Introduction
Cervical cancer (CC) is the fourth most common
female malignancy worldwide (1,2).
More than half a million women are diagnosed with CC, and the
disease results in over 300,000 deaths worldwide each year
(1,2). CC is a complex disease. In addition
to infection with the well-studied human papillomavirus (HPV),
numerous other factors can contribute to the tumourigenesis and
progression of neoplasms (3,4).
Aberrant genes, multiparity and smoking may impact the risk of CC
(3,4). Several studies have demonstrated that
the products of IFN-stimulated genes (ISGs) serve a pivotal role in
HPV infection (5,6). The pathogenesis of CC is still
unclear and probably involves the aberrant expression of numerous
oncogenes and tumour suppressors (4). Therefore, an extended understanding
of the gene expression program in CC will help us fight this
devastating disease.
RNA sequencing (RNA-seq) has been used to rapidly
generate a large amount of data on the gene expression landscape of
CC (7–10). Differential expression (DE)
analysis is the most common and straightforward analysis for a gene
expression dataset. Using DE analysis, a set of highly upregulated
and downregulated genes can be identified by comparing primary
tumour tissue (TT) and paracarcinoma tissue (TP). Although these
studies have provided significant findings for CC, most studies
have lacked normal tissue (NC) samples as a reference to allow the
analysis of tumour progression of CC from NC to TP to TT (7–10).
In the present study, RNA-seq was performed to
analyse genes that are dysregulated in CC. The relationship between
aberrant RNA expression profiles and disease states was also
analysed. Furthermore, Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) analyses were conducted to annotate the
differentially expressed genes (DEGs). The present study further
analysed the tumour evolution trajectory using pseudo-time
analysis. Finally, a cell assay was performed to examine the effect
of ISG15 ubiquitin-like modifier (ISG15 inhibition and
overexpression) on HeLa cells. The results of the present study
could guide targeted drug research for CC therapies.
Materials and methods
Samples
Samples (Table I)
from female patients (age range, 27–70 years; mean age, 50.36±13.26
years) with cervical cancer surgery were stored in a −80°C freezer.
The inclusion criteria were as follows: i) The pathological
diagnosis was clearly cervical squamous cell carcinoma; ii)
clinical data were complete, including age, tumour clinical stage,
grade, pathological type, infiltration and lymph node metastasis
(11); iii) no history of physical
therapy or preoperative chemoradiotherapy; and iv) clinical FIGO
(2018) stage IB1 to IIA2 (12).
Exclusion criteria were as follows: i) Case data were incomplete;
ii) combined immune diseases and other malignant tumours; iii) a
history of adjuvant chemotherapy and radiotherapy before surgery;
iv) the use of pathological case data without the informed consent
of the patient; v) pathological diagnosis was of another type of
cervical cancer; and vi) the surgical method did not meet the
requirements. The experiment was performed by comparative
sequencing analysis using RNA-seq data from TT, TP (2 cm ≤ TP ≥0.5
cm from the tumour tissue) and NC (≥2 cm away from the tumour
tissue) samples collected from 14 patients with CC who underwent
surgery at Pudong New Area People's Hospital Affiliated to Shanghai
Health University (Shanghai, China) between January 2019 and June
2020. All patients had CC at stages IB1-IIA2 (based on the Criteria
for Clinical Staging of Cervical Carcinoma 2018 Modified Version of
the International Federation of Gynecology and Obstetrics) and
underwent wide hysterectomy and lymphadenectomy in the pelvic
cavity. These patients were divided into the high ISG15 expression
group and the low ISG15 expression group according to the median
ISG15 gene expression level in TT tissue. The clinical data of
these patients, such as age, BMI and tumor markers, were then
collected.
 | Table I.Association between ISG15
expression and clinicopathological features in cervical cancer. |
Table I.
Association between ISG15
expression and clinicopathological features in cervical cancer.
|
Characteristics | High ISG15
expression (n=7) | Low ISG15
expression (n=7) | P-value (Fisher's
exact test) | P-value (unpaired
t-test) |
|---|
| Age, years | 53.86±14.05 | 46.85±12.39 |
| 0.34 |
| BMI | 26.27±2.16 | 23.65±3.65 |
| 0.25 |
| Staging, n (%) |
|
| 0.0047 |
|
| I | 0 (0.0) | 6 (85.7) |
|
|
| II | 7 (100.0) | 1 (14.3) |
|
|
| Tumour size, n
(%) |
|
| >0.99 |
|
| ≥4
cm | 2 (28.6) | 3 (42.9) |
|
|
| <4
cm | 5 (71.4) | 4 (57.1) |
|
|
| Depth of
infiltration, n (%) |
|
| >0.99 |
|
|
≥1/2 | 5 (71.4) | 4 (57.1) |
|
|
|
<1/2 | 2 (28.6) | 3 (42.9) |
|
|
| Tumour markers |
|
|
|
|
| SCC
antigen, ng/ml | 10.43±18.44 | 1.42±1.33 |
| 0.26 |
| CA125,
U/ml | 5.69±7.29 | 7.20±3.77 |
| 0.66 |
| CA199,
U/ml | 18.66±18.22 | 13.85±7.96 |
| 0.56 |
| AFP,
ng/ml | 11.58±11.59 | 5.30±5.23 |
| 0.25 |
| CEA,
ng/ml | 7.04±5.03 | 2.79±3.25 |
| 0.10 |
The study was approved by the Ethical Committee of
the People's Hospital of Shanghai Pudong New District
(PRYLL-QKJW1703&PKJ2021-Y21; Shanghai, China). Written informed
consent was provided by each of the patients or their guardians (if
the patient was under the legal age to provide consent or otherwise
incapacitated or deceased), and all procedures were conducted
according to the Helsinki Ethical Principles for medical
research.
RNA-seq and data analysis
A TRIzol® kit (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to purify the RNA from the samples. For
RNA extraction, tissues were lysed with 1 ml TRIzol reagent per
50–100 mg sample. For RNA extraction, the cells were first
precipitated by centrifugation at 500 × g (4°C) for 5 min and then
1 ml TRIzol was added every 5–10×106 cells, and the
mixture was agitated 2–3 times to ensure complete cell lysis. The
TRIzol lysate was transferred into EP tubes and placed at room
temperature for 5 min. A total of 0.2 ml chloroform was added for
every 1 ml TRIzol, followed by agitation for 15 sec, being placed
at room temperature for 2–3 min, and then centrifugation at 12,000
× g (2–8°C) for 15 min. The upper aqueous phase was removed and
placed in a new EP tube with an equal volume of isopropanol, and
then placed at room temperature for 10 min, before centrifugation
at 12,000 × g (2–8°C) for 10 min. The supernatant was discarded and
washed with 1 ml 75% ethanol per 1 ml TRIzol, mixed by vortex,
centrifuged at 7,500 g (2–8°C) for 5 min, and then the supernatant
was discarded. The precipitated RNA was allowed to dissolve the RNA
precipitate with RNase-free water at room temperature after natural
drying. Reverse transcription of RNA samples was used to prepare
cDNA using the SmartScribe™ Reverse Transcriptase kit
(Takara Bio Inc.). VANTS DNA Clean Beads (Vazyme Biotech, Co.,
Ltd.) was used to sort cDNA products, and TruePrep DNA Library Prep
Kit V2 for Illumina (Vazyme Biotech, Co., Ltd.) was used to obtain
200- to 1,000-bp fragments for preparing the cDNA library. All
libraries were quantified using a 2100 Bioanalyzer and pooled at
1:1 at 2 nM for HiSeq 150 bp paired-end sequencing (Illumina,
Inc.).
Data pre-processing, principal component analysis
(PCA), and hierarchical clustering analysis were performed in
R-4.12 (r-project.org) using the ‘base’ function
and ‘stat’ package (version 4.1.2). DEGs were defined based on fold
change (FC) and statistical significance using t-tests (FC >1.5
or <0.67; P<0.05). KEGG (https://www.genome.jp/kegg) pathway and GO (http://geneontology.org/) analyses were performed to
identify biological functions associated with DEGs in Metascape
(http://metascape.org/). The Cancer Genome Atlas
(TCGA; http://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
survival analysis was performed using OncoLnc (oncolnc.org). Co-expression networks were built by
computing the correlation coefficient of DEGs (cor >0.8) and
mapped using Cytoscape software (version 3.6.1; http://cytoscape.org/). Pseudo-time trajectory
analysis was implemented using the Monocle package (version 2.22.0;
http://bioconductor.org/packages/release/bioc/html/monocle.html)
in R to investigate the evolution of CC. To explore the
interactions of the top evolutionary trajectory genes, the genes
were submitted to Search Tool for the Retrieval of Interacting
Genes/Proteins (version 11.5; http://string.embl.de/) to build protein-protein
interaction (PPI) networks.
Reverse transcription-quantitative PCR
(RT-qPCR)
A TRIzol kit (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to purify the RNA from the tissue or cells. ReverTra
Ace® qPCR RT Kit (Toyobo Life Science) was used at 42°C
for 18 min and 98°C 5 min for the reverse transcription of RNA to
cDNA. The primers were designed and synthesized by General Biotech
Co., Ltd. RT-qPCR was performed using the KAPA SYBR Green SuperMix
PCR kit (Kapa Biosystems; Roche Diagnostics) and the AriaMx
Real-Time PCR System (Agilent Technologies, Inc.). Reaction
conditions were as follows: 95°C for 3 min, 95°C for 10 sec and
60°C for 30 sec, for 40 cycles. Differences in gene expression were
calculated using the relative standard curve and comparative
threshold cycle method (2−ΔΔCq) (13), and RT-qPCR was used to verify the
expression of GAPDH. The following primers were used: GAPDH
forward, 5′-GCAAGTTCAACGGCACAGTCA-3′ and reverse,
5′-ACGACATACTCAGCACCAGCAT-3′; and ISG15 forward,
5′-ACAGCCATGGGCTGGGA-3′ and reverse,
5′-GTTCGTCGCATTTGTCCACC-3′.
Immunohistochemistry (IHC)
Samples were fixed in 10% Neutral Formalin Fix
Solution (BBI Solutions), embedded in paraffin, and cut into 4-µm
thick tissue sections. For antigen retrieval, tissue sections were
placed in antigen retrieval buffer (pH 9.0; Wuhan Servicebio
Technology, Co., Ltd.) for antigen retrieval in a microwave oven on
medium power for 8 min until boiling, then cooled for 8 min and
then switched to medium-low power for 7 min. After natural cooling,
the slides were placed in PBS (pH 7.4; Wuhan Servicebio Technology,
Co., Ltd.) and washed 3 times on a destaining shaker. Next, 3%
hydrogen peroxide solution was incubated with the sample at room
temperature for 25 min to block endogenous peroxidase, followed by
blocking with 3% BSA (Sangon Biotech, Co., Ltd.) at room
temperature for 30 min. The tissue sections were incubated with
anti-ISG15 antibody (dilution, 1:100; cat. no. DF6316; Affinity
Biosciences) at room temperature (RT) for 1 h. The HRP-labelled
goat-anti-rabbit (dilution, 1:200; cat. no. G1213; Wuhan Servicebio
Technology, Co., Ltd.) antibody was used for incubation at RT for 1
h. After the 3,3′-diaminobenzidine application for 5–10 min, the
staining was observed under a fluorescence microscope (CKX53;
Olympus Corporation). The intensity was scored by eye as follows:
0, negative; 1, weak; 2, moderate; and 3, strong.
Transfection
Synthesized ISG15 overexpression vector (500
ng/µl), small interfering RNA (siRNA; 20 pmol/µl), and scrambled
siRNA controls (20 pmol/µl) were designed and synthesized by
Shanghai GenePharma Co., Ltd. A total of 1×106
cells/well were seeded into 6-well plates and transfected with
ISG15 overexpression and siRNA using HilyMax Reagent (Dojindo
Laboratories, Inc.) according to the manufacturer's instructions.
