Introduction
Female breast cancer (BC) is the second most
diagnosed malignancy worldwide (1), accounting for over two million cases
each year in the USA (2). It is
also the cause of 1 in 6 deaths due to all cancer types (1). The incidence rate of BC is
continually increasing by ~0.5% per year (3). According to the 2020 GLOBOCAN data,
the age-standardized rates for both the incidence and mortality of
BC in Saudi women were 28.8 and 8.4%, respectively (4). However, these rates continue to
increase each year (3). Notably,
the incidence numbers of BC in Saudi Arabia increased by 186% from
2004 to 2016 (5). It is considered
that this steady increase in BC cases, particularly in Saudi
Arabia, may be attributed to several factors related to lack of
awareness, delayed diagnosis, aging populations and the unhealthy
lifestyle choices related to low levels of physical activity, poor
dietary habits and smoking (6,7).
To date, the main treatment modality implemented for
patients with BC is surgery followed by chemotherapy, radiotherapy,
hormonal therapy and/or immuno-/targeted therapy to prevent the
recurrence of the disease (8).
Despite the invasive aspects of the majority of these therapeutic
interventions and their associated side effects, these treatments
are still not sufficient to effectively cure this disease and
prevent relapse. For instance, chemotherapy is a harmful procedure
that has been shown to induce drug resistance in cancer cells.
Moreover, hormonal therapy may lead to bone loss and thus, to
frailty and fractures (9,10).
Several laudable studies have been performed to
investigate BC and identify potential biomarkers. Only a few of
these therapeutic targets have already been studied in clinical
trials to evaluate the response to treatment and disease
progression, such as estrogen receptor (ER) (11), progesterone receptor (PR) (12), human epidermal growth factor
receptor 2 (HER2) (13), nuclear
protein Ki67 (Ki67) (14),
urokinase-type plasminogen activator (15), tumor protein p53 (p53) (16), cyclin E (17) and neuropeptide substance P
(18). Recent developments in
cancer immunotherapy hold promise as possible treatment options for
a variety of cancer types, including BC (19). For instance, programmed
death-ligand 1 was the first immune checkpoint blockade drug
approved by the US Food and Drug Administration (FDA) for the
treatment of patients with triple-negative metastatic BC in 2019
(20). Other types of
immunotherapies are currently available, including checkpoint
blockade, adoptive cellular therapy and cancer vaccinology
(21). Thus far, targeted
anticancer therapies are not yet able to cure BC as single agents,
which entails their combination with chemotherapy and/or
radiotherapy to obtain improved survival outcomes. Furthermore,
they were unable to provide reduced cytotoxicity on non-target
tissues and to eradicate the cancer stem cells in the tumor mass
(22,23). These hurdles to deliver effective
and personalized BC therapeutics can be explained by the molecular
pathophysiology of this aggressive tumor that remains poorly
understood.
BC is a heterogeneous disease with several clinical
subtypes, cancer stem cells niches, molecular signatures and
oncologic signaling pathways. This complexity is further amplified
by additional contributing factors, including late-stage detection,
an intricate network of crosstalks and signaling pathways, drug
resistance and a higher recurrence of the disease. These challenges
combined with the heavy burden of this malignancy on patients,
their families and the healthcare system, are urging scientists to
perform research focusing on the identification of effective
diagnostic biomarkers that may pave the way towards better
theranostics. In this context, signaling lymphocytic activation
molecule F7 (SLAMF7; previously known as CS1, CD2 subset1, CRACC
and CD319) is a glycosylated cell surface protein that has been
found to play a critical role in immune cell functions (24). This transmembrane receptor is
located on the long arm of chromosome 1 (1q23.3) and is a member of
the signaling lymphocyte activation molecule SLAM family (25). Studies have indicated that SLAMF7
has a unique pattern that is ubiquitously expressed in several
cancer types, while its expression in normal cells is restricted to
selected immune cell types, mainly natural killer (NK) cells and
mature dendritic cells, but not on normal hematopoietic stem cells
or other lymphocytes/normal tissues (24,26,27).
These properties have made SLAMF7 a promising therapeutic target
given its ability to activate NK cells specifically in tumor cells
and boost their immunogenic cell death (phagocytosis, apoptosis,
immune cells activation, cell signaling and gene expression),
particularly when triggered by an antibody or a natural ligand
(28–30). This SLAMF7-driven interaction has
been shown to result in an increase in NK cytotoxicity, macrophage
super activation and an inflammatory cytokine storm in rheumatoid
arthritis (31,32). Of note, the monoclonal antibody,
elotuzumab, has been demonstrated to specifically target SLAMF7,
which is abundantly expressed on multiple myeloma (MM) cells. It
has been shown to attract NK cells and to exert anticancer effects
via antibody-dependent cell-mediated cytotoxicity (ADCC) in MM
cells (26,33,34).
In solid tumors, SLAMF7 has recently been shown to
be expressed in colorectal cancer cells (35–37),
ovarian and cervical cancers (38,39),
as well as in liver cancer cell lines (40) and multiple murine cancer models
(41). The promise of SLAMF7 as an
immunotherapeutic target with possible clinical outcomes in
patients with various types of tumors has rendered it the focus of
several studies (as aforementioned).
To the best of our knowledge, the present study is
the first to investigate the expression of SLAMF7 in BC. This study
aimed to assess the protein expression levels of SLAMF7 and its
correlation with clinicopathological features of BC patients to
evaluate its potential value as a prognosticator of BC.
Patients and methods
Patient series
The present retrospective study included 278 lymph
node-positive cases from 730 formalin-fixed and paraffin-embedded
(FFPE) blocks of primary BC samples retrieved from the Pathology
Department, King Abdulaziz University, Jeddah, Saudi Arabia
covering the period from January 1995 to December 2014. The
inclusion criteria included all available primary BC FFPE tissues
collected from consenting patients who had full annotated
clinicopathological data, regardless of their associated systemic
diseases status. Only primary BC cases with unavailable FFPE sample
and/or annotation data were excluded. FFPE blocks were processed
routinely with hematoxylin and eosin for the evaluation of
histopathological features, histological grading and the TNM-based
staging of the tumor. The patient clinicopathological parameters
were obtained from their medical records and are summarized in
Table I.
 | Table I.Association between SLAMF7 protein
expression patterns and the clinicopathological features of
patients with breast cancer. |
Table I.
Association between SLAMF7 protein
expression patterns and the clinicopathological features of
patients with breast cancer.
|
|
| SLAMF7 protein
expression |
|
|---|
|
|
|
|
|
|---|
| Features | No. of cases | (Low) (%) | (High) (%) | P-value |
|---|
| Age, years |
|
|
| 0.007a |
|
<50 | 143 (51.4%) | 105 (73) | 38 (27) |
|
|
>50 | 134 (48.2%) | 116 (87) | 18 (13) |
|
| Missed
data | 1 (0.4%) |
|
|
|
| Tumor invasion |
|
|
| 0.008a |
|
Negative | 3 (1.1%) | 0 (0) | 3 (100) |
|
|
Positive | 258 (92.8%) | 209 (81) | 49 (19) |
|
| Missed
data | 17 (6.1%) |
|
|
|
| ER and PR hormonal
status |
|
|
| 0.912 |
|
ER−,
PR− | 72 (25.9%) | 59 (82) | 13 (18) |
|
|
ER+,
PR+ | 126 (45.3%) | 104 (83) | 22 (17) |
|
| Missed
data | 80 (28.8) |
|
|
|
| HER2 protein
status |
|
|
| 0.255 |
|
Negative | 150 (54%) | 118 (84) | 23 (16) |
|
|
Positive | 79 (28.4%) | 70 (78) | 20 (22) |
|
|
Borderline | 13 (4.7) |
|
|
|
| Missed
data | 36 (12.9%) |
|
|
|
| ER, PR and HER2
status |
|
|
| 0.579 |
|
Triple-negative | 30 (10.8%) | 26 (87) | 4 (13) |
|
|
Triple-positive | 42 (15.1%) | 33 (79) | 9 (21) |
|
| Missed
data | 206 (74.1%) |
|
|
|
| Vascular
invasion |
|
|
| 0.050 |
| Negative | 84 (30.2%) | 64 (76) | 20 (24) |
|
|
Positive | 126 (45.3%) | 109 (86) | 17 (14) |
|
| Missed
data | 68 (24.5%) |
|
|
|
| Tumor margin |
|
|
| 0.713 |
|
Negative | 222 (79.9%) | 182 (82) | 40 (18) |
|
|
Positive | 29 (10.4%) | 23 (79) | 6 (21) |
|
| Missed
data | 27 (9.7%) |
|
|
|
| Tumor size, cm |
|
|
| 0.298 |
|
0-3 | 76 (27.3%) | 56 (74) | 20 (26) |
|
|
3-6 | 136 (48.9%) | 112 (82) | 24 (18) |
|
|
>7 | 40 (14.4%) | 33 (83) | 7 (17) |
|
| Missed
data | 26 (9.4%) |
|
|
|
| Tumor grade |
|
|
| 0.844 |
| Grade
1 | 34 (12.2%) | 27 (79) | 7 (21) |
|
| Grade
2 | 135 (48.6%) | 110 (82) | 25 (18) |
|
| Grade
3 | 81 (29.1%) | 64 (79) | 17 (21) |
|
| Missed
data | 28 (10.1%) |
|
|
|
| Histopathological
type |
|
|
| 0.466 |
|
Invasive ductal | 258 (92.8%) | 207 (80) | 51 (20) |
|
|
Other | 18 (6.5%) | 13 (72) | 5 (28) |
|
| Missed
data | 2 (0.7%) |
|
|
|
| Disease
recurrence |
|
|
| 0.542 |
|
Yes | 40 (14.4%) | 34 (85) | 6 (15) |
|
| No | 112 (40.3%) | 90 (80) | 22 (20) |
|
| Missed
data | 126 (45.3%) |
|
|
|
| Status at end
point |
|
|
| 0.338 |
|
Succumbed to the disease | 25 (9%) | 18 (72) | 7 (28) |
|
|
Alive | 66 (23.7%) | 53 (80) | 13 (20) |
|
| Missed
data | 187 (67.3%) |
|
|
|
Treatment and follow-up
All (100%) consenting patients with BC were
subjected to surgery, i.e., lumpectomy, radical or modified radical
mastectomy with axillary clearance. Post-operative early adjuvant
systemic therapy in the form of chemotherapy, radiotherapy and
hormonal therapy was administered to 77, 60 and 25% of the
patients, respectively. Following treatment, the patients were
observed at 6–12-month intervals until mortality or the end of the
follow-up period in June, 2016 (date of data collection). The mean
follow-up time for the whole series was 37 months (range, 1–252
months). During the follow-up, patients were subjected to repeated
clinical examinations and bone isotope scan, chest and
abdominal-pelvic CAT scans were performed whenever needed. In most
instances, the causes of death were obvious on clinical grounds
alone. The autopsy was not performed in any case.
Tissue microarray (TMA) and
immunohistochemistry (IHC)
A total of 730 BC FFPE blocks were used to construct
a TMA as previously described (42). TMA slides were utilized for the
evaluation of SLAMF7 expression pattern using IHC with
anti-SLAMF7/CS1 primary antibody (1:100 dilution; cat. no.
ab202840, Abcam). Anti-SLAMF7 primary antibody was applied
manually. Staining and processing were performed as previously
described (43). Briefly, a fully
automated protocol was designed to include deparaffinization with
EZ Prep (Ventana Medical Systems, Inc.; Roche Diagnostics) at 75°C
and incubation with the anti-SLAMF7/CS1 primary antibody for 1 h at
37°C. Staining and processing were thereafter performed with the
ready-to-use iView DAB Detection kit (Ventana Medical Systems,
Inc.; Roche Diagnostics) which contains a pre-diluted secondary
antibody solution that is processed by the automated staining
system (Ventana Medical Systems, Inc.; Roche Diagnostics) Ventana
BenchMark XT for 1 h at 37°C. Counterstaining was performed using
hematoxylin II (Ventana Medical Systems, Inc.; Roche Diagnostics)
at room temperature for 4 min and Bluing Reagent (Ventana Medical
Systems, Inc.; Roche Diagnostics) for 4 min. The stained TMA slides
were washed with water and mild detergent followed by 3 min of
several successive immersions into alcohol buffer at increasing
concentrations (70, 95 and 100%). Tissue-Tek glass mounting medium
was applied to each slide and covered with a glass coverslip.
Evaluation of SLAMF7 staining
intensity
The expression of SLAMF7 in the tumor tissue was
assessed in a manner blinded to the clinical data using a Nikon
light microscope (Model no. 6132, Nikon Corporation) at a
magnification of ×40. The tumor cells which exhibited cytoplasmic
staining were graded into four categories as follows: 0, Negative,
no detectable staining; 1+, weak, yet detectable staining; 2+,
moderate, clearly positive yet still weak; 3+, heavy staining,
intense. As previously described (44,45),
the cytoplasmic index score was calculated where both the intensity
of the staining and the fraction of positively stained cells were
taken into account using the following formula: I=0 × f0 + 1 × f1 +
2 × f2 + 3 × f3, where (I) is the staining index and (f0-f3) are
the fractions of the cells showing a defined level of staining
intensity (from 0 to +3). Theoretically, the index scores could
vary between 0 and 300. The expression patterns were imaged and
digitized using a Coolsnap Pro Color camera and
ImagePro® Plus software (Media Cybernetics, Inc.).
Statistical analysis
Fischer's exact test was used to assess the
significance of the association between different categorical
variables. Univariate survival analysis for the outcome measure
[disease-specific survival (DSS) and disease-free survival (DFS)]
was based on the Kaplan-Meier method, with the log-rank
(Mantel-Cox) comparison test. