Introduction
Autophagy, already defined as an intracellular
catabolic phenomenon, is considered to be involved in many
pathophysiological processes, such as infection, autoimmune
disease, neurodegenerative disorders, aging, cell death, and cancer
(1–4). In the neoplastic field, it is well
established that autophagy may exert a dual role, suppressing or
contributing to tumorigenesis (5–10).
During the autophagic process, many important autophagy-related
proteins (ATGs) are involved either in its induction or the
assembly, formation, and degradation of autophagosomes (11–14).
Among the ATGs, a multifunctional protein considered autophagy
adaptor is represented by p62, also named sequestosome 1 (SQSTM 1)
(15,16); in particular, this protein may
directly interact with microtubule-associated protein light chain 3
(LC3), and further, it may be specifically degraded by autophagy
(15). Contrastingly, a defective
autophagic phenomenon may produce a p62 upregulation in human
tumors (17,18). Some reports have documented an
evident p62 expression in pancreatic, hepatocellular, mammary, and
oral squamous carcinomas, in which aggressive clinicopathological
features and poor prognosis have been referred (16–20).
In the light of these observations, it has been hypothesized that
p62 may promote the progression of cancer by repressing the
apoptotic resistance and generating reactive oxygen species (ROS),
thus, enhancing cell proliferation, tumorigenesis, and metastasis
(20–23).
In the central nervous system (CNS), p62 has been
mainly investigated in neurodegenerative disorders, such as
Parkinson's and Alzheimer's diseases, as mitochondrial dysfunction
is implicated in the pathogenesis of these disorders (24–26).
Although previous reports have hypothesized the action of SQSTM1 as
a regulator of mitochondrial function (27,28),
additional studies on cell lines have raised some doubts concerning
the exact role of p62 (29,30).
Moreover, the function of p62 in the progression of glioma is not
fully understood, even if in glioma stem-like cells, p62 should
regulate invasion by modulating energy metabolism and affecting
mitochondrial function (31,32).
However, some data about p62 level in different glial neoplastic
samples have been reported (15,33,34);
remarkably, an increase in p62 expression has been progressively
detected from low- to high-grade gliomas with prognostic value
(15,33,34),
although no correlation with isocitrate dehydrogenase (IDH)
mutation status has been documented (15). In detail, a high p62
immunohistochemical expression has been reported in 34/81 primary
high-grade gliomas and these patients had a lower mean of three
years of overall survival (33).
Moreover, a p62 immunoreactivity has been documented in 55/96
primary high grade (III and IV) gliomas with a positive correlation
with overall survival and the proteins
O6-methylguanine-DNA methyltransferase (MGMT) and
telomerase reverse transcriptase (TERT) promoter (34).
Glioblastoma (GBM) represents the most aggressive
entity in CNS tumors, in which the IDH-wild-type constitutes over
90% of GBM, with a median overall survival (OS) ranging from 12 to
18 months and the 5-year survival of about 5% (35–37).
The gold standard for the treatment of newly diagnosed GBM consists
of maximal surgical exeresis, followed by concurrent
radiotherapy/temozolomide (TMZ) and six-monthly cycles of adjuvant
TMZ (38–40). If the tumor progresses after
first-line therapy, a recurrent GBM (rGBM) occurs which makes the
treatment a challenge, although many new drugs have been tested for
their efficacy (40).
MGMT, a DNA repair protein, removes the alkylation
at the O6 position of guanine which is the most
cytotoxic lesion induced by alkylating agent chemotherapy, such as
nitrosoureas or temozolomide (TMZ) (8,9).
However, some studies have compared the methylation status of the
promoter for DNA repair protein MGMT in newly diagnosed tumors with
matched recurrence samples after TMZ treatment (41–43).
Low-level expression of MGMT protein impairs their ability to
repair DNA. Hyper-methylation of MGMT gene promoter might result in
silencing gene expression and further down-regulate protein
concentrations (42,43). Few studies have analyzed if the
MGMT methylation status of GBM might change during the disease
course, with conflicting data and variable rates of change (5–40%)
(41,42,44).
Nevertheless, it is unclear whether this transition from methylated
to unmethylated and vice versa in GBM recurrent tumors may be a
result of TMZ treatment on MGMT status or due to the selection for
a more drug-tolerant clone, or a mixture of both processes
(43,45).
In the light of the above-mentioned well-known
information concerning p62 immunoexpression and MGMT status, we
have thought to perform as novelty an analysis regarding p62
immunohistochemical pattern in a cohort of IDH1/2 wild-type GBM,
either in primary or recurrent, to verify if its expression is
maintained or changed in relation to a potential association with
relapse-free survival (RFS) or overall survival (OS). Additionally,
the relationship between MGMT status and p62 immunoexpression in
primary and corresponding recurrent IDH1/2 wild-type GBM has been
analyzed, considering the rate of change of both parameters.
Materials and methods
Ethics approval
The study was conducted in accordance with Good
Clinical Practice guidelines and the Declaration of Helsinki (1975,
revised in 2013); its retrospective nature did not require any
informed consent, although written informed consent had been
obtained from each patient before surgical procedures. The clinical
information had been retrieved from the patients' medical records
and pathology reports. Patients' initials or other personal
identifiers did not appear in any image. Finally, all samples were
anonymized before histology and immunohistochemistry. Formal
ethical approval was obtained from the Catania 1 Ethics Committee
(Catania, Italy; protocol code: 166/2015/PO;17/12/2015).
Case selection
From the archives of the Department of Human
Pathology of Adult and Evolutive Age (University of Messina,
Messina, Italy) and the Department of Medical, Surgical Sciences,
and Advanced Technologies ‘G.F. Ingrassia’ (University of Catania,
Catania, Italy), 40 consecutive patients (26 men, 14 women; mean
age, 55.85 years; range, 35–73 years) surgically treated for naȉve
IDH1/2 wild-type GBM were included in the present analysis.
Initially, during routine pathology diagnostics, IDH1/2 status was
analyzed by immunohistochemistry utilizing mouse monoclonal
antibody IDH1 R132H (work dilution 1:50, clone H09, Dianova GmbH,
catalogue n. 075874). Furtherly, the IDH1/2 wild type status on the
same casuistry was verified utilizing IDH1/2 mutation detection kit
for real-time PCR (EntroGen, product code IDH-RT38). For all cases,
primary as well as recurrent neoplasms were available and
histologically reviewed by two independent observers according to
World Health Organization (WHO) 2016 criteria. Clinical
characteristics of each patient, including age, sex, MGMT promoter
methylation status assessed by quantitative polymerase chain
reaction, disease-free interval, and overall survival were
available from the medical records of our institution.
Immunohistochemistry
For immunohistochemical procedures, 5-micron thick
sections obtained from corresponding tissue blocks were
deparaffinized, then washed in descending alcohol scale, treated
with 3% hydrogen peroxide for 10 min, washed again in deionized
water for three times, and incubated with normal sheep serum to
prevent unspecific adherence of serum proteins for 30 min at room
temperature. Subsequently, sections were washed with deionized
water and incubated for 30 min at 37°C with commercially obtained
against primary anti-human antisera mouse monoclonal
anti-SQSTM1/p62 antibody (work dilution 1:200, clone 2C11, Abcam,
catalogue n. ab 56416). Next, the sections were washed three times
with PBS and incubated with a biotinylated goat anti-mouse IgG
secondary antibody (1:300; Abcam, catalogue n. ab7064) for 20 min
at room temperature, subsequently incubated with horseradish
peroxidase-labeled secondary antibody for 30 min and developed with
diaminobenzidine tetrahydrochloride and counterstained with
hematoxylin using the ULTRA Staining system (Ventana Medical
Systems). Negative controls were obtained by omitting the specific
antisera and substituting PBS for the primary antibody. The
assessment of p62 immunoreactivity was evaluated according to the
intensity and percentage of positively stained cells, as elsewhere
reported (2,4). The cytoplasmic and nuclear
immunostaining intensity was rated as follows: 0, negative; 1,
weak; and 2, strong. The percentage of positively stained cells was
graded as follows: grade 0, 0–5%; grade 1, >5–25%; grade 2,
>25–50%; grade 3, >50–75%; and grade 4, >75–100% for all
antibodies. The immunoreactive score was calculated by adding the
staining intensity score and the percentage score of positively
stained cells (0–6). Tumors with an immunoreactive score of 0–3
were classified as negative, and those with a score of 4–6 were
classified as positive. The immunohistochemical staining samples
were independently scored by two pathologists (AI and GT), who were
blinded to patient outcomes and other clinical findings, using a
Zeiss Axioskop microscope (Carl Zeiss Microscopy GmbH) at 40×
objective magnification. The interobserver agreement for p62
immunohistochemistry staining had a kappa value ranging from
0.73-0.80 (substantial agreement). One patient was considered p62
positive if primary or corresponding recurrent GBM showed protein
expression.
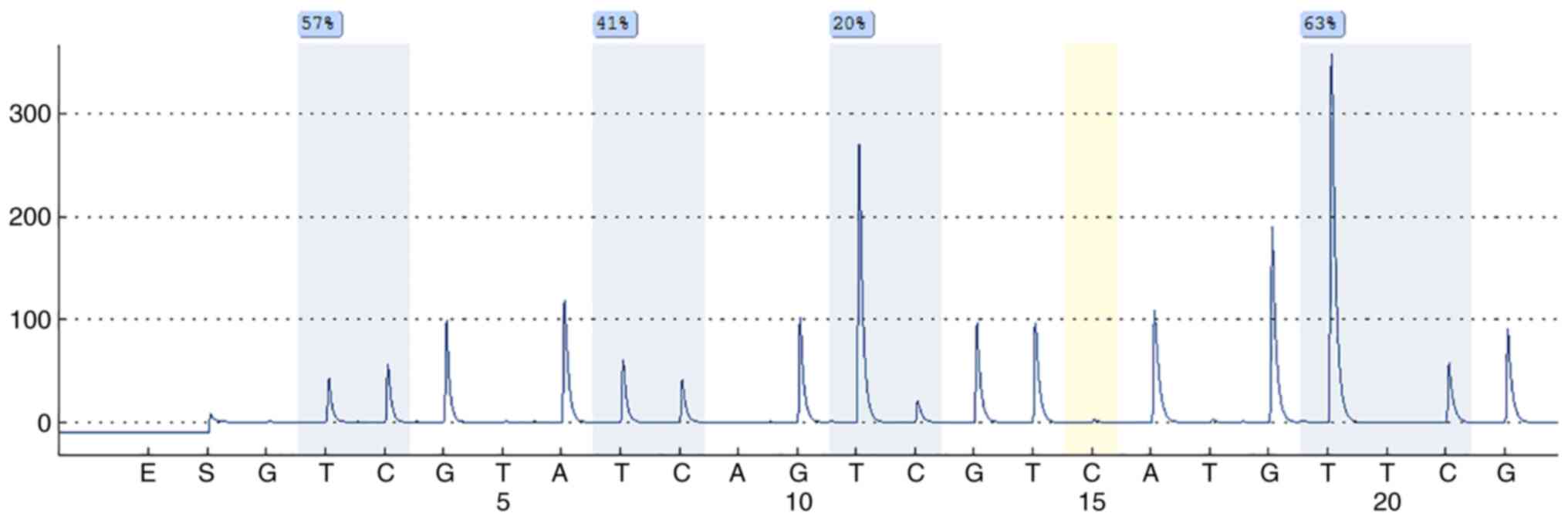
MGMT pyrosequencing analysis
The MGMT analysis was done on the DNA extracted from
paraffin-embedded tumor samples after bisulfite treatment and PCR
amplification with primers specific for exon 1 of MGMT.
Preliminarily, unmethylated cytosine residues were converted to
uracil with bisulfite treatment of 500 ng DNA using the Epi Tect
Bisulfite Kit (Qiagen) and the QiaCube automated purification
system (Qiagen) according to the manufacturer's recommendation. The
Therascreen MGMT Pyro Kit and the PyroMark Q24 system (Qiagen) were
used to assess the methylation status of the MGMT gene promoter.
Briefly, bisulfite-converted genomic DNA was amplified by PCR, the
amplicons were immobilized on streptavidin beads, and
single-stranded DNA was prepared, sequenced, and finally analyzed
on the PyroMark Q24 System. The cut-off frequency for accepting
methylation as positive was determined as elsewhere reported
(46).
Statistical analysis
Statistical evaluation was performed using the SPSS
version 13.0 software package (SPSS, Inc.). The association between
p62 expression in GBM patients and clinicopathological features
(age, sex, tumor site, MGMT status) was analyzed using the
Chi-square (χ2) or Fisher exact test. Cancer-specific
survival analysis was performed by the Kaplan-Meier method, and for
comparison of the survival curves, the Mantel-Cox log-rank test was
