Introduction
Colorectal cancer (CRC) is ranked the fourth most
deadly cancer worldwide, accounting for ~10% of all diagnosed
cancer cases and ~10% of cancer-related deaths (1). The treatment options for CRC have
been enriched over the decades, with substantial improvements to
techniques and a deeper understanding of CRC pathogenesis, which
lead to the improvement of overall survival (OS). However, since
CRC is not symptomatic until it reaches an advanced stage, and
there are high occurrence rates of metastasis, recurrence and drug
resistance, the lethality of CRC is yet to be adequately reduced
(2). Furthermore, although disease
screening using biomarkers has been implemented worldwide to
increase the early detection of CRC, there is still a lack of a
convincing test that accurately forecasts disease condition or
prognosis for patients with CRC. Therefore, constant exploration of
novel and reliable biomarkers for CRC monitoring is essential to
improve the outcomes of patients with the disease.
C-X-C motif chemokine ligands (CXCLs) are small
proteins with a cysteine-containing motif (C represents cysteine
and X represents any amino acid) near the N-terminal. The CXCLs are
key molecules that attract leukocytes to the inflammation sites,
and they bind to the corresponding CXC receptors (CXCRs) to trigger
internalization and transduction of downstream signaling pathways
(3). A growing body of evidence
has shown that CXCLs are involved in the development of a number of
malignancies. For example, CXCL1 and CXCL2 facilitate cell survival
and metastasis in breast cancer and predict poor OS in gastric
cancer (4,5). CXCL8 mediates the initiation and
development of prostate cancer, lung cancer and melanoma (6), and CXCL13 is associated with an
advanced disease stage, and poor OS and disease-free survival rates
of clear cell renal cell carcinoma (7). CXCL14 attenuates tumor progression in
squamous cell carcinoma, while predicting poor survival in breast
cancer (8,9). As shown by these studies, CXCLs
present potential as biomarkers for tumor progression and prognosis
in various cancer types, although the clinical implications of
these CXCLs in CRC have not been fully studied yet. According to
the existing evidence, we hypothesize that these CXCLs may be of
clinical value for the disease management and prognosis of CRC. In
the present study, the expression levels of tumor CXCL1, CXCL2,
CXCL8, CXCL13 and CXCL14 were detected, and their associations with
clinicopathological features and survival profile were further
assessed in patients with CRC.
Materials and methods
Patients
The present study retrospectively reviewed the cases
of 232 patients with primary CRC who underwent resection in The
First Hospital of Jilin University (Changchun, China) between
January 2012 and December 2014. All patients were initially
confirmed with primary CRC by histopathology. The age range of the
cohort was 18–80 years old. The patients were eligible if they had
well-preserved tumor tissue specimens, and complete pre-operation
tumor features and survival data, and if they were without distant
metastases, did not have recurrent or secondary CRC, had no history
of hematological malignancies or other solid tumors and had not
received neoadjuvant therapy before resection. Ethical approval for
the study was obtained from the Institutional Review Board of The
First Hospital of Jilin University (approval no. 2018-413). The
First Hospital of Jilin University provided access to the database
used in this study. All patients or their family members provided
written informed consent.
Data and sample collection
The demographic data (including the age and sex) and
preoperative tumor features [including World Health Organization
pathological grade (10), tumor
size, T stage, N stage and American Joint Committee on Cancer
Tumor-Node-Metastasis (TNM) stage (11)] were collected from the database of
The First Hospital of Jilin University. The formalin-fixed
paraffin-embedded (FFPE) tumor tissue specimens were acquired from
the Department of Pathology of The First Hospital of Jilin
University. Furthermore, the FFPE normal colon tissues were
available for 30 patients of the aforementioned 232 patients with
CRC, which were also obtained from the Department of Pathology of
The First Hospital of Jilin University.
Immunohistochemistry (IHC) assay
All tumor tissue specimens and normal colon tissues
were cut into 4-µm sections, and then the sections were
deparaffinized in 65°C overnight, washed with xylene (Sangon
Biotech, Co., Ltd.), rehydrated in a descending ethanol serials and
underwent antigen retrieval. After that, 10% goat serum
(MilliporeSigma) (30 min, room temperature) and 0.3%
H2O2 (10 min, room temperature) were added to
the sections for the blocking of non-specific binding and
peroxidase activity. Subsequently, primary antibodies (CXCL1 rabbit
polyclonal antibody; 1:100; cat. no. PA5-86508; CXCL2 recombinant
rabbit monoclonal antibody; 1:20; cat. no. 701126; CXCL8 rabbit
polyclonal antibody; 1:500; cat. no. PA5-85428; CXCL13 rabbit
polyclonal antibody; 1:500; cat. no. PA5-28827; and CXCL14 rabbit
polyclonal antibody; 1:500; cat. no. PA5-28820) (all Invitrogen;
Thermo Fisher Scientific, Inc.) were added and incubated at 4°C
overnight. The next day, horseradish peroxidase-conjugated goat
anti-rabbit IgG (H+L) secondary antibody (1:10,000; cat. no. 31460;
Invitrogen; Thermo Fisher Scientific, Inc.) was added and incubated
at 37°C for 60 min. Finally, the tissue sections were stained with
diaminobenzidine (MilliporeSigma) and counterstained with
hematoxylin (MilliporeSigma) (2 min, room temperature). The IHC
staining result was observed on a Nikon ECLIPSE E200 microscope
(Nikon Corporation) and assessed by staining intensity and staining
density of positive cells, as previously described (12). Based on the total IHC score
(staining intensity score × staining density score; score range,
0–12), the expression of CXCL1, CXCL2, CXCL8, CXCL13 and CXCL14 was
categorized as low expression (IHC score ≤3) and high expression
(IHC score >3) (12).
