Introduction
An aging population means that cancers continue to
grow as an overall health concern for numerous nations, including
China. Pancreatic cancer is rapidly becoming one of the most common
malignant tumour types in China and patients usually have poor
prognosis (1). The most profound
issues surrounding the prognosis of these patients include the lack
of any early diagnostic methods and effective treatments for this
condition (2).
This means there is a global focus on improving
patient survival while improving overall quality of life, with the
majority of studies clearly prioritising diagnosis, therapy and
risk stratification as areas of importance. This means that the
number of novel molecular biomarkers associated with pancreatic
cancer continues to increase with the family of biomarkers
expanding to include specific mutations and abnormal gene
expression (3). DNA methylation
(4), cell-free DNA (5) and exosomes (6) have also all been identified as
potential research hotspots; however, despite this growth, there is
still only a small number of markers that may be used in the
clinical setting to support pancreatic cancer management. This
means that the current clinical diagnosis and treatment continues
to rely on imaging and tumour biomarker assays for identification.
However, these examinations fail to achieve early diagnosis of
pancreatic cancer, making the discovery of novel biomarkers with
high sensitivity and specificity (7) a top priority for the field as a
whole. This means that it is important to explore all novel
biomarkers associated with the occurrence and development of
pancreatic cancer. This has been aided by the continuous
development of bioinformatics and the establishment of several
public databases, which enable an increasing number of researchers
to use bioinformatics analysis to evaluate the molecular genetic
data from this disease and identify potential molecular markers for
clinical application.
One example for this is the application of weighted
gene co-expression network analysis (WGCNA) across several cancer
types to identify potential molecular drivers in these conditions
(7). In the present study, the
expression profile array data and WGCNA were combined to identify
potential molecular biomarkers for pancreatic cancer in an effort
to provide novel diagnostic targets. Furthermore, the
differentially expressed microRNAs (miRNAs/miRs) in clinical
samples were experimentally verified, allowing for an added layer
of validation, unique from similar gene expression omnibus
(GEO)-based evaluations (7).
Materials and methods
Data retrieval and analysis
The miRNA expression profile array dataset GSE85589
was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds/) and serum miRNA
expression data were obtained for 88 patients with pancreatic
cancer and 19 healthy controls. Differentially expressed miRNAs
were screened using R software and differential expression was
considered significant when the absolute value of
log2FoldChange and false discovery rate (FDR) reached ≥1
and ≤0.05, respectively.
WGCNA
Serum miRNA expression data from 88 patients with
pancreatic cancer were obtained from GSE85589 and WGCNA analysis
was performed as previously described (8).
cDNA synthesis and quantitative PCR
(qPCR)
A total of 2 ml serum was obtained from each of the
10 healthy controls (6 males, 4 females) and 16 patients with
pancreatic ductal adenocarcinoma (PDAC) (8 males, 8 females)
undergoing treatment at the Shanxi Tumor Hospital (Taiyuan, China)
between January 2018 and January 2020. The serum samples of healthy
control subjects without tumors were collected from the Health
Examination Center, Shanxi Tumor Hospital (Taiyuan, China). Serum
samples of patients were collected from patients newly diagnosed
with PDAC and with no history of other malignancies. The healthy
controls and patients with pancreatic cancer were matched in terms
of sex and age. Total RNA was extracted according to a previous
report (9). RNA was then
reverse-transcribed using a First-Strand cDNA synthesis kit
(Agilent Technologies, Inc.) according to the manufacturer's
protocol. This cDNA was then amplified by qPCR using
SYBR® Green PCR master mix (Applied Biosystems; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The experiment was performed in an ABI 7500 PCR machine (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and the thermocycling
conditions as follows: 95°C for 10 min, and 40 cycles of 95°C for
15 sec and 60°C for 1 min. U6 was used as an internal control. The
relative miRNA expression levels were then determined by comparing
the fold change between patients with pancreatic cancer and healthy
controls using the 2−ΔΔCq method (10). These experiments were performed in
triplicate and the primer sequences were as follows: miR-4668-5p
forward, 5′-TCGGCAGGAGGGAAAAAAAAAA-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACAT-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Statistical analysis
SPSS software (version 21.0; IBM Corporation) was
used in each of the statistical evaluations and miR-4668-5p
expression was compared between patients with pancreatic cancer and
healthy individuals using an unpaired t-test. Differentially
expressed miRNAs were screened using R software via the ‘impute’
and ‘limma’ packages. P<0.05 was considered to indicate a
statistically significant difference.
