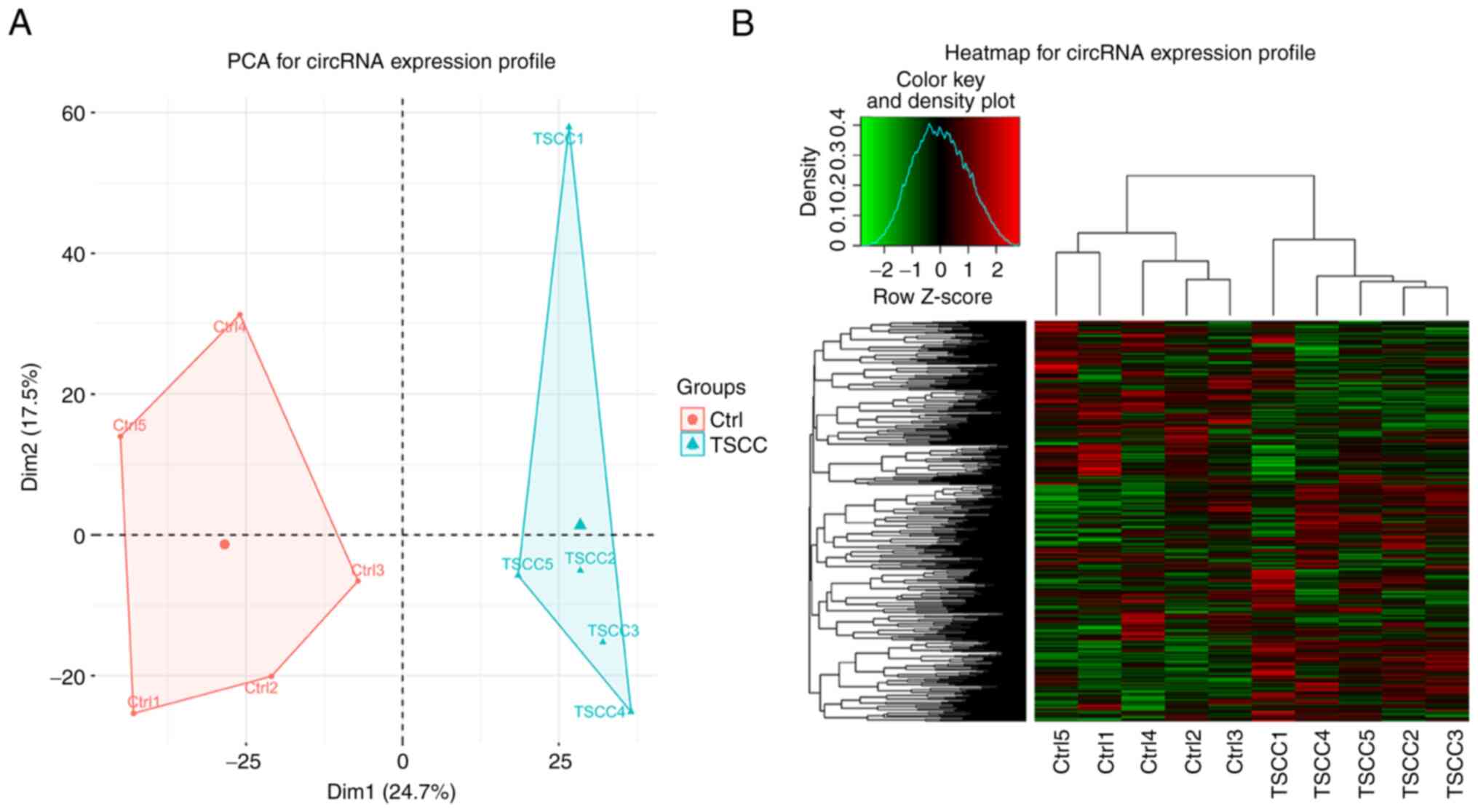
PCA and heatmap analysis for the
evaluation of the circRNA expression profile
The PCA tool was used to describe protein dynamics
and PCA analysis indicated that the DEcircRNAs detected in the TSCC
and Ctrl tissues were distributed in two separate parts, suggesting
that a clear segregation of the circRNA expression profile was
noted between TSCC and Ctrl tissues (Fig. 1A). In the row clustering step, the
TSCC tissues were all grouped together to the right side
considering their similar DEcircRNA patterns, while the Ctrl
tissues were all grouped into the left cluster since their
DEcircRNA patterns were more similar, suggesting that circRNA
expression profile could differentiate TSCC tissues from Ctrl
tissues (Fig. 1B).
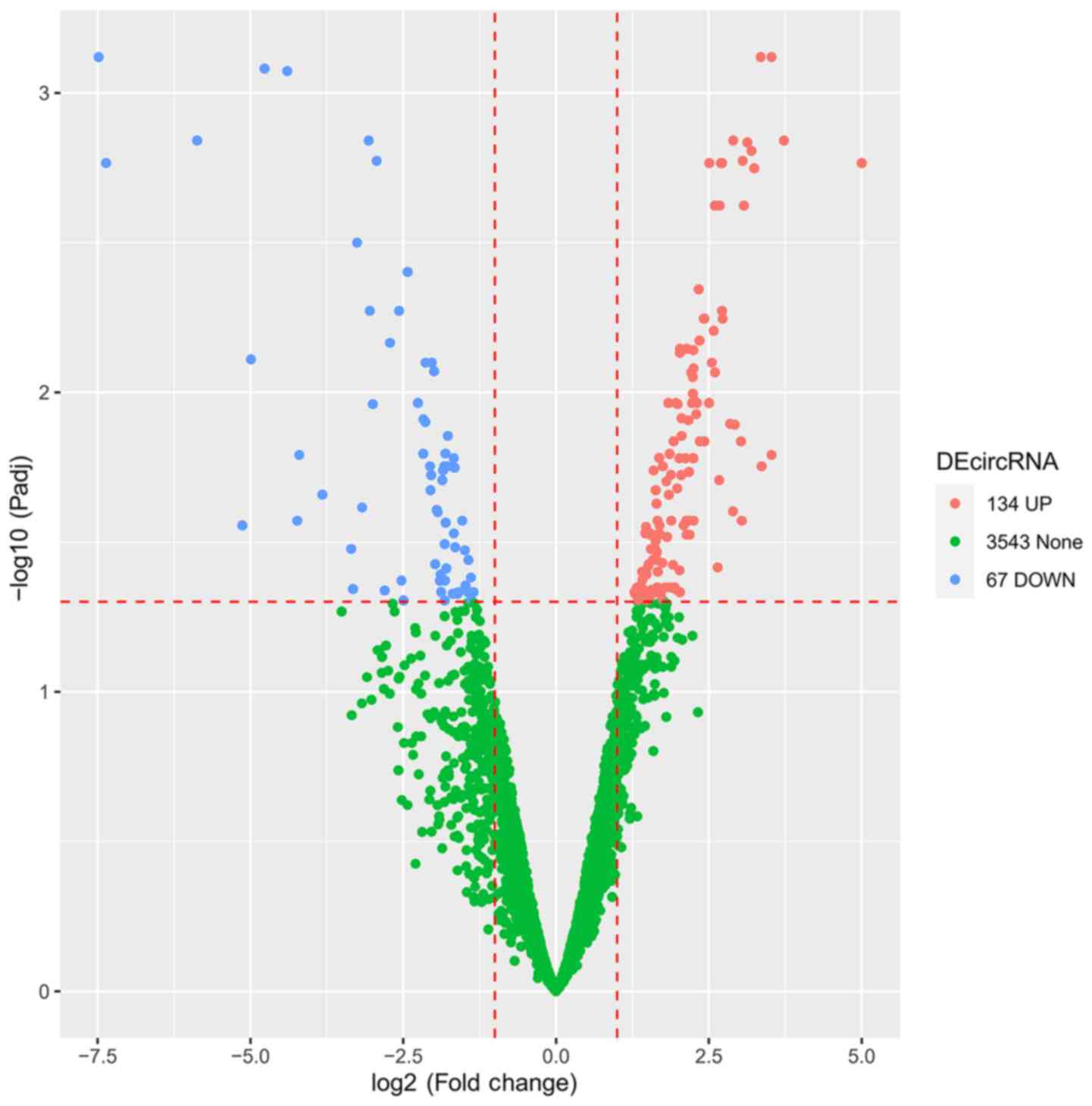
Volcano plot for the circRNA
expression profile
A total of 3,744 circRNAs were detected in more than
half of the samples, and were included in the subsequent analysis.
The volcano plot indicated 134 upregulated and 67 downregulated
DEcircRNAs. A total of 3,543 circRNAs with unaltered expression
levels were noted in the TSCC tissues compared with the Ctrl
tissues (Fig. 2).
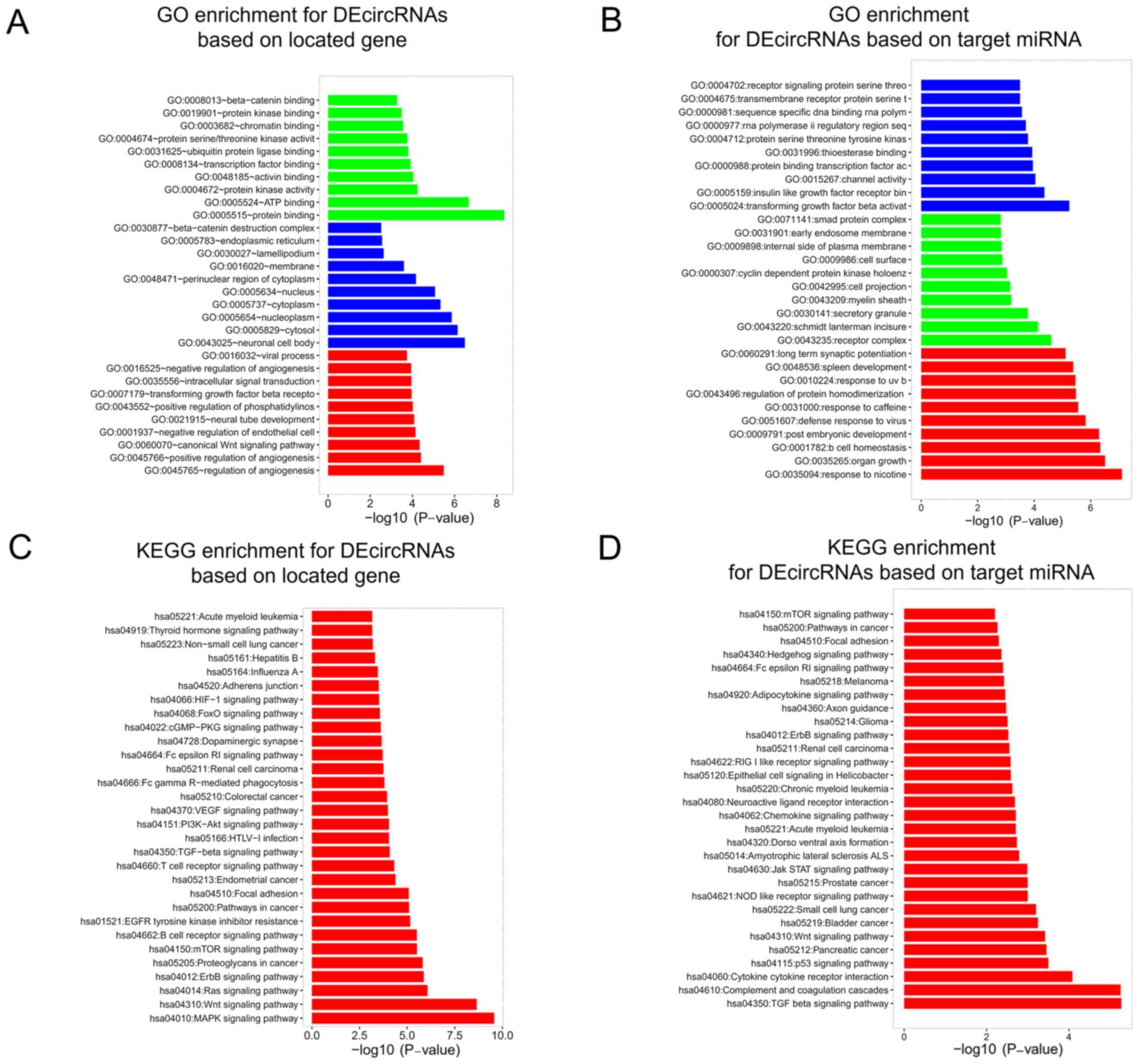
GO and KEGG enrichment analysis of
DEcircRNAs
GO and KEGG enrichment analysis was performed. GO
enrichment analysis was performed based on specific gene functions
and the data indicated that DEcircRNAs were enriched in molecular
functions, including ‘protein binding’, ‘ATP binding’ and ‘protein
kinase activity’, cellular components, including ‘neuronal cell
body’, ‘cytosol’ and ‘nucleoplasm’, and biological processes,
including ‘regulation of angiogenesis’, ‘positive regulation of
angiogenesis’ and ‘canonical Wnt signaling pathway’ based on
significance (Fig. 3A). In
addition, GO enrichment analysis was performed based on target
miRNAs and the results indicated that DEcircRNAs were enriched in
molecular functions including ‘TGF-β activation’, ‘insulin like
growth receptor binding’ and ‘channel activity’, and cellular
components including ‘receptor complex’, ‘Schmidt-Lanterman
incisures’ and ‘secretory granule’, and biological processes,
including ‘response to nicotine’, ‘organ growth’ and ‘B cell
homeostasis’ based on significance (Fig. 3B). KEGG enrichment analysis was
performed for DEcircRNAs based on specific gene functions and the
data demonstrated that DEcircRNAs were enriched in the ‘MAPK
signaling pathway’, ‘Wnt signaling pathway’ and ‘Ras signaling
pathway’ based on significance (Fig.