used. A multivariate analysis (Cox regression model) was utilized
to determine the independent effects of variables on overall
survival. P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinicopathological parameters, as well as
immunohistochemical data on p62 expression, are summarized in
Table I. In our cohort, the p62
immunoexpression was found in the nucleus and cytoplasm of
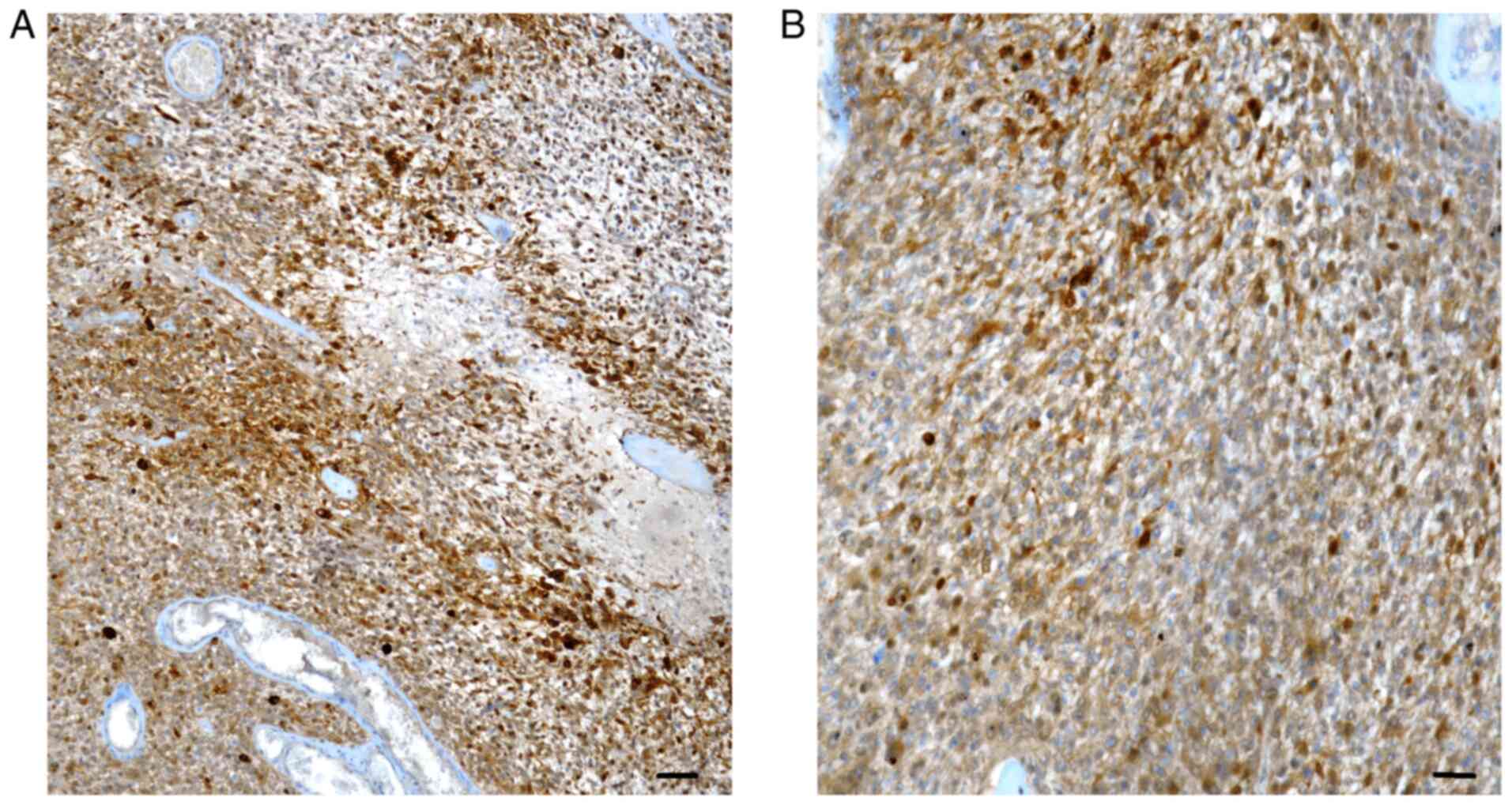
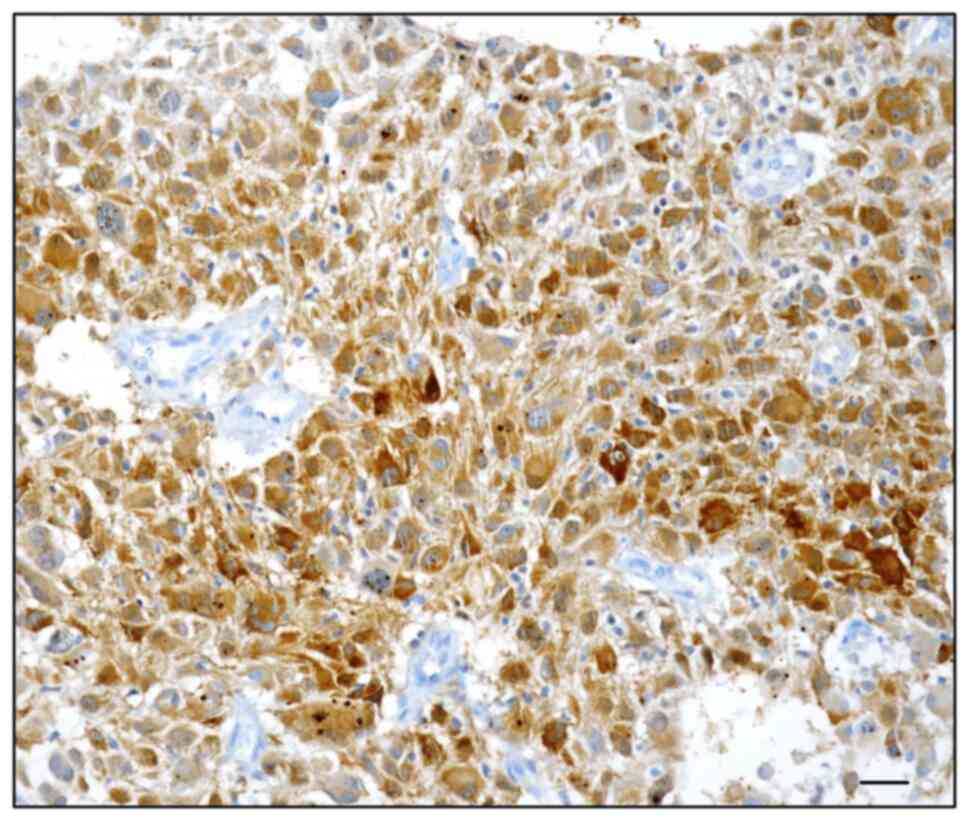
neoplastic elements in 18/40 (45%) primary (Fig. 1A and B) and 22/40 (55%) recurrent
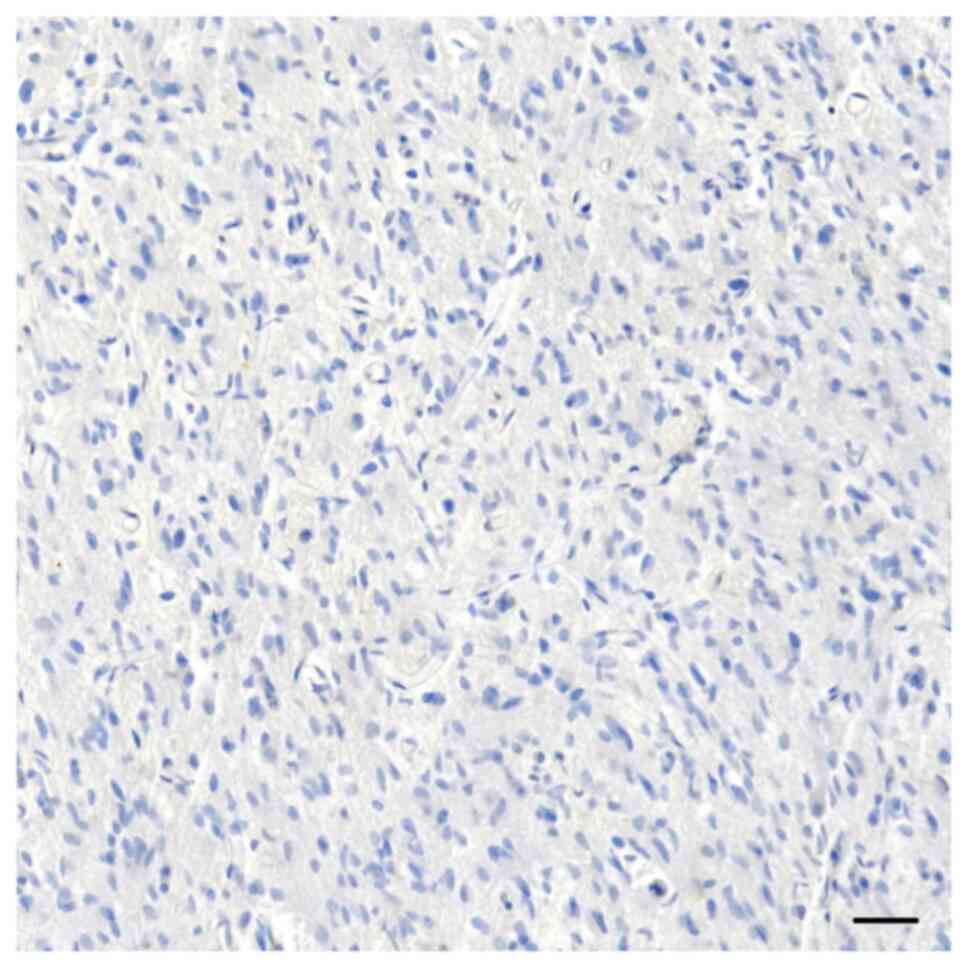
GBM (Fig. 2). By contrast, 22
primary GBM, as well as 18 recurrent GBM were consistently
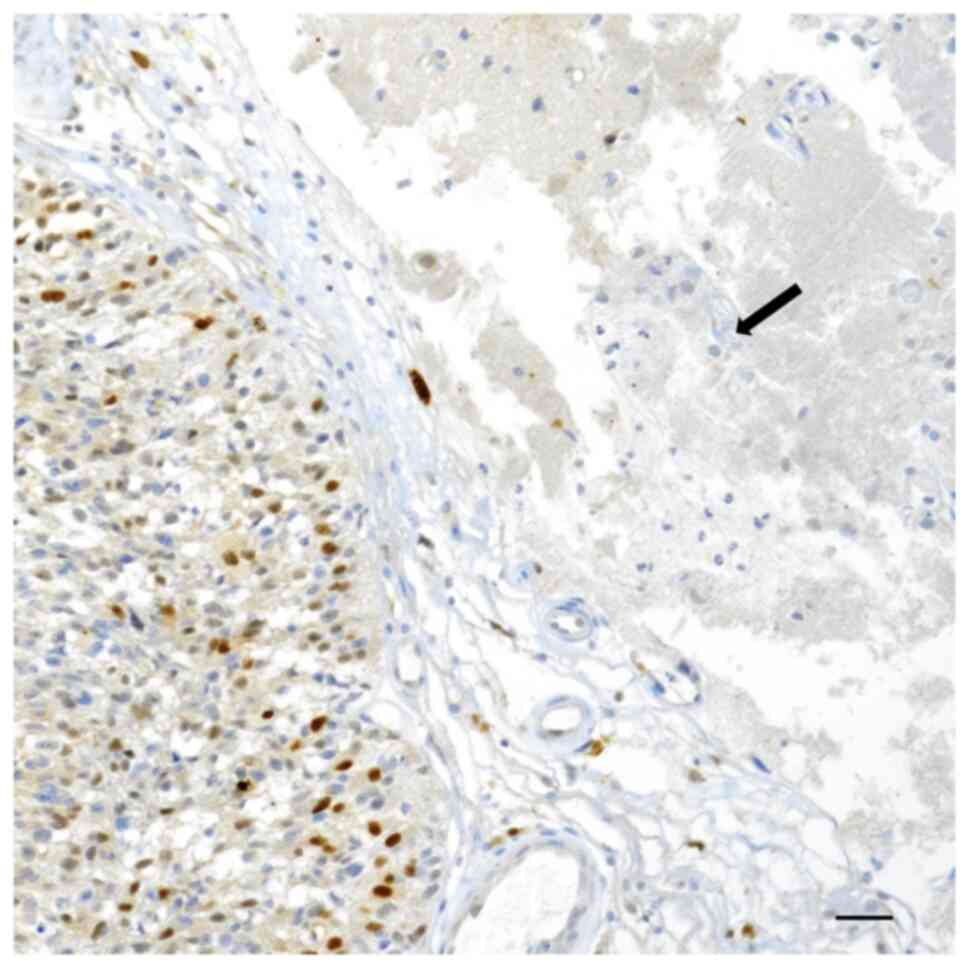
unstained (Fig. 3). Moreover,
healthy normal nervous tissue neighboring GBM exhibited a constant
p62 negative immunostaining (Fig.
4).
 | Table I.Clinicopathological parameters in
relation to p62 expression in 40 glioblastoma patients. |
Table I.
Clinicopathological parameters in
relation to p62 expression in 40 glioblastoma patients.
| Parameter | No. | p62 expression
(%) | P-value |
|---|
| Sex |
|
| NS |
|
Male | 26 | 18 (69.2) |
|
|
Female | 14 | 9 (64.3) |
|
| Tumour site |
|
| NS |
|
Frontal | 10 | 7 (70) |
|
|
Parietal | 9 | 4 (44.4) |
|
|
Fronto-parietal | 5 | 3 (60) |
|
|
Temporal | 16 | 13 (81.3) |
|
| MGMT promoter
methylation status |
|
| <0.001 |
|
Methylated | 18 | 18 (100) |
|
|
Unmethylated | 22 | 9 (40.9) |
|
Table II showed
the concordance, either negative or positive, respectively in 13/40
(32.5%) and 13/40 (32.5%); moreover, a discordant p62
immunoreactivity was found in 14/40 (35%), of which in 5/40 (12.5%)
a change from positive to negative was encountered, while in 9/40
(22.5%) a variation from negative to positive was found (Table II). In particular, the additional
Table III offered p62 detailed
ID score for all cases analyzed, either primary or recurrent.