Results
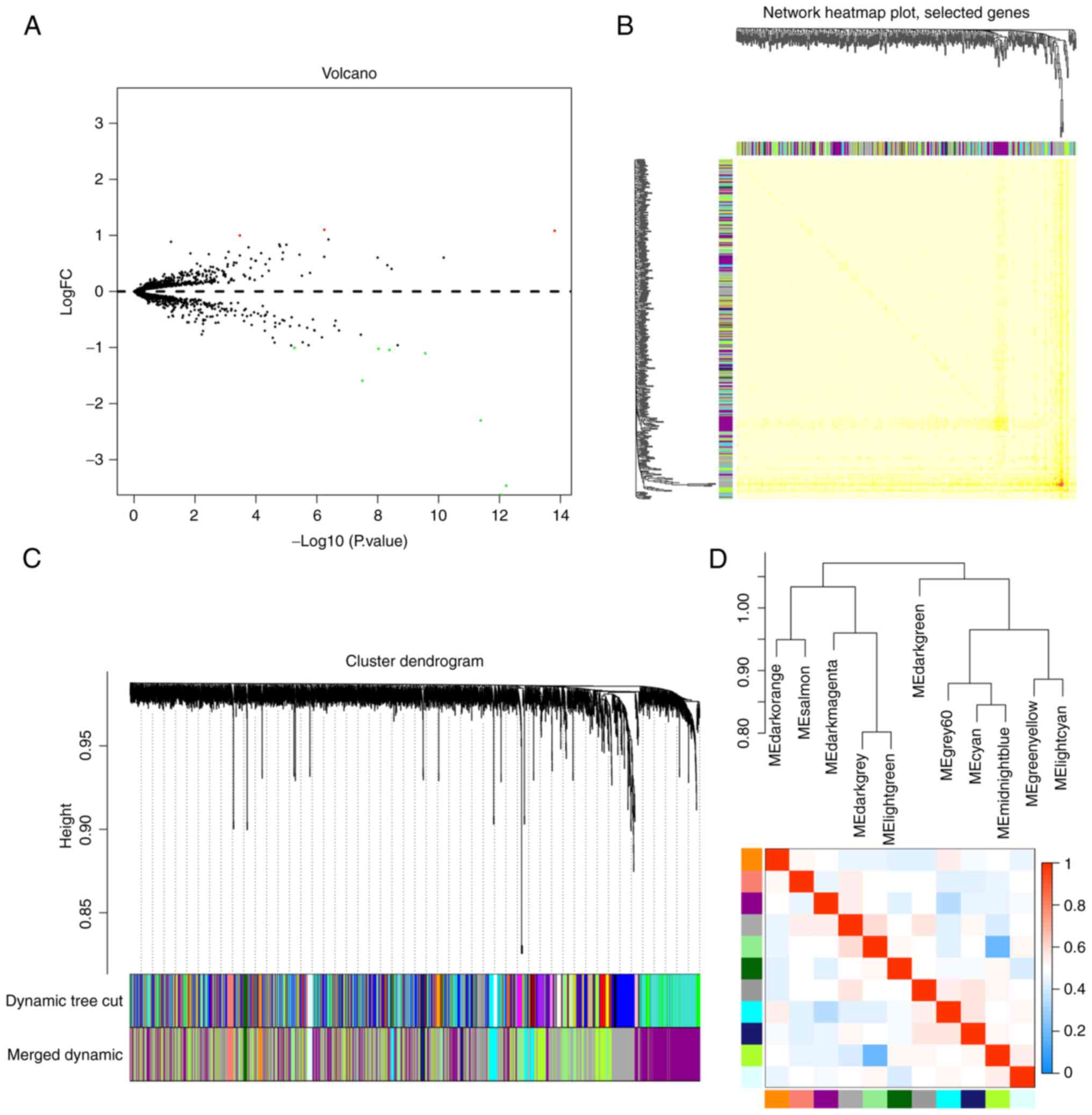
Screening identified 11 dysregulated
miRNAs in patients with pancreatic cancer
Serum miRNA expression levels in patients with
pancreatic cancer (n=88) were compared with those of a healthy
control cohort (n=19) in the GEO dataset (Fig. 1A). The comparisons identified 11
clearly dysregulated miRNAs in these samples, including miR-155-5p,
miR-4668-5p, miR-3613-3p, miR-3201, miR-548ac, miR-486-5p,
miR-548a-3p, miR-8084, miR-455-3p, miR-6068 and miR-1246. Of these,
miR-4668-5p, miR-3613-3p, miR-3201, miR-548ac, miR-486-5p,
miR-548a-3p, miR-8084, miR-6068 and miR-1246 were upregulated in
the serum of patients with pancreatic cancer, whereas miR-155-5p
and miR-455-3p were downregulated (FDR ≤0.05; Table I).