Transfection was performed using 120 µl Opti-MEM (Gibco; Thermo
Fisher Scientific, Inc.), 80 pmol (4 µl) overexpression or siRNA,
and 12 µl HilyMAX reagent at RT for 15 min. The plasmid or
siRNA-Hilymax complex was added to the cells, then incubated at
37°C for 6 h to complete transfection. The construction of
transfected cell models was validated using RT-qPCR as
aforementioned at 48 h after transfection.
The following ISG15 siRNA sequences and ISG15
overexpression were used: ISG15 siRNA sense (5′-3′),
GCACCAGCAUGAAGACAUATT and antisense (5′-3′), UAUGUCUUCAUGCUGGUGCTT;
scrambled siRNA control sense (5′-3′), UUCUCCGAACGUGUCACGUTT and
antisense (5′-3′), ACGUGACACGUUCGGAGAATT; ISG15
overexpression:
ATGGGCTGGGACCTGACGGTGAAGATGCTGGCGGGCAACGAATTCCAGGTGTCCCTGAGCAGCTCCATGTCGGTGTCAGAGCTGAAGGCGCAGATCACCCAGAAGATCGGCGTGCACGCCTTCCAGCAGCGTCTGGCTGTCCACCCGAGCGGTGTGGCGCTGCAGGACAGGGTCCCCCTTGCCAGCCAGGGCCTGGGCCCCGGCAGCACGGTCCTGCTGGTGGTGGACAAATGCGACGAACCTCTGAGCATCCTGGTGAGGAATAACAAGGGCCGCAGCAGCACCTACGAGGTACGGCTGACGCAGACCGTGGCCCACCTGAAGCAGCAAGTGAGCGGGCTGGAGGGTGTGCAGGACGACCTGTTCTGGCTGACCTTCGAGGGGAAGCCCCTGGAGGACCAGCTCCCGCTGGGGGAGTACGGCCTCAAGCCCCTGAGCACCGTGTTCATGAATCTGCGCCTGCGGGGAGGCGGCACAGAGCCTGGCGGGCGGAGCTAA.
Western blotting
Total protein was extracted from CC tissues using
RIPA buffer containing protease and phosphatase inhibitor cocktail
(both from Sigma-Aldrich; Merck KGaA). The protein concentration
was determined using a BCA Protein Assay kit (Beyotime Institute of
Biotechnology). Proteins (50 µg/lane) were separated on 10%
SDS-PAGE and transferred to PVDF membranes. The membranes were
blocked with 5% skimmed milk and 0.1% Tween-20 in Tris-buffered
saline for 2 h at RT. Membranes were probed at 4°C overnight with
primary antibodies against ISG15 (dilution, 1:5,000; cat. no.
ab133346; Abcam) and GAPDH (dilution, 1:2,500; cat. no. ab9485;
Abcam). After washing in PBST (0.5% Tween-20) three times, the
blots were incubated with HRP-conjugated anti-rabbit secondary
antibodies (dilution, 1:5,000; cat. no. ab205718; Abcam) for 1 h at
RT. The signal was visualized using ECL western blotting detection
reagents (Tanon Science and Technology Co., Ltd.). Protein
expression levels were quantified using ImageJ software (version
1.50; National Institutes of Health).
CCK8 proliferation assay
The transfected cells were placed at a density of
1×104 cells per well on a 96-well cell culture plate,
which was placed in a 37°C cell culture incubator and incubated
overnight. The old culture medium was removed from the 96-well
plate, and 100 µl DMEM (Gibco; Thermo Fisher Scientific, Inc.) was
added to each well. The change in proliferation capacity of each
group of cells was detected by the CCK-8 method at 0, 24, 48 and 72
h after the liquid change. The old solution was discarded and 95 µl
medium and 5 µl Cell Counting Kit-8 (CCK-8) reagent (Dojindo
Laboratories, Inc.) was added to each well of the 96-well plate to
be tested and incubated at 37°C for 1–3 h. The OD of each well was
measured at 450 nm using a microplate reader.
Wound-healing assay
The transfected cells (serum starved) were placed at
a density of 2×105 cells per well on a 24-well cell
culture plate, which was placed in a 37°C cell culture incubator
and incubated overnight. When they had reached 80% confluence, a
10-µl pipette tip was used to scratch a line on the cell layer. The
cells that had migrated into the wound area were imaged under a
fluorescence microscope (CKX53; Olympus Corporation) at 0 and 24 h.
The distance migrated by the cell monolayer to close the wounded
area during this time period was measured, Results were expressed
as a migration index, i.e., the distance migrated by the
ISG15-siRNA or ISG15-OE group relative to the distance migrated by
control group.
Transwell assay
A Transwell chamber (24 well; 8.0 µm pores) was used
for the invasion assay. The matrigel was diluted in serum-free
medium (Matrigel:serum-free medium, 1:8) at 4°C, and then the
diluted Matrigel was added into the wells (upper chamber) for
incubation at 37°C for 4 h. HeLa (5×104) cells were
seeded into the prepared upper chamber containing DMEM without
serum at 37°C for 30 min, and 500 µl complete medium (DMEM and 10%
FBS) was added to the lower chamber, following routine procedures.
