Introduction
Myeloid leukemia is a type of hematopoietic stem
cell malignant tumor, with differentiation disorder, uncontrolled
proliferation, or the inability of terminal differentiation of
primitive cells to retain malignant proliferation ability and
accounting for ~15% of new cases of adult leukemia (1,2). It
has been confirmed that the occurrence of myeloid leukemia is
associated with certain gene mutations, abnormal gene expression,
epigenetic disorders or abnormal expression of non-coding RNA
(3–6). Compared with traditional
chemotherapy, induced differentiation therapy has become an ideal
method for the treatment of leukemia due to its non-toxic side
effects (7,8). However, so far, only patients with
acute promyelocytic leukemia could get the complete remission
induced by differentiation-inducing drugs such as all-trans
retinoic acid; other types of leukemia have not benefited from them
(9–11). Therefore, it is necessary to
actively explore new intervention targets and corresponding
targeted drugs on the basis of in-depth exploration of the key
mechanisms of leukemia differentiation disorders. It is the
superiority of the aforementioned induced differentiation therapy
that has made differentiation induction a research hotspot in
recent years (12–15).
Expression of early growth response-1 (Egr-1) is a
member of the early growth response protein family, which has been
considered to be of great significance in a variety of
physiological processes and has been extensively studied (16,17),
especially in cell proliferation, angiogenesis, invasion and immune
response of tumors (18,19). Egr-1 can act as a transcriptional
regulator by combining the C2H2 type zinc finger with the DNA motif
of the 5′-GCG(T/G)GGGCG-3′ sequence. Regardless of the methylation
status of cytosine, it can bind to double-stranded target DNA and
the target DNA that does not bind to cytosine is oxidized to
5-formylcytosine or 5-carboxycytosine (20,21).
As it is an important part of certain signal pathways in the
process of cell signal transduction, it can mediate the coupling of
intracellular signal cascades and regulate the transcription and
transcription of a number of downstream long-term response genes
that determine cell karyotype changes (16,17).
To a certain extent, the role of Egr-1 in cell proliferation and
differentiation is heterogeneous, especially in normal somatic
cells and malignant tumor cells. For example, in normal somatic
cells, Egr-1 is in a dormant state and the expression level is very
low or even not expressed in the normal state. However, when the
cell is stimulated by some physical and chemical factors, the rapid
activation of Egr-1 allows cells to enter the proliferation phase
from the resting phase, which in turn leads to cell proliferation
(22,23). In tumor cells, the role of Egr-1 is
more complex and can be expressed as an oncogene or tumor
suppressor gene in different types of tumor, for example, Egr-1
promotes the malignant behaviors of LC cells (24), circCSPP1-miR-520h-Egr-1 activation
axis lead to the progression of prostate tumor (25), and Egr-1 as a potential oncogene
that promotes cell proliferation and defines Egr-1 as a new
molecular target in DLBCL non-Hodgkin lymphomar (26). As far as proliferation and
differentiation of leukemia cells are concerned, although there are
some studies associating it with the inhibition of proliferation
and induction of differentiation of leukemia cells (27,28),
its role in the committed differentiation of leukemia cells into
monocyte/macrophages is rarely reported.
micro (mi)RNAs are non-coding small RNAs with
post-transcriptional regulation. They are endogenous small RNAs
with a length of 18–24 nucleotides. Usually, they can base pair
with the 3′ untranslated region (UTR) of target mRNAs and silence
genes at the post-transcriptional level by inhibiting mRNA
translation or directly causing mRNA degradation and abnormal
expression often appears in the occurrence and development of
tumors (29). Among which the
miRNA (MiR)-let-7 family is downregulated in various types of tumor
tissues and has been widely studied as a tumor suppressor gene
(30,31). Increasing evidence shows that Let-7
also has the same properties as other miRNAs, which not only
participate in the occurrence and development of leukemia, but also
serve as a potential biomarker for the diagnosis and prognosis of
leukemia (32). However, whether
the miRNAs Let-7 is involved in the directional monocyte-macrophage
maturation and differentiation of leukemia cells remains to be
elucidated.
In the authors' previous work (data not published),
distinct changes in miR-let-7c-3p and Egr-1 expression were
detected in a PMA-induced differentiation model of K562 cells. The
present study focused on the role of the miR-let-7c-3p/Egr-1
signaling axis in the committed differentiation of K562 leukemia
cells into more mature monocytes/macrophages. The results
demonstrated that the miR-let-7c-3p/Egr-1 axis was closely
associated with the differentiation of K562 cells from leukemia
cells into more matured monocytes/macrophages induced by PMA.
Materials and methods
Materials
Human chronic myeloid leukemia cell line K562 was
purchased from Shanghai Cell Bank, Chinese Academy of Sciences. PMA
was purchased from American Sigma Company (cat. no. P1585-1MG).
Fetal bovine serum (FBS), BCA protein assay kit and SDS-PAGE gel
rapid preparation kit were purchased from Shanghai Biyuntian
Biotechnology Co., Ltd. RPMI 1640 medium was purchased from HyClone
(Cytiva). Swiss-Giemsa staining solution, double antibody, RIPA
protein lysis solution and 5X protein loading buffer were purchased
from Beijing Solarbio Science & Technology Co., Ltd. Standard
protein marker and Lipofectamine® 2000 transfection kit
were purchased from Thermo Fisher Scientific, Inc. The ECL
luminescence kit was purchased from Shandong Sparkjade Scientific
Instruments Co., Ltd. Egr-1 (cat. no. 22008-1-AP), GAPDH (cat. no.
10494-1-AP) and β-actin (cat. no. 20536-1-AP) primary antibodies
were purchased from ProteinTech Group, Inc. HRP-labeled rabbit
secondary antibody (cat. no. ZB-2301) was purchased from OriGene
Technologies, Inc. Primer design was provided by Sangon Biotech
Co., Ltd. Reverse transcription kit PrimeScript RT reagent kit with
gDNA Eraser (Perfect Real Time), chimeric fluorescence detection
kit and TB Green Premix Ex Taq™ (Tli RNaseH Plus) kit
was purchased from Takara Biotechnology Co., Ltd. The cycle kit was
purchased from Jiangsu KGI Biotechnology Co., Ltd. PE-CD11b (cat.
no. 301306) and FITC-CD14 (cat. no. 301804) fluorescent conjugated
antibodies were purchased from BioLegend, Inc. TRIzol®
reagent was purchased from Thermo Fisher Scientific. Inc.
Establishment of differentiation model
of K562 leukemia cells
K562 cells were grown in culture flasks containing
10% FBS in RPMI-1640 complete medium, cultured at 37°C, 5%
CO2. In the logarithmic growth phase, an appropriate
amount of K562 cell suspension and PMA solution were added to
96-well plates, so that the cell concentration in each well was
1×105/ml and the corresponding dose of PMA solution was
added. A total of three duplicate wells were set up in each group
and one zero-adjusting well was set up in each plate with only an
equal volume of RPMI1640 culture medium (100 µl) added and then
cultured at 37°C, 5% CO2 with saturated humidity. At 24,
48 and 72 h of culture 10 µl of CCK8 solution was added to each
well, except the blank well and incubated at 37°C for 2 h and
detected at 450 nm. According to the IC50 experimental
results of 48 h of culture, the control group (PMA 0 ng/ml) and the
experimental group (the final concentration of PMA 100 ng/ml) were
selected.
Observation of cell morphology by
Swiss-Giemsa staining
The control group with 0 ng/ml PMA and the
experiment group with 100 ng/ml PMA of K562 cells induced for 48 h
without staining were observed directly under an inverted optical
microscope. Cells of the above-mentioned control and experiment
group were collected at 48 h, washed twice with cold PBS,
resuspended with 100 µl PBS and mixed by gently pipetting to make a
cell suspension. After centrifugation at 200 × g for 3 min at 4°C,
the centrifuged smears were dried and stained with Wright-Giemsa
Stain Solution for 5 min at room temperature. The changes of cell
morphology were observed under different magnifications of the
optical microscope and the resulting images were captured.
Expression of differentiation antigen
CD11b and CD14 by flow cytometry
K562 cells were collected into flow tubes,
resuspended in PBS, washed twice by centrifugation at 200 × g for 3
min at 4°C, and adjusted to a cell concentration of
1×106 cells/ml. PBS (100 µl) was added to each tube,
followed by 5 µl PE-labeled mouse anti-human CD11b fluorescent
antibody and FITC-labeled mouse anti-human CD14 fluorescent
antibody respectively and incubated at 4°C for 30 min in the dark.
The cells were centrifuged at 200 × g for 4 min at 4°C and washed
twice with PBS to remove excess monoclonal antibody. The cells were
resuspended in 200 µl PBS and then fixed in 1% paraformaldehyde.
The expressions of CD11b and CD14 in different treatment groups
were analyzed on FACSVerse (BD Biosciences Pharmingen) Flow
cytometer. Isotypic rat IgG was also used to check for nonspecific
binding. The experiment was repeated three times.
Protein expression by western
blotting
The cells of the control group and the experimental
group were collected, washed twice with pre-cooled PBS, cells were
lysed with Protein Extraction reagent (Beijing Solarbio Science
& Technology Co., Ltd.), the total cell protein was extracted,
the protein concentration was determined by BCA method and 5×
protein loading buffer was added and boiled for denaturation at
95°C for 10 min. Protein (20 µg) was loaded for SDS-PAGE (10%)
electrophoresis, the separated protein was transferred to PVDF
membrane, blocked with 5% skimmed milk for 90 min at room
temperature and then incubated with the corresponding primary
antibody; Egr-1 (1:800), GAPDH (1:8,000) and β-actin (1:2,000) at
4°C overnight, then washed with 1X TBST for 30 min, then incubated
with goat anti-rabbit IgG secondary antibody (1:20,000) at room
temperature for 90 min and finally washed for 30 min, and proteins
were detected using an ECL kit (Sparkjade ECL super, ED0015-B,
Shandong Sparkjade Scientific Instruments Co., Ltd.). ImageJ
v1.51j8 was used for densitometry (National Institutes of Health).
The experiment was repeated three times.
Relationship Analysis between
miR-let-7c-3p and Egr-1
TargetScan, PITA and microRNAorg databases were used
to predict target genes of possible upstream miRNAs of Egr-1 and
intersected the predicted target miRNAs by crosstalk of the three
databases. The common target miRNAs in the three databases were
obtained, and the top 10 miRNAs were selected according to the
P-value and literature research. In addition, GeneChip miRNA 4.0
(Affymetrix Co., Ltd.) was used to detect the different miRNA
profiles between control and PMA-induced K562 cells, and 10 miRNAs
that were reduced after induced-differentiation were screened
according to the P-value.