Hematoxylin-eosin staining
The tissues, fixed in 10% formalin (Sangon Biotech,
Co., Ltd.) for 24 h at room temperature, were embedded in paraffin
and were cut into 4-µm sections. The sections were then
deparaffinized with xylene (Sangon Biotech, Co., Ltd.) and
rehydrated in a descending ethanol serials. The hematoxylin
((Sangon Biotech, Co., Ltd.) was used to stain the nuclei at room
temperature for 5 min. The cytoplasm was stained with eosin (Sangon
Biotech, Co., Ltd.) for 2 min at room temperature. The images were
taken by a Nikon ECLIPSE E200 microscope (Nikon Corporation).
Follow-up
The survival data were obtained from the patient
follow-up documents. According to the survival data, the last
follow-up date was December 31, 2018, and the median follow-up
duration was 56.0 months (range, 1.0-84.0 months). The OS time was
calculated from the date of resection to the date of death.
Human Protein Atlas Database
validation
The expression of CXCLs and CXCRs were re-assessed
using the Human Protein Atlas Database (www.proteinatlas.org), derived from The Cancer Genome
Atlas (TCGA) database. In detail, the IHC score for 597 patients
for CXCL1, CXCL2, CXCL8, CXCL13, CXCL14 are shown in Fig. S1. Besides, the survival data were
downloaded from TCGA database for subsequent analysis of the
correlation between CXCL1, CXCL2, CXCL8, CXCL13, CXCL14, CXCR1,
CXCR2, CXCR3 and CXCR5 and survival, which are shown in Figs. S1 and S2 (available from www.proteinatlas.org).
Statistical analysis
The descriptive analysis of continuous variables is
expressed as mean ± standard deviation (SD), and the descriptive
analysis of categorical variables is displayed as count
(percentage). The correlations among CXCL1, CXCL2, CXCL8, CXCL13
and CXCL14 were determined using Spearman's correlation analysis.
The comparison of quantitative data with a normal distribution
(including age and tumor size) was determined using an unpaired
Student's t-test. The comparison of an unordered categorical
variable (including sex) was assessed by χ2 test. The
comparison of ordered categorical variables (including pathological
grade, T stage, N stage and TNM stage) was performed using
Wilcoxon's rank sum test. Comparisons between the tumor tissue and
normal colon tissue with regard to CXCL1, CXCL2, CXCL8, CXCL13 and
CXCL14 expression were achieved by the paired t-test. OS was
displayed using Kaplan-Meier curves, and comparisons of OS between
two groups were determined by log-rank test. Multivariate logistic
regression analysis was performed to combine CXCL1, CXCL2, CXCL8,
CXCL13 and CXCL14 as CXCLs, and the calculation formula is shown in
Table SI. Receiver operative
curves were used for analyzing the ability of CXCLs to distinguish
between tumor tissue and normal colon tissue. Factors affecting OS
were analyzed by univariate and backward stepwise multivariate
Cox's proportional hazard regression model. Statistical analyses
were performed using SPSS (version 22.0; IBM Corp.), and figures
were plotted using GraphPad Prism (version 7.00; GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinical characteristics of patients
with CRC
The mean age ± SD for the cohort was 65.2±10.7
years, and the median age (range) was 67.5 years (39.0-80.0 years).
The sex composition was 106 (45.7%) females and 126 (54.3%) males.
A total of 34 (14.7%), 166 (71.6%) and 32 (13.8%) patients were in
pathological grades G1, G2 and G3, respectively. The mean tumor
size was 4.4±1.2 cm, and for the tumor stage, the number of
patients at TNM stage I, II and III was 30 (12.9%), 109 (47.0%) and
93 (40.1%), respectively. Other detailed clinical characteristics
are shown in Table I.
 | Table I.Clinical characteristics of patients
with colorectal cancer (n=232). |
Table I.
Clinical characteristics of patients
with colorectal cancer (n=232).
| Characteristic | Value |
|---|
| Mean age ± SD,
years | 65.2±10.7 |
| Sex, n (%) |
|
|
Female | 106 (45.7) |
| Male | 126 (54.3) |
| Pathological grade, n
(%) |
|
| G1 | 34 (14.7) |
| G2 | 166 (71.6) |
| G3 | 32 (13.8) |
| Mean tumor size ± SD,
cm | 4.4±1.2 |
| T stage, n (%) |
|
| T1 | 5 (2.2) |
| T2 | 25 (10.8) |
| T3 | 199 (85.8) |
| T4 | 3 (1.3) |
| N stage, n (%) |
|
| N0 | 139 (59.9) |
| N1 | 61 (26.3) |
| N2 | 32 (13.8) |
| TNM stage, n (%) |
|
| I | 30 (12.9) |
| II | 109 (47.0) |
| III | 93 (40.1) |
Expression of CXCL1, CXCL2, CXCL8,
CXCL13 and CXCL14 in CRC tumor tissues and normal colon
tissues
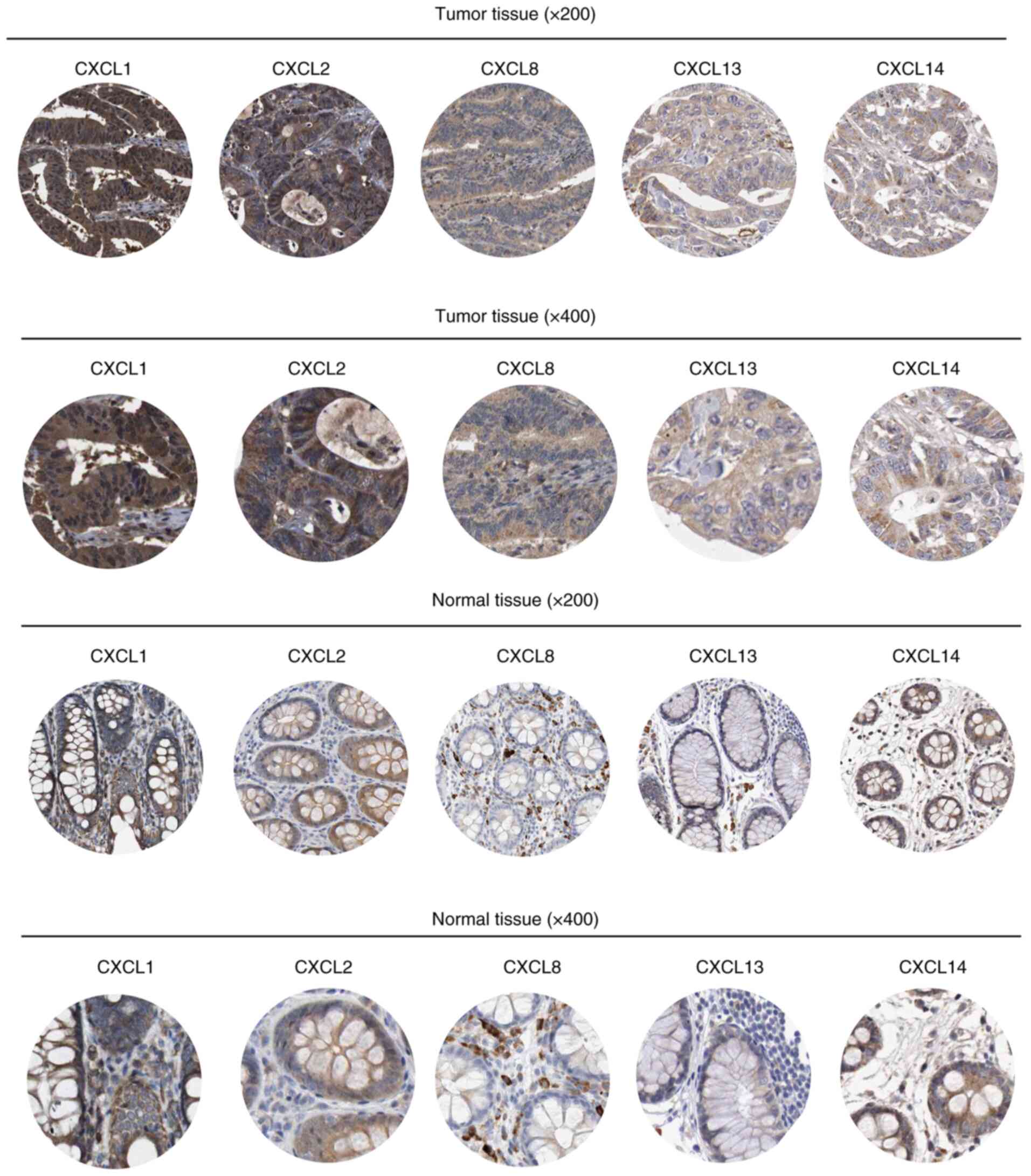
Representative staining of CXCL1, CXCL2, CXCL8,
CXCL13 and CXCL14 in tumor tissues and normal colon tissues is
shown in Fig. 1. In addition, the
tumor tissues and normal colon tissues stained using
hematoxylin-eosin staining are shown in Fig. S3. Compared with the normal colon
tissue, the tumor tissue exhibited elevated levels of CXCL2 and
CXCL8 expression (both P<0.01), but similar levels of CXCL1,
CXCL13 and CXCL14 expression (all P>0.05) (Table SII). The combination of CXCL1,
CXCL2, CXCL8, CXCL13 and CXCL14 (referred to as CXCLs) had a
certain ability for distinguishing the CRC tumor tissues from the
normal colon tissues (Fig. S4).
The correlations among CXCL1, CXCL2, CXCL8, CXCL13 and CXCL14 are
shown in Table SIII. All CXCLs
were associated with each other (all P<0.05), with the exception
of CXCL14 and CXCL8, and CXCL14 and CXCL13 (both P>0.05).
Comparison of CXCL1, CXCL2, CXCL8,
CXCL13 and CXCL14 between CRC patients with different
clinicopathological features
In patients with CRC, those with high CXCL1
expression exhibited a larger tumor size (P=0.009), and advanced T
stage (P=0.004), N stage (P=0.015) and TNM stage (P=0.006).