After 48 h, cells invading the lower chamber were collected, fixed
with 5% glutaraldehyde for 10 min at room temperature, stained with
0.5% crystal violet for 15 min at room temperature, and then
counted under a microscope (CKX53; Olympus Corporation).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software (version 7.0; GraphPad Software, Inc.), and
quantitative data are presented as the mean ± standard deviation of
three independent experiments. The difference in the levels of
ISG15 expression was calculated using Fisher's exact test. One-way
ANOVA followed by Tukey's post hoc test was used for comparisons
among groups. Differences between high ISG15 expression and low
ISG15 expression groups were analyzed using an unpaired t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Transcriptome sequencing data
analysis
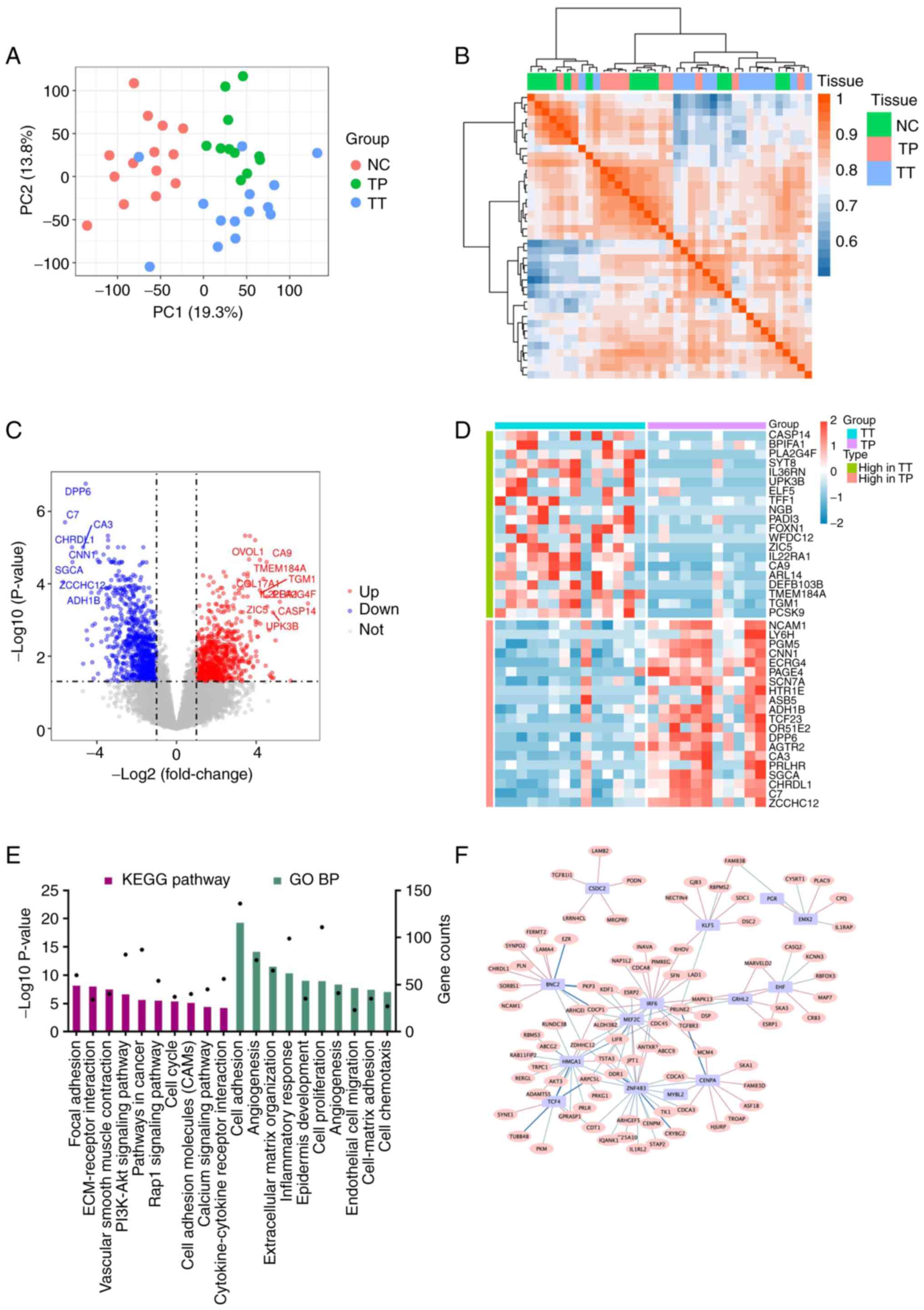
The PCA diagram shows an obvious distinction between
TT, TP and NC tissue (Fig. 1A and
B). To investigate transcriptomic differences between TT and
TP, the DEGs between the two groups were analysed. DEG analysis
revealed that there were 1,405 upregulated DEGs and 1,256
downregulated DEGs in the TT group compared with the TP group
(Fig. 1C and D). Furthermore, KEGG
pathway and GO analyses of the DEGs were performed. KEGG analysis
demonstrated that the pathways such as ‘focal adhesion’,
‘ECM-receptor interaction’, ‘vascular smooth muscle contraction’,
‘PI3K-Akt signalling pathway’ and ‘pathways in cancer’ were
enriched, among others. Additionally, GO analysis revealed the
functional enrichment of pathways such as ‘cell adhesion’, among
others (Fig. 1E). The co-network
analysis indicated that transcription factors, such as myocyte
enhancer factor 2C, interferon regulatory factor 6 and high
mobility group AT-hook 1, have regulatory effects on DEGs (Fig. 1F).
RNA-seq data reflect disease states
and stages
A graph was plotted based on a discriminative
dimensionality reduction tree analysis using the Monocle package to
demonstrate the developmental path of CC. The trajectory had a tree
structure with NC as the root state. The graph depicts the tree
best fitted to the CC developmental trajectory. The solid black
line represents the main route of the minimal spanning tree
constructed, which exhibited the backbone and order of CC
development along a pseudo-temporal continuum (NC-TP-TT; Fig. 2A). The top turning point-enriched
genes, such as ISG15, acyl-CoA thioesterase 7 and ribosomal
protein L22, are shown in Fig. 2B.
A PPI network was constructed from these genes, and it was revealed
that ISG15 had a significant regulatory role (Fig. 2C). Functional analysis revealed
significant enrichment of the ‘cell adhesion molecules (CAMs)’
pathway and ‘cell adhesion’ (Fig.
2D).