Gene expression by reverse
transcription-quantitative (RT-q) PCR
When the K562 cells were cultured to the logarithmic
growth phase, the cells (1×106) were collected and the
total RNA was extracted by TRIzol® (Thermo Fisher
Scientific, Inc.) method and the total RNA concentration was
measured by an ultra-trace nucleic acid and protein analyzer. cDNA
was synthesized according to the instructions of the PrimeScript RT
reagent kit with gDNA Eraser (Perfect Real Time) reverse
transcription kit. For reverse transcription, samples were
incubated in an Eppendorf PCR system at 42°C for 30 min, then at
90°C for 5 min and at 5°C for 5 min. cDNA was used as a template
for PCR amplification. The sense primer of miR-let-7c-3p was
5′-GCGCGCTGTACAACCTTCTAG-3′, the antisense primer was
5′-AGTGCAGGGTCCGAGGTATT-3′; the U6 sense primer was
5′-AGAGAAGATTAGCATGGCCCCTG-3′, Antisense is
5′-AGTGCAGGGTCCGAGGTATT-3′; Egr-1 sense primer was
5′-AGCAGCAGCAGCACCTTCAAC-3′, antisense was
5′-CCACCAGCACCTTCTCGTTGTTC-3′; GAPDH sense primer is
5′-CAACTTTGGTATCGTGGAAGG-3′, antisense was
5′-GCCATCACGCCAGAGTTTC-3′. The real-time fluorescence quantitative
amplification reaction was performed according to the instructions
of the TB Green® Premix Ex Taq (Tli RNaseH Plus) kit,
PCR was conducted with the following conditions: 10 sec at 95°C; 40
cycles of 5 sec at 60°C and 10 sec at 72°C; 34 sec at 60°C, and the
relative quantitative analysis was performed using the
2-ΔΔCq method (33).
The experiment was repeated three times.
Dual-luciferase reporter gene analysis
between miR-let-7c-3p and Egr-1
Using the bioinformatics prediction website
(http://www.targetscan.org) to predict
the binding fragments of Egr-1 and miR-let-7c-3p, pmirGLO-Egr-1-wt
wild plasmid vector and pmirGLO-Egr-1-mut plasmid vector were
constructed (Jinan Boshng Biotechnology Co., Ltd.) respectively,
and cells co-transfected by transfection reagent kit (jetPRIME;
Polyplus-transfection SA) with the above Egr-1 wild plasmid, Egr-1
mutant plasmid and miR-let-7c-3p mimics (sequence:
CUGUACAACCUUCUAGCUUUCC) or mimics NC (sequence:
UUCUCCGAACGUGUCACGUTT). The luciferase activity was measured by
Dual-Luciferase Reporter Assay System (Envision; PerkinElmer, Inc.)
48 h following culture and Renilla luciferase was used as an
internal control
siRNA interference
An appropriate amount of short interfering RNA
(siRNA) and its corresponding negative control were mixed with the
transfection reagent to form a transfection complex, which was
added to the 6-well plate that had been seeded with cells and
cultured at 37°C for 48 h for subsequent experiments. Egr-1
siRNA-1: 5′-CCAUGGACAACUACCCUAATT-3′, Egr-1 siRNA-2:
5′-GCCUAGUGAGCAUGACCAATT-3′ and Egr-1 siRNA-3:
5′-GCUGUCACCAACUCCUUCATT-3′ synthesized by Shanghai BioSune Co.,
Ltd. were selected. According to the preliminary experimental
results, the Egr-1 siRNA-1 with the most obvious interference
effect (5′-CCAUGGACAACUACCCUAATT-3′) was selected as the target
siRNA (hereinafter referred to as siEgr-1). As to the si-RNA
interference experiment, there were four groups. In addition to the
above 100 µg/l PMA experimental group and 0 µg/l PMA control group,
the 100 µg/l PMA experimental group was transfected with si-ctrl at
the same time as the PMA + si-Ctrl group. The 100 µg/l PMA
experimental group was transfected with siEgr-1 at the same time as
PMA + si-Egr-1 group.
Expression regulation of Egr-1 by
miR-let-7c-3p mimic and inhibitor
miR-let-7c-3p mimic and inhibitor were synthesized
by Shanghai GenePharma Co., Ltd. and their sequences were
5′-CUGUACAACCUUCUAGCUUUCC-3′, 5′-GGAAAGCUAGAAGGUUGUACAG-3′,
corresponding NCs were: 5′-UUCUCCGAACGUGUCACGUTT-3′ and
5′-GGAAAGCUAGAAGGUUGUACAG-3′, respectively. The conventional
transfection method of Lipofectamine® 2000 (Thermo
Fisher Scientific, Inc.) was used. Following transfection, RT-qPCR
and western blotting were used to detect the expression changes of
miR-let-7c-3p and downstream Egr-1 protein expression,
respectively. iii) The effect of miR-let-7c-3p on the expression of
differentiation antigens in K562 cells: In the above experiments of
transfection of miR-let-7c-3p inhibitor, the expression changes of
K562 cell differentiation antigens CD11b and CD14 were detected at
the same time and let-7c-3p inhibitor NC was used as a control. The
expression levels of differentiation antigens in the 100 µg/l PMA
experimental group and the 0 µg/l PMA control group were also
observed.
Statistical analysis
GraphPad Prism 8 software (GraphPad Software, Inc.)
was used for data processing and Shapiro-Wilk (S-W) normal
distribution was used for quantitative data. Each experiment was
repeated three times and the measurement data were expressed as
mean ± standard deviation. Comparisons between two groups were
performed using independent samples t-test and ANOVA was used for
multiple-group comparisons. The Bonferroni test was used as the
post-hoc test for the one-way ANOVA test. Pearson analysis was used
for the correlation between miR-let-7c-3p and Egr-1. All data were
analyzed by two-tailed test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Changes in cell morphology and level
of proliferation
The in vitro growth characteristics of the
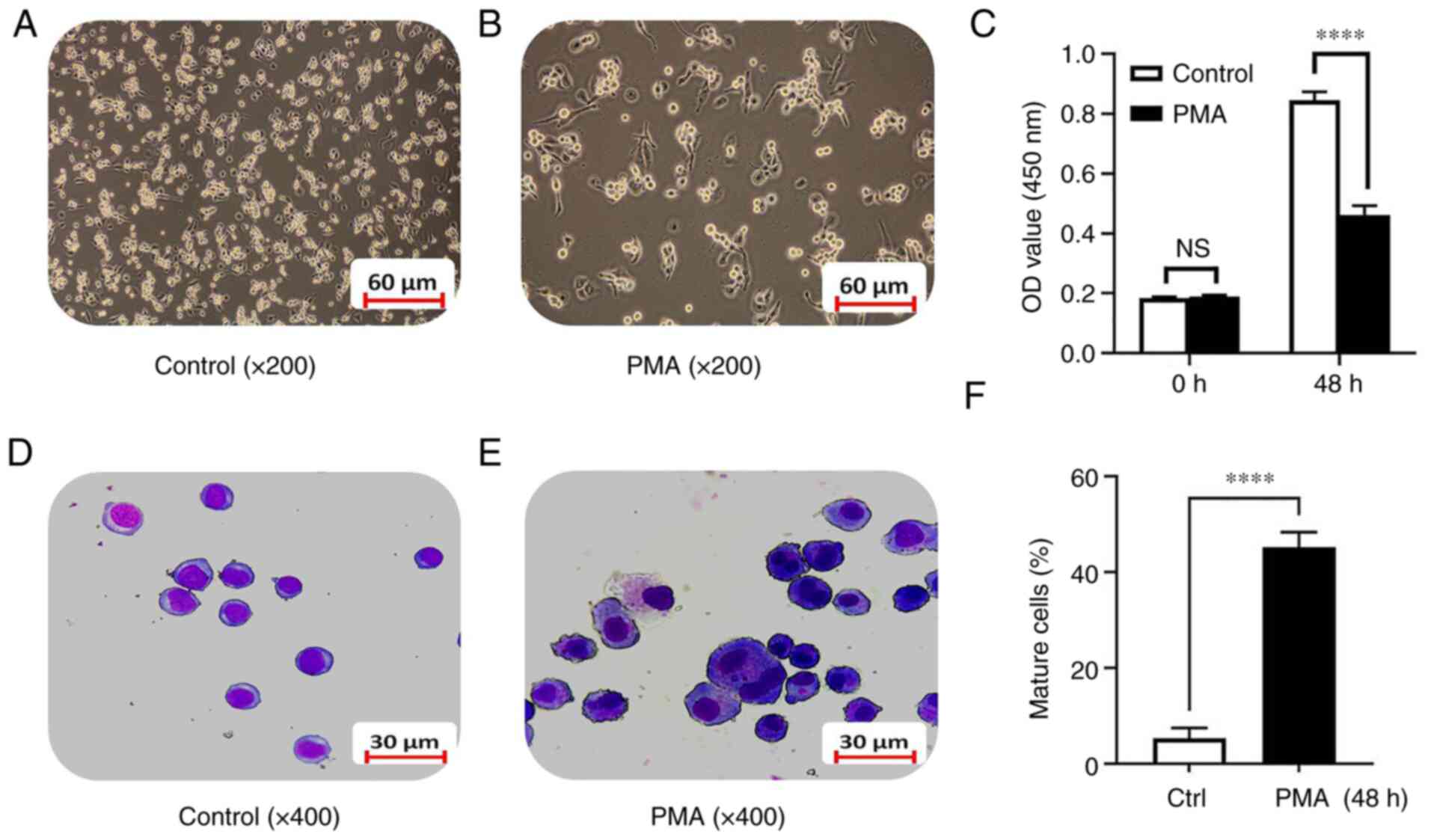
K562 cell line were directly observed under an inverted microscope.
The results showed that, compared with the control group, after
exposure to 100 ng/ml of PMA for 48 h, the K562 cells density was
significantly reduced and the cells grew from a suspension state to
an adherent state gradually (Fig. 1A
and B). The CCK8 experiment confirmed that there was no
significant difference between the PMA group and the control group
before differentiation induction. After 48 h of differentiation
induction, the proliferation ability of the PMA-induced
differentiation group was significantly decreased compared with the
control group (0.85±0.03 vs. 0.46±0.03; t=16.05; P<0.0001;
Fig. 1C). Swiss-Giemsa staining
showed that the cell volume after PMA-induced differentiation for
48 h increased significantly compared with the control group and
the cytoplasmic volume increased, the nuclear-cytoplasmic ratio
decreased and the nuclei became smaller. There was a trend towards
monocyte-macrophage differentiation (Fig. 1D and E) and the number of more
matured monocyte-macrophages increased (5.34±2.12 vs. 45.21±3.18;
t=18.07; P<0.0001; Fig. 1F).
The results indicated that the model of leukemia cell line
differentiation into monocytes/macrophages was successfully
established.
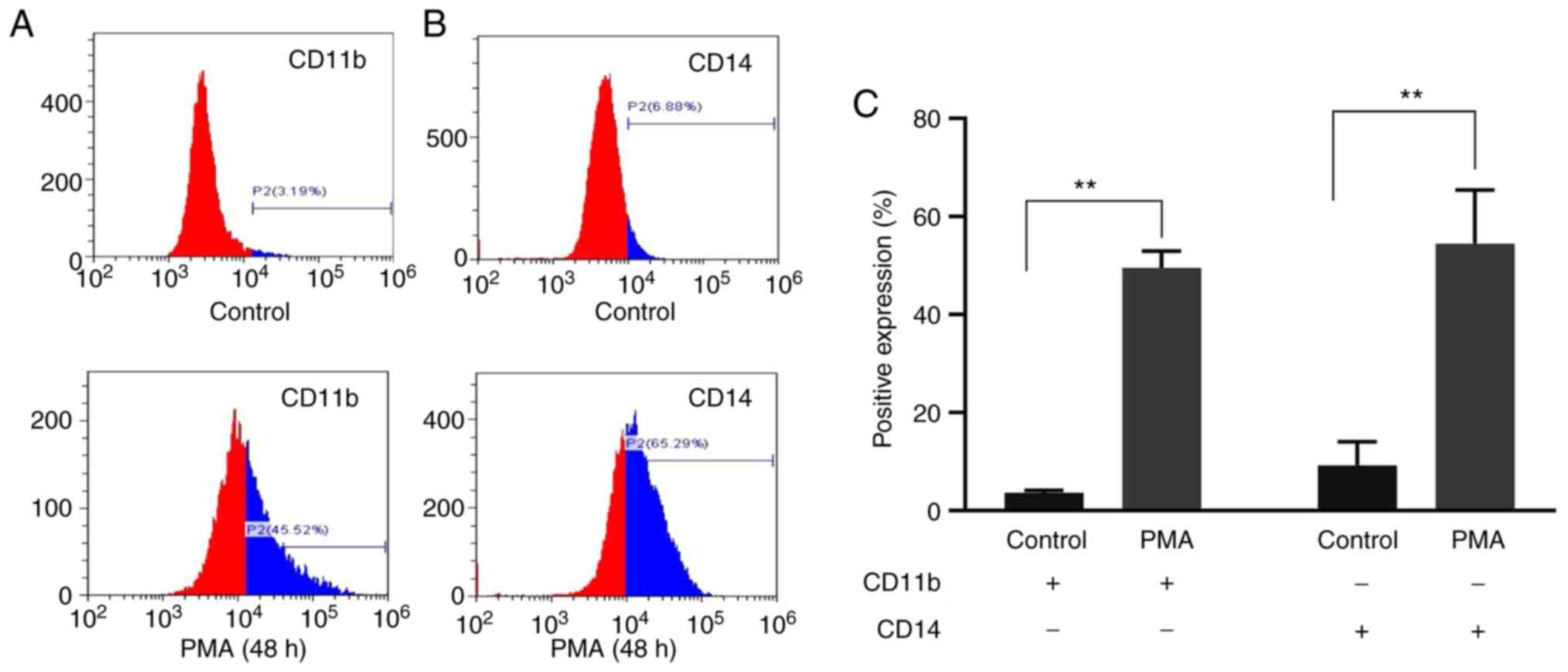
CD11b and CD14 differentiation
antigens before and after exposure to PMA
To further validate the committed differentiation of
leukemia cells into monocytes/macrophages, the present study
examined the expression of monocyte/macrophage-specific surface
markers CD11b and CD14 in K562 cells treated with PMA for 48 h. The
results of flow cytometry showed that the expression of CD11b in
the PMA-induced group was significantly higher compared with that
in the control group (49.47±3.48 vs. 3.54±0.54; t=24.070; P=0.002)
and CD14 in the PMA-induced group was significantly higher compared
with that in the control group (59.84±5.26 vs. 6.79±0.66; t=16.670;
P=0.004; Fig. 2). The results
showed that PMA could induce K562 cells to differentiate into
monocytes/macrophages.
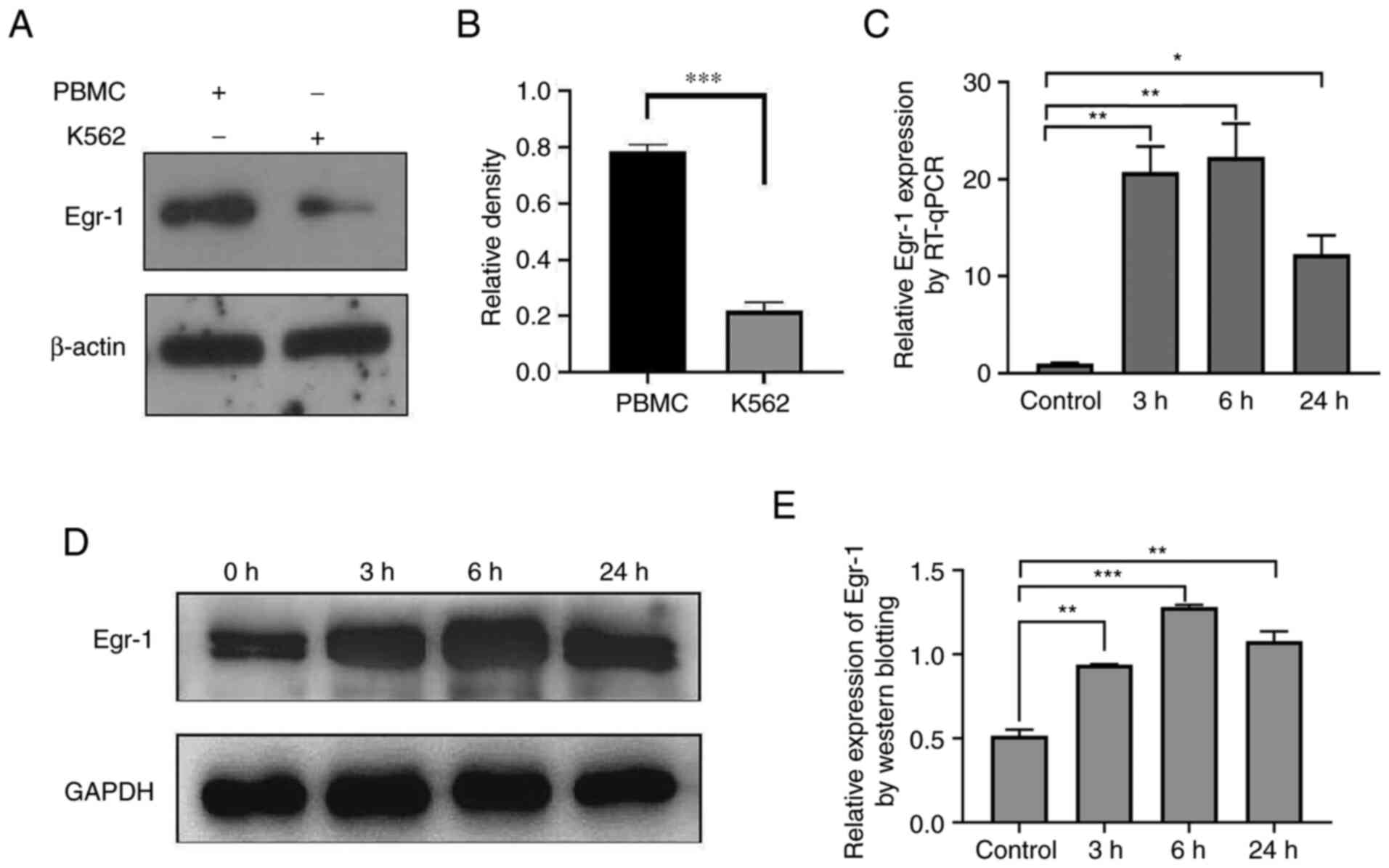
Expression of Egr-1 in K562 cells
before and after exposure to PMA
The expression level of Egr-1 in normal human
peripheral blood mononuclear cells and K562 cells was detected by
western blotting and the results showed that the expression of
Egr-1 protein in K562 cells was significantly lower compared with
that in normal controls (Fig. 3A and
B). The gene expression of Egr-1 gene was detected at the
indicated different time points (Fig.
3C) and it was also found that PMA induced an increase in the
expression of Egr-1 gene in K562 cells and the protein expression
of Egr-1 was also significantly increased (Fig. 3D and E), which contributed to the
induced differentiation from leukemia cells into
monocytes/macrophages in vitro.
