Introduction
Hepatocellular carcinoma (HCC) is a common type of
malignant tumor of the digestive system, accounting for ~90% of
liver cancer cases (1). The
incidence rate of HCC has increased in recent years worldwide, with
an estimated incidence of >1 million cases by 2025 (2). It is generally accepted that multiple
risk factors may result in HCC development, such as hepatitis B and
C viral infection, cirrhosis, non-alcoholic steatohepatitis,
dietary toxins, aflatoxins and aristolochic acid (3,4). To
date, the mainstay curative treatments for patients with HCC remain
surgery, including hepatic resection and liver transplantation
(5). In addition, transarterial
chemoembolization has been the most widely used method in standard
therapy for intermediate-stage HCC over the past two decades
(6). Although significant advances
have been made in the therapeutic methods for HCC, the discovery of
useful molecular markers for HCC therapy is an urgent requirement
due to the current lack thereof (7). It is also necessary to develop novel
therapeutic targets for the treatment of patients with HCC.
Pumilio homolog 2 (PUM2) is an RNA-binding protein
that serves as a translation repressor (8). PUM2 regulates the translation or
stability of certain mRNAs by binding directly to their 3′
untranslated region (UTR) (9). A
previous study reported that PUM2 was able to bind to the 3′UTR of
sirtuin 1 (SIRT1) mRNA to inhibit SIRT1 expression in a model of
hypoxia/reoxygenation-induced cardiomyocyte injury (10). Furthermore, PUM2 suppressed kinesin
family member 18A to affect proliferation, apoptosis and the cell
cycle of human male germ cell lines (11). In addition, PUM2 expression was
indicated to be upregulated in several types of human tumor, such
as osteosarcoma (12). It was
reported that PUM2 overexpression significantly promoted the
degradation of insulinoma-associated protein 1 mRNA and inhibited
its protein expression in MCF-7 and MDA-MB-231 cells (13). In addition, Wang et al
(14) revealed that PUM2
accelerated cell proliferation and migration by targeting the 3′UTR
of B-cell translocation gene (BTG)1 mRNA in glioblastoma cells.
However, the roles of PUM2 in HCC development have remained
elusive. In the present study, the biological roles of PUM2 and its
potential mechanism of action were investigated in HCC.
Materials and methods
Bioinformatics analysis
The mRNA levels of PUM2 were analyzed in the UALCAN
database (http://ualcan.path.uab.edu) and the
significance of differences was estimated by using Student's
unpaired t-test (15). Gene
Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn) was used to analyze the
overall survival of patients with HCC (method: Overall survival;
Group cutoff: Median; Cut-off high value and low value was set to
50%; hazard ratio: Yes; confidence interval: 95%; axis units:
Months) (16). StarBase
(http://starbase.sysu.edu.cn/) was used
to predict the interactions between PUM2 and targeting RNAs
(17). RNA-Protein Interaction
Prediction (RPISeq; http://pridb.gdcb.iastate.edu/RPISeq/index.html)
was used to confirm the interaction between PUM2 and the 3′-UTR of
BTG3 and analyze the interaction probability (RPISeq predictions
are based on Random Forest or Support Vector Machine classifiers
trained and tested on 2 non-redundant benchmark datasets of
RNA-protein interactions, RPI2241 and RPI369, extracted from PRIDB,
a comprehensive database of RNA-protein complexes extracted from
the PDB) (18).
Cell culture
The HCC cell lines HCC36, HCC-T, HCC-M and HHS-89
were purchased from the American Type Culture Collection. The HCC
cell line Huh-7 and human the hepatocyte cell line HHL5 (used as
the control cell line) were obtained from the Health Science
Research Resources Bank. The cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS; Hyclone;
Cytiva), 100 U/ml penicillin and 100 µg/ml streptomycin (both
Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere
with 5% CO2 at 37°C.
Cell transfection
The specific short hairpin RNA (shRNA) targeting
PUM2 (shRNA-PUM2-1: 5′-GCAATATAGTGTTGTATAA-3′; shRNA-PUM2-2:
5′-CATAGTTGTTGACTGTTAA-3′), the specific shRNA targeting BTG3
(shRNA-BTG3-1: 5′-GATTATGTATGGAGAGAAA-3′; shRNA-BTG3-2:
5′-GATTAATCCTCACATGTTA-3′) and the corresponding control shRNA
(shRNA-NC: 5′-CCGGCAACAAGATGAAGAGCACCAACTC-3′) were synthesized
with pRNA-U6.1/Hygro vector as the plasmid backbone by Shanghai
Integrated Biotech Solutions. These recombinant nucleotides were
transfected into Huh-7 cells using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Following 48 h of incubation, the
cells were collected for subsequent experiments.
Cell counting kit-8 (CCK-8) assay
Following transfection, Huh-7 cells were placed in
96-well plates (2×104 cells/well) and cultured in DMEM
with 10% FBS at 37°C. Following incubation for 24, 48 and 72 h, 10
µl CCK-8 solution (Beyotime Institute of Biotechnology) was added
to each well and the cells were incubated for 2 h. Finally, the
absorbance of each well was detected at 450 nm with a microplate
reader (RT-3001; Thermo Fisher Scientific, Inc.).
Colony-formation assay
The cells were seeded in 6-well plates at 500
cells/well and incubated in DMEM with 10% FBS at 37°C. Following
incubation for two weeks, the plates were fixed with 4%
paraformaldehyde for 15 min at a room temperature and stained with
0.5% crystal violet (Wako Pure Chemical Industries, Ltd.) for 30
min at room temperature. The colonies were imaged and counted by
light microscopy (Olympus Corporation). The number of colonies,
defined as >50 cells/colony, was counted.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNAs were extracted from Huh-7 cells by
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) in accordance with the manufacturer's protocol (19). The concentrations of the RNA
samples were detected using NanoDrop® 3000 (Thermo
Fisher Scientific, Inc.). Subsequently, a cDNA synthesis kit
(PrimeScript RT Master Mix; Takara Bio, Inc.) was used to reverse
transcribe 2 µg RNA into cDNA following the manufacturer's
protocol. The reaction mixture was incubated at 25°C for 5 min,
42°C for 30 min and 85°C for 5 min, and then kept at 4°C for 5 min.