Therefore, analyzing the p62 expression in primary and recurrent
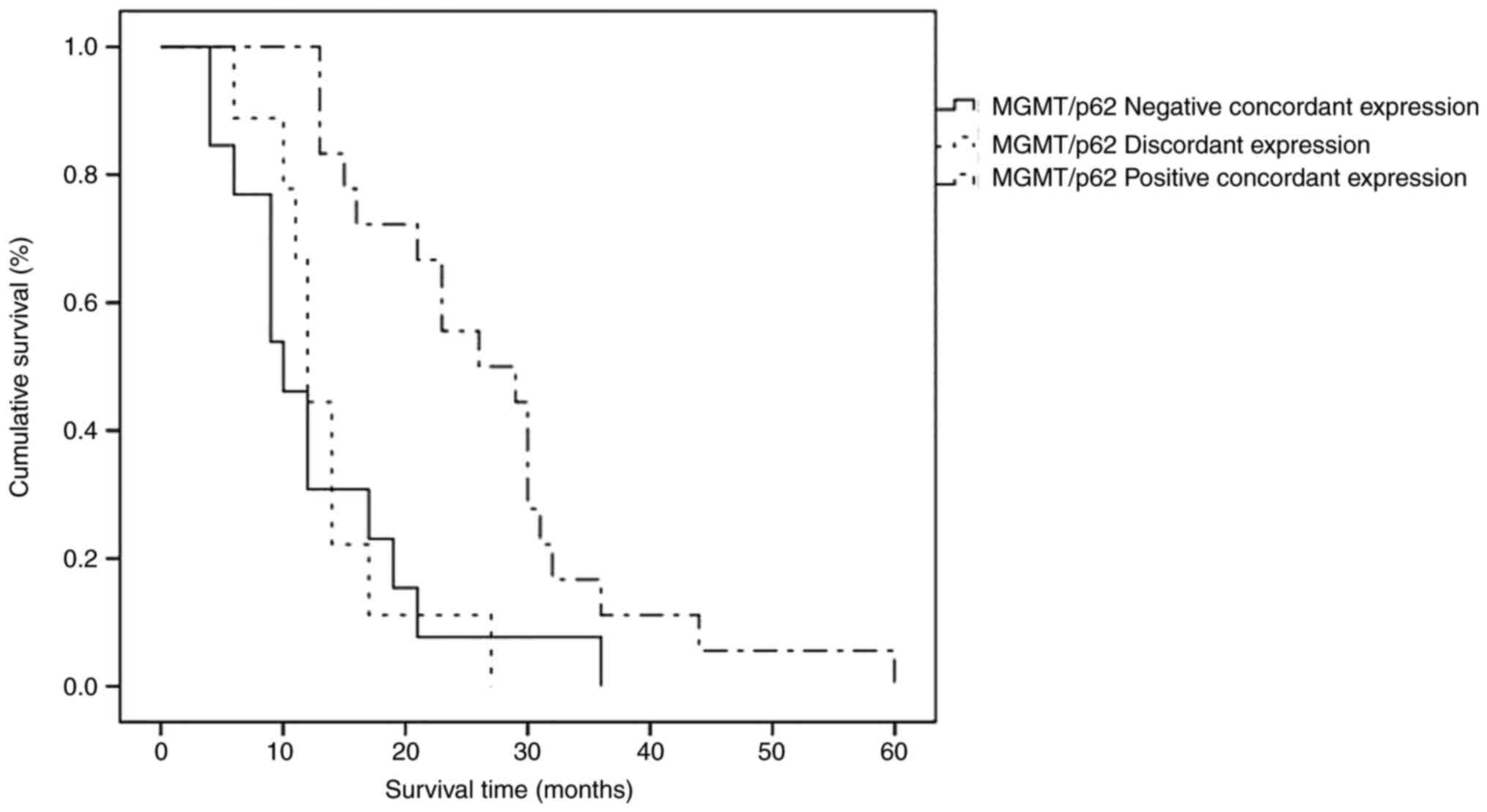
GBMs, three subgroups may be identified: Positive concordant,
negative concordant and discordant; the difference among them
(χ2=6.814) was statistically significant (P=0.033). It
merged that the most favorable prognosis was achieved when the same
GBM case was positively concordant for both parameters, in
comparison to other groups (χ2=14.538), exhibiting a
significant statistical value (P=0.001), as shown in survival
curves performed by the Kaplan-Mayer method (Fig. 5).
 | Table II.Sub-grouping for p62
immunoreactivity. |
Table II.
Sub-grouping for p62
immunoreactivity.
| Number of
cases | Primary GBM | Recurrent GBM |
|---|
| 13a | p62 +ve | p62 +ve |
| 13b | p62 -ve | p62 -ve |
| 14c | 5 p62 +ve | p62 -ve |
|
| 9 p62 -ve | p62 +ve |
 | Table III.Detailed information concerning p62
immunoreactive score either in primary or recurrent GBM. |
Table III.
Detailed information concerning p62
immunoreactive score either in primary or recurrent GBM.
| Case nr. | Age, years | Sex | Location | ID score
p62_primary | ID
scorep62_recurrence |
|---|
| 1 | 61 | F | Temporal | 0 | 0 |
| 2 | 62 | M | Parietal | 1 | 1 |
| 3 | 51 | F | Temporal | 5 | 2 |
| 4 | 64 | F | Temporal | 6 | 1 |
| 5 | 70 | F | Frontal | 0 | 0 |
| 6 | 54 | M | Temporal | 4 | 4 |
| 7 | 39 | M | Frontal | 0 | 0 |
| 8 | 53 | M |
Fronto-parietal | 6 | 6 |
| 9 | 55 | F | Temporal | 5 | 4 |
| 10 | 53 | M | Temporal | 6 | 5 |
| 11 | 62 | M |
Fronto-parietal | 2 | 2 |
| 12 | 35 | M | Frontal | 5 | 5 |
| 13 | 62 | M | Parietal | 4 | 0 |
| 14 | 61 | M | Temporal | 6 | 5 |
| 15 | 63 | M | Temporal | 4 | 5 |
| 16 | 49 | F | Frontal | 1 | 4 |
| 17 | 49 | M | Temporal | 5 | 5 |
| 18 | 52 | M | Parietal | 2 | 5 |
| 19 | 49 | M | Frontal | 1 | 4 |
| 20 | 49 | M | Temporal | 0 | 5 |
| 21 | 57 | F | Temporal | 0 | 0 |
| 22 | 70 | M | Parietal | 0 | 0 |
| 23 | 73 | M | Temporal | 1 | 6 |
| 24 | 47 | M |
Fronto-parietal | 4 | 4 |
| 25 | 57 | M | Parietal | 5 | 6 |
| 26 | 37 | F | Temporal | 4 | 0 |
| 27 | 70 | F | Temporal | 0 | 0 |
| 28 | 50 | M | Frontal | 6 | 6 |
| 29 | 65 | M | Parietal | 1 | 1 |
| 30 | 65 | M | Temporal | 2 | 6 |
| 31 | 66 | F | Frontal | 5 | 5 |
| 32 | 59 | M | Temporal | 0 | 5 |
| 33 | 66 | F | Parietal | 0 | 4 |
| 34 | 45 | M | Frontal | 1 | 1 |
| 35 | 52 | F | Parietal | 0 | 0 |
| 36 | 41 | M | Frontal | 6 | 6 |
| 37 | 52 | M | Parietal | 1 | 0 |
| 38 | 65 | M |
Fronto-parietal | 0 | 0 |
| 39 | 48 | F |
Fronto-parietal | 0 | 5 |
| 40 | 56 | F | Frontal | 4 | 0 |
Concerning MGMT status 18/40 cases showed a
methylated profile (Fig. 6). In a
univariate analysis of GBM patients, MGMT promoter methylation
status (χ2=14.517) and p62 expression
(χ2=6.590) showed a significant P-value (Table IV). By multivariate survival
analysis, only MGMT promoter methylation status emerged as an
independent prognostic parameter (Table V).
 | Table IV.Prognostic parameters examined in
glioblastoma cases: A univariate analysis of cancer-specific
mortality by Mantel-Cox log-rank test. |
Table IV.
Prognostic parameters examined in
glioblastoma cases: A univariate analysis of cancer-specific
mortality by Mantel-Cox log-rank test.
| Variable | χ2 | df | P-value |
|---|
| MGMT methylation
status | 14.517 | 1 | <0.001 |
| p62 expression | 6.590 | 1 | 0.010 |
 | Table V.Multivariate survival analysis by Cox
regression model in glioblastoma patients. |
Table V.
Multivariate survival analysis by Cox
regression model in glioblastoma patients.
| Variable | β | SE | Exp(β) RR | CI 95% Exp(β) | P-value |
|---|
| MGMT | 0.612 | 0.174 | 1.843 | 1.311-2.592 | <0.001 |
| methylation |
|
|
|
|
|
| status |
|
|
|
|
|
| p62 | - | - | - | - | 0.822 |
| expression |
|
|
|
|
|
Discussion
In the present pilot study, we have analyzed the
immunohistochemical expression of p62 in a cohort of GBM,
considering each patient as positive when this autophagic protein
was indifferently revealed in primary and/or corresponding
recurrent GBM samples. We have found a p62 immunoreaction in the
nucleus and cytoplasm of neoplastic elements in 45% of GMB primary
and 55% recurrent cases. However, a variable rate of p62
immunostaining has been elsewhere reported in primary high-grade
gliomas (33,34); specifically, the reported positive
percentage ranged from 42 to 57% (33,34).