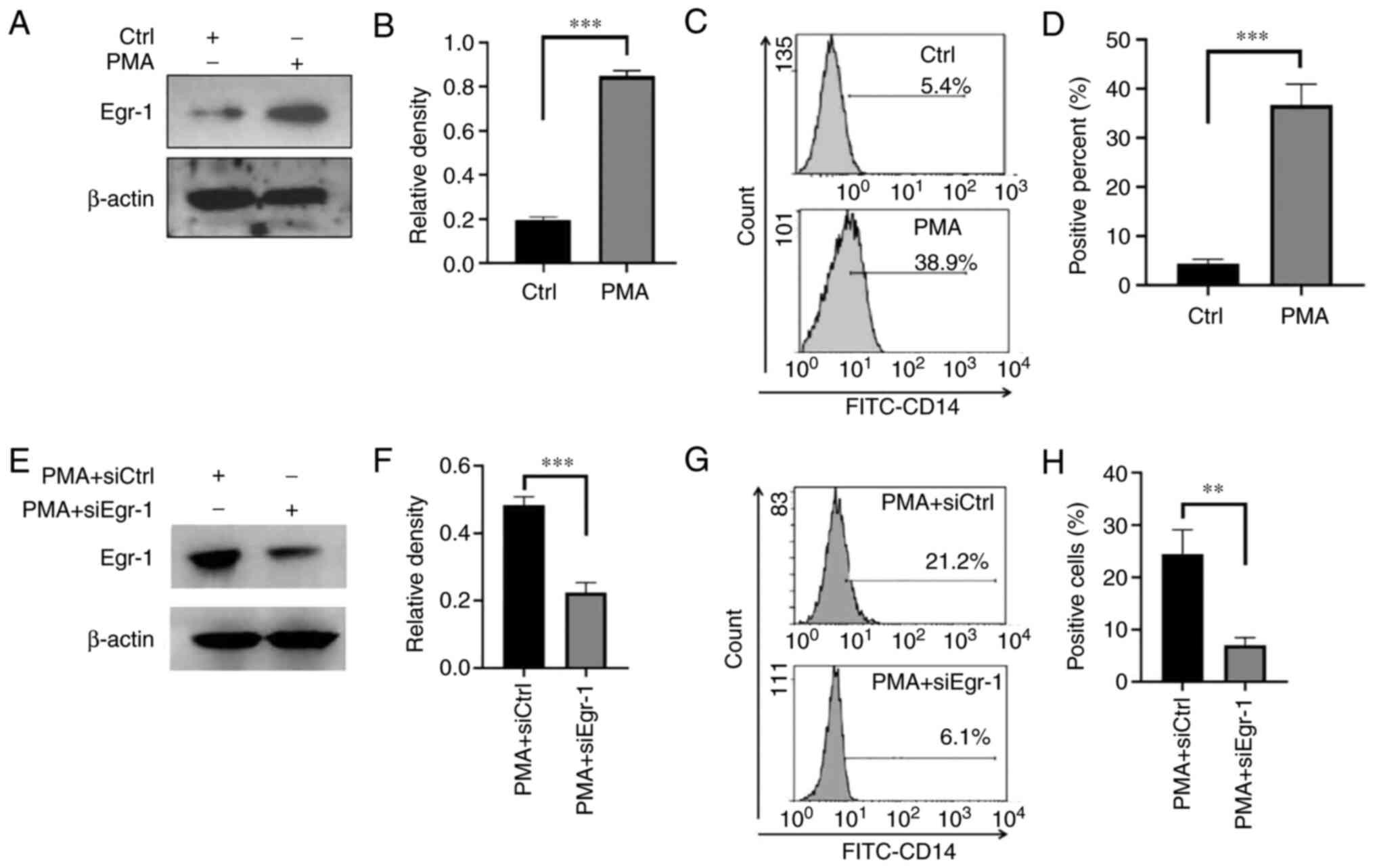
Effect of siRNA-Egr-1 on the
differentiation of K562 cells induced by PMA
The expression changes of Egr-1 in K562 cells before
and after PMA-induced differentiation were detected. The results
confirmed that compared with the control group, the expression of
Egr-1 was significantly increased after PMA induction (0.19±0.02
vs. 0.85±0.03; t=24.800; P<0.001; Fig. 4A and B). It was found that this
elevated expression of Egr-1 protein was accompanied by an elevated
expression level of K562 cell differentiation antigen CD14
(4.30±1.01 vs. 36.67±4.31; t=12.66; P=0.0002; Fig. 4C and D). Compared with the PMA
alone group, the Egr-1 protein expression in the PMA + siEgr-1
co-action group was decreased (0.22±0.03 vs. 0.48±0.03; t=11.380;
P<0.001; Fig. 4E and F) and the
expression of the differentiation antigen CD14 was significantly
decreased (7.03±1.45 vs. 24.40±4.70; t=6.113; P=0.004; Fig. 4G and H).
Target relationship between
miR-let-7c-3p and Egr-1
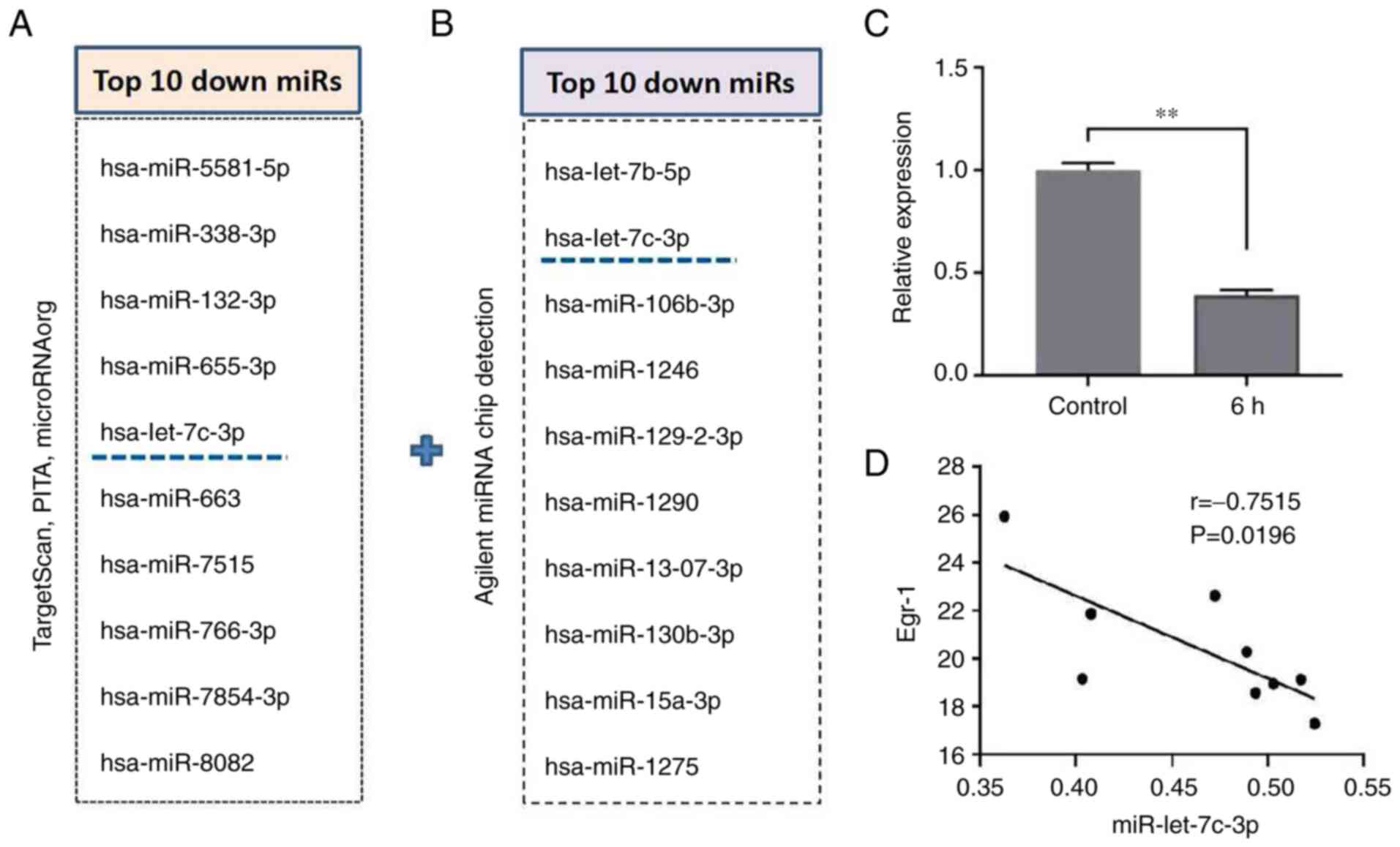
In the preliminary experiments, TargetScan, PITA and
microRNAorg databases were used to predict target genes of possible
upstream miRNAs of Egr-1 and intersected the predicted target
miRNAs by crosstalk of the three databases. According to the
results of target gene prediction, the common target miRNAs in the
three databases were obtained. After sorting according to the
P-value and literature research, the top 10 miRNAs were selected
(Fig. 5A). In addition, following
Agilent miRNA Chip detection, 10 miRNAs that were reduced after
induced-differentiation were screened according to the P-value
(Fig. 5B). Then the cross-analysis
of the database analysis and the actual detection results of the
chip was performed and it was found that miR-let-7c-3p was the only
candidate miRNA. RT-qPCR results showed that the expression level
of miR-let-7c-3p in the PMA-induced group was significantly lower
than that in the control group (1.00±0.04 vs. 0.39±0.03; t=20.040;
P=0.003; Fig. 5C). This indicated
that in the process of PMA-induced differentiation of K562 cells,
the expression level of miR-let-7c-3p was decreased. The three
different time points at which K562 cells were induced to
differentiate were randomly selected and three replicate samples
were observed. Following Pearson correlation analysis, it was found
that the changes of miR-let-7c-3p and Egr-1 were negatively
correlated (Fig. 5D).
