Introduction
Ovarian cancer (OC) is a gynecological cancer which
contributes to a large number of deaths each year in industrialized
countries; in 2020, it was estimated that there were 21,750 new
cases and 13,940 associated deaths in the United States (1). It is estimated that 80% of patients
with OC are eligible for the gold standard treatment of aggressive
surgical debulking and platinum-based chemotherapy (2) and that 70% of these will develop
platinum resistance and fatal disease following the long-term use
of platinum (3). The outcomes of
patients with platinum-resistant OC are poor, with a median overall
survival (OS) rate of <12 months (4).
Resistance to platinum-based chemotherapy is a major
clinical challenge in the treatment of OC, which results in a high
mortality-to-incidence ratio (5).
Epithelial OC (EOC) presents at an advanced stage globally and is
the most common cause of gynecological cancer-associated mortality
(6). In recent years, there have
been notable achievements in the development of treatments of EOC,
which have been validated by landmark clinical trials, such as a
combination of surgery and systemic therapy, targeted therapy,
chemotherapy and maximal surgical effort, of which the latter
remains the mainstay (7).
Therefore, overcoming platinum resistance is key to improving the
prognosis of patients with EOC.
Numerous mechanisms and biological pathways
underlying platinum resistance are being investigated. It has been
reported that cisplatin functions by covalently binding to the DNA
of tumor cells to form platinum-DNA adducts and induces cell
apoptosis (8,9). One established mechanism for
cisplatin resistance is evasion of cell apoptosis following
long-term use of cisplatin (10),
which results in resistance to cisplatin (11,12).
Integrated genomic analysis of EOC reported one of the most common
focal amplifications to be in the 8q24 region containing MYC
(13). MYCC, MYCN and MYCL nuclear
proteins are members of the Myc family that bind to and control
~15% of the human genome (14). It
has been reported that downregulation of MYCN does not influence
other MYC members such as MYCC and MYCL (15). MYC belongs to the Myc
proto-oncogene family, which encodes basic helix-loop-helix/leucine
zipper transcription factors, and functions in numerous types of
human malignancy (14), including
lung cancer (16) and mammary
adenocarcinomas (17). Moreover,
the Myc signaling pathway is one of the most commonly activated
oncogenic pathways in human malignancy (18). Furthermore, previous studies have
reported that Myc-mediated transcriptional networks are under tight
regulation in normal cells and control numerous cellular processes,
such as metabolic processes and cell proliferation, differentiation
and apoptosis (14,19,20).
As a member of the MYC family, MYCN is a type of short-lived
transcription factor that is dysregulated in numerous types of
human cancer (21), serves as a
therapeutic target (22) and is
associated with poor clinical outcome in multiple types of cancer
(23). It has been reported that
amplification of MYCN is associated with an aggressive phenotype
and a poor prognosis in neuroblastoma; relapse of
platinum-resistant neuroblastoma is the primary cause of mortality
in patients with MYCN amplification (24). MYCN overexpression is significantly
associated with poor outcomes in breast cancer (25). However, MYCN contributes to
cisplatin sensitization in acute myelogenous leukemia (26).
The function of MYCN in EOC and chemotherapeutic
resistance remains unclear. Therefore, the present study assessed
the role of MYCN in EOC chemotherapeutic resistance.
Materials and methods
Bioinformatics analysis
The datasets used in the present study are available
from The Cancer Genome Atlas (TCGA) database
(tcga-data.nci.nih.gov/tcga) under TCGA-OV project (13). The analysis of the OS of patients
receiving platin-based therapy was performed using Kaplan-Meier
Plotter software
(kmplot.com/analysis/index.php?p=service&cancer=ovar) with auto
select best cut-off.
The GSE114206 dataset (27), which contains mRNA expression
profiles of 12 patients with EOC (cisplatin-resistant patients,
n=6; cisplatin-sensitive patients, n=6) was obtained from Gene
Expression Omnibus (GEO) database (ncbi.nlm.nih.gov/geo/).
RNA sequencing data of 300 patients with OC (TCGA
pan-cancer project) was downloaded from cBioportal
(cbioportal.org/) (13). The
co-expressed genes, assessed using Spearman's correlation analysis,
were subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG)
(https://www.genome.jp/kegg/) analysis
using DAVID software version 2.0 (https://david.ncifcrf.gov/). For gene set enrichment
analysis (GSEA), the expression matrix was separated according to
the median expression of MYCN. The expression matrix was used for
Hallmark gene set enrichment using GSEA software (V.4.1.0)
(gsea-msigdb.org/gsea/index.jsp).
Tissue samples
In total, 26 female patients with EOC who underwent
primary surgery followed by cisplatin-based chemotherapy at the
First Affiliated Hospital of Chongqing Medical University
(Chongqing, China) between 2015 and 2019 were enrolled in the
present study. None of these patients received radiotherapy before
the surgery. The subtypes were assessed using histological
examination performed by pathologists. A total of 22 patients
(84.6%) were assessed as being serous and 4 (15.4%) were assessed
as having mucinous EOC. Following primary chemotherapy, patients
who relapsed within 6 months were assigned to the
cisplatin-resistant group (n=13; mean age, 56 years; range, 37–70
years) and those who relapsed after 6 months or did not relapse
were assigned to the cisplatin-sensitive group (n=13; mean age, 51
years; age range, 42–68 years). All patients provided written
informed consent prior to inclusion in the study. The present study
was approved by the Institutional Ethics Committee of The First
Affiliated Hospital of Chongqing Medical University (Approval No.
TFAHCQMU-2021-010). The characteristics of patients with EOC are
presented in Table I.
 | Table I.Characteristics of patients with
epithelial ovarian cancer. |
Table I.
Characteristics of patients with
epithelial ovarian cancer.
|
| Chemotherapy |
|
|---|
|
|
|
|
|---|
| Characteristic | Sensitive,
n=13.0 | Resistant,
n=13 | P-value |
|---|
| Median age, years
(range) | 51.0
(42.0-68.0) | 56.0
(37.0-70.0) | 0.198 |
| Histology |
|
| 0.296 |
| Serous
(%) | 10.0 (76.9) | 12.0 (92.3) |
|
|
Mucinous (%) | 3.0 (23.1) | 1.0 (7.7) |
|
| FIGO stage |
|
| 0.187 |
| I
(%) | 2.0 (15.4) | 0.0 (0.0) |
|
| II
(%) | 3.0 (23.1) | 1.0 (7.7) |
|
| III
(%) | 6.0 (46.1) | 11.0 (84.6) |
|
| IV
(%) | 2.0 (15.4) | 1.0 (7.7) |
|
| Grade |
|
| 0.500 |
| 1/2
(%) | 5.0 (38.5%) | 6.0 (46.2%) |
|
| 3
(%) | 8.0 (61.5%) | 7.0 (53.8%) |
|
| Median CA125 at
diagnosis, U/ml (range) | 474.0
(46.0-1,483.0) | 884.0
(28.5-3949) | 0.215 |
Immunohistochemical analysis
Samples were fixed by 4% PFA solution, embedded by
paraffin at 4°C overnight, sliced into 4 µm sections, and incubated
at 60°C for 30 min. Following deparaffinization by xylene I and
xylene II (each for 20 min) at room temperature, rehydration by
alcohol series (100, 95%, 85%, and 75%), antigen retrieval by
citric acid repair solution at oven for 5 min and endogenous
peroxidase inhibition by 3%H2O2 at room
temperature for 10 min, serous EOC sample slides were incubated
with anti-MYCN antibody (1:100; cat. no. 10159-2-AP; ProteinTech
Group, Inc.) at 4°C overnight. Slides were incubated with
goat-anti-rabbit horseradish peroxidase-conjugated secondary
antibodies (1:50; PR30009; ProteinTech Group, Inc.) for 1 h at room
temperature, followed by assessment of peroxidase activity using
diaminobenzidine for 10 min at room temperature. The tissue
sections were visualized using a light microscope (40×). The
statistical analysis was performed using histochemistry score
(H-score) as previously reported (28).
Cells and cell culture
The human EOC SK-OV-3 cell line, which is commonly
used in the study of cisplatin-resistant in serous EOC (29,30),
was purchased from Jiangsu KeyGEN BioTECH Co., Ltd. The human EOC
cisplatin-resistant SK-OV-3/DDP cell line was purchased from
Shanghai Chuan Qiu Biotechnology Co., Ltd. Cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% fetal bovine serum (PAN-Biotech GmbH) and 1%
penicillin/streptomycin (Beyotime Institute of Biotechnology) in an
incubator at 37°C with 5% CO2.
Cisplatin treatment
The SK-OV-3 and SK-OV-3/DDP cells were treated with
a range of concentrations of cisplatin (0, 5 and 10 µM) for 24 h at
37°C.
Lentivirus construction and
infection
The short hairpin (sh)RNA MYCN (LV-sh-MYCN;
5′-GCAGAAACCACAACATCCTGG-3′), negative control (LV-sh-NC;
5′-TTCTCCGAACGTGTCACGT-3′), MYCN-overexpressing (LV-MYCN) and NC
lentivirus (LV-NC with a scrambled sequence) were purchased from
Shanghai GenePharma Co., Ltd. The sequences were ligated into
plko.1-puro plasmid. The lentivirus was packaged by transfection of
2 µg plko.1-puro, 1 µg psPAX2 and 2 µg pMD2.G into 293 cells for 24
h. The supernatant was collected for harvesting lentivirus
particles. All lentiviruses contained GFP and puromycin resistance
genes. At 72 h post-transduction, cells (MOI=10) were selected
using puromycin (2 µg/ml) and maintained using puromycin (1 µg/ml)
(Beyotime Institute of Biotechnology). The transfection efficiency
in SK-OV-3 cells and SK-OV-3/DDP cells was assessed using western
blotting.
Western blotting
SK-OV-3 and SK-OV-3/DDP cells were treated with
cisplatin (0, 5 and 10 µM) for 24 h at 37°C and LV-sh-MYCN- SK-OV-3
cells and LV-MYCN SK-OV-3/DDP cells were treated with cisplatin (0
and 10 µM) for 24 h at 37°C, then harvested using PBS and lysed
using RIPA buffer [Roche Diagnostics (Shanghai) Co., Ltd.] with
protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). The
protein concentration was evaluated using the BCA method (Beyotime
Institute of Biotechnology). The extracted proteins (20 µg/lane)
were separated using 10% (MYCN, p53 and β-actin) or 12% (Bax and
Bcl2) SDS-PAGE, transferred to a PVDF membrane. Following blocking
using 5% skimmed milk for 2 h at room temperature, PVDF membranes
were incubated with primary antibodies overnight at 4°C and goat
anti-rabbit (1:1,000; cat. no. 7074; Cell Signaling Technology,
Inc.) and anti-mouse (1:1,000; cat. no. 7076; Cell Signaling
Technology, Inc.) horseradish peroxidase-conjugated secondary
antibodies (Cell Signaling Technology, Inc.) for 1 h at 37°C. The
proteins were visualized using chemiluminescence (ECL Plus Western
Blotting Detection system, Thermo Fisher). Primary antibodies were
as follows: MYCN (1:1,000; cat. no. 10159-2-AP; ProteinTech Group,
Inc.), MYCC (1:1,000; cat. no. 10828-1-AP; ProteinTech Group,
Inc.), MYCL1 (1:1,000; cat no. PA5-109998; Thermo Fisher
Scientific), β-actin (1:1,000; cat. no. 8457; Cell Signaling
Technology, Inc.), p53 (1:1,000; cat. no. 2527; Cell Signaling
Technology, Inc.), Bcl2 (1:1,000, cat. no. 15071; Cell Signaling
Technology, Inc.) and Bax (1:1,000, cat. no. 5023; Cell Signaling
Technology, Inc.). ImageJ software (version 1.8.0; National
Institutes of Health) was used for densitometric analysis of the
bands.
Cell Counting Kit-8 (CCK-8) assay
LV-sh-MYCN SK-OV-3 and LV-MYCN SK-OV-3/DDP cells
(1×104 cells/well) were seeded into a 96-well plate and
treated with 0, 2, 4, 6, 8, 10 and 15 µM cisplatin (Sigma-Aldrich;
Merck KGaA) for 24 h at 37°C. Cell viability was determined using
CCK-8 assay (Abcam), for which the cells were incubated at 37°C for
1 h. The absorbance was measured at 450 nm using an Infinite M200
PRO spectrophotometer (Tecan Group, Ltd.).
Apoptosis analysis
The number of apoptotic cells was quantified using
Annexin V-FITC/propidium iodide (PI) staining. LV-sh-MYCN SK-OV-3
and LV-MYCN SK-OV-3/DDP cells (1×105 cells/well) were
incubated in 6-well plates overnight at 37°C and treated with
cisplatin (0 and 10 µM) for 24 h at 37°C. Following centrifugation
at 2,000 × g for 3 min at room temperature, cells were suspended in
100 µl PBS, then mixed with 5 µl Annexin V-FITC (Beckman Coulter,
Inc.) and 5 µl PI (Beckman Coulter, Inc.) prior to incubation for
15 min in the dark at room temperature. Cells at an early stage
(FITC+/PI−) and late stage
(FITC+/PI+) were assessed as being apoptotic.
The apoptotic cell percentage was assessed using a BD FACSCalibur™
flow cytometer (BD Biosciences). Data were analyzed using FlowJo
software (version 7.6.3; FlowJo LLC).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0.1 (GraphPad Software, Inc.). Fisher's exact test was used
for the analysis of histology, FIGO stage and grade. Unpaired
Student's t test was used for analysis of age and CA125. H scores
are presented as the median + interquartile range; all other data
are from at least 3 independent experimental repeats, presented as
the mean ± standard deviation. Comparisons between 2 groups were
performed using Mann-Whitney test; for comparisons of ≥3 groups,
Kruskal-Wallis followed by Dunn's post hoc test was used. P<0.05
was considered to indicate a statistically significant
difference.
Results
High MYCN expression is positively
associated with greater OS in patients receiving platin-based
therapy
The data of 424 patients (low MYCN group, n=212;
high MYCN group, n=212) obtained from TCGA database were assessed
using OS analysis, which demonstrated that high expression of MYCN
was associated with greater OS (Fig.
1A). As platin-based therapy was the first-line therapy
approach for patients with OC, OS analysis was performed in
patients receiving platin-based therapy, which indicated that high
expression of MYCN was associated with a prolonged OS (Fig. 1B). The GSE114206 dataset, which
contains mRNA expression profiles of 12 patients with EOC
(cisplatin-resistant patients, n=6; cisplatin-sensitive patients,
n=6), was obtained from the GEO database. A heatmap of the top 50
differentially expressed genes demonstrated that MYCN was increased
in cisplatin-sensitive patients compared with cisplatin-resistant
patients (Fig. 1C). The
immunohistochemistry assessment of the tumor tissue collected in
the present study demonstrated that MYCN protein expression in
patients with cisplatin-sensitive EOC was significantly higher than
that in cisplatin-resistant EOC (Fig.