3C). Furthermore, KEGG enrichment analysis was performed for
DEcircRNAs based on target miRNAs. The data demonstrated that
DEcircRNAs were enriched in the ‘TGF beta signaling pathway’,
‘Complement and coagulation cascades’ and ‘Cytokine cytokine
receptor interaction’ based on significance (Fig. 3D).
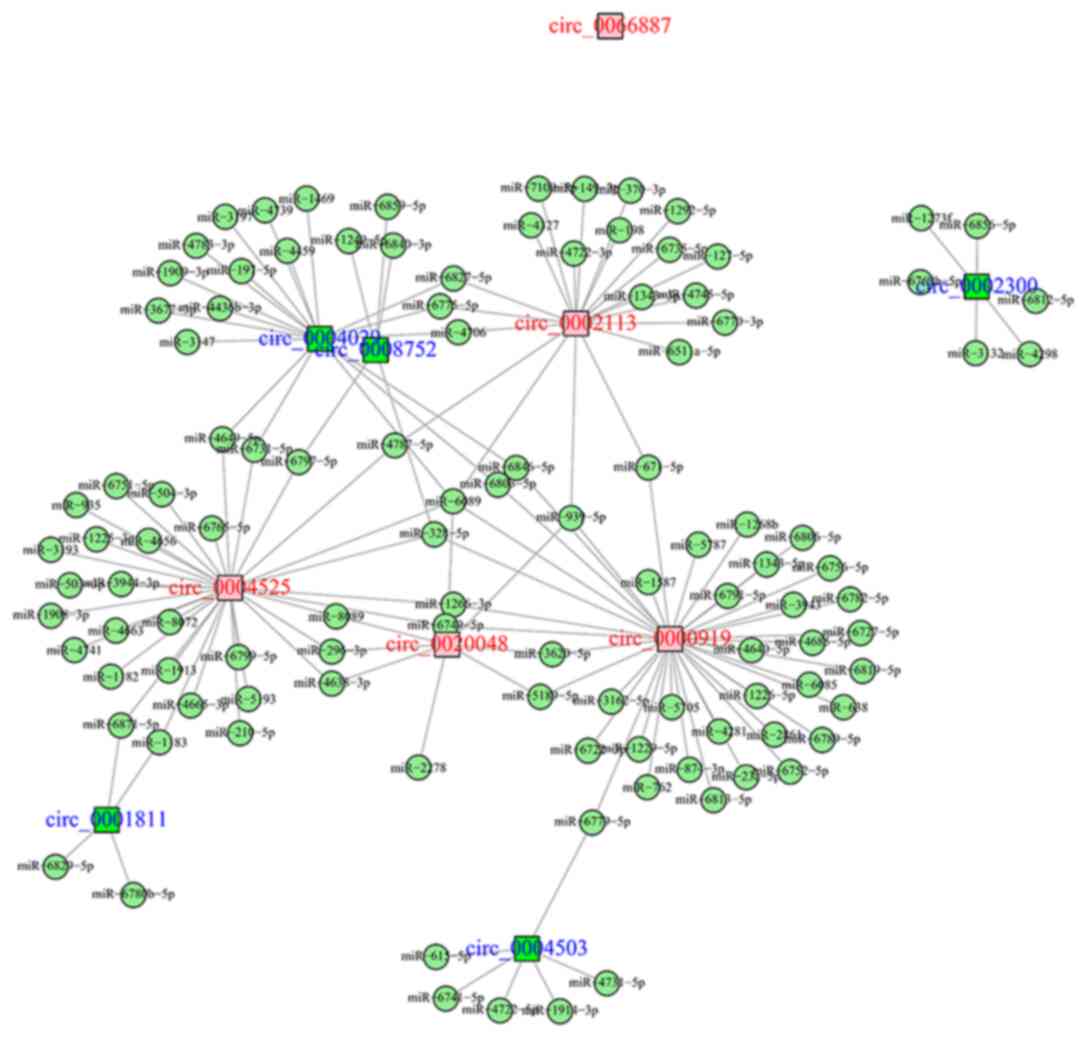
Regulatory network of circRNA-miRNA
interactions
The top 10 candidate DEcircRNAs comprised the top
five upregulated and the top five downregulated DEcircRNAs, which
were detected in five TSCC and Ctrl tissues. These circRNAs were
screened based on the log2FC value (Table II). A circRNA-miRNA regulatory
network of the top 10 DEcircRNAs was established using the miRanda
database (Fig. 4). circ_0066887
was predicted not to bind to a miRNA target, whereas the following
DEcircRNAs demonstrated multiple (≥3) target miRNAs: circ_0020048,
circ_0000919, circ_0004525, circ_0002113, circ_0004029,
circ_0004503, circ_0008752, circ_0002300 and circ_0001811.
 | Table II.Top 10 DEcircRNAs, including top five
downregulated DEcircRNAs and top five upregulated DEcircRNAs. |
Table II.
