Introduction
Acute leukemia is a malignant clonal disease
originating from progenitor or multi-potential progenitor cells
(1). Acute leukemia may be
classified into either myeloid or lymphoid lineages according to
the expression of several key antigens (2). Congenital and neonatal leukemia occur
rarely and are associated with high mortality rates; they may be
stratified into morphologically or genetically defined subtypes
(1). The etiology, pathogenesis
and biology have historically been poorly comprehended, reflected
in the ambiguity of classification and nomenclature (3). Acute myeloid leukemia (AML) at birth
indicates genetic abnormalities and possibly intrauterine exposure
to radiation, drugs or other toxins (3). An estimated 21,450 new cases of AML
occurred in the US in 2019 and the 5-year survival for patients
with AML is 28.3%. Females have a higher risk of developing infant
leukemia than males (4). AML
diagnosis may be made either from a peripheral blood sample or bone
marrow biopsy, depending on white blood count and circulating
blasts. Flow cytometric (FCM) analysis is helpful in the diagnosis
of AML and useful after treatment to evaluate AML persistence
(5). Specific chromosomal
rearrangements and certain site mutations have been identified in
congenital leukemia. Infants diagnosed with congenital leukemia
require thorough investigative workup and extensive supportive
care. Although the prognosis is poor, the recent use of
high-intensity multiagent chemotherapy regimens has produced
promising results (6). The present
study reported on a neonate who presented with massive hepatomegaly
and various neonatal diseases. Based on ultra-deep sequencing, the
patient was finally diagnosed with AFF1-KMT2A fusion-positive
AML.
Case report
Clinical presentation
In March 2021, a pregnant 24-year-old female patient
was admitted to Shaoxing Women and Children's Hospital (Shaoxing,
China) at 31 weeks of gestation due to reduced fetal movement for 3
days. Due to fetal distress in the uterus, a female premature
infant weighing ~1,600 g was delivered by Caesarean section with
IIIrd degree amniotic fluid contamination. The newborn was in a
comatose state with no milk-suckling, no bowel movements, no
autonomous respiration, neonatal pneumonia and non-traumatic
intracranial hemorrhage with neurological symptoms.
Laboratory findings
Hematologic exami-nation revealed that the number of
white blood cells (WBC) was increased to 617.57×109/l
[normal range (NR), 15.0-20.0×109/l], accompanied with a
red blood cell count of 3.31×1012/l (NR,
5.0-6.4×1012/l), in addition to a hemoglobin content of
75 g/l (NR, 180–190 g/l), platelet count of 64×109/l
(NR, 203–653×109/l), and a percentage of monocytes,
lymphocytes, neutrophil granulocytes, eosinophil granulocytes and
basophil granulocytes of 48.6% (NR, 3–10%), 45.6% (NR, 40–60%),
3.1% (NR, 31.0-40.0%), 0% (NR, 0.4-8.0%) and 2.7% (NR, 0–1%),
respectively.
Arterial blood gas analysis indicated hypoxemia and
acidosis. The concentration of potassium in the blood had reached
as high as 7.70 mmol/l (NR, 3.5-5.3 mmol/l). The pH value was 7.05
(NR, 7.35-7.45), and the arterial lactic acid concentration was 18
mmol/l. Biochemistry analysis of serum indicated a sharp increase
to various degrees in aspartate aminotransferase, adenosine
deaminase, alkaline phosphatase, γ-glutamyltransferase, lactic
dehydrogenase, total bilirubin total, direct bilirubin and indirect
bilirubin, which suggested severe liver damage. Their values were
342 U/l (NR, 13–35 U/l), 141 U/l (NR, 4–18 U/l), 560 U/l (NR,
48–406 U/l), 427 U/l (NR, 7–45 U/l), 8,795 U/l (NR, 120–250 U/l),
46.6 µmol/l (NR, 5.0-21.0 µmol/l), 26.6 µmol/l (NR, <3.4 µmol/l)
and 20.0 µmol/l (NR, 1.0-16.0 µmol/l), respectively.