The expression of Egr-1 by
miR-let-7c-3p mimic and inhibitor
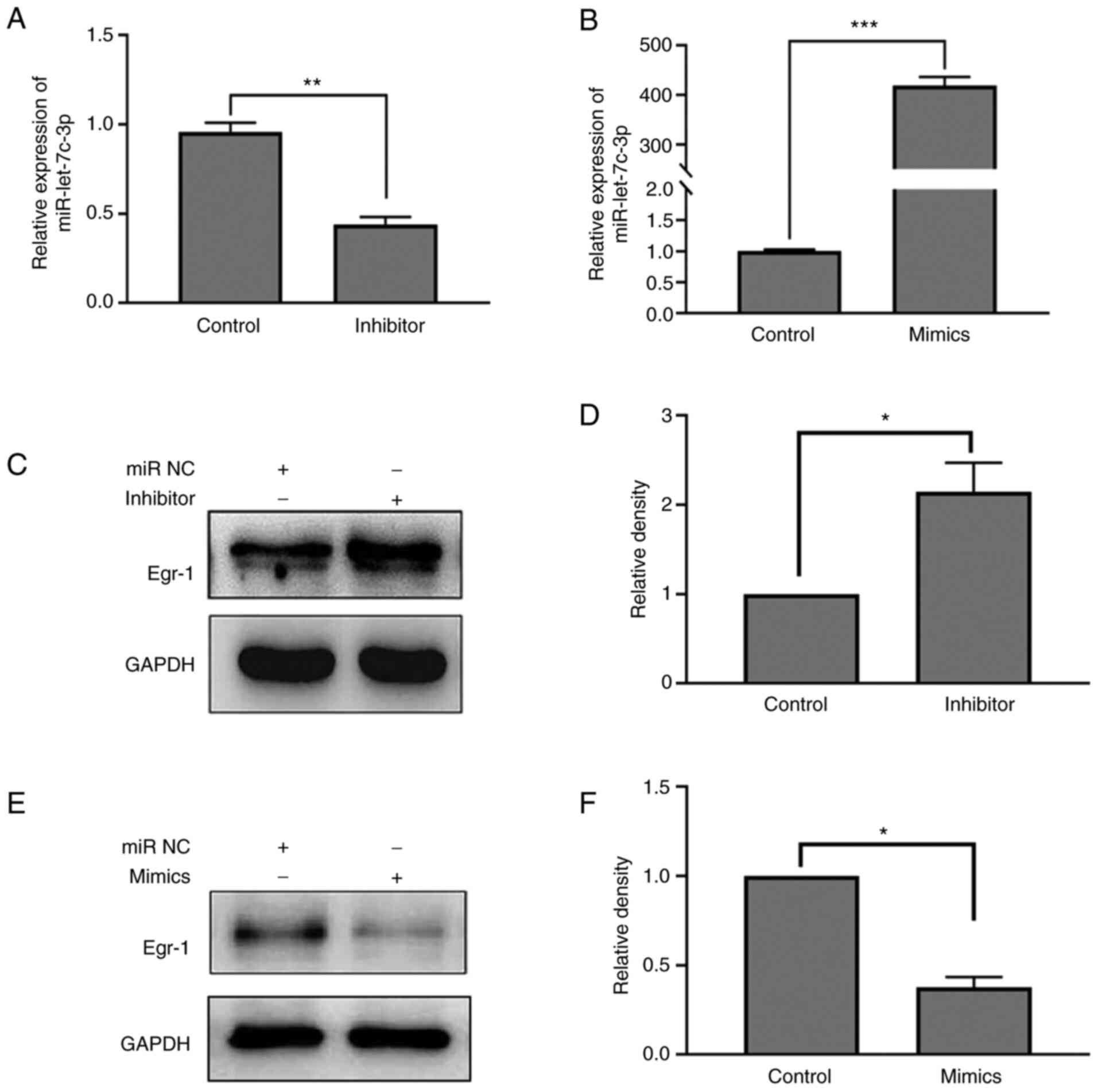
The regulatory effect of miR-let-7c-3p mimic and
inhibitor on the expression of miR-let-7c-3p were first verified
and the results confirmed that the expression level of
miR-let-7c-3p in the miR-let-7c-3p inhibitor group was
significantly lower compared with that in its negative control
group (0.44±0.42 vs. 0.96±0.05; t=12.870; P=0.006). The expression
level of miR-let-7c-3p in the miR-let-7c-3p mimic group was
significantly higher compared with that in the negative control
group (418.80±17.33 vs. 1.01±0.02; t=41.760; P<0.001; Fig. 6A and B). On the regulation of Egr-1
expression by miR-let-7c-3p, the results of western blotting showed
that the expression of Egr-1 was significantly increased following
transfection of miR-let-7c-3p inhibitor as compared with the
control (0.83±0.12 vs. 0.39±0.00; t=6.315; P=0.024; Fig. 6C and D), while the expression of
Egr-1 was significantly decreased following transfection of
miR-let-7c-3p mimic compared with the control group (0.18±0.01 vs.
0.48±0.06; t=7.809; P=0.016; Fig. 6E
and F).
Targeted binding and regulation of
Egr-1 by miR-let-7c-3p
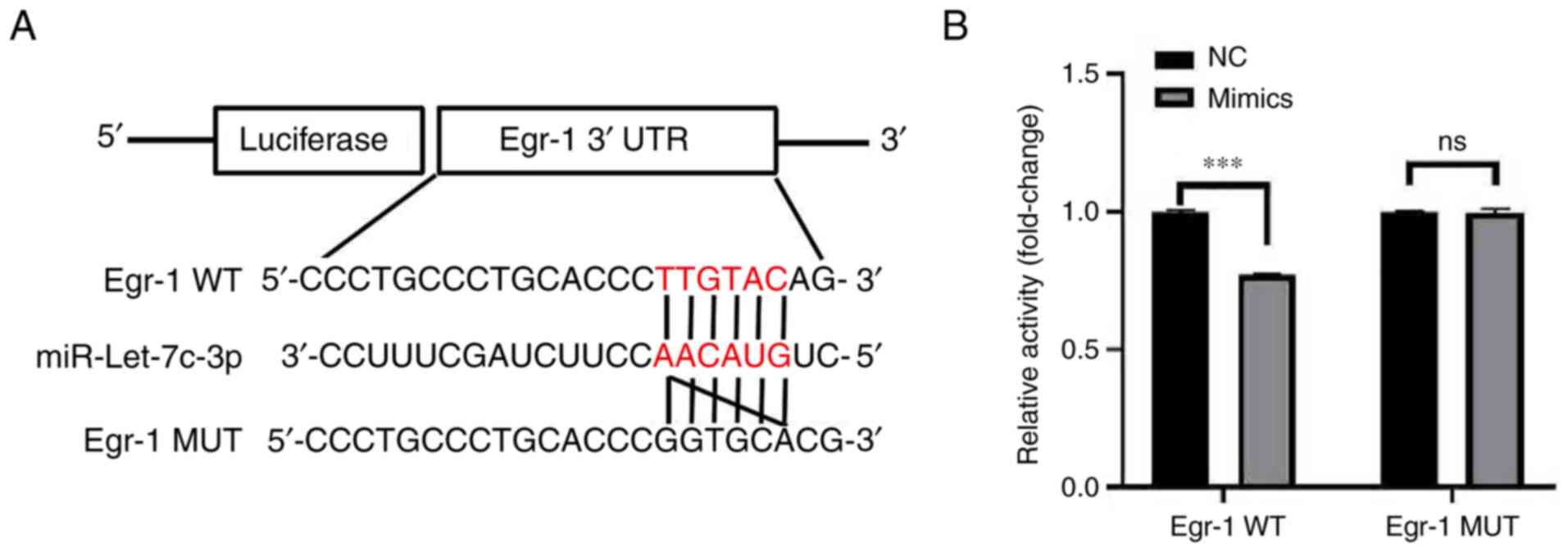
Further analysis of the StarBase database revealed a
binding site between miR-let-7c-3p and Egr-1, while the TargetScan
database predicted a pairing site between miR-let-7c-3p and Egr-1
(Fig. 7A). The results of dual
luciferase reporter gene showed that the luciferase activity in the
co-transfected mimic and Egr-1 WT groups was significantly lower
than that in the co-transfected mimic and Egr-1 MUT groups
(0.77±0.01 vs. 1.00±0.01; t=27.582; P<0.001; Fig. 7B). The dual-luciferase reporter
gene assay confirmed that there was a targeted binding activity
regulatory relationship between miR-let-7c-3p and Egr-1, indicating
that miR-let-7c-3p can target Egr-1.
The effect of miR-let-7c-3p on
PMA-induced differentiation of K562 cells
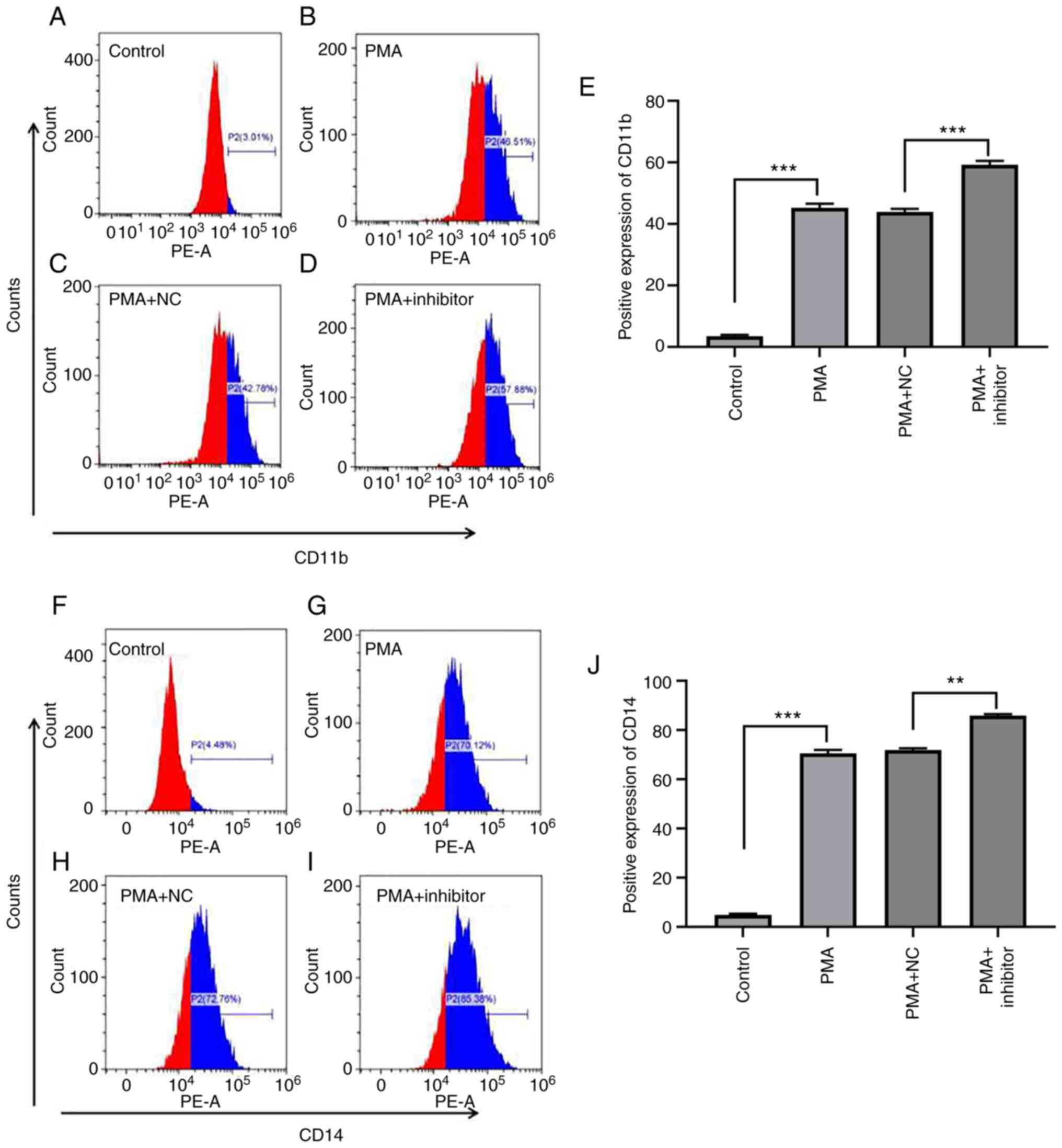
To explore the effect of miR-let-7c-3p on
PMA-induced differentiation of K562 cells, K562 cells were
transfected with miR-let-7c-3p inhibitor and its negative control,
treated with PMA (100 ng/ml) for 48 h to induce differentiation and
the expression of cell surface markers CD11b and CD14 in each group
was detected by flow cytometry. The results, as shown in Fig. 8, demonstrated that PMA clearly
induced the committed differentiation of K562 cells, which was
manifested as increased CD11b expression; for example, the
expression levels of CD11b in the control group, PMA group, PMA +
NC group and PMA + inhibitor group were 3.44±0.43%, 45.14±1.40%,
43.91±1.00% and 59.22±1.28%, respectively (Fig. 8A-E). The difference in ANOVA
analysis for CD11b between groups was statistically significant
(F=1460.318, P<0.001), and multiple Bonferroni test analysis
showed that, except for PMA and PMA+NC with P=0.238, the remaining
P-values were all <0.001, which was in line with the expected
results. The difference was statistically significant in ANOVA
analysis for CD14 between groups (F=5163.346, P<0.001), multiple
Bonferroni test analysis found that except for PMA and PMA+NC were
0.544, the remaining P-values were all <0.001, which was in line
with the expected results. Among them, the expression level of
CD11b in the PMA group was significantly higher than that in the
control group (t=39.570; P<0.001), the expression level of CD11b
in the PMA + inhibitor group was significantly higher than that in
the PMA + NC group (t=37.350; P<0.001; Fig. 8E), The expression level of CD11b in
PMA + inhibitor group was significantly higher than that in PMA
group (t=9.147; P=0.012).