Patients with high CXCL2 expression presented with advanced N stage
(P=0.016) and TNM stage (P=0.015). Patients with high CXCL8
expression presented with an advanced T stage (P=0.027) and TNM
stage (P=0.041), and those with high CXCL13 expression presented
with a higher T stage (P=0.003), N stage (P=0.001) and TNM stage
(P=0.001) (Table II). However,
there were no differences with regard to clinical characteristics
among patients with CRC with different levels of CXCL14 expression
(all P>0.05).
 | Table II.Comparison between CXCL1, CXCL2,
CXCL8, CXCL13 and CXCL14 expression levels in patients with
colorectal cancer with regard to different clinical
characteristics. |
Table II.
Comparison between CXCL1, CXCL2,
CXCL8, CXCL13 and CXCL14 expression levels in patients with
colorectal cancer with regard to different clinical
characteristics.
|
| CXCL1 expression | CXCL2 expression | CXCL8 expression | CXCL13
expression | CXCL14
expression |
|---|
|
|
|
|
|
|
|
|---|
| Items | High (n=119) | Low (n=119) | P-value | High (n=139) | Low (n=93) | P-value | High (n=103) | Low (n=129) | P-value | High (n=95) | Low (n=137) | P-value | High (n=72) | Low (n=160) | P-value |
|---|
| Mean age ± SD,
years | 64.4±10.2 | 66.0±11.2 | 0.241 | 64.9±10.6 | 65.6±11.0 | 0.648 | 65.0±10.7 | 65.4±10.8 | 0.786 | 63.8±10.4 | 66.2±10.9 | 0.097 | 64.1±9.7 | 65.7±11.2 | 0.289 |
| Sex, n (%) |
|
| 0.052 |
|
| 0.345 |
|
| 0.179 |
|
| 0.147 |
|
| 0.549 |
|
Female | 47 (39.5) | 59 (52.2) |
| 60 (43.2) | 46 (49.5) |
| 42 (40.8) | 64 (49.6) |
| 38 (40.0) | 68 (49.6) |
| 35 (48.6) | 71 (44.4) |
|
|
Male | 72 (60.5) | 54 (47.8) |
| 79 (56.8) | 47 (50.5) |
| 61 (59.2) | 65 (50.4) |
| 57 (60.0) | 69 (50.4) |
| 37 (51.4) | 89 (55.6) |
|
| Pathological grade,
n (%) |
|
| 0.218 |
|
| 0.072 |
|
| 0.338 |
|
| 0.090 |
|
| 0.483 |
| G1 | 16 (13.5) | 18 (15.9) |
| 18 (12.9) | 16 (17.2) |
| 14 (13.6) | 20 (15.5) |
| 13 (13.7) | 21 (15.3) |
| 7 (9.7) | 27 (16.9) |
|
| G2 | 83 (69.7) | 83 (73.5) |
| 97 (69.8) | 69 (74.2) |
| 72 (69.9) | 94 (72.9) |
| 63 (66.3) | 103 (75.2) |
| 56 (77.8) | 110 (68.7) |
|
| G3 | 20 (16.8) | 12 (10.6) |
| 24 (17.3) | 8 (8.6) |