These values are greatly superimposable with ours in primary and
recurrent GBM. In addition, our data confirm that an increase in
p62 protein was detected in about 50% of GBM cases analyzed, with a
concordant rate of 65% between primary and recurrent GBM.
Interestingly, a discordant p62 immunoreactivity was found in 35%
of GBM cases, although a variation from negative to positive and
vice versa has been documented. The occurrence of changes in
biomarker expression in tumors represents biological evidence
frequently observed in oncology. As largely documented in the
literature, a lack of concordance of oncogene expression (i.e.,
HER2) has been reported between primary and metastatic/recurrent
neoplasias, such as breast and gastric cancer (47–49);
therefore, a similar phenomenon may also be suggested in brain
gliomas. However, to explain the documented change in biomarker
expression, many different mechanisms have been hypothesized such
as intratumoral heterogeneity, clone selection promoted by
cytotoxic treatments, and lastly analytical bias (47–49).
A progressive p62 enhancement moving from the WHO
grade II to grade IV, as previously elsewhere suggested (15,33).
Moreover, p62 overexpression has been reported also in glioma cell
lines and no difference in p62 expression between IDH wild-type or
IDH mutated groups was reported, suggesting that p62 function may
be considered independent of IDH status (15,33).
Consequently, it can be argued that p62 overexpression stimulates
the classical autophagic pathway, allowing GBM cell survival by
antagonizing apoptosis and producing drug resistance to proteasome
inhibitors (17,50,51).
Alternatively, an accumulation of the autophagy substrate p62 may
reveal a defective process that cannot degrade its substrates.
Therefore, p62 may act as a tumor promoter in glioma cells not only
by the regulation of autophagy but also by interfering with
proliferation, migration, and Temozolomide resistance (15).
The 2016 classification by WHO of brain tumors
introduced new molecular markers in high-grade gliomas, such as
MGMT methylation, IDH1, TP53, and TERT promoter mutation (52); this approach may represent a
crucial point in the neoplastic strategy treatment, predicting the
sensitivity of gliomas to chemotherapy as well as the prognosis
(53–56). In the present paper, we have
combined the p62 expression and MGMT promoter methylation status to
evaluate if an association between these two parameters may be
appreciable; in detail, in relation to this point, three groups may
be identified: negative concordant, positive concordant and
discordant. Taking into consideration the suggestion that MGMT
promoter methylation presence has been considered as an independent
favorable prognostic factor in GBM, we have documented the
achievement of the most favorable prognosis when the same GBM case
was positively concordant for both p62 expression and MGMT
methylated status. Interestingly, this association is further
emphasized by the comparative analysis of primary and corresponding
recurrent GBM in relation to MGMT methylation. Therefore, a
significant association between these latter two parameters should
be hypothesized, similarly to that elsewhere reported (34). On the other hand, the univariate
analysis allowed us to identify MGMT promoter methylation status as
well as p62 expression as significant prognostic factors able to
define GBM long survivors, although only MGMT methylation emerged
as an independent marker in multivariate analysis. These data
confirm recent findings that have demonstrated a worse prognostic
behavior in GBM patients with high levels of autophagy-related
genes and MGMT promoter unmethylated (57). However, autophagy can have a tumor
suppressor function in GBM destroying damaging unfolded proteins,
oncogenic protein substrates, and injured organelles (58,59).
Recently, it has been reported that elevated levels of ATGs were
linked to better survival in glioma patients (60–62).
In particular, higher AKT and mTOR hyperphosphorylation has been
reported in high-grade gliomas in comparison to low-grade ones
(63,64). It has been suggested that mTOR
signaling pathway activation is associated with autophagy
inhibition, supporting the glioma stem cell proliferation, tumor
infiltration, and therapeutic resistance (65,66).
Although the relationship between autophagy and
programmed cell death is not fully elucidated, we may hypothesize
that the capability to repair DNA damage should be reduced by a
methylated MGMT status, and therefore, autophagy and apoptosis may
interact with each other through several pathways. However, the
coexistence observed by us of p62 expression and MGMT profile in
GBM needs to be analyzed further in their putative prognostic role,
since only a few data are available on the association between
autophagy and other synchronized mutations and therefore, in the
future, an extensive study on a larger cohort should be carried out
as the next step.
Acknowledgements
The authors would like to thank Professor Sandra De
Dominici for her assistance in reviewing the English style and
grammar of the manuscript.
Funding
This research was funded by grants from the Italian Minister of
Research and University (FFABR ANVUR 2021).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AI and GT designed the project and wrote the paper.
CP, GB, AG, GMVB and PV contributed to data collection and
analysis. RC and GG analyzed data and critically reviewed/edited
the manuscript. AI and GT confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki, and approved by the Catania 1
Ethics Committee (Catania, Italy; protocol code:
166/2015/PO;17/12/2015).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ieni A, Cardia R, Giuffrè G, Rigoli L,
Caruso RA and Tuccari G: Immunohistochemical expression of
autophagy-related proteins in advanced tubular gastric
adenocarcinomas and its implications. Cancers (Basel). 11:3892019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Broggi G, Ieni A, Russo D, Varricchio S,
Puzzo L, Russo A, Reibaldi M, Longo A, Tuccari G, Staibano S and
Caltabiano R: The macro-autophagy-related protein beclin-1
immunohistochemical expression correlates with tumor cell type and
clinical behavior of uveal melanoma. Front Oncol. 10:5898492020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ieni A, Pizzimenti C, Giuffrè G, Caruso RA
and Tuccari G: Autophagy-related prognostic signature in HER2
positive gastric carcinomas. Curr Mol Med. 22:809–818. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eskelinen EL: The dual role of autophagy
in cancer. Curr Opin Pharmacol. 11:294–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chmurska A, Matczak K and Marczak A: Two
faces of autophagy in the struggle against cancer. Int J Mol Sci.
22:29812021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Verma AK, Bharti PS, Rafat S, Bhatt D,
Goyal Y, Pandey KK, Ranjan S, Almatroodi SA, Alsahli MA, Rahmani
AH, et al: Autophagy paradox of cancer: Role, regulation, and
duality. Oxid Med Cell Longev. 2021:88325412021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gerada C and Ryan KM: Autophagy, the
innate immune response and cancer. Mol Oncol. 14:1913–1929. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yun CW, Jeon J, Go G, Lee JH and Lee SH:
The dual role of autophagy in cancer development and a therapeutic
strategy for cancer by targeting autophagy. Int J Mol Sci.
22:1792020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Amaravadi RK, Kimmelman AC and Debnath J:
Targeting autophagy in cancer: Recent advances and future
directions. Cancer Discov. 9:1167–1181. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang CW and Klionsky DJ: The molecular
mechanism of autophagy. Mol Med. 9:65–76. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mizushimaa N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell1. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng Y and Klionsky DJ: Autophagy
regulates DNA repair through SQSTM1/p62. Autophagy. 13:995–996.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Metur SP and Klionsky DJ: Autophagy under
construction: Insights from in vitro reconstitution of
autophagosome nucleation. Autophagy. 17:383–384. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Deng D, Luo K, Liu H, Nie X, Xue L, Wang
R, Xu Y, Cui J, Shao N and Zhi F: p62 acts as an oncogene and is
targeted by miR-124-3p in glioma. Cancer Cell Int. 19:2802019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chao X, Ni HM and Ding W: An unexpected
tumor suppressor role of SQSTM1/p62 in liver tumorigenesis.