1D and E).
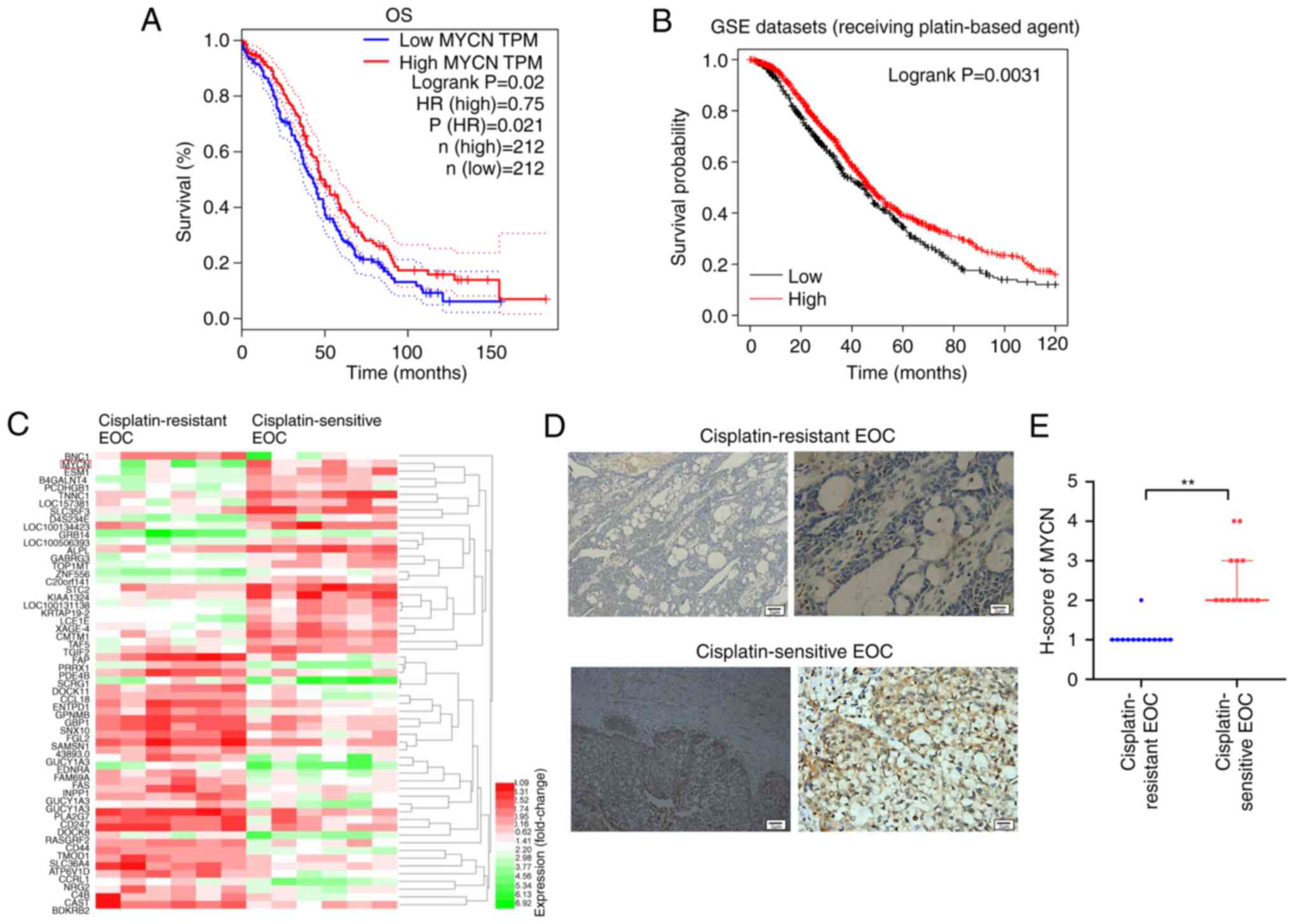 | Figure 1.Bioinformatics analysis. (A) OS
analysis of The Cancer Genome Atlas dataset using Kaplan-Meier
plotter (low MYCN group, n=212; high MYCN group, n=212). (B) OS
analysis of GSEA datasets (patients with OC receiving platin-based
therapy) using the Kaplan-Meier plotter (low, n=474; high, n=935).
(C) Gene expression profiling of GSE114206 dataset was performed
using microarray analysis (cisplatin-resistant patients, n=6;
cisplatin-sensitive patients, n=6). (D) MYCN protein expression in
tumor tissue from patients with cisplatin-resistant and -sensitive
EOC assessed using immunohistochemistry (n=13). (E) Statistical
analysis of MYCN protein expression levels according to the
H-score. Scale bar=2 µm. **P<0.01. OS, overall
survival; TPM, Transcripts Per Million; HR, hazard ratio; GSEA,
gene set enrichment analysis; EOC, epithelial ovarian cancer;
H-score, histochemistry score. |
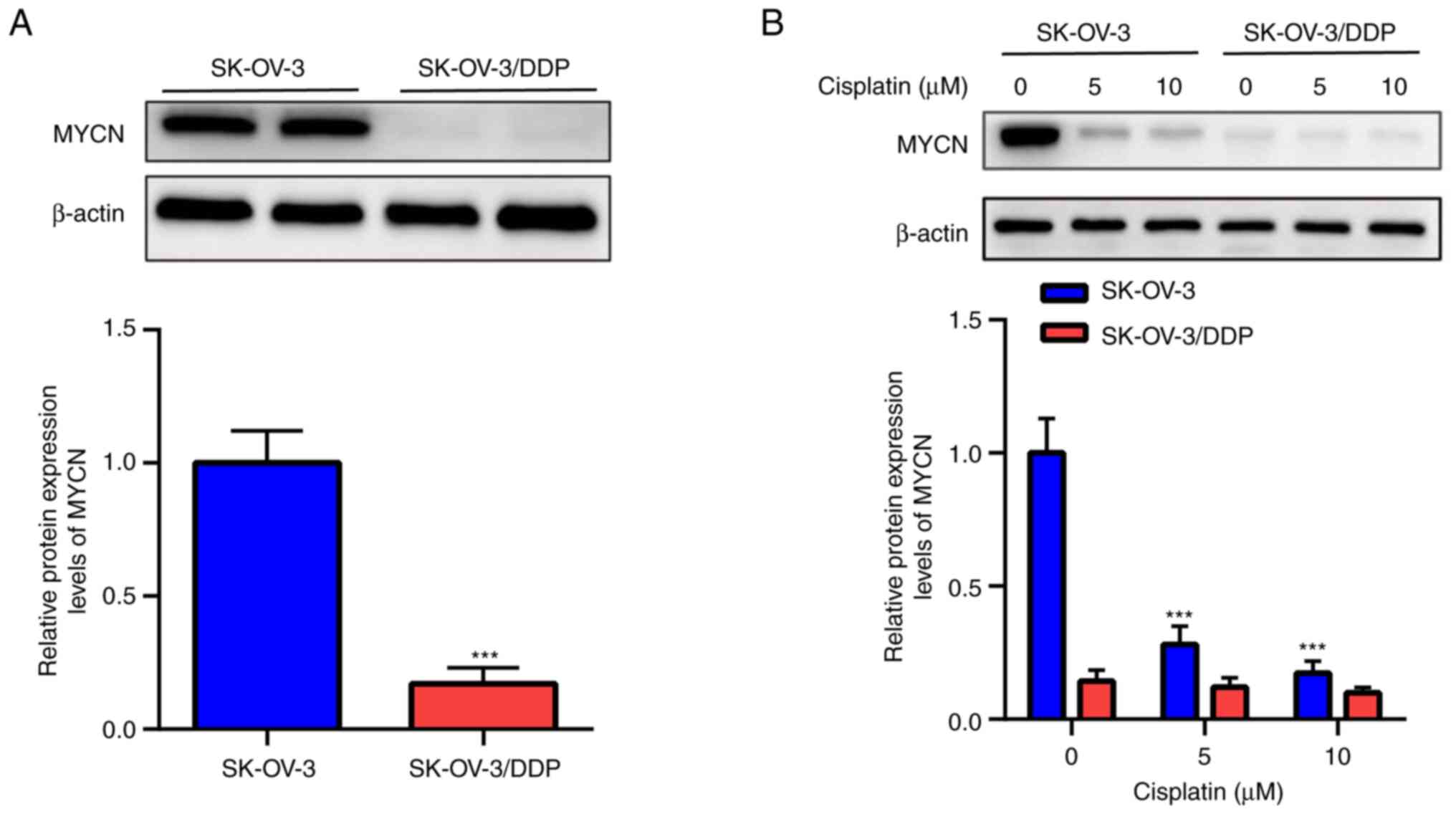
Low MYCN protein expression levels are
positively associated with cisplatin-resistance
Western blotting demonstrated that MYCN expression
was significantly lower in SK-OV-3/DDP compared with SK-OV-3 cells
(Fig. 2A). Moreover, cisplatin (0,
5 and 10 µM) significantly decreased MYCN protein expression in a
dose-dependent manner in SK-OV-3 cells but not in SK-OV-3/DDP cells
(Fig. 2B).