 | Table I.Differentially expressed miRNAs from
the gene expression omnibus database. |
Table I.
Differentially expressed miRNAs from
the gene expression omnibus database.
| miRNA |
|Log2FC| | Average
expression | t | P-value | Adjusted P-value |
|---|
|
miR-155-5pa | 1.08 | 0.85 | 8.86 |
1.55×10−14 |
4.00×10−11 |
|
miR-4668-5pb | 3.46 | 6.48 | −8.15 |
6.05×10−13 |
7.80×10−10 |
|
miR-3613-3pb | 3.63 | 6.58 | −8.07 |
9.27×10−13 |
7.96×10−10 |
| miR-3201b | 2.30 | 2.59 | −7.78 |
4.14×10−12 |
2.67×10−9 |
|
miR-548acb | 1.10 | 1.91 | −6.94 |
2.76×10−10 |
1.19×10−7 |
|
miR-486-5pb | 1.04 | 6.62 | −6.39 |
4.08×10−9 |
1.17×10−6 |
|
miR-548a-3pb | 1.02 | 2.09 | −6.21 |
9.44×10−9 |
2.18×10−6 |
| miR-8084b | 1.59 | 2.14 | −5.95 |
3.19×10−8 |
6.32×10−6 |
|
miR-455-3pa | 1.10 | 4.71 | 5.31 |
5.67×10−7 |
7.39×10−5 |
| miR-6068b | 1.00 | 3.02 | −4.78 |
5.44×10−6 |
4.38×10−4 |
| miR-1246b | 1.00 | 4.86 | 3.70 |
3.39×10−4 |
1.07×10−2 |
miR-4668-5p is associated with a
multitude of modules during WGCNA
The WGCNA package provides R functions for weighted
correlation network analysis, which allowed the investigation of
the co-expression patterns of the various miRNAs identified in the
88 patient samples. These evaluations identified 11 clusters
(modules) of highly correlated genes, which were highlighted as
follows: Dark orange, salmon, dark magenta, dark grey, light green,
dark green, grey 60, cyan, midnight blue, green yellow and light
cyan. A heat map of these miRNAs was also generated (Fig. 1B-D) and most of the previously
identified miRNAs were grouped into the dark grey, dark magenta and
light green modules (Table II).
The module membership value for each of these miRNAs was then
calculated and they were used to identify the most significantly
linked modules in this data set (P≤0.05; Table II). Of note, these evaluations
identified miR-4668-5p as the transcript with the highest number of
associations, with this transcript linked to various modules
including the dark magenta, dark grey, light green, grey 60 and
green yellow modules. Therefore, it was suggested that miR-4668-5p
may serve as a potential molecular biomarker for pancreatic
cancer.
 | Table II.Models and MM value of these
differentially expressed miRNAs. |
Table II.
Models and MM value of these
differentially expressed miRNAs.