Amplification of the cDNA was performed by real-time qPCR using the
SYBR Premix Ex Taq™ II kit (Takara Bio, Inc.) following
the manufacturer's protocol in an ABI PRISM 7900 Real-Time system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR
program was 95°C for 3 min, followed by 35 cycles of denaturation
at 95°C for 30 sec, annealing at 60°C for 30 sec and extension at
72°C for 1 min. A final extension step at 72°C for 7 min was
performed in each PCR assay. The primer sequences for PCR were as
follows: PUM2 forward, 5′-GGGAATGGGAGAGACCATTCAA-3′ and reverse,
5′-AGGATTAGGAAGAGGCCCCA-3′; BTG3 forward,
5′-TCCACCTCTTCCAATGTGGC-3′ and reverse, 5′-TCCGGTCACAATGCATTCCA-3′;
GAPDH forward, 5′-GGGAAACTGTGGCGTGAT-3′ and reverse,
5′-GAGTGGGTGTCGCTGTTGA-3′. The relative expression levels of the
target gene were calculated by the relative quantification
(2−ΔΔCq) method (20)
and GAPDH mRNA levels were used for normalization.
Terminal deoxynucleotidyl
transferase-mediated dUTP nick-end labeling (TUNEL) assay
A TUNEL assay was performed to evaluate Huh-7 cell
apoptosis using an apoptosis detection kit (cat. no. 11684795910;
Roche Diagnostics) in accordance with the manufacturer's
guidelines. The transfected cells were fixed with 4%
paraformaldehyde for 10 min at 4°C and incubated with proteinase K
(Beijing Solarbio Science & Technology Co., Ltd.) for 15 min at
37°C. Subsequently, the cells were placed in 3%
H2O2 for 15 min at room temperature and
stained with the reagents from the TUNEL kit, followed by
counterstaining with DAPI for 10 min at room temperature. The
labeled cells were observed using fluorescence microscopy (Olympus
Corporation; magnification, ×200). The number of TUNEL-positive
(green) and DAPI-positive cells (blue nuclear stain) was visually
counted and at least 10 fields per section were examined. The
percentage of apoptotic cells was calculated as (number of
TUNEL-positive cells/total number of cells) ×100%.
Luciferase reporter assay
To obtain a 3′-UTR-luciferase reporter plasmid, the
3′UTR of BTG3 was amplified using PCR from genomic DNA of the human
HCC cell line Huh-7 (Invitrogen; Thermo Fisher Scientific, Inc.)
and cloned into the XhoI/NotI sites of the psiCHECK-2
vector (Promega Corporation) following digestion with XhoI
and NotI (Beyotime Institute of Biotechnology).
Amplification of the cDNA was performed by real-time qPCR using the
SYBR Premix Ex Taq™ II kit (Takara Bio, Inc.) following
the manufacturer's protocol in an ABI PRISM 7900 Real-Time system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
amplification condition was 95°C for 3 min, followed by 35 cycles
of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec
and extension at 72°C for 1 min, prior to a final extension step at
72°C for 7 min. The 3′-UTR-luciferase reporter plasmids and the
short hairpin (sh)RNA-PUM2 or negative control (NC) sequences were
co-transfected into Huh-7 cells using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. Following transfection, the cells were
incubated for 48 h and the dual-luciferase assay system (Promega
Corporation) was used to measure firefly and Renilla
luciferase activity levels.
RNA-binding protein
immunoprecipitation (RIP) assay
The interaction between PUM2 and BTG3 was identified
by an RIP assay using the EZ-Magna RIP™ RNA-Binding
Protein Immunoprecipitation Kit (cat. no. 17-701; MilliporeSigma)
according to the manufacturer's instructions. The cells were lysed
in complete RIP lysis buffer and the protein extract was then
prepared. Anti-PUM2 antibody (cat. no. ab92390; 1:10 dilution;
Abcam) and NC normal rabbit IgG (cat. no. NI01; 1/200 dilution;
MilliporeSigma) were incubated with the protein extract from the
lysed cells at 37°C overnight. The co-precipitated RNAs were
detected by RT-qPCR as specified above.
Western blot analysis
Total protein was extracted from cells using RIPA
buffer (Bio-Rad Laboratories, Inc.). Total protein was quantified
using a BCA assay (Beyotime Institute of Biotechnology), according
to the manufacturer's protocol. A total of 30 µg protein per lane
was loaded on 10% SDS-polyacrylamide gels and after
electrophoresis, proteins were transferred to polyvinylidene
membranes (MilliporeSigma). The membranes were blocked in 5%
non-fat milk (Beyotime Institute of Biotechnology) at room
temperature for 2 h and incubated with primary antibodies against
PUM2 (1:1,000 dilution; cat. no. ab92390), Bcl-2 (1:1,000 dilution;
cat. no. ab32124), Bax (1:1,000 dilution; cat. no. ab32503),
cleaved caspase 3 (1:500 dilution; cat. no. ab32042), caspase 3
(1:1,000 dilution; cat. no. ab32351), cleaved poly(ADP-ribose)
polymerase (PARP; 1:1,000 dilution; cat. no. ab32561), PARP
(1:1,000 dilution; cat. no. ab32138), BTG3 (1:1,000 dilution; cat.
no. ab112938) and GAPDH (1:1,000 dilution; cat. no. ab8245; all
from Abcam) overnight at 4°C. Finally, the membranes were incubated
with horseradish peroxidase-labeled anti-rabbit IgG (cat. no. 7074;
1:1,000 dilution; Cell Signaling Technology, Inc.) or anti-mouse
IgG (cat. no. 14709; 1:1,000 dilution; Cell Signaling Technology,
Inc.) at room temperature for 1 h. The protein bands were
visualized using an enhanced chemiluminescence detection system
(Amersham; Cytiva) according to the manufacturer's instructions.
The density of each band was quantified by ImageJ software
(v.1.8.0; National Institutes of Health).
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corporation). Values are expressed as the mean ±
standard deviation. Significant differences between two groups were
analyzed by Student's unpaired t-test, while differences among
multiple groups were analyzed using one-way analysis of variance
followed by Bonferroni's post-hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
PUM2 expression is upregulated in HCC
tissues and cells
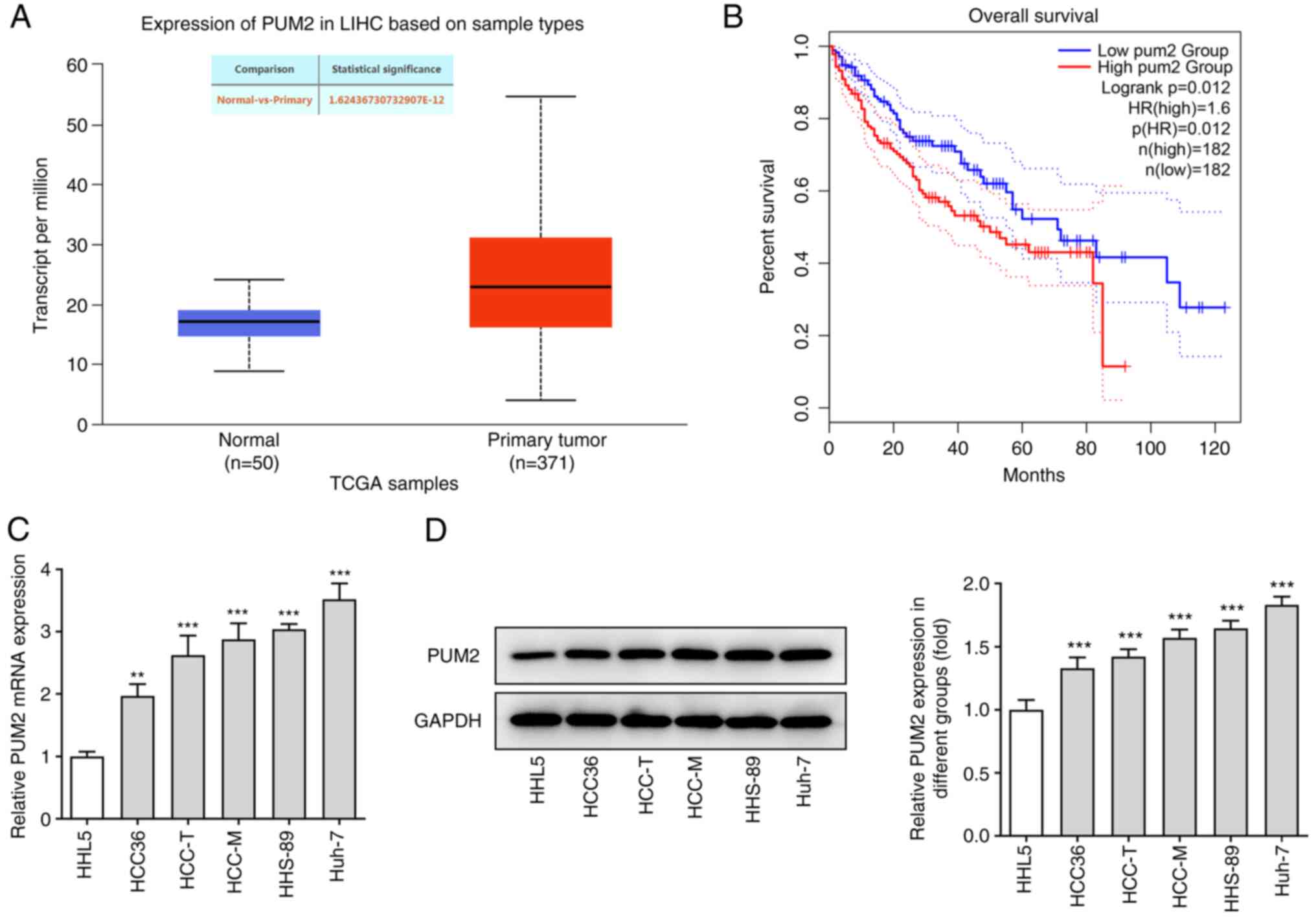
To explore the role of PUM2 in HCC development, the
expression levels of PUM2 were initially detected in a public
dataset of patients with HCC and in HCC cell lines. According to
the data obtained from the UALCAN database, PUM2 was highly
expressed in HCC tissues compared with those in the control
subjects (Fig. 1A). In addition,
analysis with the GEPIA database indicated that upregulation of
PUM2 expression was associated with poor prognosis of patients with
HCC (Fig. 1B). In addition,
RT-qPCR and western blot assays indicated significantly higher mRNA
and protein expression levels of PUM2 in HCC cell lines compared
with those in the non-cancerous control cell line. Huh-7 cells
exhibited the highest PUM2 expression level; therefore, Huh-7 cells
were selected for the subsequent experiments (Fig. 1C and D).
Inhibition of PUM2 expression reduces
cell proliferation and facilitates apoptosis in Huh-7 cells