MYCN downregulation promotes cisplatin
resistance in EOC cells
Western blotting demonstrated notable knockdown of
MYCN by LV-sh-MYCN compared with LV-sh-NC in SK-OV-3 cells
(Fig. 3A). The present study
assessed the effect of MYCN knockdown on MYCC and MYCL protein
expression levels, which demonstrated that LV-sh-MYCN did not
affect MYCC and MYCL expression compared with LV-sh-NC in SK-OV-3
cells. CCK-8 assay demonstrated that cisplatin markedly decreased
SK-OV-3 cell viability in a dose-dependent manner; furthermore,
compared with LV-sh-NC, cell viability was significantly higher in
the LV-sh-MYCN group following cisplatin treatment (Fig. 3B). Flow cytometry of SK-OV-3 cells
demonstrated that cisplatin induced significant cell apoptosis in
the LV-sh-NC group compared with the untreated LV-sh-NC group, but
there was no significant difference between cisplatin treated group
and cisplatin untreated group in the LV-sh-MYCN group. Furthermore,
compared with LV-sh-NC, cisplatin-induced cell apoptosis was
significantly decreased in the LV-sh-MYCN group. In groups without
cisplatin treatment, there was significantly decreased cell
apoptosis in the LV-sh-MYCN group compared with the LV-sh-NC group
(Fig. 3C).
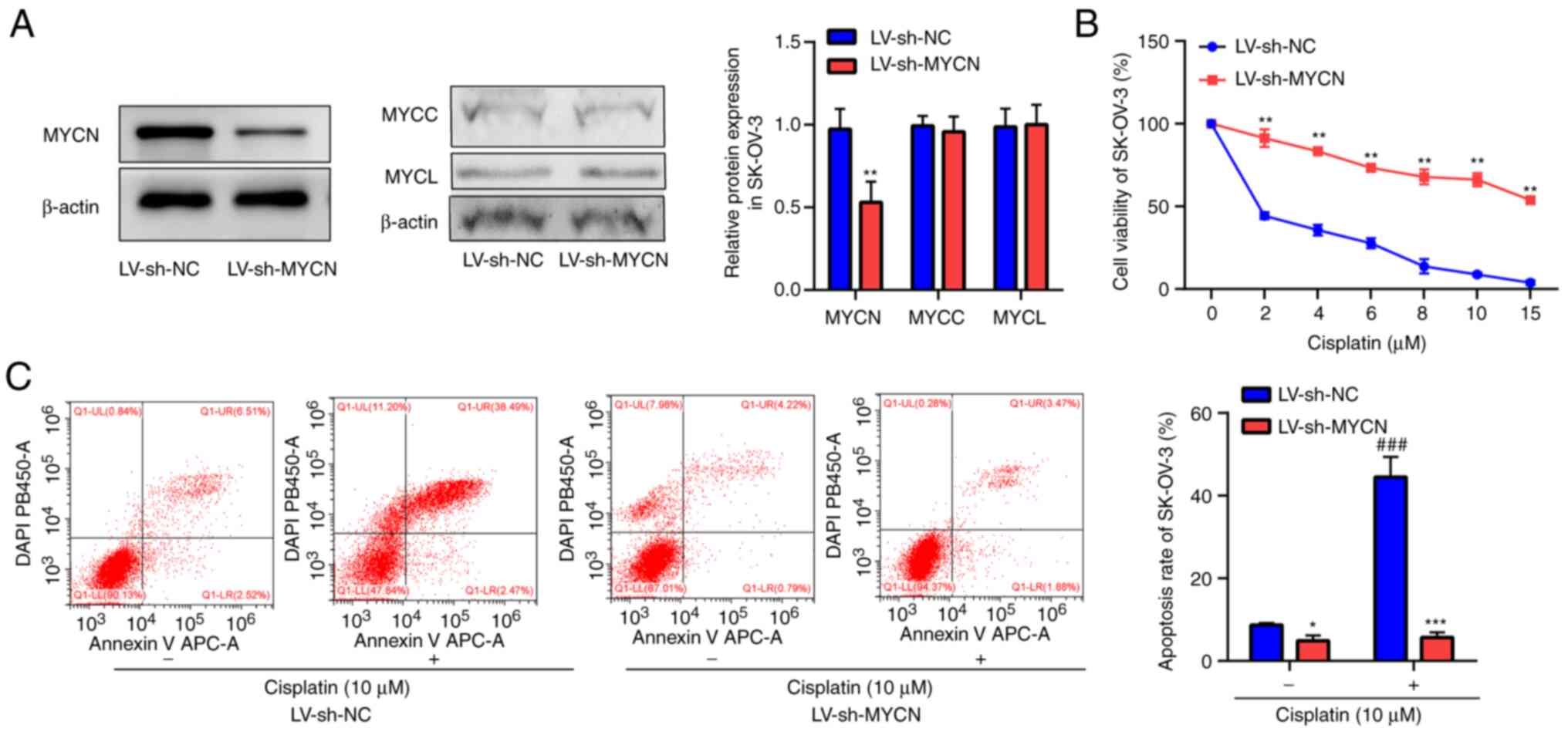 | Figure 3.Viability and apoptosis in SK-OV-3