| Model/miRNAs |
|
| MM P<0.05 |
|
|
|---|
| Darkgrey |
|
|
|
|
|
|
hsa-miR-4668-5p | Darkmagenta | Darkgrey | Lightgreen | Grey60 | Greenyellow |
|
hsa-miR-3613-3p | Darkmagenta | Darkgrey | Lightgreen | Greenyellow |
|
|
hsa-miR-6068 | Greenyellow | grey60 | Lightgreen | Darkgrey |
|
| Darkmagenta |
|
|
|
|
|
|
hsa-miR-548ac | Darkmagenta | Darkgrey | Cyan |
|
|
|
hsa-miR-3201 | Darkmagenta | Cyan |
|
|
|
|
hsa-miR-548a-3p | Darkmagenta | Darkgrey |
|
|
|
|
hsa-miR-8084 | Darkmagenta |
|
|
|
|
| Lightgreen |
|
|
|
|
|
|
hsa-miR-155-5p | Lightgreen |
|
|
|
|
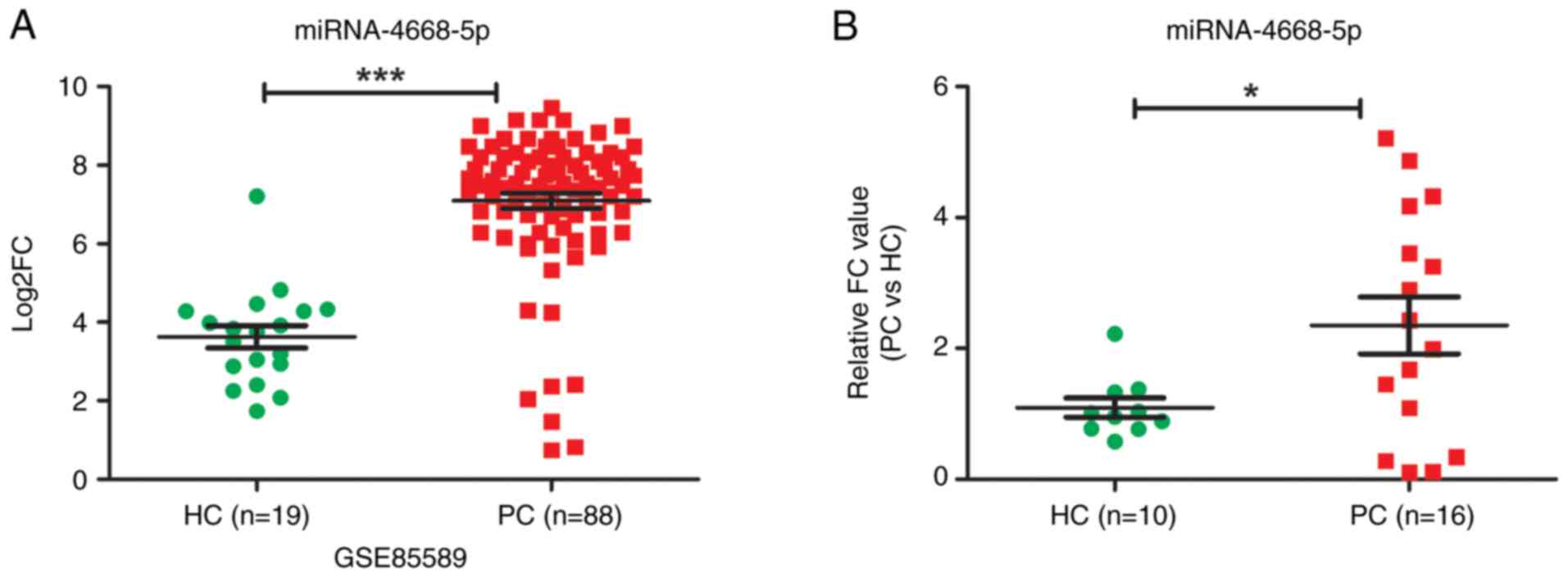
miR-4668-5p is consistently
upregulated in serum of patients with pancreatic cancer
The GEO and WGCNA analysis of the present study
suggested that miR-4668-5p may be a key marker for pancreatic
cancer. And evaluations of GSE85589 revealed a significant
upregulation of miR-4668-5p in patients with pancreatic cancer
(n=88) when compared to the healthy controls (n=19) (P≤0.0001;
Fig. 2A). Given this, the study
went on to validate the expression levels of miR-4668-5p using an
RT-qPCR assay. The results confirmed that miR-4668-5p expression
was consistently upregulated in the serum of patients with
pancreatic cancer (n=16) when compared with that of healthy
controls (n=10) (P≤0.05; Fig.
2B).
Discussion
The present study was designed to identify a subset
of co-expressed miRNAs that may help to differentiate between
pancreatic cancer and healthy tissues when applied in a liquid
biopsy setting. The study relied on the WGCNA-based evaluation of
GEO data to identify these differentially expressed transcripts and
facilitate their further evaluation. These analyses identified 11
differentially expressed miRNAs in patients with pancreatic cancer
and revealed that miR-4668-5p was likely a central
regulator/effector, as it presented with the most associations and
modular links to other miRNAs. This suggests that miR-4668-5p may
be a potential molecular biomarker for pancreatic cancer. Given
this, its upregulation was confirmed in patients with pancreatic
cancer using RT-qPCR, which validated the initial identification
supporting the notion that this transcript may be a valuable marker
for diagnosis of pancreatic cancer.