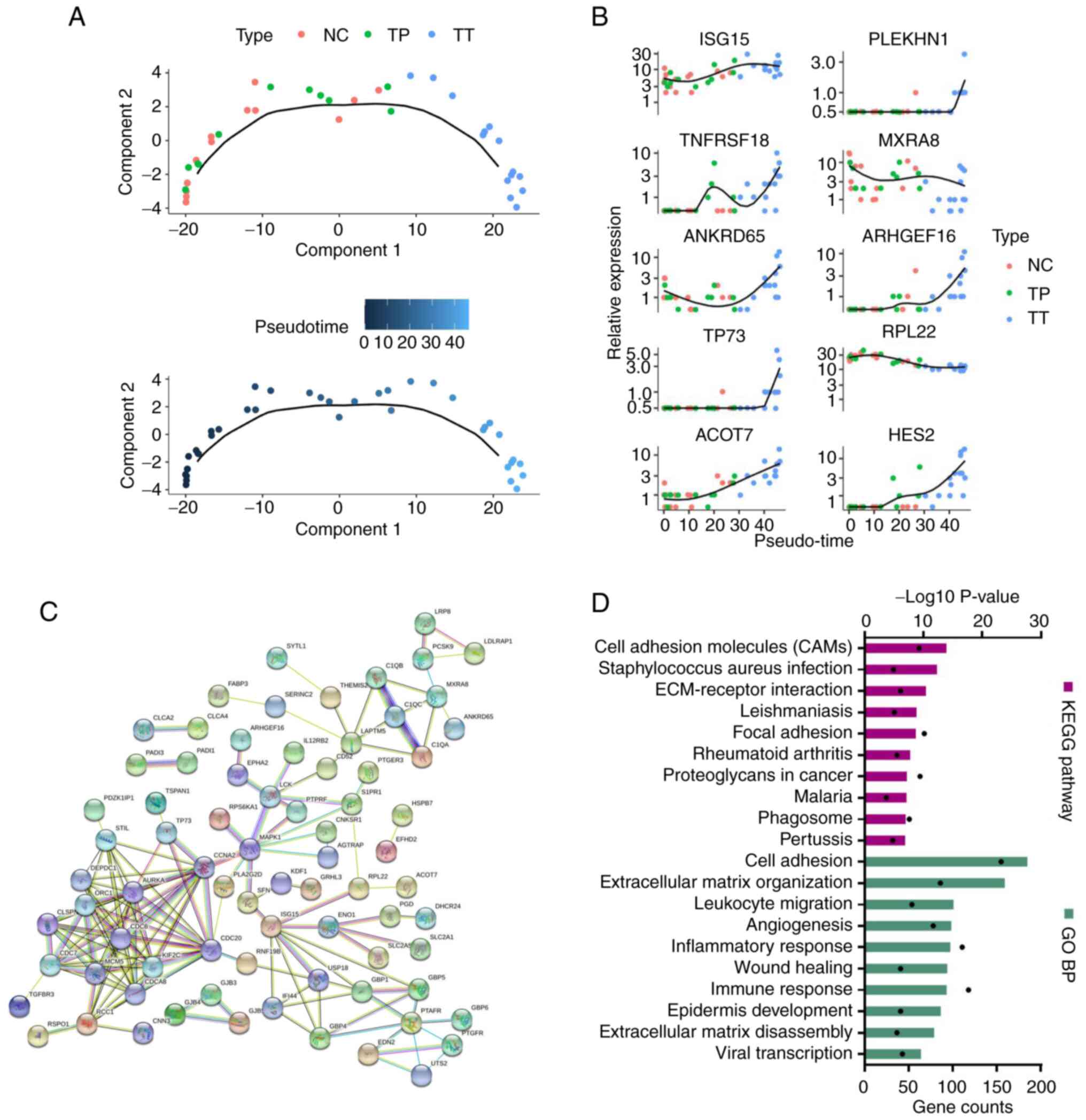 | Figure 2.RNA sequencing data reflected disease
states and stages. (A) Pseudo-time trajectory analysis of cervical
cancer development. (B) Top turning point-enriched genes, such as
ISG15, ACOT7 and RPL22, in pseudo-time trajectory
analysis. (C) Protein-protein interaction network showing that
ISG15 had a significant regulatory role. (D) KEGG pathway
and GO BP analysis of turning point-enriched genes showing P-values
(black dots) and gene counts (bars). ACOT7, acyl-CoA
thioesterase 7; ANKRD65, ankyrin repeat domain 65;
ARHGEF16, rho guanine nucleotide exchange factor 16; BP,
biological process; GO, Gene Ontology; HES2, hes family bHLH
transcription factor 2; ISG15, ISG15 ubiquitin-like
modifier; KEGG, Kyoto Encyclopedia of Genes and Genomes;
MXRA8, matrix remodeling associated 8; NC, normal tissue;
PLEKHN1, pleckstrin homology domain containing N1;
RPL22, ribosomal protein L22; TNFRSF18, TNF receptor
superfamily member 18; TP, paracarcinoma tissue; TP73,
tumour protein p73; TT, primary tumour tissue. |
Validation of the significance of
ISG15 expression in CC
TCGA survival (14)
analysis demonstrated that ISG15 expression had no
significant effect on the prognosis of patients with CC (Fig. 3A). RT-qPCR (Fig. 3B) and IHC assays (Fig. 3C) were performed to verify the
regulatory relationships among NC, TP and TT. The results revealed
that ISG15 expression was significantly higher in the TT
group compared with that in the TP group. In addition, RT-qPCR
assays demonstrated that the ISG15 expression in the TP
group was higher than that in the NC group. As shown in Table I, the associations between
ISG15 and the clinical features of CC indicated that
ISG15 expression was significantly associated with tumour
stage (P=0.0047).