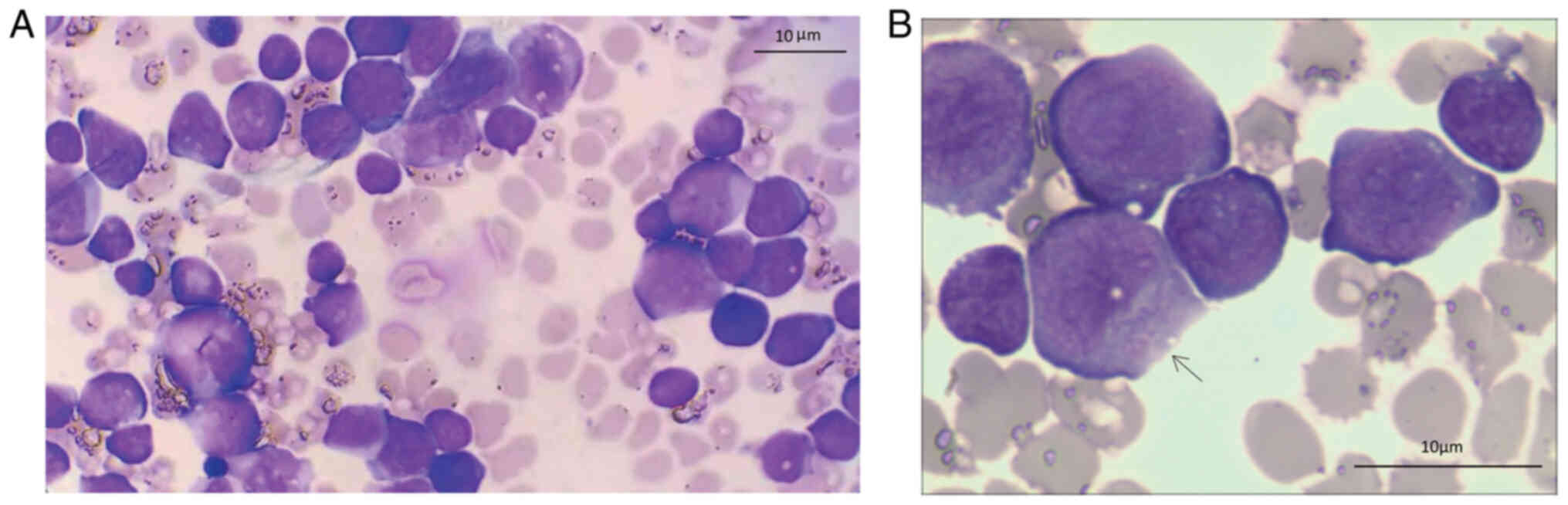
A peripheral blood smear indicating a small number
of immature cells revealed the following: Most cells, varied in
size and dyed purple on Wright staining, were circular in shape,
exhibited cytoplasm reduction, swelling of nucleus. Under oil
immersion lens of microscopy, blasts of varying sizes were observed
(Fig. 1A) and granules were seen
in the blast (labeled by black arrow) (Fig. 1B). It was concluded that the
diagnosis of the present case of neonatal leukemia was probably
AML.
Targeted panel sequencing and
bioinformatics
Sequencing was performed to determine the
pathogenesis of neonatal leukemia. Genomic DNA (gDNA) and RNA were
isolated from the patient's whole blood specimen using the QIAamp
DNA Blood Mini Kit and the PAXgene Blood RNA Kit (both from Qiagen
GmbH), respectively. For mutation analysis, gDNA of an adequate
quantity and quality was fragmented to a size ranging from 200 to
400 bp, followed by adaptor ligation. Adaptor-ligated DNA underwent
hybrid capture using a HEME mutpanel that contained 505 genes
related to hematological malignancies. For fusion analysis, a
minimum of 1 µg total RNA was subjected to rRNA depletion, followed
by canonical RNA-Seq library construction. The resulting cDNA
library was hybridized with a capture HEME-fuse panel, consisting
of 99 fusion genes. The entire capture process was performed
according to the manufacturer's protocol using reagents supplied by
Integrated DNA Technologies. The captured libraries were sequenced
with a NovaSeq 6000 (Illumina, Inc.) and 150 bp paired-end sequence
data were generated for fusion analysis. NGS service consisting 505
genes and 99 fusion genes was provided by MEDx Translational
Medicine Co. Ltd.