PMA also significantly increased the expression of
CD14 in K562 cells. The expression levels of CD14 in the control
group, PMA group, PMA + NC group and PMA + inhibitor group were
4.91±0.42%, 70.54±1.44%, 71.91±0.74% and 85.98±0.52%, respectively
(Fig. 8F-J), the difference was
statistically significant following ANOVA analysis between groups
(F=5163.346; P<0.001), multiple LSD analysis found that except
for PMA and PMA + NC (P=0.91), the remaining P-values were all
<0.001, which was in line with the expected results. The
expression level of CD14 in the PMA group was significantly higher
compared with the control group (t=94.720; P<0.001), the
expression level of CD14 in the PMA + inhibitor group was
significantly higher compared with the PMA + NC group (t=19.310;
P=0.003) and the expression of CD14 in the PMA + inhibitor group
was significantly higher compared with the PMA group (t=18.660;
P=0.003; Fig. 8J). The results
indicated that reduction of the expression of miR-let-7c-3p could
promote the directed differentiation of PMA-induced leukemia.
Discussion
Myeloid leukemia is a type of hematopoietic stem
cell malignant tumor with the characteristics of differentiation
disorder and uncontrolled proliferation (1,2). At
present, the treatment of leukemia is mainly based on the
combination of chemotherapy and targeted therapy, but there are
obvious side effects and a high recurrence rate. The
differentiation induction therapy represented by all-trans retinoic
acid is one of the best methods for the treatment of leukemia
(9–11); in that, the leukemia cells are
induced to differentiate into mature promyelocytes by all trans
retinoic acid (ATRA) and their malignant proliferation is
inhibited. It is precisely because of this selective effect on
leukemia cells that it does not affect normal hematopoietic and
immune functions, making it a research hotspot (12–15).
However, elucidating the molecular mechanism of development and
occurrence of patients with leukemia and how to find new
differentiation-inducing drugs has become a challenge in the field
of differentiation induction. At present, in addition to a number
of studies on the induction of leukemia cells into myeloid cells
(8,34–36),
there are also some reports on the induction of leukemia cells into
monocytes/macrophages. For example, Chou and Hsu (37) show that PMA can induce the
differentiation of chronic myeloid leukemia cell line K562 into
monocyte-macrophages. On this basis, the present study first used
multiple methods to verify that PMA induced K562 to differentiate
into monocytes/macrophages; it was found that the cells gradually
changed from the suspension growth to the adherent state, the cell
volume increased, the antennae were prominent, the cytoplasmic
folds increased, the nucleocytoplasmic ratio was significantly
reduced and multinucleation appeared, showing the development trend
of mature monocytes/macrophages following PMA induction. At the
same time, the monocyte/macrophage surface molecules CD11b and CD14
were significantly increased (38). CCK8 experiments also showed that
PMA could effectively inhibit the proliferation of K562 cells. All
of the above results indicated that PMA possessed a strong ability
to induce K562 to differentiate into monocytes/macrophages.
Therefore, this differentiation induction model provides a model
for studying the molecular mechanism of leukemia cells to
differentiate into monocytes/macrophages.
The Egr-1 gene, as a member of the zinc finger
structure transcription factor family, is located on human
chromosome 5q31 and encodes a DNA-binding transcription factor of
~80 kDa (39). It has been
demonstrated that Egr-1 serves an important role in cell growth,
differentiation and apoptosis (16,40–44).
At the same time, Egr-1 is closely associated with the occurrence
and development of certain diseases, for example tutor and
leukemia, however the specific signaling pathway of each of them
remains to be elucidated. It is also found so far that Egr-1 is an
important multifunctional transcriptional regulator, and various
factors can affect the expression of Egr-1. Usually, in
unstimulated cells, Egr-1 expression was difficult to detect or
only detectable at very low levels. However, Egr-1 can be
upregulated in a rapid and transient manner under the activation by
different extracellular stimuli (45–48),
such as some cytokines or differentiation inducers (39,49,50).
Egr-1 includes transactivation and repression regions, as well as
three DNA-binding zinc finger regions that recognize GC-rich
fragments of the promoter regions of target genes (51). Egr-1 has a wide range of functions,
involving the control of synaptic plasticity, wound repair,
inflammation, blood coagulation, pulmonary vascular permeability,
growth and apoptosis of a number of cells (50,52–56).
Some findings suggest that Egr-1 can reverse the disease
progression of acute myeloid leukemia by regulating changes in the
downstream target genes c-myc and E2F1 (57). In chronic myeloid leukemia, a
decrease in Egr-1 leads to an increase in the number of leukemia
stem cells in the blood, accelerating disease progression, whereas
Egr-1 serves an important role in normal hematopoietic stem cell
differentiation, quiescence and terminal differentiation of
monocyte/macrophage cells (58).
As early as the last century, some researchers identified Egr-1 as
an important differentiation response gene during
monocyte/macrophage development (59), it is greatly upregulated in HL-60
and U-937 leukemia cells following exposure to PMA. In the present
study, the expression of Egr-1 in K562 cells also showed a trend of
upregulation, which was induced by PMA. Combined with the Egr-1
interference experiment, the PMA-induced upregulation of CD14
expression was reduced by the si-Egr-1 group, as compared with that
of PMA + si-ctrl group, indicating that Egr-1 was involved in the
differentiation of leukemia K562 cells into
monocyte/macrophage.
It has been demonstrated that some important
transcriptional regulatory proteins are often regulated by
epigenetic factors, such as non-coding RNAs, at the same time that
some important transcriptional regulatory proteins are involved in
transcriptional regulation. Usually, one important pathway for
microRNAs (miRNAs) to result in gene silencing or translational
repression was mainly contributed to binding to 3′UTR of target
mRNAs (60–62). The regulatory factors of miRNA not
only participate in the occurrence and development of various human
tumors (63), but also show great
potential in disease diagnosis, prognosis judgment and targeted
therapy (64). Among them, it has
been found that miRNAs are involved in differentiation,
proliferation, apoptosis and other processes of leukemia cells, and
its relatively specific mutational and deregulated expression
profiles also have potential as diagnostic or prognostic biomarkers
(65). In addition, non-coding
RNAs also serve an important role in the chemoresistance of tumors
or leukemias. Different miRNA species serve different roles in the
formation or reversal of drug resistance. It can be used both as a
biomarker of drug resistance and as one of the targets for drug
resistance intervention (66,67).
For example, miRNA expression profiling of drug-resistant melanoma
patients and their cell lines reveals that miRNA-181a and
miRNA-181b are significantly downregulated in drug-resistant
melanoma patients and drug-resistant cell lines. Reconstruction of
miR-181a/b expression reversed the resistance of melanoma cells to
the BRAF inhibitor dabrafenib. Clinical observations show that
melanoma patients with high expression of miRNA-181a and miRNA-181b
have longer progression-free survival time (68).
miR-let-7, as the earliest discovered human miRNA,
is one of the most widely studied miRNAs. Its family members are
abnormally expressed in various malignant tumors and become a new
target for tumor prevention and treatment (32,69).
For example, in cervical cancer, nanocarriers can target
miR-let-7c-5p to inhibit tumor cells and exhibit reduced
cytotoxicity (70). The miR-let-7
family has been shown to be downregulated in various types of tumor
tissues and has been extensively studied as a tumor suppressor gene
(30,31,71).
Accumulating evidence suggests that Let-7 also has the same
properties as other miRNAs and is not only involved in the
occurrence and development of leukemia, but also a potential
biomarker for leukemia diagnosis and prognosis (32). However, it has not been elucidated
whether miRNA Let-7 is involved in leukemia cell-directed
monocyte-macrophage maturation and differentiation.
In the preliminary experiments of the present study,
it was found that some miRNAs changed significantly during the
process of differentiation of leukemia cells into mature
monocytes/macrophages induced by PMA. Further analysis of the
StarBase database revealed a binding site between miR-let-7c-3p and
Egr-1, while the TargetScan database predicted a pairing site
between miR-let-7c-3p and Egr-1. In addition, following Agilent
miRNA Chip assay, miR-let-7c-3p also reduced after
induced-differentiation by PMA with the upregulation of Egr-1 mRNA.
Furthermore, it was found that the expressions of miR-let-7c-3p and
Egr-1 showed an inverse relationship by Pearson analysis in the
differentiation process induced by PMA and that they had a nucleic
acid sequence basis for targeted binding. In addition, Egr-1 is
widely associated with miRNAs and can be regulated by miRNAs in a
variety of tumors (72,73). Therefore, it was hypothesized that
the miR-let-7c-3p signal axis may serve a role in the committed
differentiation of leukemia cells into monocytes/macrophages.