cells. (A) Relative MYCN, MYCC and MYCL protein expression levels
in LV-sh-NC and LV-sh-MYCN cells were semi-quantified using western
blotting (n=3). (B) Viability of LV-sh-NC and LV-shRNA-MYCN cells
was assessed using the Cell Counting Kit-8 assay following
treatment with cisplatin (0, 2, 4, 6, 8, 10 and 15 µM) for 24 h
(n=5). (C) Apoptosis of LV-sh-NC and LV-sh-MYCN cells was assessed
using flow cytometry assay following cisplatin (10 µM) treatment
for 24 h (n=3). *P<0.05, **P<0.01 and
***P<0.001 vs. LV-sh-NC. ###P<0.001 vs.
cisplatin (0 µM). LV, lentivirus; sh, short hairpin; NC, negative
control. |
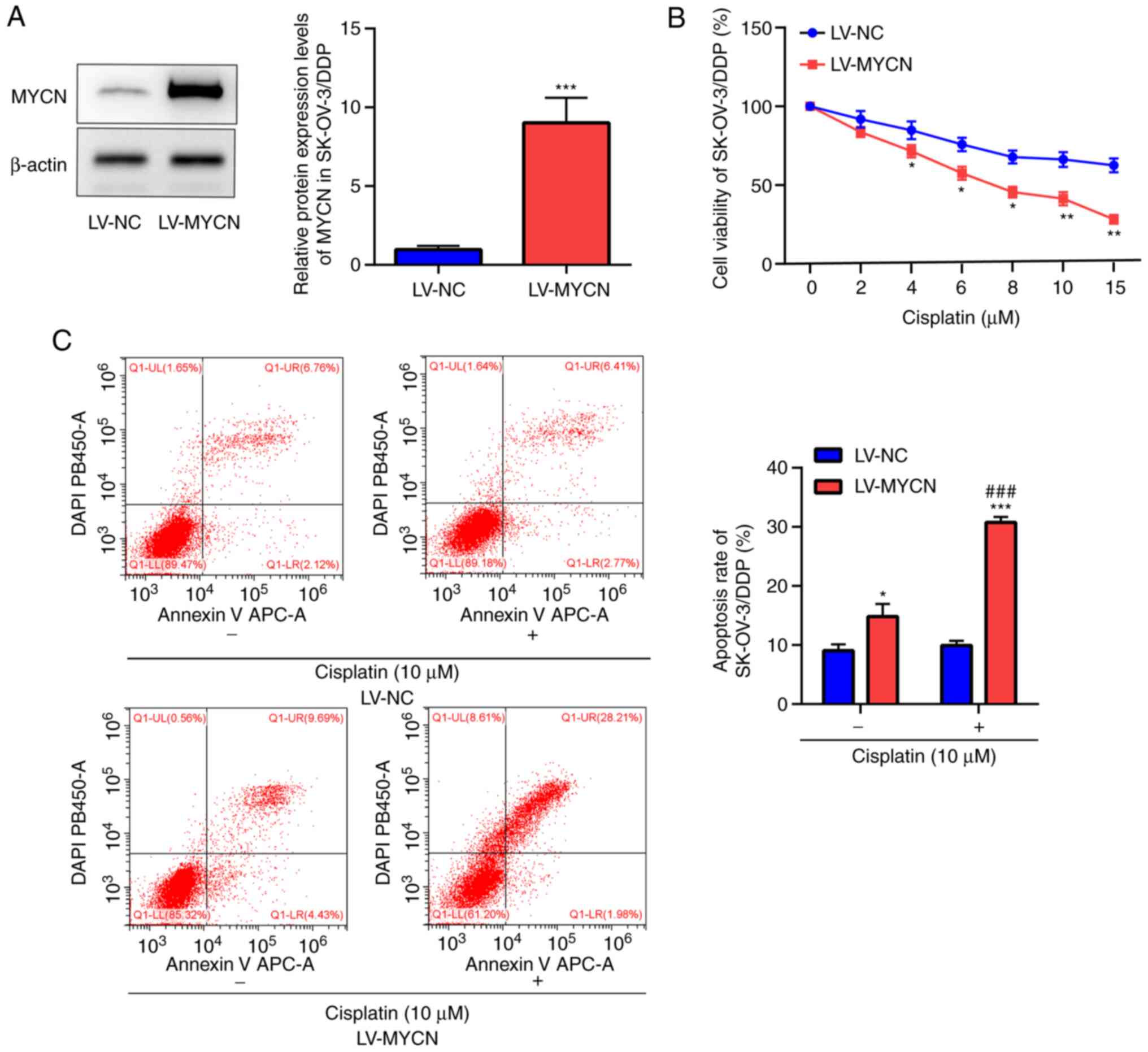
MYCN upregulation reverses cisplatin
resistance of EOC cells
Western blotting demonstrated significant
overexpression of MYCN in LV-MYCN SK-OV-3/DDP cells compared with
LV-NC in SK-OV-3/DDP cells (Fig.
4A). CCK-8 assay in the SK-OV-3/DDP cells demonstrated that
cisplatin markedly decreased cell viability in a dose-dependent
manner; moreover, compared with LV-NC, cell viability was
significantly decreased by cisplatin (≥4 µM) in the LV-MYCN group
(Fig. 4B). Flow cytometry
demonstrated that in the SK-OV-3/DDP cells, cisplatin induced
significantly increased apoptosis in the LV-MYCN group compared
with the LV-NC group; furthermore, compared with LV-NC,
cisplatin-induced cell apoptosis was significantly increased in the
LV-MYCN group. In the groups without cisplatin treatment, there was
significantly increased cell apoptosis in the LV-MYCN group
compared with the LV-NC group (Fig.
4C).