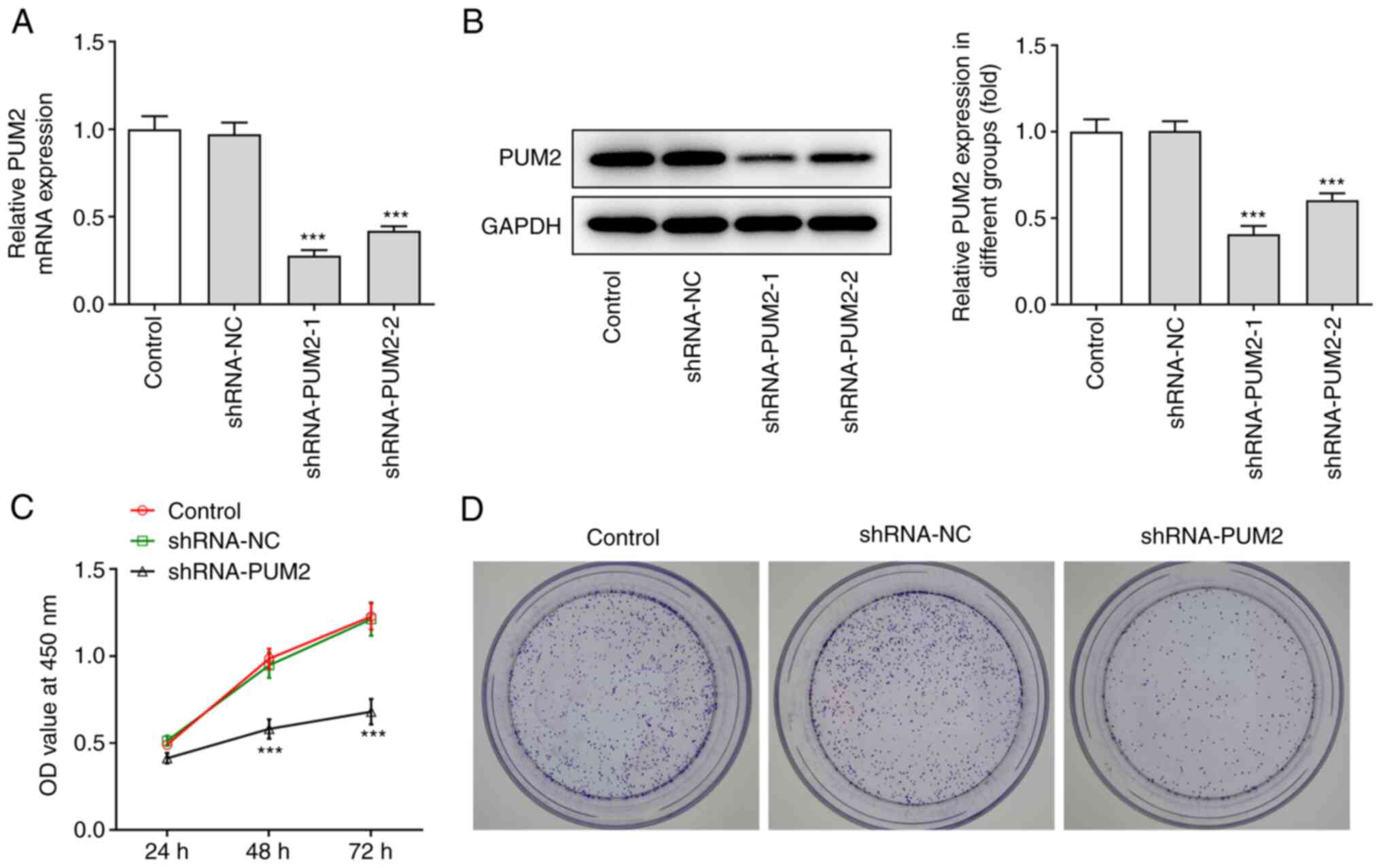
To investigate the effect of PUM2 on HCC cell
proliferation and apoptosis, specific shRNA sequences targeting
PUM2 were transfected into Huh-7 cells. The transfection efficiency
was evaluated by RT-qPCR and western blot analyses (Fig. 2A and B). The knockdown efficiency
of shRNA-PUM2-1 was better than that of shRNA-PUM2-2 and thus,
shRNA-PUM2-1 was used for the further experiments (named as
shRNA-PUM2 from here onwards). Subsequently, the CCK-8 assay was
used to assess cell proliferation. The results indicated that PUM2
silencing significantly suppressed Huh-7 cell proliferation
compared with that noted in the NC group (Fig. 2C). Furthermore, the
colony-formation assay demonstrated that the number of colonies in
PUM2-silenced cells was decreased compared with that in the NC
group (Fig. 2D). Furthermore, it
was observed that the apoptotic rate of Huh-7 cells transfected
with shRNA-PUM2 was considerably elevated. Induction of apoptosis
was accompanied with decreased Bcl-2 levels and increased levels of
Bax, cleaved caspase 3 and cleaved PARP, as determined by western
blot analysis (Fig. 3).
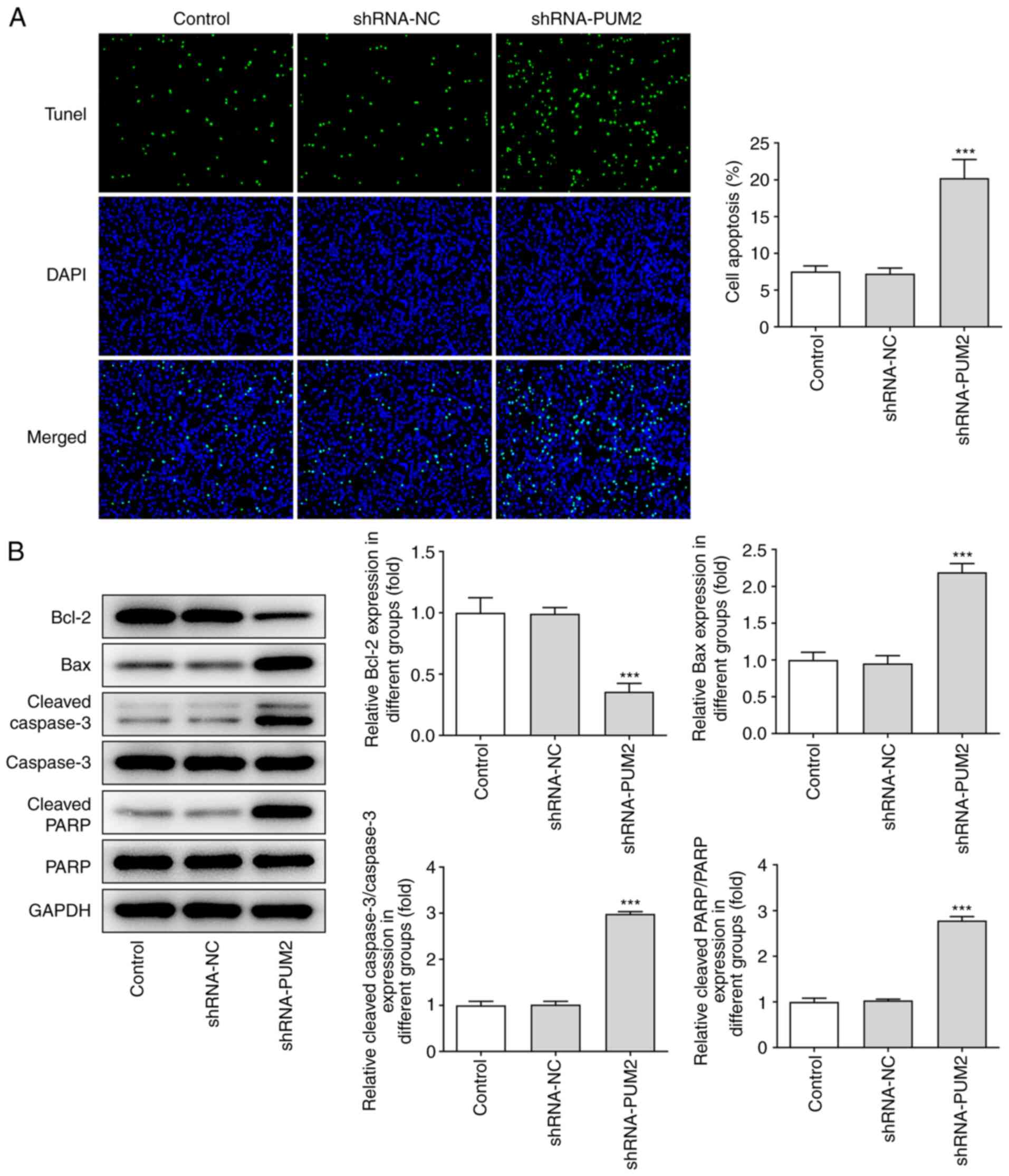 | Figure 3.PUM2 silencing represses apoptosis in
Huh-7 cells. (A) A TUNEL assay was carried out to quantify
apoptosis of Huh-7 cells transfected with shRNA-PUM2
(magnification, ×200). (B) Western blot analysis was performed to