The sequence data were aligned to the reference
human genome (GRCh37) and subjected to adaptor trimming and
sequencing quality control. Single nucleotide variants with a
variant allele fraction >1%, as well as small insertions and
deletions <50 bp in size were detected using Varscan v2.3.9.
Possible germline polymorphisms were filtered out if the allele
frequency was > 0.1% in the Genome Aggregation Database
(http://gnomad.broadinstitute.org/).
Fusion events were analyzed using STAR-FUSION v1.5 (https://github.com/STAR-Fusion/STAR-Fusion/wiki). As
presented in Fig. 2, five missense
mutations, namely MutS homolog 6 (MSH6) K854M (Fig. 2A), Rat sarcoma of NIH3T3 (NRAS)
Q61K (Fig. 2B), phospholipase C
gamma 2 (PLCG2) T396S (Fig. 2C),
tyrosine kinase 2 (TYK2) Y1080C (Fig.
2D) and Runt related transcription factor 1 (RUNX1) S424A
(Fig. 2E), as well as AF4/FMR2
family, member 1 (AFF1)-lysine methyltransferase 2A (KMT2A) fusion
(Fig. 2F), were detected. All
somatic missense site mutations are shown in Table I.
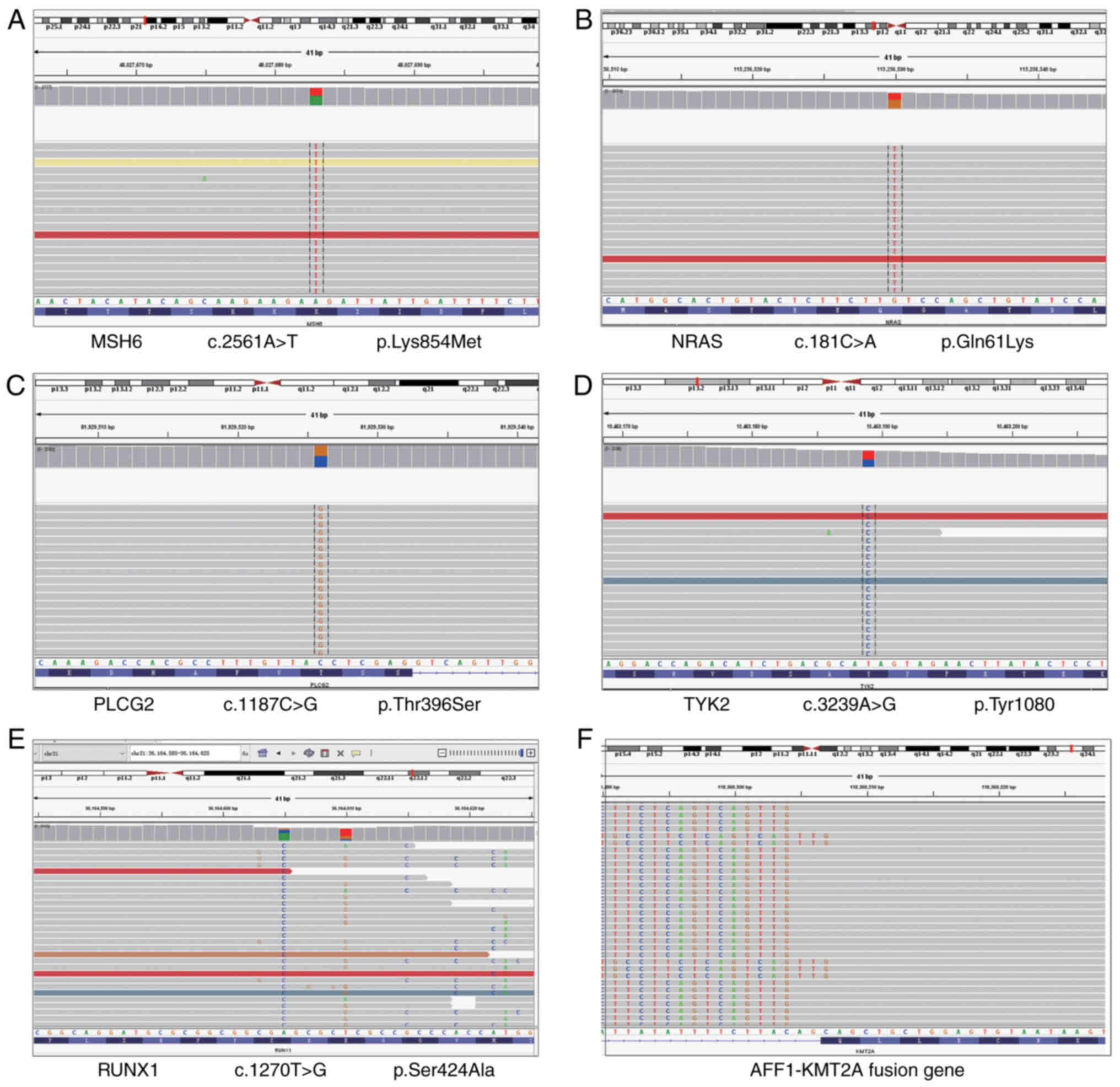 | Figure 2.Representative mutations and fusion
genes with clinical significance. (A) MSH6 K854M, (B) NRAS Q61K,
(C) PLCG2 T396S, (D) TYK2 Y1080C, (E) RUNX1 S424A and (F)
AFF1-KMT2A fusion. MSH6, MutS homolog 6; NRAS, rat sarcoma of
NIH3T3; PLCG2, phospholipase C gamma 2; TYK2, tyrosine kinase 2;
RUNX1, Runt-related transcription factor 1; AFF1, AF4/FMR2 family,
member 1; KMT2A, lysine methyltransferase 2A. |
 | Table I.List of all site somatic mutations of
the patient. |
Table I.
List of all site somatic mutations of
the patient.
| Gene | Transcript no. | Nucleotide | Amino acid | Exon location | Variation
pattern | Variation ratio
(%) |
|---|
| KDR | NM_002253.3 | c.3724A>G | p.Ile1242Val | Exon 28 | Missense
mutation | 51.54 |
| CD79B | NM_001039933.1 | c.221A>C | p.Asn74Thr | Exon 3 | Missense
mutation | 51.17 |
| NTRK3 | NM_001012338.1 | c.278C>T | p.Thr93Met | Exon 4 | Missense
mutation | 49.47 |
| PLCG2 | NM_002661.1 | c.1187C>G | p.Thr396Ser | Exon 13 | Missense
mutation | 49.20 |
| MLH3 | NM_001040108.1 | c.1879T>C | p.Phe627Leu | Exon 2 | Missense
mutation | 48.33 |