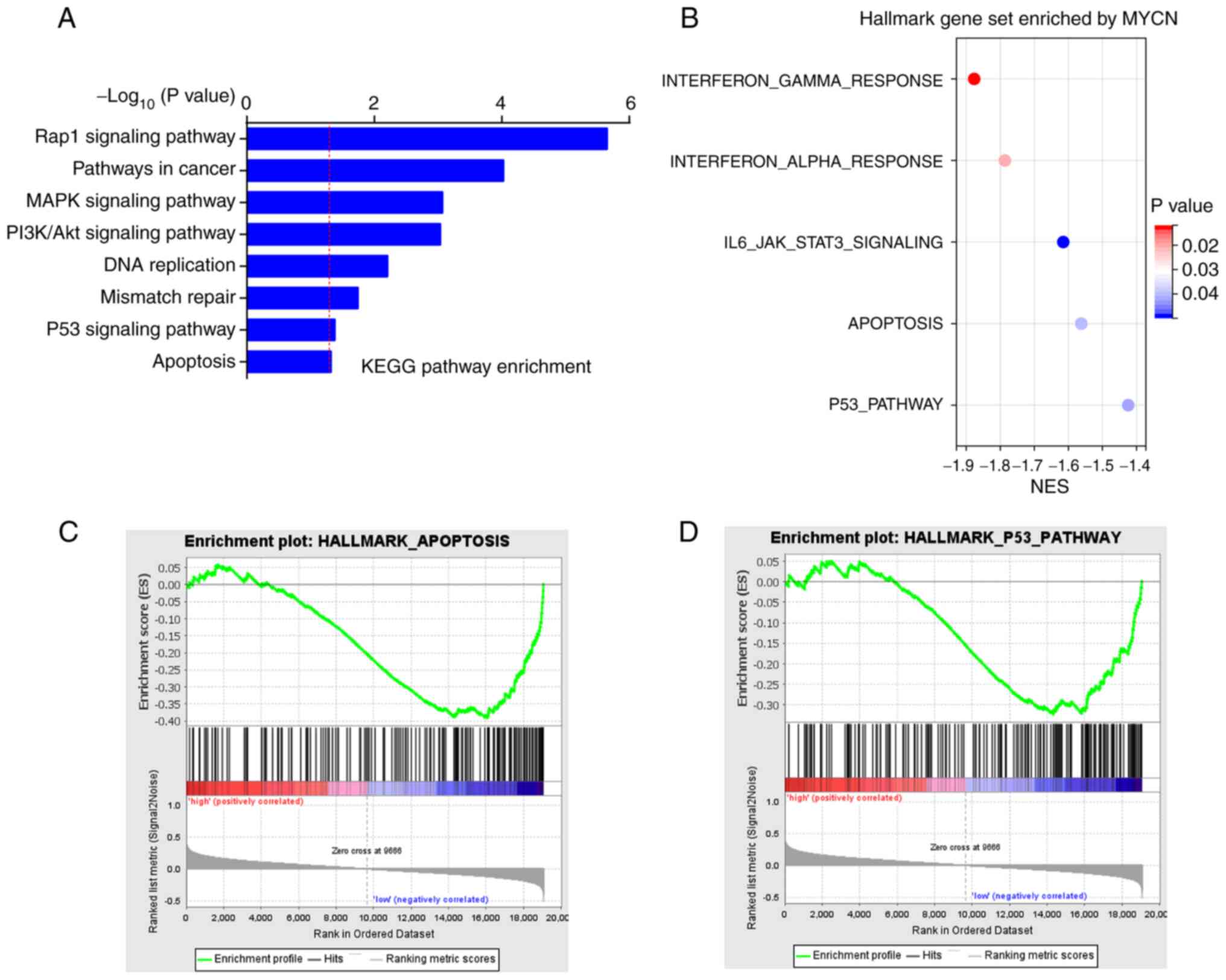
MYCN downregulation promotes cisplatin
resistance by decreasing cisplatin-induced apoptosis
KEGG enrichment analysis of co-expressed genes of
MYCN in the TCGA-OC dataset demonstrated that they were primarily
involved in pathways that contributed to chemotherapeutic
resistance and participated in cell apoptosis, such as ‘MAPK
signaling pathway’, ‘PI3K/AKT signaling pathway’ and ‘p53 signaling
pathway’ (Fig. 5A). Globally, GSEA
demonstrated that MYCN was enriched in ‘apoptosis’ and ‘p53
pathway’ in the hallmark gene set (Fig. 5B-D).
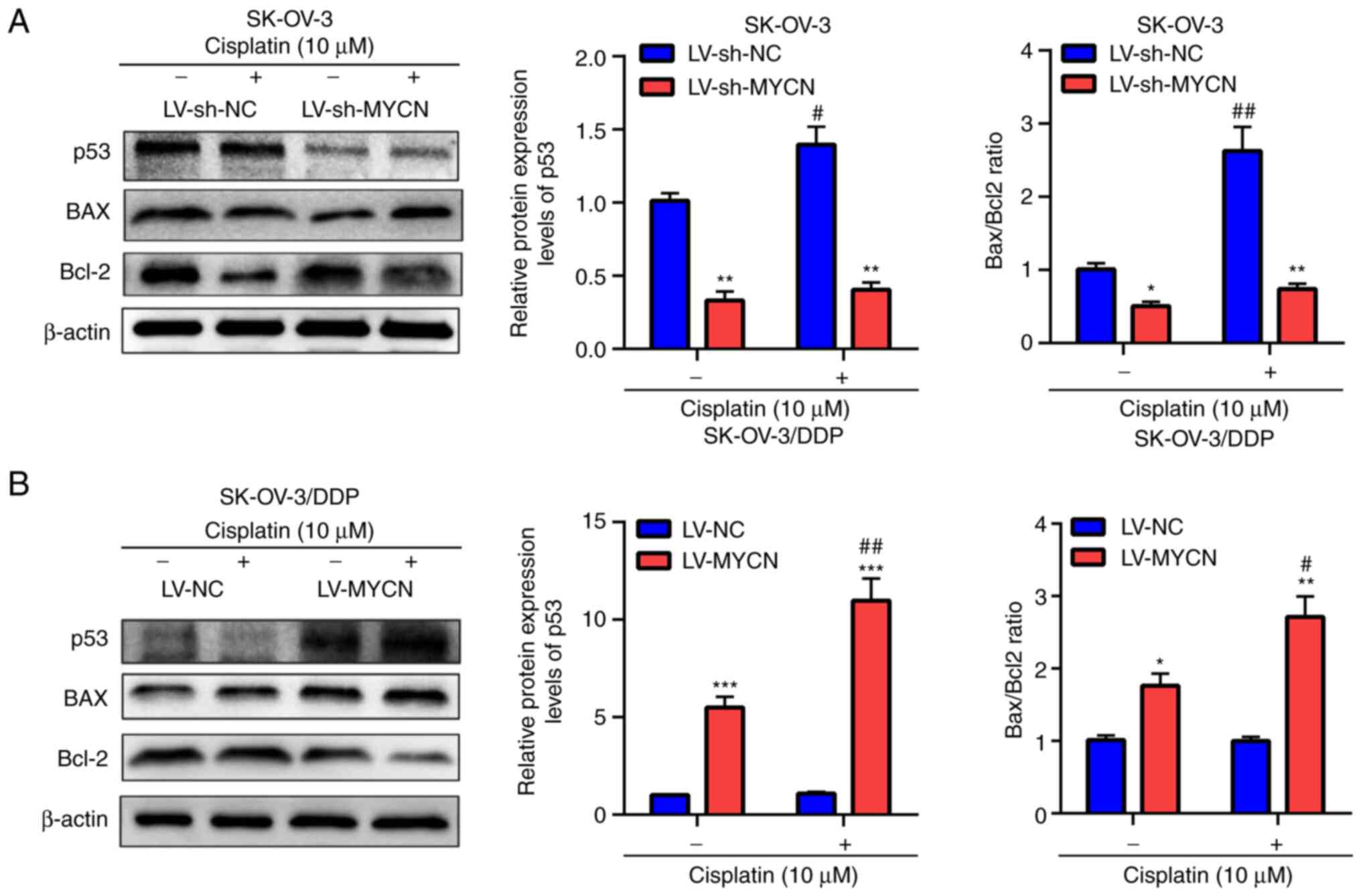
To evaluate these results, p53, BAX and Bcl-2
protein expression levels were assessed using western blotting. In
SK-OV-3 cells, treatment with cisplatin induced the significant
upregulation of p53 protein expression levels and significantly
increased the Bax/Bcl2 ratio in the LV-sh-NC group compared with
the untreated group; however no significant difference was
demonstrated in the LV-sh-MYCN group compared with the untreated
group (Fig. 6A). Furthermore,
compared with LV-sh-NC, cisplatin treatment induced significant
upregulation of p53 protein expression levels and significantly
decreased Bax/Bcl2 ratio in the LV-sh-MYCN group. Moreover, in the
groups without cisplatin treatment, there was a significantly lower
p53 protein expression and Bax/Bcl2 ratio in the LV-sh-MYCN group
compared with LV-sh-NC group (Fig.
6A). However, in the SK-OV-3/DDP cells, cisplatin treatment
induced significant upregulation of p53 protein expression levels
and significantly increased Bax/Bcl2 ratio in the LV-MYCN group
compared with the untreated group, however no significant
difference was demonstrated in the LV-NC group compared with the
untreated group. Furthermore, compared with LV-NC, cisplatin
induced significant upregulation of p53 protein expression levels
and significantly increased Bax/Bcl2 ratio in LV-MYCN group;
moreover, in the groups without cisplatin treatment, there were
significantly higher p53 protein expression levels and Bax/Bcl2
ratio in the LV-MYCN group compared with the LV-NC group (Fig. 6B).