Autophagy. 18:459–461. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang J, Li Y, Xia S, Li J, Yang Q, Ding K
and Zhang H: Sequestosome 1/p62: A multitasker in the regulation of
malignant tumor aggression (Review). Int J Oncol. 59:772021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li D, He C, Ye F, Ye E, He H, Chen G and
Zhang J: p62 overexpression promotes bone metastasis of lung
adenocarcinoma out of LC3-dependent autophagy. Front Oncol.
11:6095482021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thongchot S, Vidoni C, Ferraresi A,
Loilome W, Khuntikeo N, Sangkhamanon S, Titapun A, Isidoro C and
Namwat N: Cancer-associated fibroblast-derived IL-6 determines
unfavorable prognosis in cholangiocarcinoma by affecting
autophagy-associated chemoresponse. Cancers (Basel). 13:21342021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Umemura A, He F, Taniguchi K, Nakagawa H,
Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S,
Duran A, et al: p62, upregulated during preneoplasia, induces
hepatocellular carcinogenesis by maintaining survival of stressed
HCC-initiating cells. Cancer Cell. 29:935–948. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim JW, Jun SY, Kim JM, Oh YH, Yoon G,
Hong SM and Chung JY: Prognostic value of LC3B and p62 expression
in small intestinal adenocarcinoma. J Clin Med. 10:53982021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim HM and Koo JS: Autophagy-related
proteins are differentially expressed in adrenal cortical
tumor/pheochromocytoma and associated with patient prognosis. Int J
Mol Sci. 22:104902021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Perng DS, Hung CM, Lin HY, Morgan P, Hsu
YC, Wu TC, Hsieh PM, Yeh JH, Hsiao P, Lee CY, et al: Role of
autophagy-related protein in the prognosis of combined
hepatocellular carcinoma and cholangiocarcinoma after surgical
resection. BMC Cancer. 21:8282021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haack TB, Ignatius E, Calvo-Garrido J,
Iuso A, Isohanni P, Maffezzini C, Lönnqvist T, Suomalainen A, Gorza
M, Kremer LS, et al: Absence of the autophagy adaptor SQSTM1/p62
causes childhood-onset neurodegeneration with ataxia, dystonia, and
gaze palsy. Am J Hum Genet. 99:735–743. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Muto V, Flex E, Kupchinsky Z, Primiano G,
Galehdari H, Dehghani M, Cecchetti S, Carpentieri G, Rizza T,
Mazaheri N, et al: Biallelic SQSTM1 mutations in early-onset,
variably progressive neurodegeneration. Neurology. 91:e319–e330.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pytte J, Anderton RS, Flynn LL, Theunissen
F, Jiang L, Pitout I, James I, Mastaglia FL, Saunders AM, Bedlack
R, et al: Association of a structural variant within the SQSTM1
gene with amyotrophic lateral sclerosis. Neurol Genet. 6:e4062020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seibenhener ML, Du Y, Diaz-Meco MT, Moscat
J, Wooten MC and Wooten MW: A role for sequestosome 1/p62 in
mitochondrial dynamics, import and genome integrity. Biochim
Biophys Acta. 1833:452–459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bartolome F, Esteras N, Martin-Requero A,
Boutoleau-Bretonniere C, Vercelletto M, Gabelle A, Le Ber I, Honda
T, Dinkova-Kostova AT, Hardy J, et al: Pathogenic p62/SQSTM1
mutations impair energy metabolism through limitation of
mitochondrial substrates. Sci Rep. 7:16662017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Calvo-Garrido J, Maffezzini C, Schober FA,
Clemente P, Uhlin E, Kele M, Stranneheim H, Lesko N, Bruhn H,
Svenningsson P, et al: SQSTM1/p62-directed metabolic reprogramming
is essential for normal neurodifferentiation. Stem Cell Reports.
12:696–711. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Poon A, Saini H, Sethi S, O'Sullivan GA,
Plun-Favreau H, Wray S, Dawson LA and McCarthy JM: The role of
SQSTM1 (p62) in mitochondrial function and clearance in human
cortical neurons. Stem Cell Reports. 16:1276–1289. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Galavotti S, Bartesaghi S, Faccenda D,
Shaked-Rabi M, Sanzone S, McEvoy A, Dinsdale D, Condorelli F,
Brandner S, Campanella M, et al: The autophagy-associated factors
DRAM1 and p62 regulate cell migration and invasion in glioblastoma
stem cells. Oncogene. 32:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang YL, Li YF, Chou CH, Huang LC, Wu YP,
Kao Y and Tsai CK: Diosmin inhibits glioblastoma growth through
inhibition of autophagic flux. Int J Mol Sci. 22:104532021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang T and Wu Z: Immunohistochemical
assessment of autophagic protein LC3B and p62 levels in glioma
patients. Int J Clin Exp Pathol. 11:862–868. 2018.PubMed/NCBI
|
|
34
|
Tamrakar S, Yashiro M, Kawashima T, Uda T,
Terakawa Y, Kuwae Y, Ohsawa M and Ohata K: Clinicopathological
significance of autophagy-related proteins and its association with
genetic alterations in gliomas. Anticancer Res. 39:1233–1242. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ostrom QT, Adel Fahmideh M, Cote DJ,
Muskens IS, Schraw JM, Scheurer ME and Bondy ML: Risk factors for
childhood and adult primary brain tumors. Neuro Oncol.
21:1357–1375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ostrom QT, Truitt G, Gittleman H, Brat DJ,
Kruchko C, Wilson R and Barnholtz-Sloan JS: Relative survival after
diagnosis with a primary brain or other central nervous system
tumor in the national program of cancer registries, 2004 to 2014.
Neurooncol Pract. 7:306–312. 2020.PubMed/NCBI
|
|
37
|
Wen PY, Rodon JA, Mason W, Beck JT,
DeGroot J, Donnet V, Mills D, El-Hashimy M and Rosenthal M: Phase
I, open-label, multicentre study of buparlisib in combination with
temozolomide or with concomitant radiation therapy and temozolomide
in patients with newly diagnosed glioblastoma. ESMO Open.
5:e0006732020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Seyve A, Lozano-Sanchez F, Thomas A,
Mathon B, Tran S, Mokhtari K, Giry M, Marie Y, Capelle L, Peyre M,
et al: Initial surgical resection and long time to occurrence from
initial diagnosis are independent prognostic factors in resected
recurrent IDH wild-type glioblastoma. Clin Neurol Neurosurg.
196:1060062020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Le Rhun E and Weller M: Sex-specific
aspects of epidemiology, molecular genetics and outcome: Primary
brain tumours. ESMO Open. 5 (Suppl 4):e0010342020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Birzu C, French P, Caccese M, Cerretti G,
Idbaih A, Zagonel V and Lombardi G: Recurrent glioblastoma: From
molecular landscape to new treatment perspectives. Cancers (Basel).