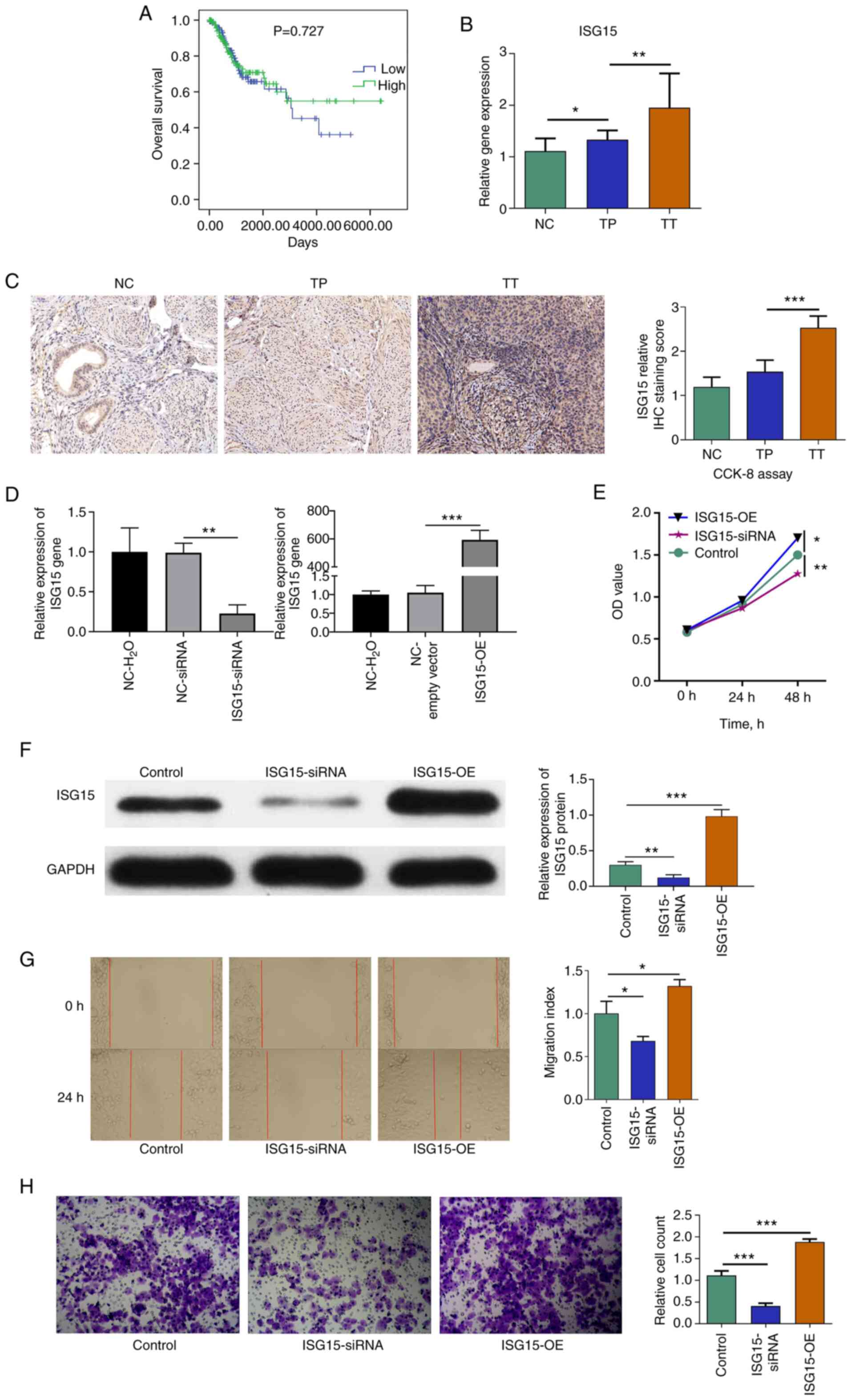 | Figure 3.ISG15 promotes the
proliferation and invasion of HeLa cells. (A) The Cancer Genome
Atlas survival analysis of ISG15. (B) Relative ISG15 expression in
cervical cancer tissues. (C) IHC staining (×200 magnification) and
ISG15 score. (D) RT-qPCR results demonstrated the success of the
ISG15 interference assay, where ISG15 expression was reduced in
cells transfected with ISG15 siRNA compared with the siRNA negative
control (P<0.01); similarly, ISG15 expression was increased in
cells transfected with the ISG15 overexpression vector compared
with the empty vector (P<0.001). (E) A cell proliferation assay
indicated that ISG15 promoted the proliferation of HeLa
cells. (F) Western blot analysis revealed a significant decrease in
ISG15 expression in the ISG15 siRNA group compared with the control
group (P<0.01) and an upregulation of ISG15 expression in the
ISG15-OE group (P<0.001). (G) Cell scratch assays (×100
magnfication) demonstrated that inhibition of ISG15
effectively inhibited the migration of HeLa cells. (H) Transwell
assays (×100 magnification) revealed that invasion was
significantly inhibited in the cells treated with
ISG15-siRNA. *P<0.05, **P<0.01,
***P<0.001. CCK-8, Cell Counting Kit-8; IHC,
immunohistochemistry; ISG15, ISG15 ubiquitin-like modifier;
NC, normal tissue; OD, optical density; siRNA, small interfering
RNA; TP, paracarcinoma tissue; TT, primary tumour tissue. |
ISG15-siRNA inhibits the proliferation
and invasion of HeLa cells
To further investigate the effect of ISG15 on
CC cell proliferation and migration, ISG15 interference
experiments were performed using HeLa cells in vitro. The
results of transfection showed that ISG15 expression was reduced in
cells transfected with ISG15 siRNA compared with the siRNA negative
control (P<0.01). Similarly, ISG15 expression was increased in
cells transfected with the ISG15 overexpression vector (ISG15-OE)
compared with the empty vector (P<0.001) (Fig. 3D). CCK-8 assays indicated that the
inhibition of ISG15 significantly reduced the proliferation of HeLa
cells (48 h; P<0.01), and overexpression of ISG15 significantly
increased the proliferation of HeLa cells (48 h; P<0.05)
compared with the control (Fig.