Discussion
The present study was based on bioinformatics
analysis, which demonstrated that patients with high MYCN
expression had greater OS. High MYCN expression was associated with
increased OS of patients receiving platin-based therapy;
immunohistochemistry of tumor tissue collected in the present study
demonstrated that there was significantly higher MYCN protein
expression levels in cisplatin-sensitive EOC than
cisplatin-resistant EOC. Therefore, it was hypothesized that MYCN
was inhibited cisplatin resistance in EOC.
A previous study of expression profile of EOC
reported that MYCN is overexpressed in C5 subtype tumors compared
with three other molecular subtypes (C1, C2 and C4) of high-grade
serous EOC (31), which indicated
its aggressive role in high-grade serous EOC. Furthermore, MYCN
overexpression has been reported to be predictive of an aggressive
phenotype and poor prognosis in neuroblastoma (23), breast cancer (24) and spinal ependymoma (32); however, MYCN contributes to
cisplatin sensitization in acute myelogenous leukemia (25). Consistent with this, the present
study demonstrated that MYCN protein expression in SK-OV-3/DDP
cells was significantly lower compared with that in SK-OV-3 cells
and cisplatin significantly decreased MYCN protein expression
levels in SK-OV-3 cells, but not in SK-OV-3/DDP cells. These
results indicated that cisplatin functioned by suppressing
expression of MYCN in EOC. However, the association between MYCN
protein expression and cisplatin-induced cell behavior in EOC is
unknown.
Apoptosis serves a key role in tissue homeostasis in
response to numerous stimuli (33); decreased apoptosis associated with
occurrence, development and drug resistance of tumors (34). Cisplatin functions by covalently
binding to the DNA of tumor cells to form platinum-DNA adducts and
induces cell apoptosis (8,9). Once the cisplatin-induced apoptotic
pathway is blocked, tumor cells acquire resistance to the
proapoptotic effects of cisplatin, thus decreasing its antitumor
efficacy (35). In the present
study, viability and apoptosis of SK-OV-3 and SK-OV-3/DDP cells
following treatment with cisplatin was assessed, which demonstrated
that the SK-OV-3 cells in which MYCN was knocked down exhibited a
significantly decreased sensitivity to cisplatin-induced cell
apoptosis compared with NC. Furthermore, SK-OV-3/DDP cells with
MYCN overexpression exhibited a significantly increased sensitivity
to cisplatin-induced cell apoptosis. Collectively, these results
demonstrated that MYCN increased cisplatin-induced apoptosis and
that apoptosis may be the primary mechanism by which MYCN inhibits
cisplatin resistance in EOC. However, the molecules that mediate
the role of MYCN in EOC remain to be elucidated.
The genes co-expressed with MYCN were primarily
involved in pathways which contributed to chemotherapeutic
resistance and participated in cell apoptosis, including ‘MAPK
signaling pathway’, ‘PI3K/AKT signaling pathway’ and ‘p53 signaling
pathway’. Globally, GSEA demonstrated that MYCN was enriched in
‘apoptosis’ and ‘p53 pathway’ in the hallmark gene sets. The tumor
suppressor p53 is a transcription factor that regulates molecules
in extrinsic (Bcl2 family) and intrinsic (mitochondrial) apoptotic
pathways (36–38). Balance of Bcl2 family members
determines whether a cell undergoes apoptosis or survival (39). Cisplatin increases p53 levels and
facilitates the apoptotic response in tumor cells (40); moreover, cisplatin activates Bax,
decreases expression of Bcl2 and shifts the Bax/Bcl2 ratio in a
pro-apoptotic direction in tumor cells (41). Furthermore, the emergence of p53
mutant cisplatin-resistant OC cells has been demonstrated following
drug exposure (42) and patients
with OC who have p53 mutations are more resistant to
cisplatin-based therapy (43). In
the present study, p53, Bax and Bcl-2 protein expression levels
were assessed using western blotting, which demonstrated that
following treatment with cisplatin, SK-OV-3 cells in which MYCN was
knocked down exhibited significantly decreased p53 protein
expression levels and Bax/Bcl2 ratio, whereas SK-OV-3/DDP cells
with overexpressed MYCN exhibited significantly increased p53
protein expression levels and Bax/Bcl2 ratio. Therefore, it was
hypothesized that MYCN affected cisplatin resistance by regulating
p53 expression and ratio of Bax/Bcl2.
In conclusion, the present study suggested that MYCN
served as a potential marker for cisplatin treatment in EOC.
Specifically, the present study demonstrated that patients with
high expression of MYCN were more sensitive to cisplatin, whereas
patients with low expression of MYCN may be resistant to cisplatin.
Furthermore, it may be hypothesized that the findings for cisplatin
may be analogous to other chemotherapeutic drugs that lead to cell
apoptosis. However, one weakness in current study is the use of
only one EOC cell line and experiments should be replicated using
another EOC cell line.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Chongqing (grant no. cstc2021jcyj-msxmX0120).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GEO repository, accession number
GSE114206.
Authors' contributions
RY, HZ and RW performed experiments and data
analysis. LX conceived and supervised the study and wrote the
manuscript. RY and LX confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided signed consent prior to their
inclusion in the present study. The present study was approved by
the Institutional Ethics Committee of the First Affiliated Hospital
of Chongqing Medical University (approval No.
TFAHCQMU-2021-010).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Markman M and Bookman MA: Second-line
treatment of ovarian cancer. Oncologist. 5:26–35. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Assis J, Pereira C, Nogueira A, Pereira D,
Carreira R and Medeiros R: Genetic variants as ovarian cancer
first-line treatment hallmarks: A systematic review and
meta-analysis. Cancer Treat Rev. 61:35–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marchetti C, De Felice F, Romito A,
Iacobelli V, Sassu CM, Corrado G, Ricci C, Scambia G and Fagotti A:
Chemotherapy resistance in epithelial ovarian cancer: Mechanisms
and emerging treatments. Semin Cancer Biol. 77:144–166. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khan MA, Vikramdeo KS, Sudan SK, Singh S,
Wilhite A, Dasgupta S, Rocconi RP and Singh AP: Platinum-resistant
ovarian cancer: From drug resistance mechanisms to liquid
biopsy-based biomarkers for disease management. Semin Cancer Biol.
77:99–109. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lheureux S, Gourley C, Vergote I and Oza
AM: Epithelial ovarian cancer. Lancet. 393:1240–1253. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kurnit KC, Fleming GF and Lengyel E:
Updates and new options in advanced epithelial ovarian cancer
treatment. Obstet Gynecol. 137:108–121. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kelland L: The resurgence of
platinum-based cancer chemotherapy. Nat Rev Cancer. 7:573–584.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang J, Wei H, Liu X, Wang N, Qi Y, Zhang
Y and Zhang S: Downregulation of phosphoglycerate dehydrogenase
inhibits proliferation and enhances cisplatin sensitivity in
cervical adenocarcinoma cells by regulating Bcl-2 and caspase-3.