Top 10 DEcircRNAs, including top five
downregulated DEcircRNAs and top five upregulated DEcircRNAs.
| circRNA | Probe name | Chr | Symbol | Log2(FC) | P-value | Padj value | Trend |
|---|
| circ_0020048 | ASCRP001058 | chr10 | TCF7L2 | 5.002 | 6.7×10-6 | 1.7×10-3 | Up |
| circ_0000919 | ASCRP000027 | chr19 | ATP13A1 | 3.728 | 3.2×10-6 | 1.4×10-3 | Up |
| circ_0004525 | ASCRP003285 | chr20 | RBCK1 | 3.526 | 5.7×10-7 | 7.6×10-4 | Up |
| circ_0066887 | ASCRP003739 | chr3 | GSK3B | 3.526 | 3.1×10-4 | 1.6×10-2 | Up |
| circ_0002113 | ASCRP003424 | chr21 | IFNGR2 | 3.364 | 3.7×10-4 | 1.8×10-2 | Up |
| circ_0004029 | ASCRP003104 | chr2 | UXS1 | −7.482 | 3.1×10-7 | 7.6×10-4 | Down |
| circ_0004503 | ASCRP004245 | chr5 | UBE2D2 | −7.361 | 7.7×10-6 | 1.7×10-3 | Down |
| circ_0008752 | ASCRP004812 | chr8 | CNOT7 | −5.871 | 2.4×10-6 | 1.4×10-3 | Down |
| circ_0002300 | ASCRP004304 | chr5 | CANX | −5.133 | 8.2×10-4 | 2.8×10-2 | Down |
| circ_0001811 | ASCRP004898 | chr8 | STAU2 | −4.991 | 7.5×10-5 | 7.8×10-3 | Down |
Validation of candidate DEcircRNA
expression in TSCC tissues
The expression levels of 10 candidate DEcircRNAs
were further assessed in the tumor and paired Ctrl tissues of 60
patients with TSCC using RT-qPCR analysis. The expression levels of
circ_0020048, circ_0000919, circ_0004525 (Fig. 5A-C; all P<0.001) and
circ_0002113 (Fig. 5E; P<0.001)
were increased in TSCC tumor tissues compared with in Ctrl tissues,
while the expression levels of circ_0004029, circ_0004503 (Fig. 5F and G; both P<0.001),
circ_0008752 (Fig. 5H; P=0.002),
circ_0002300 (Fig. 5I; P<0.001)
and circ_0001811 (Fig. 5J;
P=0.005) were decreased in TSCC tumor tissues compared with in Ctrl
tissues. The expression levels of circ_0066887 were similar between
TSCC tumor tissues and Ctrl tissues (Fig. 5D; P=0.131).
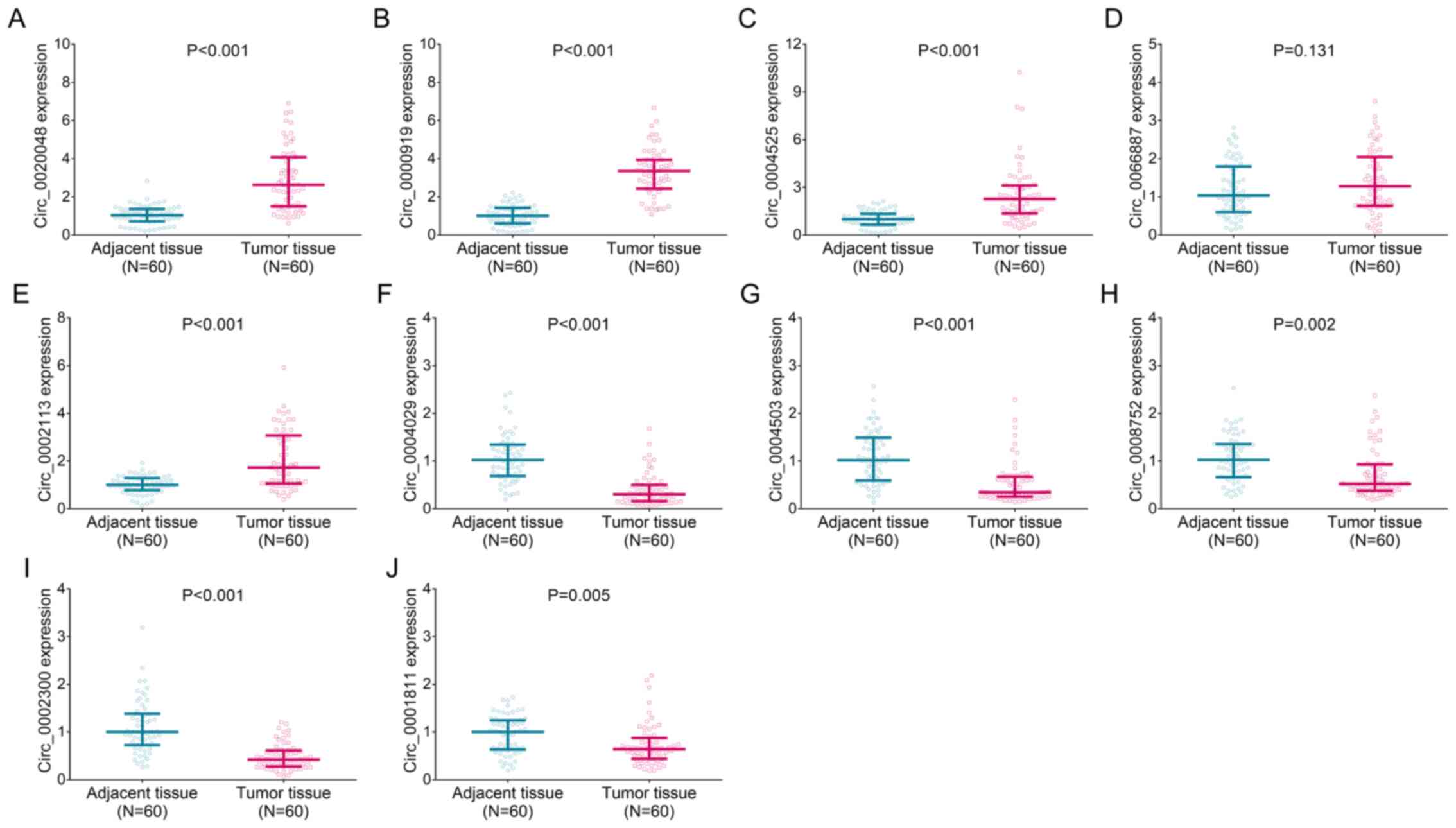 | Figure 5.Validation of candidate differentially
expressed circRNA expression levels in patients with TSCC.
Comparison of the expression levels of (A) circ_0020048, (B)
circ_0000919, (C) circ_0004525, (D) circ_0066887, (E) circ_0002113,
(F) circ_0004029, (G) circ_0004503, (H) circ_0008752, (I)
circ_0002300 and (J) circ_0001811 between 60 TSCC and adjacent
tissues. circRNA/circ, circular RNA; TSCC, tongue squamous cell
carcinoma. |
Association between the expression
levels of candidate DEcircRNAs and tumor features in patients with
TSCC
The association between the expression levels of
candidate tumor DEcircRNAs and tumor features was further detected
in patients with TSCC (Table
III). The expression levels of circ_0020048 in tumor tissues
exhibited a positive association with T stage (P=0.010) and TNM
stage (P=0.006). The expression levels of circ_0000919 exhibited a
positive association with T stage (P=0.022), N stage (P<0.001)
and TNM stage (P<0.001). In contrast to these findings, the
expression levels of circ_0004503 in the tumor tissues were
negatively associated with pathological grade (P=0.015), whereas
those of circ_0008752 were negatively associated with pathological
grade (P=0.024) and N stage (P=0.036). The expression levels of
circ_0002300 noted in the tumor samples were negatively associated
with pathological grade (P=0.048), N stage (P=0.048) and TNM stage
(P=0.021). The expression levels of other candidate DEcircRNAs were
not associated with tumor features, including pathological grade, T
stage, N stage and TNM stage (all P>0.05).
 | Table III.Associations between candidate
differentially expressed circRNAs and clinical features were
determined using Spearman's rank sum test. |
Table III.