3E). Western blot analysis revealed a significant decrease in
ISG15 expression in the ISG15-siRNA group compared with the control
group (P<0.01) and upregulation of ISG15 in the ISG15-OE group
(P<0.001) (Fig. 3F). Scratch
assays demonstrated that the inhibition of ISG15 effectively
inhibited the migration of HeLa cells (P<0.05), and
overexpression of ISG15 effectively promoted the migration of HeLa
cells (P<0.05) compared with the control (Fig. 3G). In addition, Transwell assays
revealed that invasion was significantly inhibited in the cells
treated with ISG15-siRNA (P<0.001), and invasion was
significantly promoted in the cells treated with ISG15-OE
(P<0.001) compared with the control (Fig. 3H).
Discussion
In the present study, the transcriptomic differences
among NC, TP, and TT tissues from patients with CC were analysed
and tumour progression was simulated using pseudo-time analysis. It
was observed that ISG15 served a key role in the development
of CC. The results of immunohistochemical and RT-qPCR assays
indicated that ISG15 expression was increased in TT. An
increasing trend of ISG15 expression from NC to TP to TT was
also observed, which suggests that higher expression of
ISG15 was closely associated with the malignant evolution of
CC tissues. HeLa cell experiments demonstrated that
ISG15-siRNA inhibited cell proliferation and invasion.
ISG15 is one of the genes that are most
highly induced in response to viral infection (15). A study has demonstrated that
ISGylation of the HPV L1 capsid protein has a dominant inhibitory
effect on the infectivity of HPV16 pseudoviruses (15). Cannella et al (16) observed higher ISG15
expression in patients with CC with low-risk HPV infection than in
those with high-risk HPV. Pierangeli et al (17) found that ISG15 expression was
higher in low-risk HPV infection samples than in HPV-negative
samples, while high-risk HPV infections had a low ISG15 level.
Rajkumar et al (18) found
that ISG15 was upregulated in patients with CC compared with normal
controls, which was confirmed in the present study, suggesting that
ISG15 may serve an important role in the progression of CC.
Several studies have provided evidence that
ISG15 and its conjugation are involved in cancer
pathogenesis (19–22). ISG15 is secreted or released
by various cell types, including fibroblasts, neutrophils,
monocytes, and lymphocytes. ISG15 has emerged as a pivotal
regulator in diverse cellular processes, including proliferation,
apoptosis, and DNA repair. The receptor for extracellular
ISG15 has been identified as leukocyte function-associated
antigen-1 (LFA-1), an adhesion molecule of the integrin family
composed of an αL and β2 subunit (23,24).
LFA-1 binding to intercellular adhesion molecule 1 is critical in
homing leukocytes to sites of inflammation (23,24).
Elevated ISG15 expression has been demonstrated to be
required for tumourigenic phenotypes in triple-negative breast
cancer (TNBC) possessing inactivated p53 and ADP ribosylation
factor, suggesting the potential role of ISG15 in TNBC
development and progression (25).
High levels of ISG15 accelerate the invasion of breast
cancer cells and are associated with poor prognosis in patients
with breast cancer (25–28). ISG15 and enzymes involved in
ISGylation promote the proliferation and migration of
hepatocellular carcinoma (HCC) by stabilizing apoptosis inhibitors
(21,29). Furthermore, the expression of
ISG15 is higher in HBV-related HCC tissues than in
non-tumour tissues (21,29). The expression of ISG15 has
been demonstrated to be elevated in oesophageal squamous cell
carcinoma (ESCC) and to be closely associated with clinical
outcomes, indicating that ISG15 may be used as a prognostic
biomarker in patients with ESCC (30).
The present results indicated that ISG15 promotes CC
cell proliferation and invasion. ISG15 may act as an
oncogene in the tumourigenesis of CC. Taken together, these data
demonstrate that ISG15 serves pivotal roles in cancer cell
proliferation and apoptotic cell death.
The limitation of the present study was the small
number of patient cases enrolled. Survival analysis could not be
performed because of the small sample size. However, ISG15
expression had no significant effect on the prognosis of patients
with CC in TCGA data. In conclusion, the present study demonstrated
that ISG15 was upregulated in CC tumour tissue and
positively associated with the development of CC. ISG15 may
be a potential anticancer drug target for future CC-targeted drug
discovery.
Acknowledgements
Not applicable.
Funding
The present study was supported by Science and Technology
Development Fund of Shanghai Pudong New Area (grant nos.
PKJ2017-Y34 and PKJ2021-Y21).
Availability of data and materials
The datasets generated and/or analysed during the
current study are available in the GEO database repository
(GSE192804, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE192804).
All other data used and/or analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
PT, LS, FL and BY designed the research, performed
the analyses and wrote the paper. YS, YW and YY performed
experiments. PT and BY confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Ethics
Committee of the Pudong New Area People's Hospital Affiliated to
Shanghai Health University (PRYLL-QKJW1703; Shanghai, China).
Written informed consent to participate in the study was obtained
from each patient before they entered the study.
Patient consent for publication
Patient consent for publication was covered by the
informed consent document.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cohen PA, Jhingran A, Oaknin A and Denny
L: Cervical cancer. Lancet. 393:169–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li H, Wu X and Cheng X: Advances in
diagnosis and treatment of metastatic cervical cancer. J Gynecol
Oncol. 27:e432016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang H, Zhao Y, Chen M and Cui J:
Identification of novel long non-coding and circular RNAs in human
papillomavirus-mediated cervical cancer. Front Microbiol.
8:17202017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Choi YJ and Park JS: Clinical significance
of human papillomavirus genotyping. J Gynecol Oncol. 27:e212016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stanley MA: Epithelial cell responses to
infection with human papillomavirus. Clin Microbiol Rev.