| 17 (16.5) | 15 (11.6) |
| 19 (20.0) | 13 (9.5) |
| 9 (12.5) | 23 (14.4) |
|
| Mean tumor size ±
SD, cm | 4.6±1.3 | 4.2±1.1 | 0.009 | 4.5±1.3 | 4.3±1.2 | 0.241 | 4.6±1.3 | 4.3±1.2 | 0.064 | 4.6±1.4 | 4.3±1.1 | 0.056 | 4.4±1.3 | 4.4±1.2 | 0.900 |
| T stage, n (%) |
|
| 0.004 |
|
| 0.056 |
|
| 0.027 |
|
| 0.003 |
|
| 0.862 |
| T1 | 1 (0.8) | 4 (3.5) |
| 2 (1.4) | 3 (3.2) |
| 1 (1.0) | 4 (3,1) |
| 0 (0.0) | 5 (3.6) |
| 1 (1.4) | 4 (2.5) |
|
| T2 | 8 (6.8) | 17 (15.1) |
| 12 (8.6) | 13 (14.0) |
| 7 (6.8) | 18 (13.9) |
| 6 (6.3) | 19 (13.9) |
| 8 (11.1) | 17 (10.6) |
|
| T3 | 107 (89.9) | 92 (81.4) |
| 122 (87.8) | 77 (82.8) |
| 93 (90.3) | 106 (82.2) |
| 86 (90.5) | 113 (82.5) |
| 62 (86.1) | 137 (85.6) |
|
| T4 | 3 (2.5) | 0 (0.0) |
| 3 (2.2) | 0 (0.0) |
| 2 (1.9) | 1 (0.8) |
| 3 (3.2) | 0 (0.0) |
| 1 (1.4) | 2 (1.3) |
|
| N stage, n (%) |
|
| 0.015 |
|
| 0.016 |
|
| 0.140 |
|
| 0.001 |
|
| 0.579 |
| N0 | 63 (52.9) | 76 (67.3) |
| 75 (54.0) | 64 (68.8) |
| 56 (54.4) | 83 (64.3) |
| 46 (48.4) | 93 (67.9) |
| 45 (62.5) | 94 (58.8) |
|
| N1 | 34 (28.6) | 27 (23.9) |
| 40 (28.8) | 21 (22.6) |
| 31 (30.1) | 30 (23.3) |
| 28 (29.5) | 33 (24.1) |
| 18 (25.0) | 43 (26.8) |
|
| N2 | 22 (18.5) | 10 (8.8) |
| 24 (17.2) | 8 (8.6) |
| 16 (15.5) | 16 (12.4) |
| 21 (22.1) | 11 (8.0) |
| 9 (12.5) | 23 (14.4) |
|
| TNM stage, n
(%) |
|
| 0.006 |
|
| 0.015 |
|
| 0.041 |
|
| 0.001 |
|
| 0.699 |
| I | 9 (7.6) | 21 (18.6) |
| 14 (10.1) | 16 (17.2) |
| 8 (7.8) | 22 (17.0) |
| 6 (6.3) | 24 (17.5) |
| 9 (12.5) | 21 (13.1) |
|
| II | 54 (45.4) | 55 (48.7) |
| 61 (43,9) | 48 (51.6) |
| 48 (46.6) | 61 (47.3) |
| 40 (42.1) | 69 (50.4) |
| 36 (50.0) | 73 (45.6) |
|
|
III | 56 (47.0) | 37 (32.7) |
| 64 (46.0) | 29 (31.2) |
| 47 (45.6) | 46 (35,7) |
| 49 (51.6) | 44 (32.1) |
| 27 (37.5) | 66 (41.3) |
|
Associations between CXCL1, CXCL2,
CXCL8, CXCL13 and CXCL14 expression levels and OS in patients with
CRC
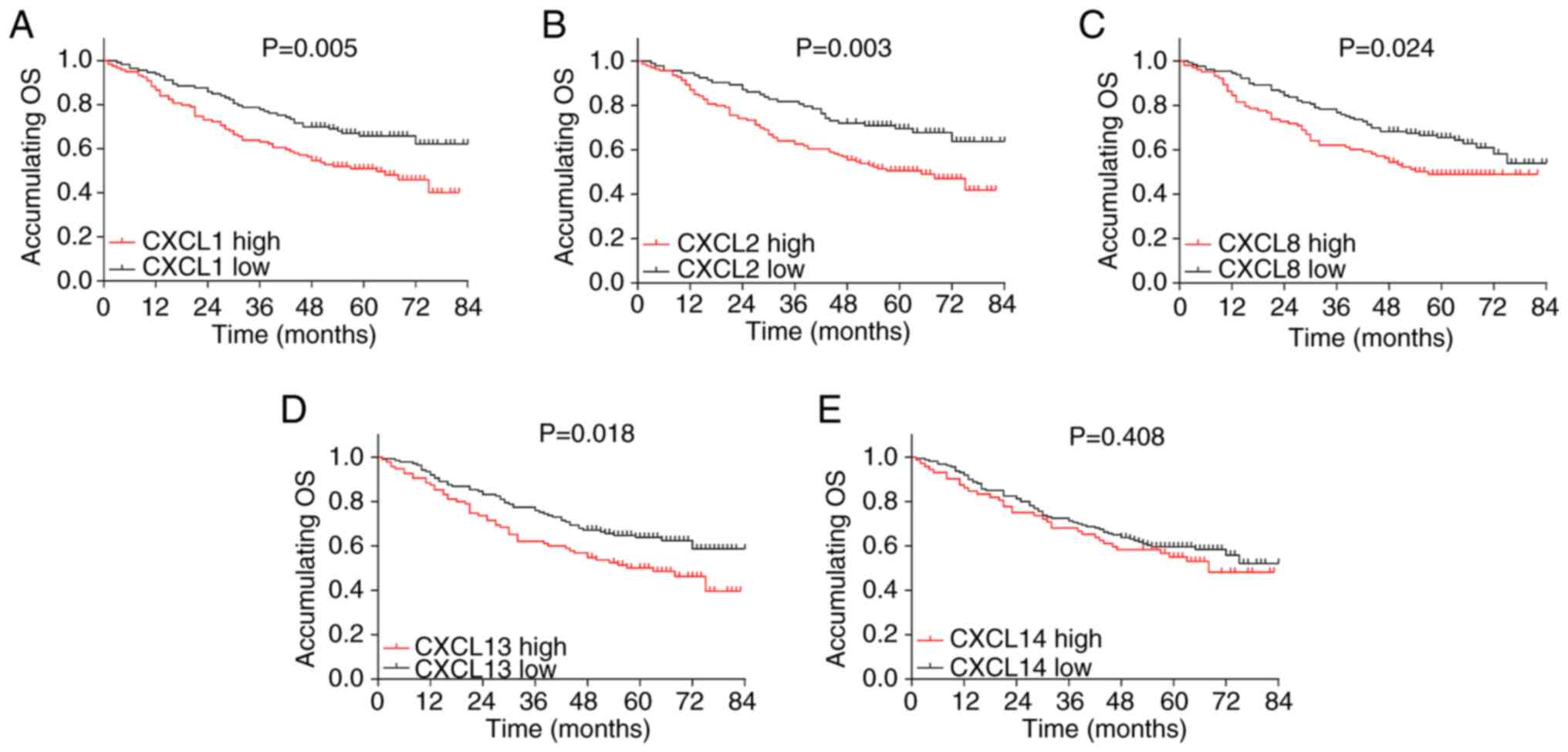
High CXCL1 expression (P=0.005) (Fig. 2A), high CXCL2 expression (P=0.003)
(Fig. 2B), high CXCL8 expression
(P=0.024) (Fig. 2C) and high
CXCL13 expression (P=0.018) (Fig.