Associations between candidate
differentially expressed circRNAs and clinical features were
determined using Spearman's rank sum test.
|
Characteristics | circ_0020048,
median (IQR) | circ_0000919,
median (IQR) | circ_0004525,
median (IQR) | circ_0066887,
median (IQR) | circ_0002113,
median (IQR) | circ_0004029,
median (IQR) | circ_0004503,
median (IQR) | circ_0008752,
median (IQR) | circ_0002300,
median (IQR) | circ_0001811,
median (IQR) |
|---|
| Pathological
grade |
|
|
|
|
|
|
|
|
|
|
| G1 | 1.898 | 2.853 | 2.502 | 1.100 | 1.280 | 0.384 | 0.561 | 0.800 | 0.502 | 0.509 |
|
| (1.117-2.779) | (1.389-3.597) | (1.548-3.206) | (0.563-2.137) | (0.986-2.806) | (0.193-0.918) | (0.278-1.190) | (0.419-1.465) | (0.376-1.047) | (0.265-0.793) |
| G2 | 3.310 | 3.360 | 1.781 | 1.449 | 1.729 | 0.281 | 0.359 | 0.553 | 0.414 | 0.667 |
|
| (2.290-4.836) | (2.554-4.295) | (1.159-2.980) | (0.771-2.213) | (0.981-3.207) | (0.151-0.477) | (0.285-0.712) | (0.420-0.833) | (0.279-0.580) | (0.525-0.919) |
| G3 | 2.231 | 3.410 | 2.260 | 1.068 | 1.868 | 0.306 | 0.250 | 0.387 | 0.310 | 0.650 |
|
| (1.335-3.644) | (2.499-4.287) | (1.281-3.844) | (0.799-2.002) | (1.390-3.051) | (0.100-0.536) | (0.194-0.391) | (0.309-0.488) | (0.236-0.590) | (0.421-0.924) |
|
P-value | 0.539 | 0.100 | 0.628 | 0.844 | 0.249 | 0.224 | 0.015 | 0.024 | 0.048 | 0.334 |
| T stage |
|
|
|
|
|
|
|
|
|
|
| T1 | 1.497 | 2.907 | 1.897 | 1.503 | 1.729 | 0.400 | 0.322 | 0.685 | 0.545 | 0.740 |
|
| (1.165-2.448) | (2.008-3.189) | (1.175-2.514) | (0.576-2.372) | (0.751-2.043) | (0.184-0.756) | (0.286-0.851) | (0.465-0.895) | (0.407-1.035) | (0.583-1.408) |
| T2 | 2.855 | 3.392 | 2.310 | 0.982 | 1.772 | 0.306 | 0.349 | 0.491 | 0.413 | 0.642 |
|
| (1.501-4.160) | (2.323-4.069) | (1.332-3.095) | (0.757-1.649) | (1.073-3.215) | (0.145-0.504) | (0.266-0.612) | (0.375-1.091) | (0.269-0.580) | (0.463-0.959) |
| T3 | 3.678 | 3.740 | 2.371 | 1.830 | 1.602 | 0.257 | 0.292 | 0.523 | 0.322 | 0.518 |
|
| (2.014-4.647) | (3.157-4.410) | (1.390-3.666) | (0.970-2.869) | (1.093-3.209) | (0.179-0.443) | (0.209-0.649) | (0.321-0.750) | (0.190-0.660) | (0.336-0.673) |
|
P-value | 0.010 | 0.022 | 0.386 | 0.299 | 0.485 | 0.288 | 0.312 | 0.472 | 0.084 | 0.081 |
| N stage |
|
|
|
|
|
|
|
|
|
|
| N0 | 2.379 | 2.907 | 2.260 | 1.218 | 1.772 | 0.322 | 0.349 | 0.591 | 0.447 | 0.650 |
|
| (1.260-4.094) | (2.211-3.634) | (1.308-2.977) | (0.750-2.053) | (0.957-3.125) | (0.166-0.575) | (0.286-0.697) | (0.400-0.949) | (0.310-0.634) | (0.479-1.065) |
| N1 | 3.310 | 3.946 | 1.493 | 1.488 | 1.659 | 0.249 | 0.250 | 0.441 | 0.365 | 0.542 |
|
| (2.086-4.170) | (3.558-4.679) | (1.047-4.052) | (1.108-2.463) | (1.064-2.949) | (0.147-0.558) | (0.203-0.831) | (0.343-0.730) | (0.241-0.627) | (0.339-0.680) |
| N2 | 2.922 | 4.006 | 3.047 | 0.830 | 1.866 | 0.179 | 0.412 | 0.319 | 0.251 | 0.689 |
|
| (2.815-4.320) | (3.217-5.404) | (2.065-4.453) | (0.369-1.817) | (1.267-3.345) | (0.141-0.252) | (0.263-0.571) | (0.198-0.447) | (0.226-0.283) | (0.354-0.737) |
|
P-value | 0.082 | <0.001 | 0.316 | 0.402 | 0.845 | 0.221 | 0.235 | 0.036 | 0.048 | 0.138 |
| TNM stage |
|
|
|
|
|
|
|
|
|
|
| I | 1.497 | 2.907 | 1.897 | 1.503 | 1.729 | 0.400 | 0.322 | 0.685 | 0.545 | 0.740 |
|
| (1.165-2.448) | (2.008-3.189) | (1.175-2.514) | (0.576-2.372) | (0.751-2.043) | (0.184-0.756) | (0.286-0.851) | (0.465-0.895) | (0.407-1.035) | (0.583-1.408) |
| II | 2.639 | 2.967 | 2.281 | 0.977 | 1.772 | 0.322 | 0.349 | 0.567 | 0.434 | 0.642 |
|
| (1.263-4.908) | (2.211-3.639) | (1.308-3.045) | (0.750-1.743) | (0.957-3.324) | (0.154-0.505) | (0.272-0.614) | (0.399-1.422) | (0.265-0.619) | (0.447-1.065) |
|
III | 3.350 | 3.939 | 2.227 | 1.644 | 1.694 | 0.286 | 0.271 | 0.482 | 0.399 | 0.554 |
|
| (2.159-4.263) | (3.394-4.547) | (1.204-3.864) | (1.128-2.694) | (1.064-3.058) | (0.158-0.512) | (0.204-0.762) | (0.353-0.710) | (0.272-0.593) | (0.341-0.683) |
| IV | 2.922 | 4.006 | 3.047 | 0.830 | 1.866 | 0.179 | 0.412 | 0.319 | 0.251 | 0.689 |
|
| (2.815-4.320) | (3.217-5.404) | (2.065-4.453) | (0.369-1.817) | (1.267-3.345) | (0.141-0.252) | (0.263-0.571) | (0.198-0.447) | (0.226-0.283) | (0.354-0.737) |
|
P-value | 0.006 | <0.001 | 0.183 | 0.524 | 0.355 | 0.140 | 0.321 | 0.062 | 0.021 | 0.118 |
Association between the expression
levels of candidate DEcircRNAs and prognosis of patients with
TSCC
To explore the potential prognostic value of the top
10 candidate DEcircRNAs in TSCC, the association of their
corresponding expression levels with OS time was assessed. The
higher expression levels of circ_0000919 in tumor tissues were
associated with decreased OS time in patients with TSCC (Fig. 6B; P=0.041), while no association
was observed between the expression levels of the other DEcircRNAs,
including circ_0020048 (Fig. 6A),
circ_0004525 circ_0066887, circ_0002113, circ_0004029,
circ_0004503, circ_0008752, circ_0002300 and circ_0001811 (Fig. 6C-J), and the OS time of patients
with TSCC (all P>0.05).