| KCNT1 | NM_020822.1 | c.3139G>A | p.Val1047Ile | Exon 27 | Missense
mutation | 47.66 |
| TYK2 | NM_003331.4 | c.3239A>G | p.Tyr1080Cys | Exon 23 | Missense
mutation | 47.62 |
| CROCC | NM_014675.4 | c.3635G>A | p.Arg1212His | Exon 24 | Missense
mutation | 47.26 |
| GRIK4 | NM_014619.4 | c.664T>G | p.Ser222Ala | Exon 7 | Missense
mutation | 46.96 |
| DLC1 | NM_182643.2 | c.1664T>C | p.Val555Ala | Exon 9 | Missense
mutation | 46.04 |
| KRT79 | NM_175834.2 | c.266G>A | p.Gly89Asp | Exon 1 | Missense
mutation | 45.85 |
| MLLT10 | NM_004641.3 | c.2552C>T | p.Thr851Ile | exon2 1 | Missense
mutation | 45.68 |
| NSD1 | NM_022455.4 | c.1852A>G | p.Lys618Glu | Exon 5 | Missense
mutation | 45.58 |
| MSH6 | NM_000179.2 | c.2561A>T | p.Lys854Met | Exon 4 | Missense
mutation | 43.96 |
| NRAS | NM_002524.4 | c.181C>A | p.Gln61Lys | Exon 3 | Missense
mutation | 43.65 |
| HYDIN | NM_001270974.1 | c.11803C>T | p.Gln3935* | Exon 70 | Nonsense
mutation | 43.65 |
| CSMD3 | NM_198123.1 | c.7520A>G | p.Lys2507Arg | Exon 48 | Missense
mutation | 42.53 |
| CACNA1G | NM_018896.4 | c.4382G>A | p.Arg1461Gln | Exon 23 | Missense
mutation | 42.32 |
| CACNA1B | NM_000718.3 | c.2990C>T | p.Thr997Met | Exon 19 | Missense
mutation | 22.14 |
| RUNX1 | NM_001754.1 | c.1270T>G | p.Ser424Ala | Exon 9 | Missense
mutation | 11.25 |
| MLLT1 | NM_005934.3 | c.805A>C | p.Lys269Gln | Exon 6 | Missense
mutation | 3.85 |
Interventions, outcomes and
lessons
All of the tests and clinical manifestation
indicated that the newborn was suffering severe asphyxia, shock,
neonatal pneumonia, leukemia, hypoxic-ischemic encephalopathy,
intracranial haemorrhage, hypoglycaemia and acidosis. The patient
was shifted to salvage chemotherapy with blood pressure and blood
sugar maintenance, as well as oxygen supply to relieve seizures,
cerebral edema and syndrome of the brain stem. Intubation and
ventilator support were administered after parental informed
consent. In spite of accurate corresponding emergency rescue
measures implemented in a timely manner, such as correction of
acidosis and anemia, as well as vitamin K1 and calcium gluconate
injection, the neonate died two and a half days after birth. FCM
and bone marrow biopsy were all missed. In general, FCM analysis of
peripheral blood and bone marrow biopsy may be used for making a
definite diagnosis of leukemia; low age and a high initial WBC
count are high-risk factors (6).
Combined analysis of morphology, immunology, cytogenetics and
molecular biology for leukemia typing were insufficient. The
remaining blood samples were subjected to deep sequencing for
analysis of site mutations and fusion genes. The Institutional
Review Board of Shaoxing Women and Children's Hospital (Shaoxing,
China) approved this retrospective case study.
Discussion
Due to reduced fetal movement for 3 days and fetal
distress in the uterus, the mother was admitted to hospital at 31
weeks of gestation and gave birth to a premature female infant by
Caesarean delivery. According to the hospitalization records, the
expectant mother denied any history of exposure to toxins or
radiation and prenatal genetic testing indicated a low risk. From
the first trimester on, 0.4 mg folic acid was supplemented daily.
The results of regular prenatal detection and three-dimensional
ultrasonic imaging revealed that maternal nutrition and fetal
development were all normal until 3 days prior to hospitalization
due to decreased fetal movement.
The infant was born prematurely with a body weight
of 1.6 kg and IIIrd degree amniotic fluid contamination. The
neonate was weak due to numerous types of neonatal disease, as
mentioned earlier. Of note, the infant's WBC in the peripheral
blood outdistanced the normal range, particularly the monocyte
count, with a sharp increase. Follow-up routine peripheral blood
smear indicated that both monocytes and lymphocytes were all
atypical and immature. Hyperactive monocyte proliferation suggested
a high probability of monocytic leukemia. Children presenting with
multiple leukemias were more likely to suffer from genetic
predisposition. In order to determine the cause of the pathology of
the present case, gene detection of the hematologic tumor, was
performed and 505 genes and 75 fused genes were analysed using
targeted panel sequencing. K854M of MSH6, Q61K of NRAS, T396S of
PLCG2, Y1080C of TYK2, S424A of RUNX1 and AFF1-KMT2A fusion were
detected.
K854M in the MSH6 gene was reported to be associated
with hereditary nonpolyposis colorectal cancer (7). In a recent study, NRAS mutations were
discovered in 13% of patients with AML (152 of 1,149), and Q61K and
Q61R substitutions of NRAS frequently occurred (8). Mutations in PLCG2 are found in most
patients with chronic lymphocytic leukemia and have been assumed to
be the causative drivers of ibrutinib resistance (9). TYK2 is a member of the Janus kinase
family involved in cytokine signal transduction in immune and
haematopoietic cells. TYK2 variants were found in 25.8% of cases of
B-acute lymphoblastic leukemia, which is the most frequent
childhood cancer and accounts for 25% of adult acute leukemias
(10). The RUNX1 gene, a member of
the transcription factor family, has a critical role in myeloid
differentiation and hematopoietic stem cell emergence and
regulation (11). Mutations in
RUNX1 were detected in 9.1% of AML and 13.9% of myelodysplastic
syndrome cases (12).
A 3,969 aa nuclear protein encoded by
KMT2A/mixed–lineage leukemia (MLL) is divided into two parts
through proteolysis by Taspase1 and then dimerizes to generate the
functional unit, which is essential for normal hematopoiesis
(13). MLL rearrangements (MLL-r)
originated in utero is a devastating malignancy with a
dismal prognosis, which exhibits a clear correlation with age. It
accounts for ~70% of acute leukemias in infants. MLL-r occurs in 5%
of childhood ALL cases, 70–80% of ALL in infants, 15–20% of
childhood AML and 50% of infant AML cases. MLL-r leukemia has long
been suspected to originate from an uncommitted precursor (14). MLL-r results in the fusion of the
N-terminus of MLL with the C-terminus of a partner. A total of 79
different MLL partner genes have now been identified. In infant
ALL, 4 partner genes account for 93% of cases: AF4 (49%),
eleven-nineteen leukemia (22%), AF9 (17%) and AF10 (5%). In infant
AML, 3 partner genes account for 66% of cases: AF9 (22%), AF10
(27%) and ELL (17%) (4). MLL and
AF4 are fused in a balanced recombination event to cause the
generation of the two fusion genes MLL-AF4 and AF4-MLL. Fusion
proteins bind directly to their target gene and upregulate gene
expression by increasing H3K79me2 through disruptor of telomeric
silencing 1-like recruitment (15). The most frequent rearrangement is
MLL-AF4, which is relatively common in ALL but uncommon in AML
(16).
The age at diagnosis is an important predictor of
prognosis, regardless of the therapeutic approach (1). Infant leukemia refers to acute
leukemia diagnosed prior to 1 year of age. Infant leukemia is rare
but requires much attention from clinicians to due to aggressive
clinical presentation and poor prognosis (1). The patient of the present study was
prematurely delivered at our hospital and diagnosed with a series
of severe neonatal illnesses of the central nervous system,
respiratory system and hematologic system. According to the
progress notes, prenatal screening indicated no Down's syndrome
(DS) and other congenital genetic defects in the first trimester.
Pediatric patients with DS have an increased risk of both ALL and
AML (17). Children are vulnerable
to the neurotoxic effects of chemicals, radioactive exposure,
certain elements and heavy metals, air pollutants and amniotic
fluid contamination, particularly in the prenatal period. The
mother denied any history of the above-mentioned items, and the
mother and her fetus were healthy in the early pregnancy. The
etiology of infant leukemia is not fully clarified and cannot be
fully explained; single-gene and chromosomal defects are only
partially responsible for the pathology of the present case. In the
present study, sequencing was used to gain insight into the
cellular and molecular factors that drive neonatal leukemia. Based
on all the available data, AFF1-KMT2A fusion may be the key factor,
while missense mutations at other sites may have contributed to the
pathogenesis. Contaminated amniotic fluid and poor living
conditions may also contribute to progression and exacerbation. All
mothers-to-be must avoid contact with any harmful factors mentioned
above.
Acknowledgements
Not applicable.
Funding
The study was supported by the Science Technology Department of
Zhejiang Province, China (grant no. LGF22H190009) and the Health
Commission of Zhejiang Province, China (grant nos. 2020KY325,
2022KY411 and 2020RC128).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BQ and JD designed the study and obtained funding
support. BQ, XD and JD performed the research; BQ analyzed data and
wrote the manuscript. BQ, XD and JD confirm the authenticity of all
the raw data All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study involving human participants was reviewed
and approved by the Institutional Ethics Committee of Shaoxing
Maternity and Child Health Care Hospital (Shaoxing, China).