2D) were associated with poor OS in patients with CRC, whereas
no association was observed between CXCL14 expression level and OS
(P=0.408) (Fig. 2E).
Factors affecting OS in patients with
CRC
In total, 101 patients died during the follow-up
period. Of these, 92 patients died of cancer or its related causes
and 9 patients died from other causes, including 8 patient deaths
due to complications of their disease and 1 patient death from an
accident (a fall causing a head injury). High CXCL1 (P=0.006,
HR=1.756), high CXCL2 (P=0.004, HR=1.883), high CXCL8 (P=0.025,
HR=1.561) and high CXCL13 (P=0.019, HR=1.593) expression levels, as
well as higher pathological grade (P<0.001, HR=2.166), greater
tumor size (P=0.023, HR=1.657) and advanced TNM stage (P<0.001,
HR=1.826) were associated with a lower OS rate in the patients with
CRC (Table III). Backward
stepwise multivariate Cox's regression further illustrated that
CXCL1 high expression (P=0.043, HR=1.563), higher pathological
grade (P<0.001, HR=2.191), greater tumor size (P=0.003,
HR=1.975) and advanced TNM stage (P=0.001, HR=1.662) were
independent predictive factors for poor OS rate in patients with
CRC. All the enrolled patients in the study received a surgical
resection with curative intent. After the surgical resection, 220
patients achieved an R0 resection. An analysis was performed for R0
resection status and OS. The results showed that patients with an
R0 resection exhibited a prolonged accumulating OS time compared
with those patients who did not achieve an R0 resection (P=0.001;
Fig. S5).
 | Table III.Factors affecting OS. |
Table III.
Factors affecting OS.
|
| Cox's proportional
hazard regression |
|---|
|
|
|
|---|
|
|
|
| 95% CI |
|---|
|
|
|
|
|
|---|
| Factor | P-value | HR | Lower | Higher |
|---|
| Univariate Cox's
regression |
| CXCL1
high | 0.006 | 1.756 | 1.176 | 2.623 |
| CXCL2
high | 0.004 | 1.883 | 1.228 | 2.888 |
| CXCL8
high | 0.025 | 1.561 | 1.056 | 2.308 |
| CXCL13
high | 0.019 | 1.593 | 1.078 | 2.354 |
| CXCL14
high | 0.410 | 1.189 | 0.787 | 1.797 |
| Age
(>60 years) | 0.991 | 0.998 | 0.665 | 1.496 |
|
Male | 0.409 | 1.180 | 0.797 | 1.747 |
|
Pathological grade | <0.001 | 2.166 | 1.495 | 3.139 |
| Tumor
size (>5 cm) | 0.023 | 1.657 | 1.071 | 2.562 |
| TNM
stage | <0.001 | 1.826 | 1.337 | 2.492 |
| Backward stepwise
multivariate Cox's regression |
| CXCL1
high | 0.043 | 1.563 | 1.013 | 2.411 |
|
Pathological grade | <0.001 | 2.191 | 1.505 | 3.190 |
| Tumor
size (>5 cm) | 0.003 | 1.975 | 1.263 | 3.090 |
| TNM
stage | 0.001 | 1.662 | 1.226 | 2.255 |
Validation of CXCL expression and
correlation with survival in CRC patients
The expression of CXCLs in CRC was re-assessed using
the Human Protein Atlas Database and divided into high and low
expression according to the median expression value (fragments per
kilobase per million) (Fig. S1A, C,
E, G and I). High CXCL1, CXCL2, CXCL8, CXCL13 and CXCL14
expression was associated with poor 5-year survival in patients
with CRC (all P<0.05) (Fig. S1B,
D, F, H and J). Furthermore, the associations of the CXCRs with
survival were also determined, which showed that only high CXCR1
expression was associated with prolonged OS time (P=0.035), while
CXCR2, CXCR3 and CXCR5 were not associated with OS (all P>0.05)
(Fig. S2A-D).
Discussion
The present study found that CXCL1, CXCL2, CXCL8,
CXCL13 and CXCL14 were sufficiently expressed in CRC tissues and
they were closely correlated with each other, with the exception of
CXCL8 and CXCL14, and CXCL14 and CXCL13. Most importantly, CXCL1,
CXCL2, CXCL8, CXCL13, but not CXCL14, were associated with advanced
tumor features and poor OS in patients with CRC.
CXCLs are known to be associated with tumor
formation and metastasis (4,5,7–9,13–15).
In the pathology of CRC, CXCL1, CXCL2, CXCL8, CXCL13 and CXCL14
have been shown to modulate tumor progression, such as the
tumor-specific immune response, angiogenesis and metastasis
(5,7–9,13–18),
whereas their associations with clinicopathological features in
patients with CRC are obscure (19). The high level of CXCLs may activate
the carcinoma-associated fibroblasts that promote cancer cell
growth, migration and invasion, leading to lymph node metastasis
and a higher TNM stage of CRC (8,18).
From another prospective, CXCL13 activates the Wnt/β-catenin
pathway and the production of IL-12, IL-17 and IgG4, and CXCL1,
CXCL2 and CXCL8 activate the NF-κB pathway, both of which are
contributors for the tumorigenesis and metastasis of CRC (17,20).