13:472020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brandes AA, Franceschi E, Tosoni A,
Bartolini S, Bacci A, Agati R, Ghimenton C, Turazzi S, Talacchi A,
Skrap M, et al: O(6)-methylguanine DNA-methyltransferase
methylation status can change between first surgery for newly
diagnosed glioblastoma and second surgery for recurrence: Clinical
implications. Neuro Oncol. 12:283–288. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brandes AA, Franceschi E, Paccapelo A,
Tallini G, De Biase D, Ghimenton C, Danieli D, Zunarelli E, Lanza
G, Silini EM, et al: Role of MGMT methylation status at time of
diagnosis and recurrence for patients with glioblastoma: Clinical
implications. Oncologist. 22:432–437. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Storey K, Leder K, Hawkins-Daarud A,
Swanson K, Ahmed AU, Rockne RC and Foo J: Glioblastoma recurrence
and the role of O6-methylguanine-DNA methyltransferase
promoter methylation. JCO Clin Cancer Inform. 3:1–12. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Felsberg J, Thon N, Eigenbrod S, Hentschel
B, Sabel MC, Westphal M, Schackert G, Kreth FW, Pietsch T, Löffler
M, et al: Promoter methylation and expression of MGMT and the DNA
mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary
and recurrent glioblastomas. Int J Cancer. 129:659–670. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Álvarez-Torres MDM, Fuster-García E,
Balaña C, Puig J and García-Gómez JM: Lack of benefit of extending
temozolomide treatment in patients with high vascular glioblastoma
with methylated MGMT. Cancers (Basel). 13:54202021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Brigliadori G, Foca F, Dall'Agata M,
Rengucci C, Melegari E, Cerasoli S, Amadori D, Calistri D and Faedi
M: Defining the cutoff value of MGMT gene promoter methylation and
its predictive capacity in glioblastoma. J Neurooncol. 128:333–339.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ieni A, Barresi V, Caltabiano R, Caleo A,
Bonetti LR, Lanzafame S, Zeppa P, Caruso RA and Tuccari G:
Discordance rate of HER2 status in primary gastric carcinomas and
synchronous lymph node metastases: A multicenter retrospective
analysis. Int J Mol Sci. 15:22331–22341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ieni A, Barresi V, Caltabiano R, Cascone
AM, Del Sordo R, Cabibi D, Zeppa P, Lanzafame S, Sidoni A, Franco V
and Tuccari G: Discordance rate of HER2 status in primary breast
carcinomas versus synchronous axillary lymph node metastases: A
multicenter retrospective investigation. Onco Targets Ther.
7:1267–1272. 2014.PubMed/NCBI
|
|
49
|
Ieni A, Cardia R, Pizzimenti C, Zeppa P
and Tuccari G: HER2 heterogeneity in personalized therapy of
gastro-oesophageal malignancies: An overview by different
methodologies. J Pers Med. 10:102020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zeng RX, Zhang YB, Fan Y and Wu GL:
p62/SQSTM1 is involved in caspase-8 associated cell death induced
by proteasome inhibitor MG132 in U87MG cells. Cell Biol Int.
38:1221–1226. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ivankovic D, Chau KY, Schapira AH and Gegg
ME: Mitochondrial and lysosomal biogenesis are activated following
PINK1/parkin-mediated mitophagy. J Neurochem. 136:388–402. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Śledzińska P, Bebyn MG, Furtak J,
Kowalewski J and Lewandowska MA: Prognostic and predictive
biomarkers in gliomas. Int J Mol Sci. 22:103732021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Brandner S, McAleenan A, Kelly C, Spiga F,
Cheng HY, Dawson S, Schmidt L, Faulkner CL, Wragg C, Jefferies S,
et al: MGMT promoter methylation testing to predict overall
survival in people with glioblastoma treated with temozolomide: A
comprehensive meta-analysis based on a cochrane systematic review.
Neuro Oncol. 23:1457–1469. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Broggi G, Salvatorelli L, Barbagallo D,
Certo F, Altieri R, Tirrò E, Massimino M, Vigneri P, Guadagno E,
Maugeri G, et al: Diagnostic utility of the immunohistochemical
expression of serine and arginine rich splicing factor 1 (SRSF1) in
the differential diagnosis of adult gliomas. Cancers (Basel).
13:20862021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Certo F, Altieri R, Maione M, Schonauer C,
Sortino G, Fiumanò G, Tirrò E, Massimino M, Broggi G, Vigneri P, et
al: FLAIRectomy in supramarginal resection of glioblastoma
correlates with clinical outcome and survival analysis: A
prospective, single institution, case series. Oper Neurosurg
(Hagerstown). 20:151–163. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Stella M, Falzone L, Caponnetto A, Gattuso
G, Barbagallo C, Battaglia R, Mirabella F, Broggi G, Altieri R,
Certo F, et al: Serum extracellular vesicle-derived circHIPK3 and
circSMARCA5 Are two novel diagnostic biomarkers for glioblastoma
multiforme. Pharmaceuticals (Basel). 14:6182021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang QW, Liu HJ, Zhao Z, Zhang Y, Wang Z,
Jiang T and Bao ZS: Prognostic correlation of autophagy-related
gene expression-based risk signature in patients with glioblastoma.
Onco Targets Ther. 13:95–107. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Khan I, Baig MH, Mahfooz S, Rahim M,
Karacam B, Elbasan EB, Ulasov I, Dong JJ and Hatiboglu MA:
Deciphering the role of autophagy in treatment of resistance
mechanisms in glioblastoma. Int J Mol Sci. 22:13182021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Batara DCR, Choi MC, Shin HU, Kim H and
Kim SH: Friend or foe: Paradoxical roles of autophagy in
gliomagenesis. Cells. 10:14112021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Shukla S, Patric IR, Patil V, Shwetha SD,
Hegde AS, Chandramouli BA, Arivazhagan A, Santosh V and
Somasundaram K: Methylation silencing of ULK2, an autophagy gene,
is essential for astrocyte transformation and tumor growth. J Biol
Chem. 289:22306–22318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Miracco C, Cosci E, Oliveri G, Luzi P,
Pacenti L, Monciatti I, Mannucci S, De Nisi MC, Toscano M,
Malagnino V, et al: Protein and mRNA expression of autophagy gene
beclin 1 in human brain tumours. Int J Oncol. 30:429–436.
2007.PubMed/NCBI
|
|
62
|
Aoki H, Kondo Y, Aldape K, Yamamoto A,
Iwado E, Yokoyama T, Hollingsworth EF, Kobayashi R, Hess K,
Shinojima N, et al: Monitoring autophagy in glioblastoma with
antibody against isoform B of human microtubule-associated protein
1 light chain 3. Autophagy. 4:467–475. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mecca C, Giambanco I, Donato R and Arcuri
C: Targeting mTOR in glioblastoma: Rationale and
preclinical/clinical evidence. Dis Markers. 2018:92304792018.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li XY, Zhang LQ, Zhang XG, Li X, Ren YB,
Ma XY, Li XG and Wang LX: Association between AKT/mTOR signalling
pathway and malignancy grade of human gliomas. J Neurooncol.
103:453–458. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jhanwar-Uniyal M, Jeevan D, Neil J,
Shannon C, Albert L and Murali R: Deconstructing mTOR complexes in
regulation of glioblastoma multiforme and its stem cells. Adv Biol
Regul. 53:202–210. 2013. View Article : Google Scholar : PubMed/NCBI
|