detect the protein levels of Bcl-2, Bax, caspase3, cleaved
caspase3, PARP and cleaved PARP. Values are expressed as the mean ±
standard deviation. ***P<0.001 vs. shRNA-NC. PUM2, Pumilio
homolog 2; NC, negative control; shRNA, short hairpin RNA; TUNEL,
terminal deoxynucleotidyl transferase-mediated dUTP nick-end
labeling; PARP, poly(ADP ribose) polymerase. |
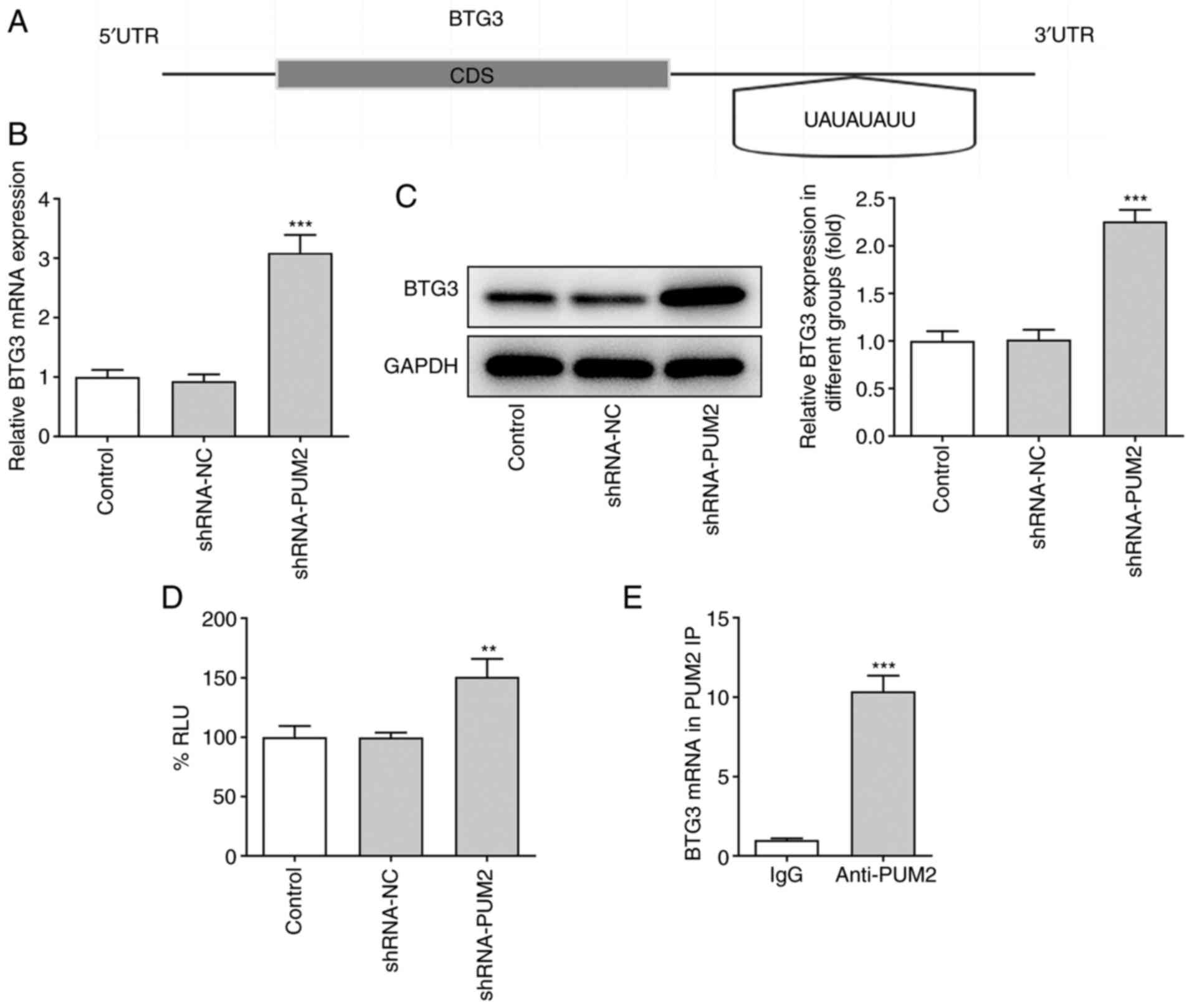
PUM2 binds directly to the 3′UTR of
BTG3
The mechanisms underlying the regulatory role of
PUM2 were further explored in HCC cells. By using the starBase
database, PUM2 was predicted to bind to several RNAs. Among these
RNAs, PUM2 protein was predicted to bind to the 3′-UTR of BTG3 and
the interaction probability was 0.99 according to the RPISeq
website. Furthermore, the binding sequence of the PUM2 and BTG3
3′UTR was predicted by Ensembl (Fig.
4A). Furthermore, the mRNA and protein levels of BTG3 were both
increased following shRNA-mediated inhibition of PUM2 expression in
Huh-7 cells (Fig. 4B and C). A
luciferase reporter assay was performed and the data indicated that
the luciferase activity of the 3′UTR of BTG3 was significantly
increased by PUM2 silencing, while no apparent changes were noted
in the luciferase activity in the shRNA-NC group as compared with
the control group (Fig. 4D). In
addition, the results of the RIP assay were able to verify the
combination of PUM2 and BTG3 (Fig.
4E).
PUM2 silencing represses cell
proliferation and promotes apoptosis of Huh-7 cells by targeting
BTG3
To assess the role of BTG3 in PUM2-mediated HCC
progression, BTG3 expression was silenced in Huh-7 cells. RT-qPCR
analysis indicated a significant decrease in BTG3 expression
following transfection with shRNA-BTG3-1 or −2. shRNA-BTG3-1
exhibited improved knockdown efficiency; therefore, shRNA-BTG3-1
(denoted as shRNA-BTG3 from here onwards) was selected for the
subsequent assays (Fig. 5A). The
CCK-8 assay results indicated that BTG3 silencing reduced the
optical density values of Huh-7 cells transfected with shRNA-PUM2
(Fig. 5B). In addition, depletion
of BTG3 inhibited the reduction of the number of colonies by
downregulation of PUM2 (Fig. 5C).
Furthermore, the TUNEL assay indicated a marked decrease in the
apoptotic rate of Huh-7 cells co-transfected with shRNA-PUM2 and
shRNA-BTG3 compared with that in cells transfected with shRNA-PUM2
(Fig. 5D and E). In addition,
western blot analysis indicated that BTG3 silencing reversed the
effects of PUM2 silencing on the protein levels of Bcl-2, Bax,
cleaved caspase 3 and cleaved PARP in transfected Huh-7 cells
(Fig. 5F and G).
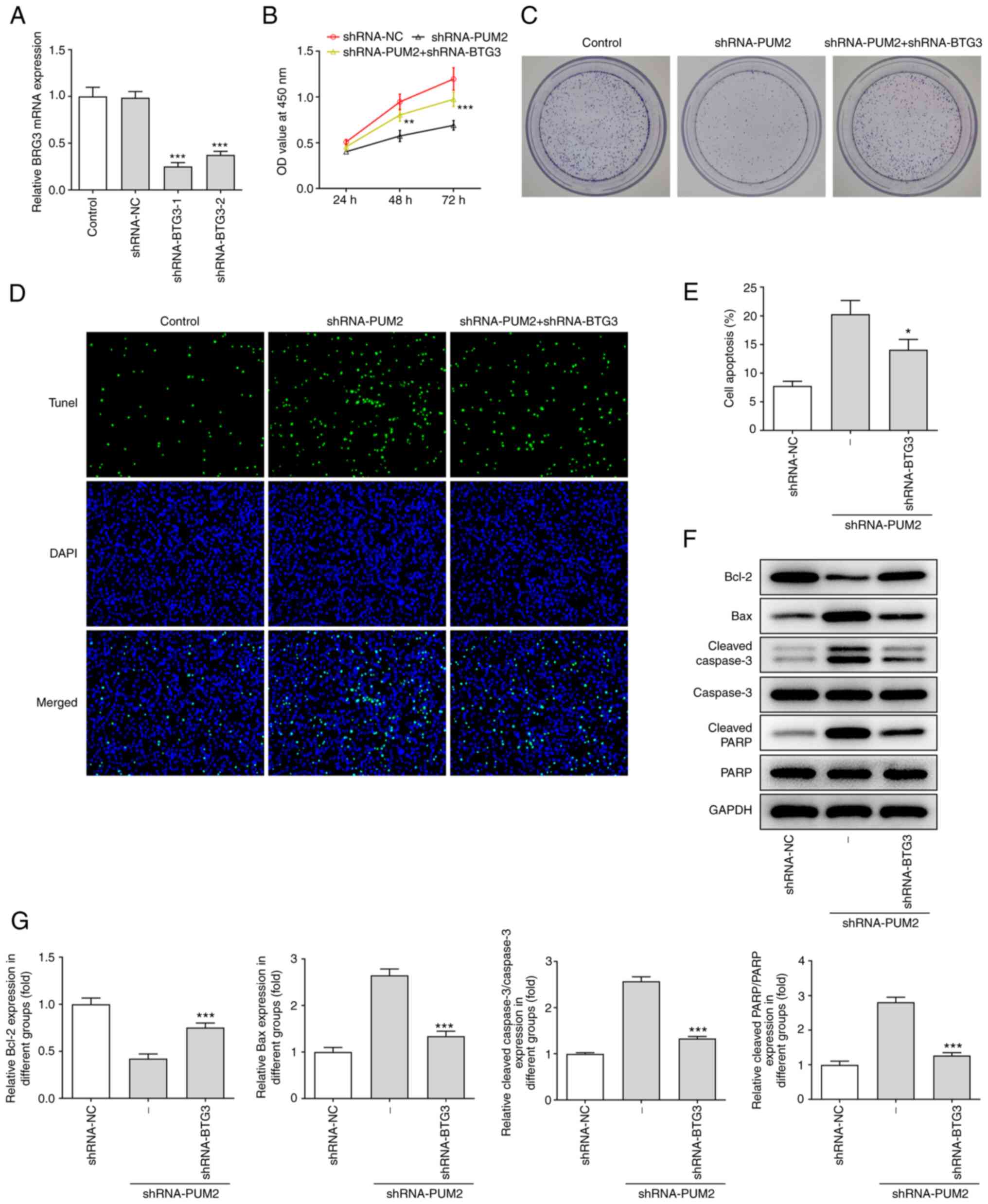 | Figure 5.PUM2 silencing inhibits Huh-7 cell
proliferation and promotes apoptosis by targeting BTG3. (A) mRNA
expression of BTG3 in Huh-7 cells after BTG3 was silenced were
detected by reverse transcription-quantitative PCR. (B) A Cell
Counting Kit-8 assay and (C) colony-formation assay were used to
examine cell proliferation. (D and E) A TUNEL assay was employed to
determine apoptosis in Huh-7 cells transfected with shRNA-BTG3. (D)
Representative images of TUNEL staining (magnification, ×200) and
(E) quantified results. (F and G) Western blot analysis was
utilized to evaluate the protein level of Bcl-2, Bax, caspase3,
cleaved caspase3, PARP and cleaved PARP. (F) Representative western
blots and (G) quantified protein levels. Values are expressed as
the mean ± standard deviation. *P<0.05, **P<0.01,
***P<0.001 vs. shRNA-NC in A or vs. shRNA-PUM2 in B-G. PUM2,
Pumilio homolog 2; NC, negative control; shRNA, short hairpin RNA;
PARP, poly(ADP ribose) polymerase; TUNEL, terminal deoxynucleotidyl
transferase-mediated dUTP nick-end labeling; OD, optical density;
BTG3, B-cell translocation gene 3. |
Discussion
HCC is a common malignant tumor type of the
digestive system with high morbidity and mortality (21). A high degree of malignancy and
distal metastasis is observed in the majority of patients with HCC
at diagnosis, leading to poor prognosis (22). In recent years, the development of
diagnostic and therapeutic applications for HCC has improved the
prognosis of HCC to a certain extent; however, the 5-year survival
rate of patients with HCC remains <26% (23,24).