With regard to CXCL14, its role in cancer is controversial, being
tumor suppressive in squamous cell carcinoma, but tumor promotive
in breast cancer (8,9). Therefore, no association between
CXCL14 and any clinical characteristics was observed in the
patients with CRC in the present study. Considering these results,
the detection of CXCL levels may assist pathological assessment in
clinical settings.
Overall, the upregulation of CXCLs is associated
with a poor prognosis in cancer. Specifically, CXCL1 and CXCL2 are
independent predictive factors for poor OS in patients with gastric
cancer (5), CXCL8 is closely
associated with unfavorable survival in papillary thyroid carcinoma
(21), CXCL13 predicts poor OS and
disease-free survival in clear cell renal cell carcinoma, as well
as the recurrence of hepatocellular carcinoma after hepatectomy
(16), and CXCL14 accelerates cell
growth in breast cancer, and induces drug resistance and
metastasis, which leads to a poor prognosis for patients with
breast cancer (7,16). Regarding CRC, although previous
studies have reported the influence of these CXCLs on tumor
initiation and development, the prognostic value of these CXCLs
towards CRC has not been fully investigated and needs further
validation (2,3,19).
The present study observed that CXCL1, CXCL2, CXCL8 and CXCL13, but
not CXCL14, predicted poor OS, and that CXCL1 was an independent
predictive factor for unfavorable OS in patients with CRC.
Moreover, the negative association of high CXCL expression with
poor 5-year survival was observed via analysis using the Human
Protein Atlas Database. These results regarding prognosis were in
accordance with an existing study indicating that CXCL1, CXCL2,
CXCL8 and CXCL13 were predictive factors for poor survival in
patients with colorectal cancer (3). There are several explanations for
these CXCLs being able to predict poor OS: i) CXCL1, CXCL2 and
CXCL8 are ligands binding to receptor CXCR2, and are proangiogenic
and facilitate chemoresistance under chemotherapeutic drugs in
various cancer types. Therefore, they are associated with poor
survival in patients with CRC. ii) CXCL13 binds to CXCR5 and
regulates lymphocyte migration, promotes inflammation, and promotes
CRC cell growth, invasion and metastasis via the PI3K/AKT pathway,
which leads to poor survival (22). CXCL14 was not associated with
clinical characteristics of the patients with CRC in the present
study, and considering that the role of CXCL14 in cancer is
controversial, it is predictable that CXCL14 was not associated
with the survival of these patients. Due to the accessibility of
CXCL expression levels, the evaluation of CXCLs might be of
clinical value for identifying patients at risk of a poor prognosis
in order for appropriate treatment approaches. One unneglectable
limitation in the present study needed to be clarified: The study
was retrospective, and when it was performed, 101 patients had
already died; therefore the consent was signed by the family
members on behalf of the patients..
In this study, the associations of key CXCLs with
clinical characteristics and survival in patients with CRC were
explored; however, there were still several restrictions. Above
all, since this was a small-scale study with limited samples
recruited from a single geographic area, the results might be
subjected to selection bias. Further large-scale and multi-center
investigation is necessary to validate the findings. Secondly,
although CXCLs have been studied for their potential as CRC
prognostic markers in this study, the molecular mechanisms of CXCLs
in CRC were not investigated. In this study, the expression of
CXCLs was detected by IHC, and the cutoff value for CXCL high and
low expression was an IHC score of 3. However, the optimal cutoff
for CXCLs should be determined by Youden index or the Maxstat
method, or the χ2 test, which was not performed since
control data was lacking. Furthermore, the association of CXCLs
with prognosis in patients with CRC could be validated with more
thorough analysis, such as use of nomograms, or using an additional
cohort from a second hospital assessed with IHC. This study was a
retrospective study; therefore, its inherent limitation should not
be neglected such as patient selection bias and insufficient data.
Even though the multivariate Cox's regression analysis was
performed to eliminate the potential cofounding factors such as
age, the range of the enrolled patients was large (mean age,
65.2±10.7 years; range, 39–80 years), which may affect the
generalization of the results. Fresh frozen tissues were not stored
for use in this study; therefore, the RNA level of the CXCLs was
not determined in the present study. Finally, the CXCR levels in
patients with CRC and their associations with survival should be
determined in further studies.
In conclusion, the present study found that CXCL1,
CXCL2, CXCL8 and CXCL13, but not CXCL14, were associated with worse
tumor features and unfavorable OS in patients with CRC. This may be
of potential in assisting predictive and individualized CRC
treatments.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and LW made substantial contributions to
conception and design. XL, JT, YZ, PZ, DS and LW collected and
analyzed the data. XL, JT, YZ, PZ and DS were involved in drafting
the manuscript and revising it critically for important
intellectual content. XL and LW confirm the authenticity of all the
raw data. All authors given final approval of the version to be
published, and agreed to be accountable for all aspects of the work
in ensuring that questions related to the accuracy or integrity of
any part of the work are appropriately investigated and resolved.
All authors have read and approved the manuscript.
Ethics approval and consent to
participate
Ethical approval for the study was obtained from the
Institutional Review Board of The First Hospital of Jilin
University (approval no. 2018-413). All patients or their family
members provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CXCL
|
C-X-C motif chemokine ligand
|
|
CRC
|
colorectal cancer
|
|
IHC
|
immunohistochemistry
|
|
OS
|
overall survival
|
|
SD
|
standard deviation
|
References
|
1
|
Dekker E, Tanis PJ, Vleugels JL, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oh HH and Joo YE: Novel biomarkers for the
diagnosis and prognosis of colorectal cancer. Intest Res.
18:168–183. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cabrero-de Las Heras S and
Martínez-Balibrea E: CXC family of chemokines as prognostic or
predictive biomarkers and possible drug targets in colorectal
cancer. World J Gastroenterol. 24:4738–4749. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Acharyya S, Oskarsson T, Vanharanta S,
Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N,
Seshan VE, et al: A CXCL1 paracrine network links cancer
chemoresistance and metastasis. Cell. 150:165–178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kasashima H, Yashiro M, Nakamae H, Masuda
G, Kinoshita H, Morisaki T, Fukuoka T, Hasegawa T, Nakane T, Hino
M, et al: Clinicopathologic significance of the CXCL1-CXCR2 axis in
the tumor microenvironment of gastric carcinoma. PLoS One.
12:e01786352017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T,
Chen Y, Han X and Wu K: The CXCL8-CXCR1/2 pathways in cancer.
Cytokine Growth Factor Rev. 31:61–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng Z, Cai Y, Chen H, Chen Z, Zhu D,
Zhong Q and Xie W: CXCL13/CXCR5 axis predicts poor prognosis and
promotes progression through PI3K/AKT/mTOR pathway in clear cell
renal cell carcinoma. Front Oncol. 8:6822019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Zhang J, Sun X, Su Q and You C:
Down-regulation of miR-29b in carcinoma associated fibroblasts
promotes cell growth and metastasis of breast cancer. Oncotarget.
8:39559–39570. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kondo T, Ozawa S, Ikoma T, Yang XY,
Kanamori K, Suzuki K, Iwabuchi H, Maehata Y, Miyamoto C, Taguchi T,
et al: Expression of the chemokine CXCL14 and cetuximab-dependent
tumour suppression in head and neck squamous cell carcinoma.
Oncogenesis. 5:e2402016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA;
WHO Classification of Tumours Editorial Board, : The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tong GJ, Zhang GY, Liu J, Zheng ZZ, Chen
Y, Niu PP and Xu XT: Comparison of the eighth version of the
american joint committee on cancer manual to the seventh version
for colorectal cancer: A retrospective review of our data. World J
Clin Oncol. 9:148–161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu H, Jin C, Zhu Q, Liu T, Ke B, Li A and
Zhang T: Dysregulated expressions of PTEN, NF-κB, WWP2, p53 and
c-Myc in different subtypes of B cell lymphoma and reactive
follicular hyperplasia. Am J Transl Res. 11:1092–1101.
2019.PubMed/NCBI
|
|
13
|
Ding J, Xu K, Zhang J, Lin B, Wang Y, Yin
S, Xie H, Zhou L and Zheng S: Overexpression of CXCL2 inhibits cell
proliferation and promotes apoptosis in hepatocellular carcinoma.
BMB Rep. 51:630–635. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sunaga N, Kaira K, Tomizawa Y, Shimizu K,
Imai H, Takahashi G, Kakegawa S, Ohtaki Y, Nagashima T and Kasahara
N: Clinicopathological and prognostic significance of interleukin-8
expression and its relationship to KRAS mutation in lung
adenocarcinoma. Br J Cancer. 110:2047–2053. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Uehara H, Troncoso P, Johnston D, Bucana
CD, Dinney C, Dong Z, Fidler IJ and Pettaway CA: Expression of
interleukin-8 gene in radical prostatectomy specimens is associated
with advanced pathologic stage. Prostate. 64:40–49. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu T, Ruan H, Song Z, Cao Q, Wang K, Bao
L, Liu D, Tong J, Yang H, Chen K and Zhang X: Identification of
CXCL13 as a potential biomarker in clear cell renal cell carcinoma
via comprehensive bioinformatics analysis. Biomed Pharmacother.
118:1092642019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li C, Kang D, Sun X, Liu Y, Wang J and Gao
P: The effect of C-X-C motif chemokine 13 on hepatocellular
carcinoma associates with Wnt signaling. Biomed Res Int.
2015:3454132015.PubMed/NCBI
|
|
18
|
Mishra P, Banerjee D and Ben-Baruch A:
Chemokines at the crossroads of tumor-fibroblast interactions that
promote malignancy. J Leukoc Biol. 89:31–39. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Verbeke H, Struyf S, Laureys G and Van
Damme J: The expression and role of CXC chemokines in colorectal
cancer. Cytokine Growth Factor Rev. 22:345–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ruiz de Porras V, Bystrup S,
Martínez-Cardús A, Pluvinet R, Sumoy L, Howells L, James MI, Iwuji
C, Manzano JL, Layos L, et al: Curcumin mediates
oxaliplatin-acquired resistance reversion in colorectal cancer cell
lines through modulation of CXC-Chemokine/NF-κB signalling pathway.
Sci Rep. 6:246752016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li X, He J, Zhou M, Cao Y, Jin Y and Zou
Q: Identification and validation of core genes involved in the
development of papillary thyroid carcinoma via bioinformatics
analysis. Int J Genomics. 2019:58949262019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu Z, Zhang X, Guo H, Fu L, Pan G and Sun
Y: CXCL13-CXCR5 axis promotes the growth and invasion of colon
cancer cells via PI3K/AKT pathway. Mol Cell Biochem. 400:287–295.
2015. View Article : Google Scholar : PubMed/NCBI
|