It has been indicated that patients with
incidentally diagnosed pancreatic cancer have a better prognosis
than those who are diagnosed upon developing symptoms (11), which explains the clear benefits of
early diagnosis in improving prognosis. Although there are numerous
novel diagnostic methods, liquid biopsy continues to be a firm
favourite, as this method is largely non-invasive and is designed
to be convenient, economical and minimally traumatic (12). However, liquid biopsy relies on the
identification of disease-specific biomarkers. To date, no
clinically useful biomarkers have been reported for pancreatic
cancer. Given this, in the present study the non-coding RNA profile
array for patients with pancreatic cancer was downloaded from the
GEO database and a weighted screening of the various differentially
expressed miRNAs was performed in this dataset. The present
evaluation identified 11 differentially expressed miRNAs in these
samples, which were then further narrowed down to the key
transcripts using WGCNA. Of note, the present evaluations included
the construction of a co-expression network and modules, designed
to increase the current understanding of the host-pathogen
relationship. WGCNA has gradually been used to construct various
co-expression networks (13),
particularly for miRNAs. One example of this is the identification
of several hub miRNAs associated with prognosis in colorectal
cancer (14,15). Here, WGCNA was used to construct
various modules and co-expression networks with the aim of
identifying particularly well-connected transcripts, as a proxy for
their importance in this pathology. These evaluations reduced the
target miRNAs from 11 to just one transcript miR-4668-5p. This
suggests that this transcript is likely key to this pathology and
thus is a solid target for liquid biopsy development in the
future.
Dysregulated miR-4668-5p expression has been
observed in the peripheral blood and tissues of numerous diseases,
including hepatocellular carcinoma, where it is indicated that this
miRNA may have a role in cancer progression, vascular invasion and
immune surveillance, as well as the potential coordination of other
upregulated miRNAs (16).
miR-4668-5p has also been observed to be dysregulated in gastric
cancer (17) and dedifferentiated
liposarcoma tissues (18) when
compared to healthy controls. Although abnormal miR-4668-5p
expression has been observed in several malignant tumour types, the
molecular mechanism involved in the occurrence and progression of
these tumours remains elusive. Relevant research is currently
limited and more work is required in the future. From a clinical
perspective, the relationship between miRNA expression, clinical
stage and survival time of patients with pancreatic cancer will be
investigated in a future study. A study of the effects of miRNA
overexpression on the biological behaviour of malignant tumour
cells in vivo and in vitro may also be performed with
the aim to uncover the target genes and regulatory pathways
impacted by this transcript with a focus on understanding its role
in tumour progression. It is also worth noting that miR-4668-5p is
also dysregulated in various diseases of the nervous system,
including Alzheimer's disease (10) and mesial temporal lobe epilepsy
with hippocampal sclerosis (19).
However, studies evaluating miR-4668-5p expression in pancreatic
cancer remain limited.
Despite its clear contribution, it is important to
note that the present study does have certain limitations,
including the fact that it is better to combine miRNA expression
and clinical data, such as those on overall survival, clinical
stage, etc., when evaluating the diagnostic/prognostic value. In
addition, the potential mechanism underlying miR-4668-5p-mediated
regulation of its target genes and their functions remain undefined
and should be prioritised moving forward. Finally, these
observations require to be confirmed in larger disease cohorts to
determine the diagnostic capacity.
In conclusion, the present study was the first to
indicate that miR-4668-5p is a differentially expressed miRNA
consistently upregulated in the serum of patients with pancreatic
cancer and the first to suggest that this transcript may be a
potential diagnostic marker for pancreatic cancer. The present data
may help facilitate the development of novel diagnostics for this
condition in the future.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The public datasets analysed are available from the
sources stated above. Apart from that, data sharing is not
applicable to this article, as no datasets were generated during
the current study.