Patient consent for publication
Written informed consent was provided by the
infant's mother.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brown P, Pieters R and Biondi A: How I
treat infant leukemia. Blood. 133:205–214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arber DA, Orazi A, Hasserjian R, Thiele J,
Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M and Vardiman JW:
The 2016 revision to the World Health Organization classification
of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Deschler B and Lubbert M: Acute myeloid
leukemia: Epidemiology and etiology. Cancer. 107:2099–2107. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brown P: Treatment of infant leukemias:
Challenge and promise. Hematology Am Soc Hematol Educ Program.
2013:596–600. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Newell LF and Cook RJ: Advances in acute
myeloid leukemia. BMJ. 375:n20262021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tasian SK, Loh ML and Hunger SP: Childhood
acute lymphoblastic leukemia: Integrating genomics into therapy.
Cancer. 121:3577–3590. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Doss CG and Sethumadhavan R: Investigation
on the role of nsSNPs in HNPCC genes-a bioinformatics approach. J
Biomed Sci. 16:422009. View Article : Google Scholar
|
|
8
|
Wang S, Wu Z, Li T, Li Y, Wang W, Hao Q,
Xie X, Wan D, Jiang Z, Wang C and Liu Y: Mutational spectrum and
prognosis in NRAS-mutated acute myeloid leukemia. Sci Rep.
10:121522020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lampson BL and Brown JR: Are BTK and PLCG2
mutations necessary and sufficient for ibrutinib resistance in
chronic lymphocytic leukemia? Expert Rev Hematol. 11:185–194. 2018.
View Article : Google Scholar
|
|
10
|
Turrubiartes-Martinez E, Bodega-Mayor I,
Delgado-Wicke P, Molina-Jiménez F, Casique-Aguirre D,
González-Andrade M, Rapado I, Camós M, Díaz-de-Heredia C, Barragán
E, et al: TYK2 Variants in B-Acute Lymphoblastic Leukaemia. Genes
(Basel). 11:14342020. View Article : Google Scholar
|
|
11
|
Haferlach T, Nagata Y, Grossmann V, Okuno
Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T,
et al: Landscape of genetic lesions in 944 patients with
myelodysplastic syndromes. Leukemia. 28:241–247. 2014. View Article : Google Scholar
|
|
12
|
Wang K, Zhou F, Cai X, Chao H, Zhang R and
Chen S: Mutational landscape of patients with acute myeloid
leukemia or myelodysplastic syndromes in the context of RUNX1
mutation. Hematology. 25:211–218. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kumar AR, Yao Q, Li Q, Sam TA and Kersey
JH: t(4;11) leukemias display addiction to MLL-AF4 but not to
AF4-MLL. Leuk Res. 35:305–309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen C, Yu W, Alikarami F, Qiu Q, Chen C,
Flournoy J, Gao P, Uzun Y, Fang L, Davenport JW, et al: Single-cell
multiomics reveals increased plasticity, resistant populations and
stem-cell-like blasts in KMT2A-rearranged leukemia. Blood.
139:2198–2211. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rice S, Jackson T, Crump NT, Fordham N,
Elliott N, O'Byrne S, Fanego MDML, Addy D, Crabb T, Dryden C, et
al: A human fetal liver-derived infant MLL-AF4 acute lymphoblastic
leukemia model reveals a distinct fetal gene expression program.
Nat Commun. 12:69052021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schwaller J: MLL-AF4+ infant leukemia: A
microRNA affair. Blood. 138:2014–2015. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Laetsch TW, Maude SL, Balduzzi A, Rives S,
Bittencourt H, Boyer MW, Buechner J, De Moerloose B, Qayed M,
Phillips CL, et al: Tisagenlecleucel in pediatric and young adult
patients with Down syndrome-associated relapsed/refractory acute
lymphoblastic leukemia. Leukemia. 36:1508–1515. 2022. View Article : Google Scholar
|