Therefore, it is important to further explore the pathogenesis of
HCC and identify biomarkers for evaluation of the prognosis of the
disease (25,26). In the present study, the expression
levels of the RNA binding protein PUM2 were elevated in HCC tumor
tissues and cell lines. Silencing of PUM2 inhibited HCC cell
proliferation and induced cell apoptosis. In addition, PUM2 was
demonstrated to bind to the 3′UTR of BTG3 and regulate BTG3
expression in Huh-7 cells. Downregulation of BTG3 expression
reversed the effects of PUM2 knockdown on Huh-7 cell proliferation
and apoptosis. Taken together, the present study demonstrated for
the first time, to the best of our knowledge, that PUM2 may be a
therapeutic target for HCC, which has a regulatory role by binding
to the 3′UTR of BTG3.
The RNA-binding proteins of the Pumilio and FBF (PUF
family) have crucial roles in the occurrence and development of
multiple diseases (27–29). The PUF family proteins have
regulatory roles by combining with Pumilio binding elements located
on the 3′UTR of specific mRNAs (30). PUM2 is the mammalian member of the
PUF family that has been extensively investigated in germ and stem
cells; however, its involvement in the development of various
cancer types remains to be fully elucidated (31–33).
A recent study reported that PUM2 expression was upregulated in
cervical cancer (CC) tissues, while its silencing inhibited the
viability of CC cells (34). An
additional study revealed that knockdown of PUM2 expression
significantly suppressed cell proliferation and migration, while
promoting apoptosis of the glioblastoma cell lines U87 and U251
(14). In the present study, PUM2
expression was upregulated in HCC tumor tissues and cell lines.
Furthermore, PUM2 knockdown reduced cell proliferation and induced
apoptosis of Huh-7 cells, which is consistent with the previously
published reports.
BTG3 belongs to the B-cell translocation
gene/transducer of the Erb-B2 receptor tyrosine kinase 2 protein
family (35). It has been
indicated that BTG3 serves as a tumor suppressor gene in various
cancer types, including lung adenocarcinoma (36), oral squamous cell cancer (37) and renal cell carcinoma (38). StarBase was used to predict whether
PUM2 binds to BTG3. Bioinformatics analysis also revealed a
reaction element responsible for the binding of PUM2 with the 3′UTR
of BTG. The RPISeq website predicted that the index of PUM2
required to bind to the 3′UTR of BTG3 (positive) was 0.99. In
addition, the mRNA and protein levels of BTG3 were upregulated in
Huh-7 cells following knockdown of PUM2. The binding of PUM2 to the
3′UTR of BTG3 was confirmed by luciferase reporter and RIP assays.
Ren et al (39)
demonstrated that knockdown of BTG3 expression accelerated the
proliferation, migration and invasion of gastric cancer cells. Wang
et al (40) further
reported that BTG3 reversed the effects of miR-519c-3p
overexpression to promote Hep3B cell proliferation, migration and
invasion in HCC. The results of the present study are consistent
with these findings and indicate that BTG3 silencing reverses the
suppressive effects of shRNA-PUM2 in HCC cells by promoting the
shRNA-PUM2-mediated inhibition of cell proliferation and by
reducing the cell apoptosis rate. Collectively, these findings
indicated that BTG3 acts as a tumor suppressor in HCC. The findings
also indicated that BTG3 was involved in the regulatory roles of
PUM2 in HCC.
In conclusion, the results of the present study
demonstrated that PUM2 expression was upregulated in HCC and that
it was associated with poor prognosis. PUM2 regulated cell
proliferation and apoptosis of HCC cells by directly binding to the
3′UTR of BTG3. Rescue experiments indicated that BTG3 silencing
reversed the effects of PUM2 expression knockdown on cell
proliferation and apoptosis, which implies the potential for this
protein to be used as a novel therapeutic target for the treatment
of HCC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
available from the corresponding author on reasonable request.
Authors' contributions
ZL and CL designed the study, performed the
experiments and drafted and revised the manuscript. ZL analyzed the
data and performed the literature search. ZL and CL confirmed the
authenticity of all the raw data. Both authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cree IA, Indave Ruiz BI, Zavadil J, McKay
J, Olivier M, Kozlakidis Z, Lazar AJ, Hyde C, Holdenrieder S,
Hastings R, et al: The international collaboration for cancer
classification and research. Int J Cancer. 148:560–571. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kanwal F, Kramer J, Asch SM, Chayanupatkul
M, Cao Y and El-Serag HB: Risk of hepatocellular cancer in HCV
patients treated with direct-acting antiviral agents.
Gastroenterology. 153:996–1005.e1. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Estes C, Razavi H, Loomba R, Younossi Z
and Sanyal AJ: Modeling the epidemic of nonalcoholic fatty liver
disease demonstrates an exponential increase in burden of disease.
Hepatology. 67:123–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
European Association for the Study of the
Liver, . Electronic address simpleeasloffice@easloffice.eu.
European Association for the Study of the Liver: EASL clinical
practice guidelines: Management of hepatocellular carcinoma. J
Hepatol. 69:182–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Llovet JM and Bruix J: Systematic review
of randomized trials for unresectable hepatocellular carcinoma:
Chemoembolization improves survival. Hepatology. 37:429–442. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abbastabar M, Sarfi M, Golestani A and
Khalili E: lncRNA involvement in hepatocellular carcinoma
metastasis and prognosis. EXCLI J. 17:900–913. 2018.PubMed/NCBI
|
|
8
|
White EK, Moore-Jarrett T and Ruley HE:
PUM2, a novel murine puf protein, and its consensus RNA-binding
site. RNA. 7:1855–1866. 2001.PubMed/NCBI
|
|
9
|
Fox M, Urano J and Reijo Pera RA:
Identification and characterization of RNA sequences to which human
PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind.