First, to detect the possible effect of miR-let-7c-3p on binding
3′UTR and modulating activity of Egr-1 transcription, luciferase
assay was performed and the results indicated that the
miR-let-7c-3p could bind 3′UTR of Egr-1 and modulate its activity.
The luciferase activity of the co-transfected mimic and Egr-1 WT
group was significantly lower than that of the co-transfected mimic
and Egr-1 MUT group. Furthermore, the miR's endogenous expression
in K562 cells was also demonstrated by miR-let-7c-3p mimics
transfection; the expression level of Egr-1 in miR-let-7c-3p mimic
group was significantly lower than that in the negative control
group. The expression of miR-let-7c-3p in K562 cells before and
after treatment with PMA was detected by RT-qPCR assay and the
results showed that the expression level of miR-let-7c-3p in the
induction group was significantly lower than that in the control
group, as in agreement with the effect of miR-let-7c-3p inhibitor
and the expression level of Egr-1 in the miR-let-7c-3p inhibitor
group was significantly lower compared with the negative control
group.
Some other studies (39,74,75)
report that the role of Egr-1 can be modulated by some miRNAs,
including miR-424, miR-146a, miR181a and miR-337. In the present
study, the expression of Egr-1 is regulated in K562 cells
transfected with miR-let-7c-3p mimics and inhibitor. Western
blotting showed that compared with the control group, the
expression of Egr-1 in the miR-let-7c-3p mimics transfected group
was decreased and the expression of Egr-1 in the miR-let-7c-3p
inhibitor transfected group was increased. Thus, miR-let-7c-3p can
be targeted to bind to Egr-1 and has a negative regulatory
relationship. In brief, the expression of Egr-1 and miR-let-7c-3p
was upregulated or downregulated after exposure to PMA in
vitro and miR-let-7c-3p could directly bind to the 3′UTR of
Egr-1 and modulated its promoter activity. The miR-let-7c-3p/Egr-1
signaling axis contributed to the differentiation from K562
leukemia cells into more matured monocytes/macrophage cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the ‘Twelfth Five-Year’
National Science and Technology Support Program (grant no.
2013BAI07B02), Natural Science Foundation of China (grant no.
81573467), Natural Science Foundation of Shandong (grant nos.
ZR2020QH160 and ZR2021MH080), The Foundation for Teachers' research
project of Jining Medical University (grant no. JYFC2019FKJ102) and
Science and Technology Project of Jinan Municipal Health Commission
(grant no. 2019-1-66).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GSJ, XDW, FQ, XPW and CZW made substantial
contributions to the conception and design and also critically
reviewed the study. FQ, XPW, CZW, RJS and HW performed the
experiments. FQ, SZZ, PCD and JW analyzed the data and wrote the
manuscript. GSJ and XDW confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu YH, Zhu M, Lei PP, Pan XY and Ma WN:
ND-09 inhibits chronic myeloid leukemia K562 cell growth by
regulating BCR-ABL signaling. Oncol Rep. 46:1362021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yao FY, Zhao C, Zhong FM, Qin TY, Wen F,
Li MY, Liu J, Huang B and Wang XZ: m(6)A modification of lncRNA
NEAT1 regulates chronic myelocytic leukemia progression via
miR-766-5p/CDKN1A axis. Front Oncol. 11:6796342021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cai SF and Levine RL: Genetic and
epigenetic determinants of AML pathogenesis. Semin. Hematol.
56:84–89. 2019.PubMed/NCBI
|
|
4
|
Padmakumar D, Chandraprabha VR, Gopinath
P, Vimala Devi ART, Anitha GRJ, Sreelatha MM, Padmakumar A and
Sreedharan H: A concise review on the molecular genetics of acute
myeloid leukemia. Leuk Res. 111:1067272021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhat AA, Younes SN, Raza SS, Zarif L,
Nisar S, Ahmed I, Mir R, Kumar S, Sharawat SK, Hashem S, et al:
Role of non-coding RNA networks in leukemia progression, metastasis
and drug resistance. Mol Cancer. 19:572020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schwarzer A, Emmrich S, Schmidt F, Beck D,
Ng M, Reimer C, Adams FF, Grasedieck S, Witte D, Käbler S, et al:
The non-coding RNA landscape of human hematopoiesis and leukemia.
Nat Commun. 8:2182017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
de Thé H: Differentiation therapy
revisited. Nat Rev Cancer. 18:117–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Madan V and Koeffler HP: Differentiation
therapy of myeloid leukemia: Four decades of development.
Haematologica. 106:26–38. 2021.PubMed/NCBI
|
|
9
|
Yen JH, Lin CY, Chuang CH, Chin HK, Wu MJ
and Chen PY: Nobiletin promotes megakaryocytic differentiation
through the MAPK/ERK-dependent EGR1 expression and exerts
anti-leukemic effects in human chronic myeloid leukemia (CML) K562
cells. Cells. 9:8772020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Josa-Culleré L, Madden KS, Cogswell TJ,
Jackson TR, Carter TS, Zhang D, Trevitt G, Davies SG, Vyas P, Wynne
GM, et al: A phenotypic screen identifies a compound series that
induces differentiation of acute myeloid leukemia cells in vitro
and shows antitumor effects in vivo. J Med Chem. 64:15608–15628.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pinto CA, DE Sousa Portilho AJ, Barbosa
MC, DE Moraes MEA, DE Lemos JAR, Burbano RMR and Moreira-Nunes CA:
Combined therapy of ATRA and imatinib mesylate decreases BCR-ABL
and ABCB1/MDR1 expression through cellular differentiation in a
chronic myeloid leukemia model. In Vivo. 35:2661–2667. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mishra M, Thacker G, Sharma A, Singh AK,
Upadhyay V, Sanyal S, Verma SP, Tripathi AK, Bhatt MLB and Trivedi
AK: FBW7 inhibits myeloid differentiation in acute myeloid leukemia
via GSK3-dependent ubiquitination of PU.1. Mol Cancer Res.
19:261–273. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang Q, Wang L, Ran Q, Wang J, Wang C, He
H, Li L and Qi H: Notopterol-induced apoptosis and differentiation
in human acute myeloid leukemia HL-60 cells. Drug Des Devel Ther.
13:1927–1940. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dembitz V, Lalic H, Tomic B, Smoljo T,
Batinic J, Dubravcic K, Batinic D, Bedalov A and Visnjic D:
All-trans retinoic acid induces differentiation in primary acute
myeloid leukemia blasts carrying an inversion of chromosome 16. Int
J Hematol. 115:43–53. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Minamiguchi H, Fujita H, Atsuta Y, Asou N,
Sakura T, Ueda Y, Sawa M, Dobashi N, Taniguchi Y, Suzuki R, et al:
Predictors of early death, serious hemorrhage, and differentiation
syndrome in Japanese patients with acute promyelocytic leukemia.
Ann Hematol. 99:2787–2800. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shao S, Ju M, Lei J, Lu X, Li H, Wang D
and Xia C: Egr-1 inhibits colon cancer cell proliferation,
migration and invasion via regulating CDKL1 at the transcriptional
level. Oncol Rep. 46:1692021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su B, Wang X, Sun Y, Long M, Zheng J, Wu W
and Li L: miR-30e-3p promotes cardiomyocyte autophagy and inhibits
apoptosis via regulating Egr-1 during ischemia/hypoxia. Biomed Res
Int. 2020:72312432020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du K, Wu X, Ji X, Liang N and Li Z: Early
growth response 1 promoted the invasion of glioblastoma multiforme
by elevating HMGB1. J Neurosurg Sci. Dec 9–2020.(Epub ahead of
print). View Article : Google Scholar
|
|
19
|
Chen J, Zhan Y, Xu J, Wang Y and Gao Q:
EGR1 overexpression inhibits the occurrence of preeclampsia by
binding to MicroRNA-574 promoter and upregulating GAB1. Reprod Sci.
28:1112–1121. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Squires A, Atas E and Meller A: Nanopore
sensing of individual transcription factors bound to DNA. Sci Rep.
5:116432015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thiel G and Cibelli G: Regulation of life
and death by the zinc finger transcription factor Egr-1. J Cell
Physiol. 193:287–292. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang C, Hu X, Wang L, Cheng H, Lin Y,
Pang Y, Yuan W, Cheng T and Wang J: Excessive proliferation and
impaired function of primitive hematopoietic cells in bone marrow
due to senescence post chemotherapy in a T cell acute lymphoblastic
leukemia model. J Transl Med. 13:2342015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cera AA, Cacci E, Toselli C, Cardarelli S,
Bernardi A, Gioia R, Giorgi M, Poiana G and Biagioni S: Egr-1
maintains NSC proliferation and its overexpression counteracts cell
cycle exit triggered by the withdrawal of epidermal growth factor.
Dev Neurosci. 40:223–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang L, Ren R, Yang X, Ge Y, Zhang X and
Yuan H: Oncogenic role of early growth response-1 in liver cancer
through the regulation of the microRNA-675/sestrin 3 and the
Wnt/β-catenin signaling pathway. Bioengineered. 12:5305–5322. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu J, Zhong C, Luo J, Shu F, Lv D, Liu Z,
Tan X, Wang S, Wu K, Yang T, et al: HnRNP-L-regulated
circCSPP1/miR-520h/EGR1 axis modulates autophagy and promotes
progression in prostate cancer. Mol Ther Nucleic Acids. 26:927–944.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kimpara S, Lu L, Hoang NM, Zhu F, Bates
PD, Daenthanasanmak A, Zhang S, Yang DT, Kelm A, Liu Y, et al: EGR1
addiction in diffuse large B-cell lymphoma. Mol Cancer Res.
19:1258–1269. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gibbs JD, Liebermann DA and Hoffman B:
Egr-1 abrogates the E2F-1 block in terminal myeloid differentiation
and suppresses leukemia. Oncogene. 27:98–106. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Racine RR, Manalo NA, Hall JMF, Dibas A,
Raffel GD and Mummert ME: CD44 induced enhancement of phosphatase
activity and calcium influx: Modifications of EGR-1 expression and
cell proliferation. Biochem Biophys Rep. 6:172–178. 2016.PubMed/NCBI
|
|
29
|
Zhao F, Yang Z, Gu X, Feng L, Xu M and
Zhang X: miR-92b-3p regulates cell cycle and apoptosis by targeting
CDKN1C, thereby affecting the sensitivity of colorectal cancer
cells to chemotherapeutic drugs. Cancers (Basel). 13:33232021.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kou N, Liu S, Li X, Li W, Zhong W, Gui L,
Chai S, Ren X, Na R, Zeng T and Liu H: H19 facilitates tongue
squamous cell carcinoma migration and invasion via sponging
miR-let-7. Oncol Res. 27:173–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han X, Zhang JJ, Han ZQ, Zhang HB and Wang
ZA: Let-7b attenuates cisplatin resistance and tumor growth in
gastric cancer by targeting AURKB. Cancer Gene Ther. 25:300–308.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen H, Wang J, Wang H, Liang J, Dong J,
Bai H and Jiang G: Advances in the application of Let-7 microRNAs
in the diagnosis, treatment and prognosis of leukemia. Oncol Lett.