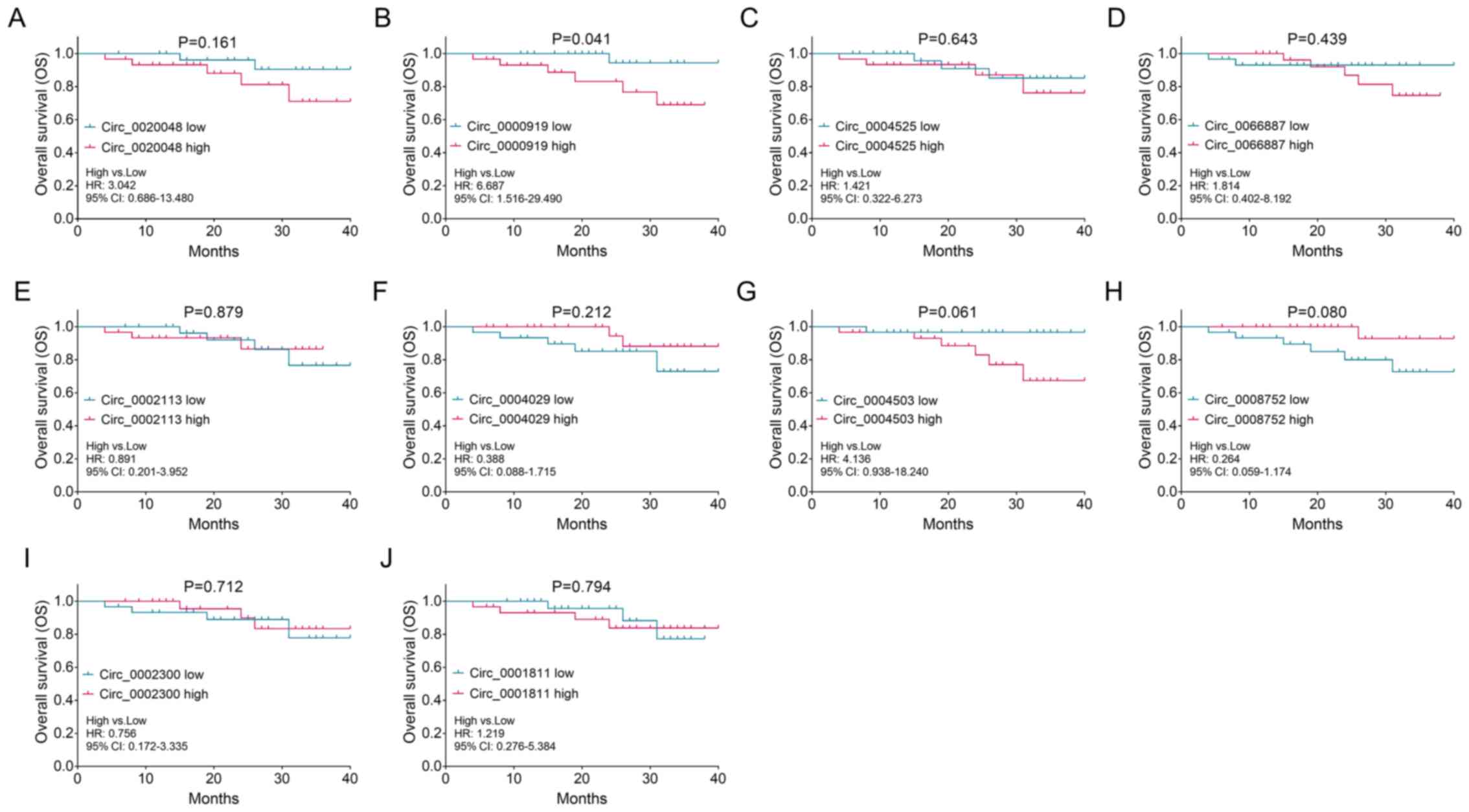 | Figure 6.Association between candidate
differentially expressed circRNAs and prognosis of patients with
TSCC. Association between the expression levels of (A)
circ_0020048, (B) circ_000919, (C) circ_0004525, (D) circ_0066887,
(E) circ_0002113, (F) circ_0004029, (G) circ_0004503, (H)
circ_0008752, (I) circ_0002300 and (J) circ_0001811 in tumors
derived from 60 patients with TSCC and overall survival. 95% CI,
95% confidence interval; circRNA/circ, circular RNA; HR, hazard
ratio; OS, overall survival; TSCC, tongue squamous cell
carcinoma. |
The data regarding the comparison of the expression
levels of the 10 candidate DEcircRNAs between tumor and Ctrl
tissues, as well as their corresponding association with tumor
features and OS time are summarized in Table IV. Among the 10 candidate
DEcircRNAs, the expression levels of circ_0000919 were increased in
TSCC tumor tissues compared with in the Ctrl tissues and associated
with higher TNM stage and low OS time of patients with TSCC,
suggesting that circ_0000919 could be used as a diagnostic and
prognostic biomarker in TSCC.
 | Table IV.Summary of study findings. |
Table IV.