25:215–222. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hong S, Mehta KP and Laimins LA:
Suppression of STAT-1 expression by human papillomaviruses is
necessary for differentiation-dependent genome amplification and
plasmid maintenance. J Virol. 85:9486–9494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Tian R, Gao H, Yang Y, Williams BRG,
Gantier MP, McMillan NAJ, Xu D, Hu Y and Gao Y: Identification of a
histone family gene signature for predicting the prognosis of
cervical cancer patients. Sci Rep. 7:164952017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brant AC, Menezes AN, Felix SP, de Almeida
LM, Sammeth M and Moreira MAM: Characterization of HPV integration,
viral gene expression and E6E7 alternative transcripts by RNA-Seq:
A descriptive study in invasive cervical cancer. Genomics.
111:1853–1861. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gong Z, Liu J, Xie X, Xu X, Wu P, Li H,
Wang Y, Li W and Xiong J: Identification of potential target genes
of USP22 via ChIP-seq and RNA-seq analysis in HeLa cells. Genet Mol
Biol. 41:488–495. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu B, Chen L, Zhang W, Li Y, Zhang Y, Gao
Y, Teng X, Zou L, Wang Q, Jia H, et al: TOP2A and CENPF are
synergistic master regulators activated in cervical cancer. BMC Med
Genomics. 13:1452020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu YP, Wang ZQ, Liang XD, Wang Y and Wang
JL: Comparative analysis of the prognosis of patients with locally
advanced cervical cancer undergoing laparoscopic or abdominal
surgery. Zhonghua Fu Chan Ke Za Zhi. 55:609–616. 2020.(In Chinese).
PubMed/NCBI
|
|
12
|
Berek JS, Matsuo K, Grubbs BH, Gaffney DK,
Lee SI, Kilcoyne A, Cheon GJ, Yoo CW, Li L, Shao Y, et al:
Multidisciplinary perspectives on newly revised 2018 FIGO staging
of cancer of the cervix uteri. J Gynecol Oncol. 30:e402019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Anaya J: OncoLnc: Linking TCGA survival
data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2:e672016.
View Article : Google Scholar
|
|
15
|
Durfee LA, Lyon N, Seo K and Huibregtse
JM: The ISG15 conjugation system broadly targets newly synthesized
proteins: Implications for the antiviral function of ISG15. Mol
Cell. 38:722–732. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cannella F, Scagnolari C, Selvaggi C,
Stentella P, Recine N, Antonelli G and Pierangeli A: Interferon
lambda 1 expression in cervical cells differs between low-risk and
high-risk human papillomavirus-positive women. Med Microbiol
Immunol. 203:177–184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pierangeli A, Degener AM, Ferreri ML, Riva
E, Rizzo B, Turriziani O, Luciani S, Scagnolari C and Antonelli G:
Interferon-induced gene expression in cervical mucosa during human
papillomavirus infection. Int J Immunopathol Pharmacol. 24:217–223.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rajkumar T, Sabitha K, Vijayalakshmi N,
Shirley S, Bose MV, Gopal G and Selvaluxmy G: Identification and
validation of genes involved in cervical tumourigenesis. BMC
Cancer. 11:802011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Desai SD, Reed RE, Burks J, Wood LM,
Pullikuth AK, Haas AL, Liu LF, Breslin JW, Meiners S and Sankar S:
ISG15 disrupts cytoskeletal architecture and promotes motility in
human breast cancer cells. Exp Biol Med (Maywood). 237:38–49. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kiessling A, Hogrefe C, Erb S, Bobach C,
Fuessel S, Wessjohann L and Seliger B: Expression, regulation and
function of the ISGylation system in prostate cancer. Oncogene.
28:2606–2620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Wang J, Zhang H, Zhu M, Chen F, Hu
Y, Liu H and Zhu H: Interferon-stimulated gene 15 (ISG15) is a
trigger for tumorigenesis and metastasis of hepatocellular
carcinoma. Oncotarget. 5:8429–8441. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wood LM, Pan ZK, Seavey MM, Muthukumaran G
and Paterson Y: The ubiquitin-like protein, ISG15, is a novel
tumor-associated antigen for cancer immunotherapy. Cancer Immunol
Immunother. 61:689–700. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Swaim CD, Scott AF, Canadeo LA and
Huibregtse JM: Extracellular ISG15 signals cytokine secretion
through the LFA-1 integrin receptor. Mol Cell. 68:581–590.e5. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
D'Cunha J, Knight E Jr, Haas AL, Truitt RL
and Borden EC: Immunoregulatory properties of ISG15, an
interferon-induced cytokine. Proc Natl Acad Sci USA. 93:211–215.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Forys JT, Kuzmicki CE, Saporita AJ,
Winkeler CL, Maggi LB Jr and Weber JD: ARF and p53 coordinate tumor
suppression of an oncogenic IFN-β-STAT1-ISG15 signaling axis. Cell
Rep. 7:514–526. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hadjivasiliou A: ISG15 implicated in
cytoskeleton disruption and promotion of breast cancer. Expert Rev
Proteomics. 9:72012.
|
|
27
|
Cerikan B and Schiebel E: DOCK6
inactivation highlights ISGylation as RHO-GTPase balancer. Cell
Cycle. 16:304–305. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burks J, Reed RE and Desai SD: ISGylation
governs the oncogenic function of Ki-Ras in breast cancer.
Oncogene. 33:794–803. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qiu X, Hong Y, Yang D, Xia M, Zhu H, Li Q,
Xie H, Wu Q, Liu C and Zuo C: ISG15 as a novel prognostic biomarker
for hepatitis B virus-related hepatocellular carcinoma. Int J Clin
Exp Med. 8:17140–17150. 2015.PubMed/NCBI
|
|
30
|
Yan W, Shih JH, Rodriguez-Canales J,
Tangrea MA, Ylaya K, Hipp J, Player A, Hu N, Goldstein AM, Taylor
PR, et al: Identification of unique expression signatures and
therapeutic targets in esophageal squamous cell carcinoma. BMC Res
Notes. 5:732012. View Article : Google Scholar : PubMed/NCBI
|