23:12022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi M, Ren X, Wang X, Wang H, Liu G, Yuan
X, Zheng S, Yu L, Pan S, Song G, et al: A novel combination of
oridonin and valproic acid in enhancement of apoptosis induction of
HL-60 leukemia cells. Int J Oncol. 48:734–746. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Song G, Shi L, Guo Y, Yu L, Wang L, Zhang
X, Li L, Han Y, Ren X, Guo Q, et al: A novel PAD4/SOX4/PU.1
signaling pathway is involved in the committed differentiation of
acute promyelocytic leukemia cells into granulocytic cells.
Oncotarget. 7:3144–3157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li L, Wang Y, Zhang X, Song G, Guo Q,
Zhang Z, Diao Y, Yin H, Liu H and Jiang G: Deubiquitinase USP48
promotes ATRA-induced granulocytic differentiation of acute
promyelocytic leukemia cells. Int J Oncol. 53:895–903.
2018.PubMed/NCBI
|
|
37
|
Chou CC and Hsu CY: Involvement of PKC in
TPA-potentiated apoptosis induction during hemin-mediated erythroid
differentiation in K562 cells. Naunyn Schmiedebergs Arch Pharmacol.
379:1–9. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang C, Guo LY, Mu D, Gong JH and Chen J:
Induction of apoptosis and erythroid differentiation of human
chronic myelogenous leukemia K562 cells by low concentrations of
lidamycin. Oncol Rep. 41:475–482. 2019.PubMed/NCBI
|
|
39
|
Tian J, Li Z, Han Y, Jiang T, Song X and
Jiang G: The progress of early growth response factor 1 and
leukemia. Intractable Rare Dis Res. 5:76–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Park YS, Kim HS, Kim JH, Choi SH, Kim DS,
Ryoo ZY, Kim JY and Lee S: NAB2-STAT6 fusion protein mediates cell
proliferation and oncogenic progression via EGR-1 regulation.
Biochem Biophys Res Commun. 526:287–292. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang M, Liao Y and Lönnerdal B: EGR-1 is
an active transcription factor in TGF-β2-mediated small intestinal
cell differentiation. J Nutr Biochem. 37:101–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Toan NK, Tai NC, Kim SA and Ahn SG:
Soluble Klotho regulates bone differentiation by upregulating
expression of the transcription factor EGR-1. FEBS Lett.
594:290–300. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Myung DS, Park YL, Kim N, Chung CY, Park
HC, Kim JS, Cho SB, Lee WS, Lee JH and Joo YE: Expression of early
growth response-1 in colorectal cancer and its relation to tumor
cell proliferation and apoptosis. Oncol Rep. 31:788–794. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zins K, Pomyje J, Hofer E, Abraham D,
Lucas T and Aharinejad S: Egr-1 upregulates Siva-1 expression and
induces cardiac fibroblast apoptosis. Int J Mol Sci. 15:1538–1553.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jeong SH, Kim HJ, Jang Y, Ryu WI, Lee H,
Kim JH, Bae HC, Choi JE, Kye YC and Son SW: Egr-1 is a key
regulator of IL-17A-induced psoriasin upregulation in psoriasis.
Exp Dermatol. 23:890–895. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yeo H, Ahn SS, Lee JY and Shin SY: EGR-1
acts as a transcriptional activator of KLK7 under IL-13
stimulation. Biochem Biophys Res Commun. 534:303–309. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Q, Salman H, Danilenko M and
Studzinski GP: Cooperation between antioxidants and
1,25-dihydroxyvitamin D3 in induction of leukemia HL60 cell
differentiation through the JNK/AP-1/Egr-1 pathway. J Cell Physiol.
204:964–974. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bestilny LJ and Riabowol KT: A role for
serine proteases in mediating phorbol ester-induced differentiation
of HL-60 cells. Exp Cell Res. 256:264–271. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu C, Calogero A, Ragona G, Adamson E and
Mercola D: EGR-1, the reluctant suppression factor: EGR-1 is known
to function in the regulation of growth, differentiation, and also
has significant tumor suppressor activity and a mechanism involving
the induction of TGF-beta1 is postulated to account for this
suppressor activity. Crit Rev Oncog. 7:101–125. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li TT, Liu MR and Pei DS: Friend or foe,
the role of EGR-1 in cancer. Med Oncol. 37:72019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gashler A and Sukhatme VP: Early growth
response protein 1 (Egr-1): Prototype of a zinc-finger family of
transcription factors. Prog Nucleic Acid Res Mol Biol. 50:191–224.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Minatohara K, Akiyoshi M and Okuno H: Role
of immediate-early genes in synaptic plasticity and neuronal
ensembles underlying the memory trace. Front Mol Neurosci.
8:782016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fan YY, Ye GH, Lin KZ, Yu LS, Wu SZ, Dong
MW, Han JG, Feng XP and Li XB: Time-dependent expression and
distribution of Egr-1 during skeletal muscle wound healing in rats.
J Mol Histol. 44:75–81. 2013.PubMed/NCBI
|
|
54
|
Ho LC, Sung JM, Shen YT, Jheng HF, Chen
SH, Tsai PJ and Tsai YS: Egr-1 deficiency protects from renal
inflammation and fibrosis. J Mol Med (Berl). 94:933–942. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xu JH, Lu SJ, Wu P, Kong LC, Ning C and Li
HY: Molecular mechanism whereby paraoxonase-2 regulates coagulation
activation through endothelial tissue factor in rat haemorrhagic
shock model. Int Wound J. 17:735–741. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yan SF, Fujita T, Lu J, Okada K, Shan Zou
Y, Mackman N, Pinsky DJ and Stern DM: Egr-1, a master switch
coordinating upregulation of divergent gene families underlying
ischemic stress. Nat Med. 6:1355–1361. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gibbs JD, Liebermann DA and Hoffman B:
Leukemia suppressor function of Egr-1 is dependent on transforming
oncogene. Leukemia. 22:1909–1916. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Maifrede S, Magimaidas A, Sha X, Mukherjee
K, Liebermann DA and Hoffman B: Loss of Egr1, a human del5q gene,
accelerates BCR-ABL driven chronic myelogenous leukemia.
Oncotarget. 8:69281–69294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nguyen HQ, Hoffman-Liebermann B and
Liebermann DA: The zinc finger transcription factor Egr-1 is
essential for and restricts differentiation along the macrophage
lineage. Cell. 72:197–209. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cannell IG, Kong YW and Bushell M: How do
microRNAs regulate gene expression? Biochem Soc Trans.
36:1224–1231. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li P, Chen Y, Juma CA, Yang C, Huang J,
Zhang X and Zeng Y: Differential inhibition of target gene
expression by human microRNAs. Cells. 8:7912019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Towler BP, Jones CI and Newbury SF:
Mechanisms of regulation of mature miRNAs. Biochem Soc Trans.
43:1208–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Smolarz B, Durczyński A, Romanowicz H,
Szyłło K and Hogendorf P: miRNAs in cancer (review of literature).
Int J Mol Sci. 23:28052022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Vishnoi A and Rani S: MiRNA biogenesis and
regulation of diseases: An overview. Methods Mol Biol. 1509:1–10.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liao Q, Wang B, Li X and Jiang G: miRNAs
in acute myeloid leukemia. Oncotarget. 8:3666–3682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hahne JC and Valeri N: Non-coding RNAs and
resistance to anticancer drugs in gastrointestinal tumors. Front
Oncol. 8:2262018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chen B, Dragomir MP, Yang C, Li Q, Horst D
and Calin GA: Targeting non-coding RNAs to overcome cancer therapy
resistance. Signal Transduct Target Ther. 7:1212022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Barbato A, Iuliano A, Volpe M, D'Alterio
R, Brillante S, Massa F, De Cegli R, Carrella S, Salati M, Russo A,
et al: Integrated genomics identifies miR-181/TFAM pathway as a
critical driver of drug resistance in melanoma. Int J Mol Sci.
22:18012021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chirshev E, Oberg KC, Ioffe YJ and
Unternaehrer JJ: Let-7 as biomarker, prognostic indicator, and
therapy for precision medicine in cancer. Clin Transl Med.
8:242019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shao L, Wang R, Sun Y, Yue Z, Sun H, Wang
X, Wang P, Sun G, Hu J, Sun H, et al: Delivery of
MicroRNA-let-7c-5p by biodegradable silica nanoparticles suppresses
human cervical carcinoma cell proliferation and migration. J Biomed
Nanotechnol. 16:1600–1611. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liu P, Qi M, Ma C, Lao G and Liu Y and Liu
Y and Liu Y: Let7a inhibits the growth of endometrial carcinoma
cells by targeting Aurora-B. FEBS Lett. 587:2523–2529. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ma S, Cheng J, Wang H, Ding N, Zhou F, Ji
R, Zhu L, Zhu C and Pan Y: A novel regulatory loop
miR-101/ANXA2/EGR1 mediates malignant characteristics of liver
cancer stem cells. Carcinogenesis. 42:93–104. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yang Y, Wu F, Zhang J, Sun R, Li F, Li Y,
Chang S, Wang L, Wang X, Liu L and Huang C: EGR1 interacts with
DNMT3L to inhibit the transcription of miR-195 and plays an
anti-apoptotic role in the development of gastric cancer. J Cell
Mol Med. 23:7372–7381. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Shen X, Tang J, Hu J, Guo L, Xing Y and Xi
T: MiR-424 regulates monocytic differentiation of human leukemia
U937 cells by directly targeting CDX2. Biotechnol Lett.
35:1799–1806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zha F, Qu X, Tang B, Li J, Wang Y, Zheng
P, Ji T, Zhu C and Bai S: Long non-coding RNA MEG3 promotes
fibrosis and inflammatory response in diabetic nephropathy via
miR-181a/Egr-1/TLR4 axis. Aging (Albany NY). 11:3716–3730. 2019.
View Article : Google Scholar : PubMed/NCBI
|