Summary of study findings.
|
|
| Significant
association |
|---|
|
|
|
|
|---|
| circRNAs | Significant
difference Tumor vs. adjacent | Pathological
grade | T stage | N stage | TNM stage | OS |
|---|
| circ_0020048 | Yes | No | Yes | No | Yes | No |
| circ_0000919 | Yes | No | Yes | Yes | Yes | Yes |
| circ_0004525 | Yes | No | No | No | No | No |
| circ_0066887 | No | No | No | No | No | No |
| circ_0002113 | Yes | No | No | No | No | No |
| circ_0004029 | Yes | No | No | No | No | No |
| circ_0004503 | Yes | Yes | No | No | No | No |
| circ_0008752 | Yes | Yes | No | Yes | No | No |
| circ_0002300 | Yes | Yes | No | Yes | Yes | No |
| circ_0001811 | Yes | No | No | No | No | No |
Discussion
circRNAs are expressed in the cytoplasm of
eukaryotic cells and display high enrichment, stability and
diversity (17,18). They also exhibit tissue- and
developmental phase-specific expression (17,18).
circRNAs possess unique structures and high specificity (8). Furthermore, they exert diverse
regulatory functions and present potential value as diagnostic and
prognostic markers in multiple cancer cells, including cancer
cells, immune cells, hematopoietic stem cells, etc. (18,19).
In addition, the application of microarray-based technologies has
increased the understanding of the diverse functions of circRNAs
(17). Their contributions in
carcinogenesis have attracted considerable attention (17). Therefore, a high number of circRNAs
and their potential functions as cancer-associated regulators have
been identified (20). However,
studies investigating the circRNA expression profile in TSCC are
limited. Specific circRNAs, including circ_0001742 and circ_081069,
are involved in the development of TSCC (11,12).
Therefore, the present study aimed to identify the comprehensive
circRNA expression profile associated with TSCC. The findings may
provide additional evidence regarding the role of circRNAs in
TSCC.
In the present study, microarray and bioinformatics
analyses were conducted in five pairs of TSCC and Ctrl tissues. The
data revealed 134 upregulated DEcircRNAs and 67 downregulated
DEcircRNAs in TSCC tissues compared with Ctrl tissues, suggesting
that these DEcircRNAs may be involved in the development of TSCC.
Additional GO and KEGG enrichment analysis demonstrated that these
DEcircRNAs were enriched in tumor-associated biological processes,
including ‘regulation of angiogenesis’ and the ‘canonical Wnt
signaling pathway’, as well as in several oncogenic signaling
pathways, such as the ‘Wnt signaling pathway’, ‘MAPK signaling
pathway’ and ‘Ras signaling pathway’. These results may be
interpreted in several ways. First, DEcircRNAs may activate
angiogenic factors, which further contribute to an active
angiogenic state and promote TSCC development. This was based on
the existing evidence that angiogenesis is the process of
generating new blood vessels required for sufficient nutrition and
oxygen supply and tumor growth (18). Second, it was hypothesized that
DEcircRNAs may regulate the oncogenic components of the Wnt
signaling pathway, such as adenomatous polyposis coli and
β-catenin, and lead to its stimulation, which promotes TSCC cell
differentiation and survival and enhances TSCC development
(21). This was based on previous
studies demonstrating that the activation of the Wnt signaling
pathway is associated with oncogenic properties of cancer stem
cells and the progression of EMT (21,22).
Third, it was suggested that DEcircRNAs may regulate the components
of the MAPK signaling pathways, leading to excessive activation of
proteins and kinases of the Ras signaling pathway, which in turn
will affect the development of TSCC. This conclusion was based on
the ability of the MAPK cascade to regulate a wide range of
cellular processes, including differentiation and apoptosis
(20). The MAPK signaling pathway
is implicated in the differentiation and survival of TSCC cells via
interaction with other signaling pathways, such as the Ras and ERK
signaling pathways (23).
Furthermore, this finding was in line with the observation that
DEcircRNAs were enriched in the Ras signaling pathway.
In the present study, the top five upregulated and
downregulated DEcircRNAs were selected as candidates for RT-qPCR
validation in a total of 60 patients with TSCC. Notably,
circ_0000919 exhibited a positive association with T stage and it
could be used to predict low OS in patients with TSCC. The results
suggested the potential of circ_0000919 as a TSCC diagnostic and
prognostic biomarker, which may aid disease screening and
monitoring of patients with TSCC. The possible explanations for
these conclusions are as follows: First, circ_0000919 may serve as
the sponge of several antitumor miRNAs, such as miR-1587 and
miR-1226-3p, which in turn may mediate TSCC cell repair capacity
and proliferation, as well as radiosensitivity of these tumors
(24,25). Therefore, it was suggested that the
expression levels of circ_0000919 were associated with increased
TNM stage and unfavorable survival due to its ability to serve as a
sponge of antitumor miRNAs. Furthermore, circ_0000919 may enhance
translation efficiency of certain TSCC stimulators or inhibit the
translation of several tumor suppressors via protein carrier
functions, thereby promoting TSCC tumor development and resulting
in poor prognosis of patients with TSCC (17).
However, the present study had several limitations.
Firstly, the sample size for candidate DEcircRNA validation was 60,
which may lead to limited statistical power. The patients were
enrolled in one hospital, which may also contribute to selection
bias. Secondly, the present study was a retrospective study and the
information regarding tumor recurrence was not recorded in detail
in the majority of patients with TSCC. Therefore, the association
of candidate DEcircRNAs with disease-free survival was not
assessed. Thirdly, in the present study, circ_0000919 was
demonstrated to have diagnostic and prognostic value as a biomarker
of TSCC. However, the detailed mechanism of the involvement of
circ_0000919 in TSCC progression requires further investigation in
in vivo and in vitro studies. Furthermore, the top five upregulated
and downregulated DEcircRNAs were selected as candidate DEcircRNAs
based on the absolute value of log2FC for further RT-qPCR
validation in 60 patients with TSCC, since these DEcircRNAs were
predicted to be novel and promising biomarkers for early screening
and follow-up of these patients. RT-qPCR analysis was not conducted
to validate all DEcircRNAs based on the microarray data of the 60
patients with TSCC to avoid unnecessary cost. Finally, in the
present study, tissue specimens were collected for detection and
the data demonstrated that circ_0000919 may be a candidate TSCC
biomarker. Considering that blood samples are more easily collected
compared with tissue samples, whether the value of circ_0000919 in
blood samples is the same as that in tissue samples requires
additional experimental exploration.