Authors' contributions
PL performed the primary bioinformatics analysis and
experiments and wrote the manuscript; ZH and HZ made substantial
contributions to data analysis and technical support. JL made
substantial contributions to conception and design, and revising
the manuscript critically for important intellectual content, and
given final approval of the version to be published, and agreed to
be accountable for all aspects of the work. PL and JL checked and
confirmed the authenticity of the raw data. All authors contributed
to the article and read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The study was approved by the ethics committee of
the Shanxi Tumour Hospital (Taiyuan, China; no. 2022JC19) and the
patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jia X, Du P, Wu K, Xu Z, Fang J, Xu X and
Lin K: Pancreatic cancer mortality in China: Characteristics and
prediction. Pancreas. 47:233–237. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGuigan A, Kelly P, Turkington RC, Jones
C, Coleman HG and McCain RS: Pancreatic cancer: A review of
clinical diagnosis, epidemiology, treatment and outcomes. World J
Gastroenterol. 24:4846–4861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Previdi MC, Carotenuto P, Zito D, Pandolfo
R and Braconi C: Noncoding RNAs as novel biomarkers in pancreatic
cancer: What do we know? Future Oncol. 13:443–453. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mishra NK and Guda C: Genome-wide DNA
methylation analysis reveals molecular subtypes of pancreatic
cancer. Oncotarget. 8:28990–29012. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vietsch EE, Graham GT, McCutcheon JN,
Javaid A, Giaccone G, Marshall JL and Wellstein A: Circulating
cell-free DNA mutation patterns in early and late stage colon and
pancreatic cancer. Cancer Genet. 218–219. 39–50. 2017.PubMed/NCBI
|
|
6
|
Zhao C, Gao F, Weng S and Liu Q:
Pancreatic cancer and associated exosomes. Cancer Biomark.
20:357–367. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chu LC, Goggins MG and Fishman EK:
Diagnosis and detection of pancreatic cancer. Cancer J. 23:333–342.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ruan QF, Jiang MJ, Ye ZQ, Zhao CL and Xie
WG: Analysis of microRNA expression profile in serum of patients
with electrical burn or thermal burn. Zhonghua Shao Shang Za Zhi.
33:37–42. 2017.(In Chinese). PubMed/NCBI
|
|
10
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou B, Xu JW, Cheng YG, Gao JY, Hu SY,
Wang L and Zhan HX: Early detection of pancreatic cancer: Where are
we now and where are we going? Int J Cancer. 141:231–241. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chu D and Park BH: Liquid biopsy:
Unlocking the potentials of cell-free DNA. Virchows Arch.
471:147–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhai X, Xue Q, Liu Q, Guo Y and Chen Z:
Colon cancer recurrence-associated genes revealed by WGCNA
co-expression network analysis. Mol Med Rep. 16:6499–6505. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yepes S, López R, Andrade RE,
Rodriguez-Urrego PA, Lopez-Kleine L and Torres MM: Co-expressed
miRNAs in gastric adenocarcinoma. Genomics. 108:93–101. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou XG, Huang XL, Liang SY, Tang SM, Wu
SK, Huang TT, Mo ZN and Wang QY: Identifying miRNA and gene modules
of colon cancer associated with pathological stage by weighted gene
co-expression network analysis. Onco Targets Ther. 11:2815–2830.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pascut D, Krmac H, Gilardi F, Patti R,
Calligaris R, Croce LS and Tiribelli C: A comparative
characterization of the circulating miRNome in whole blood and
serum of HCC patients. Sci Rep. 9:82652019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bibi F, Naseer MI, Alvi SA, Yasir M,
Jiman-Fatani AA, Sawan A, Abuzenadah AM, Al-Qahtani MH and Azhar
EI: microRNA analysis of gastric cancer patients from Saudi Arabian
population. BMC Genomics. 17 (Suppl 9):S7512016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fricke A, Cimniak AFV, Ullrich PV,
Becherer C, Bickert C, Pfeifer D, Heinz J, Stark GB, Bannasch H,
Braig D and Eisenhardt SU: Whole blood miRNA expression analysis
reveals miR-3613-3p as a potential biomarker for dedifferentiated
liposarcoma. Cancer Biomark. 22:199–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yan S, Zhang H, Xie W, Meng F, Zhang K,
Jiang Y, Zhang X and Zhang J: Altered microRNA profiles in plasma
exosomes from mesial temporal lobe epilepsy with hippocampal
sclerosis. Oncotarget. 8:4136–4146. 2017. View Article : Google Scholar : PubMed/NCBI
|