Cancer Biol Ther. 16:541–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Binju M, Amaya-Padilla MA, Wan G,
Gunosewoyo H, Suryo Rahmanto Y and Yu Y: Therapeutic inducers of
apoptosis in ovarian cancer. Cancers (Basel). 11:17862019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kartalou M and Essigmann JM: Mechanisms of
resistance to cisplatin. Mutat Res. 478:23–43. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wernyj RP and Morin PJ: Molecular
mechanisms of platinum resistance: Still searching for the
Achilles' heel. Drug Resist Updat. 7:227–232. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cancer Genome Altas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meyer N and Penn LZ: Reflecting on 25
years with MYC. Nat Rev Cancer. 8:976–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Neri F, Zippo A, Krepelova A, Cherubini A,
Rocchigiani M and Oliviero S: Myc regulates the transcription of
the PRC2 gene to control the expression of developmental genes in
embryonic stem cells. Mol Cell Biol. 32:840–851. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zajac-Kaye M: Myc oncogene: A key
component in cell cycle regulation and its implication for lung
cancer. Lung Cancer. 34 (Suppl 2):S43–S46. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Boxer RB, Jang JW, Sintasath L and Chodosh
LA: Lack of sustained regression of c-MYC-induced mammary
adenocarcinomas following brief or prolonged MYC inactivation.
Cancer Cell. 6:577–586. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bild AH, Yao G, Chang JT, Wang Q, Potti A,
Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al:
Oncogenic pathway signatures in human cancers as a guide to
targeted therapies. Nature. 439:353–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mo H, Vita M, Crespin M and Henriksson M:
Myc overexpression enhances apoptosis induced by small molecules.
Cell Cycle. 5:2191–2194. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McMahon SB: MYC and the control of
apoptosis. Cold Spring Harb Perspect Med. 4:a0144072014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koach J, Holien JK, Massudi H, Carter DR,
Ciampa OC, Herath M, Lim T, Seneviratne JA, Milazzo G, Murray JE,
et al: Drugging MYCN oncogenic signaling through the MYCN-PA2G4
binding interface. Cancer Res. 79:5652–5667. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Beltran H: The N-myc oncogene: Maximizing
its targets, regulation, and therapeutic potential. Mol Cancer Res.
12:815–822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jung M, Russell AJ, Liu B, George J, Liu
PY, Liu T, DeFazio A, Bowtell DD, Oberthuer A, London WB, et al: A
Myc activity signature predicts poor clinical outcomes in
Myc-associated cancers. Cancer Res. 77:971–981. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nakagawara A, Li Y, Izumi H, Muramori K,
Inada H and Nishi M: Neuroblastoma. Jpn J Clin Oncol. 48:214–241.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mizukami Y, Nonomura A, Takizawa T,
Noguchi M, Michigishi T, Nakamura S and Ishizaki T: N-myc protein
expression in human breast carcinoma: Prognostic implications.
Anticancer Res. 15:2899–2905. 1995.PubMed/NCBI
|
|
26
|
Huang X, Qi L, Lu W, Li Z, Li W and Li F:
MYCN contributes to the sensitization of acute myelogenous leukemia
cells to cisplatin by targeting SRY-box transcription factor 4.
Bioengineered. 2021. View Article : Google Scholar
|
|
27
|
Veskimäe K, Scaravilli M, Niininen W,
Karvonen H, Jaatinen S, Nykter M, Visakorpi T, Mäenpää J, Ungureanu
D and Staff S: Expression analysis of platinum sensitive and
resistant epithelial ovarian cancer patient samples reveals new
candidates for targeted therapies. Transl Oncol. 11:1160–1170.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schulz H, Kuhn C, Hofmann S, Mayr D,
Mahner S, Jeschke U and Schmoeckel E: Overall survival of ovarian
cancer patients is determined by expression of galectins-8 and −9.
Int J Mol Sci. 19:3232018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu H, Zou X, Lin S, Hu X and Gao J:
Effects of naringin on reversing cisplatin resistance and the
Wnt/β-catenin pathway in human ovarian cancer SKOV3/CDDP cells. J
Int Med Res. 48:3000605198878692020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zou GP, Yu CX, Shi SL, Li QG, Wang XH, Qu
XH, Yang ZJ, Yao WR, Yan DD, Jiang LP, et al: Mitochondrial
dynamics mediated by DRP1 and MFN2 contributes to cisplatin
chemoresistance in human ovarian cancer SKOV3 cells. J Cancer.
12:7358–7373. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Helland Å, Anglesio MS, George J, Cowin
PA, Johnstone CN, House CM, Sheppard KE, Etemadmoghadam D, Melnyk
N, Rustgi AK, et al: Deregulation of MYCN, LIN28B and LET7 in a
molecular subtype of aggressive high-grade serous ovarian cancers.
PLoS One. 6:e180642011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ghasemi DR, Sill M, Okonechnikov K,
Korshunov A, Yip S, Schutz PW, Scheie D, Kruse A, Harter PN,
Kastelan M, et al: MYCN amplification drives an aggressive form of
spinal ependymoma. Acta Neuropathol. 138:1075–1089. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Oliveira CB, Comunello LN, Maciel ES,
Giubel SR, Bruno AN, Chiela EC, Lenz G, Gnoatto SC, Buffon A and
Gosmann G: The inhibitory effects of phenolic and terpenoid
compounds from Baccharis trimera in Siha cells: Differences in
their activity and mechanism of action. Molecules. 18:11022–11032.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Plati J, Bucur O and Khosravi-Far R:
Dysregulation of apoptotic signaling in cancer: Molecular
mechanisms and therapeutic opportunities. J Cell Biochem.
104:1124–1149. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang H, Luo Y, Qiao T, Wu Z and Huang Z:
Luteolin sensitizes the antitumor effect of cisplatin in
drug-resistant ovarian cancer via induction of apoptosis and
inhibition of cell migration and invasion. J Ovarian Res.
11:932018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Green DR and Kroemer G: The
pathophysiology of mitochondrial cell death. Science. 305:626–629.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Czabotar PE, Lessene G, Strasser A and
Adams JM: Control of apoptosis by the BCL-2 protein family:
Implications for physiology and therapy. Nat Rev Mol Cell Biol.
15:49–63. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vousden KH: p53: Death star. Cell.
103:691–694. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pistritto G, Trisciuoglio D, Ceci C,
Garufi A and D'Orazi G: Apoptosis as anticancer mechanism: Function
and dysfunction of its modulators and targeted therapeutic
strategies. Aging (Albany NY). 8:603–619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
An SH, Kang JH, Kim DH and Lee MS: Vitamin
C increases the apoptosis via up-regulation p53 during cisplatin
treatment in human colon cancer cells. BMB Rep. 44:211–216. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li X, Mu J, Lin Y, Zhao J and Meng X:
Combination of cyanidin-3-O-glucoside and cisplatin induces
oxidative stress and apoptosis in HeLa cells by reducing activity
of endogenous antioxidants, increasing bax/bcl-2 mRNA expression
ratio, and downregulating Nrf2 expression. J Food Biochem.
45:e138062021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Righetti SC, Perego P, Corna E, Pierotti
MA and Zunino F: Emergence of p53 mutant cisplatin-resistant
ovarian carcinoma cells following drug exposure: Spontaneously
mutant selection. Cell Growth Differ. 10:473–478. 1999.PubMed/NCBI
|
|
43
|
Xie X, Lozano G and Siddik ZH:
Heterozygous p53(V172F) mutation in cisplatin-resistant human tumor
cells promotes MDM4 recruitment and decreases stability and
transactivity of p53. Oncogene. 35:4798–806. 2016. View Article : Google Scholar : PubMed/NCBI
|