In summary, the present study demonstrated an
aberrant circRNA expression profile and potential circRNA-miRNA
interactions in TSCC and further identified that circ_0000919
expression was associated with tumor features. The expression
levels of this circRNA could be used to predict unfavorable OS.
These findings suggested that circ_0000919 may be used as a
candidate biomarker for disease management of TSCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HL contributed to the study design. HL, QL, HQ, FD
and YQ made substantial contributions to acquisition and
interpretation of data. HL, QL and HQ contributed to data analysis
and presentation. HL, QL, HQ, FD and YQ were involved in drafting
the manuscript and revising it for important intellectual content.
HL and QL confirmed the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Cangzhou Medical College (Cangzhou, China) with the
approval no. ‘20190611-3’. Written informed consent was provided by
each eligible patient or his/her guardian if the patients were dead
when the clinical data was collected.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
circRNA
|
circular RNA
|
|
Ctrl
|
adjacent tissue
|
|
DEcircRNA
|
differentially expressed circRNA
|
|
EMT
|
epithelial-mesenchymal transition
|
|
FC
|
fold change
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
OS
|
overall survival
|
|
PCA
|
principal component analysis
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
TSCC
|
tongue squamous cell carcinoma
|
References
|
1
|
Kim YJ and Kim JH: Increasing incidence
and improving survival of oral tongue squamous cell carcinoma. Sci
Rep. 10:78772020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shrestha AD, Vedsted P, Kallestrup P and
Neupane D: Prevalence and incidence of oral cancer in low- and
middle-income countries: A scoping review. Eur J Cancer Care
(Engl). 29:e132072020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ohta K and Yoshimura H: Squamous cell
carcinoma of the dorsal tongue. CMAJ. 191:E13102019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Machiels JP, René Leemans C, Golusinski W,
Grau C, Licitra L and Gregoire V: EHNS Executive Board. Electronic
address. simplesecretariat@ehns.orgESMO
Guidelines Committee. Electronic address. simpleclinicalguidelines@esmo.orgESTRO
Executive Board. Electronic address. simpleinfo@estro.orgSquamous cell
carcinoma of the oral cavity, larynx, oropharynx and hypopharynx:
EHNS-ESMO-ESTRO Clinical Practice Guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 31:1462–1475. 2020. View Article : Google Scholar
|
|
5
|
Okubo M, Iwai T, Nakashima H, Koizumi T,
Oguri S, Hirota M, Mitsudo K and Tohnai I: Squamous Cell Carcinoma
of the Tongue Dorsum: Incidence and Treatment Considerations.
Indian J Otolaryngol Head Neck Surg. 69:6–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Montero PH and Patel SG: Cancer of the
oral cavity. Surg Oncol Clin N Am. 24:491–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ge P, Zhang J, Zhou L, Lv MQ, Li YX, Wang
J and Zhou DX: CircRNA expression profile and functional analysis
in testicular tissue of patients with non-obstructive azoospermia.
Reprod Biol Endocrinol. 17:1002019. View Article : Google Scholar
|
|
8
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar
|
|
9
|
Fan HY, Jiang J, Tang YJ, Liang XH and
Tang YL: circRNAs: A New Chapter in Oral Squamous Cell Carcinoma
Biology. OncoTargets Ther. 13:9071–9083. 2020. View Article : Google Scholar
|
|
10
|
Wang YF, Li BW, Sun S, Li X, Su W, Wang
ZH, Wang F, Zhang W and Yang HY: Circular RNA Expression in Oral
Squamous Cell Carcinoma. Front Oncol. 8:3982018. View Article : Google Scholar
|
|
11
|
Yao Y, Bi L and Zhang C: Circular
RNA_0001742 has potential to predict advanced tumor stage and poor
survival profiles in tongue squamous cell carcinoma management. J
Clin Lab Anal. 34:e233302020. View Article : Google Scholar
|
|
12
|
Wei T, Ye P, Yu GY and Zhang ZY: Circular
RNA expression profiling identifies specific circular RNAs in
tongue squamous cell carcinoma. Mol Med Rep. 21:1727–1738.
2020.PubMed/NCBI
|
|
13
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: the 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar
|
|
14
|
World Health Organization, . The ICD-10
classification of mental and behavioural disorders: diagnostic
criteria for research. World Health Organization; Geneva,
Switzerland: 1993, https://apps.who.int/iris/handle/10665/37108October
25–2016
|
|
15
|
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y,
Sun B, Wang Y, Wang L and Huang Q: Circular RNAs expression
profiles in human gastric cancer. Sci Rep. 7:90602017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Prats AC, David F, Diallo LH, Roussel E,
Tatin F, Garmy-Susini B and Lacazette E: Circular RNA, the Key for
Translation. Int J Mol Sci. 21:E85912020. View Article : Google Scholar
|
|
18
|
Tucker D, Zheng W, Zhang DH and Dong X:
Circular RNA and its potential as prostate cancer biomarkers. World
J Clin Oncol. 11:563–572. 2020. View Article : Google Scholar
|
|
19
|
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie
JW, Wang JB, Lin JX, Chen QY, Cao LL, et al: Circular RNA
circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to
facilitate gastric cancer invasion and metastasis. Cancer Lett.
471:38–48. 2020. View Article : Google Scholar
|
|
20
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martin-Orozco E, Sanchez-Fernandez A,
Ortiz-Parra I and Ayala-San Nicolas M: WNT Signaling in Tumors: The
Way to Evade Drugs and Immunity. Front Immunol. 10:28542019.
View Article : Google Scholar
|
|
22
|
Basu S, Cheriyamundath S and Ben-Ze'ev A:
Cell-cell adhesion: Linking Wnt/β-catenin signaling with partial
EMT and stemness traits in tumorigenesis. F1000 Res. 7:72018.
View Article : Google Scholar
|
|
23
|
Guo YJ, Pan WW, Liu SB, Shen ZF, Xu Y and
Hu LL: ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med.
19:1997–2007. 2020.PubMed/NCBI
|
|
24
|
Liu R, Shen L, Lin C, He J, Wang Q, Qi Z,
Zhang Q, Zhou M and Wang Z: MiR-1587 Regulates DNA Damage Repair
and the Radiosensitivity of CRC Cells via Targeting LIG4. Dose
Response. 18:15593258209369062020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park EJ, Jung HJ, Choi HJ, Jang HJ, Park
HJ, Nejsum LN and Kwon TH: Exosomes co-expressing AQP5-targeting
miRNAs and IL-4 receptor-binding peptide inhibit the migration of
human breast cancer cells. FASEB J. 34:3379–3398. 2020. View Article : Google Scholar : PubMed/NCBI
|