Genomics. 85:92–105. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cao Y, Liu C, Wang Q, Wang W, Tao E and
Wan L: Pum2 mediates Sirt1 mRNA decay and exacerbates
hypoxia/reoxygenation-induced cardiomyocyte apoptosis. Exp Cell
Res. 393:1120582020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smialek MJ, Kuczynska B, Ilaslan E,
Janecki DM, Sajek MP, Kusz-Zamelczyk K and Jaruzelska J: Kinesin
KIF18A is a novel PUM-regulated target promoting mitotic
progression and survival of a human male germ cell line. J Cell
Sci. 133:jcs2409862020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu R, Zhu X, Chen C, Xu R, Li Y and Xu W:
RNA-binding protein PUM2 suppresses osteosarcoma progression via
partly and competitively binding to STARD13 3′UTR with miRNAs. Cell
Prolif. 51:e125082018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tao W, Ma J, Zheng J, Liu X, Liu Y, Ruan
X, Shen S, Shao L, Chen J and Xue Y: Silencing SCAMP1-TV2 inhibited
the malignant biological behaviors of breast cancer cells by
interaction with PUM2 to facilitate INSM1 mRNA degradation. Front
Oncol. 10:6132020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Sun W, Yang J, Yang L, Li C, Liu
H, Liu X and Jiao B: PUM2 promotes glioblastoma cell proliferation
and migration via repressing BTG1 expression. Cell Struct Funct.
44:29–39. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res.
45:(W1)W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:(Database Issue). D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Muppirala UK, Honavar VG and Dobbs D:
Predicting RNA-protein interactions using only sequence
information. BMC Bioinformatics. 12:4892011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cao Y, Ren Y, Ma H, Zhou C, Liu J, Shi Q,
Feng G, Zheng C and Xiong B: Classification of hepatocellular
carcinoma diameter by statistical technology and prognostic
evaluation in patients after the combined use of transarterial
chemoembolization and radiofrequency ablation. J Cancer Res Ther.
16:356–364. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang W and Wei C: Advances in the early
diagnosis of hepatocellular carcinoma. Genes Dis. 7:308–319. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim DW, Talati C and Kim R: Hepatocellular
carcinoma (HCC): Beyond sorafenib-chemotherapy. J Gastrointest
Oncol. 8:256–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ichikawa T, Sano K and Morisaka H:
Diagnosis of pathologically early HCC with EOB-MRI: Experiences and
current consensus. Liver Cancer. 3:97–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gentile D, Donadon M, Lleo A, Aghemo A,
Roncalli M, di Tommaso L and Torzilli G: Surgical treatment of
hepatocholangiocarcinoma: A systematic review. Liver Cancer.
9:15–27. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gennarino VA, Singh RK, White JJ, De Maio
A, Han K, Kim JY, Jafar-Nejad P, di Ronza A, Kang H, Sayegh LS, et
al: Pumilio1 haploinsufficiency leads to SCA1-like
neurodegeneration by increasing wild-type Ataxin1 levels. Cell.
160:1087–1098. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen D, Zheng W, Lin A, Uyhazi K, Zhao H
and Lin H: Pumilio 1 suppresses multiple activators of p53 to
safeguard spermatogenesis. Curr Biol. 22:420–425. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Crittenden SL, Bernstein DS, Bachorik JL,
Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R,
Wickens M and Kimble J: A conserved RNA-binding protein controls
germline stem cells in Caenorhabditis elegans. Nature. 417:660–663.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nishanth MJ and Simon B: Functions,
mechanisms and regulation of Pumilio/Puf family RNA binding
proteins: A comprehensive review. Mol Biol Rep. 47:785–807. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moore FL, Jaruzelska J, Fox MS, Urano J,
Firpo MT, Turek PJ, Dorfman DM and Pera RA: Human Pumilio-2 is
expressed in embryonic stem cells and germ cells and interacts with
DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc Natl Acad
Sci USA. 100:538–543. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shigunov P, Sotelo-Silveira J, Kuligovski
C, de Aguiar AM, Rebelatto CK, Moutinho JA, Brofman PS, Krieger MA,
Goldenberg S, Munroe D, et al: PUMILIO-2 is involved in the
positive regulation of cellular proliferation in human
adipose-derived stem cells. Stem Cells Dev. 21:217–227. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee MH, Wu X and Zhu Y: RNA-binding
protein PUM2 regulates mesenchymal stem cell fate via repression of
JAK2 and RUNX2 mRNAs. J Cell Physiol. 235:3874–3885. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Duan W, Nian L, Qiao J and Liu NN: LncRNA
TUG1 aggravates the progression of cervical cancer by binding PUM2.
Eur Rev Med Pharmacol Sci. 23:8211–8218. 2019.PubMed/NCBI
|
|
35
|
Matsuda S, Rouault J, Magaud J and Berthet
C: In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS
Lett. 497:67–72. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yoneda M, Suzuki T, Nakamura T, Ajima R,
Yoshida Y, Kakuta S, Katsuko S, Iwakura Y, Shibutani M, Mitsumori
K, et al: Deficiency of antiproliferative family protein Ana
correlates with development of lung adenocarcinoma. Cancer Sci.
100:225–232. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamamoto N, Uzawa K, Yakushiji T,
Shibahara T, Noma H and Tanzawa H: Analysis of the ANA gene as a
candidate for the chromosome 21q oral cancer susceptibility locus.
Br J Cancer. 84:754–759. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu L, Liu S, Duan Q, Chen L, Wu T, Qian
H, Yang S, Xin D, He Z and Guo Y: MicroRNA-142-5p promotes cell
growth and migration in renal cell carcinoma by targeting BTG3. Am
J Transl Res. 9:2394–2402. 2017.PubMed/NCBI
|
|
39
|
Ren XL, Zhu XH, Li XM, Li YL, Wang JM, Wu
PX, Lv ZB, Ma WH, Liao WT, Wang W, et al: Down-regulation of BTG3
promotes cell proliferation, migration and invasion and predicts
survival in gastric cancer. J Cancer Res Clin Oncol. 141:397–405.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang L, Mo H, Jiang Y, Wang Y, Sun L, Yao
B, Chen T, Liu R, Li Q, Liu Q and Yin G: MicroRNA-519c-3p promotes
tumor growth and metastasis of hepatocellular carcinoma by
targeting BTG3. Biomed Pharmacother. 118:1092672019. View Article : Google Scholar : PubMed/NCBI
|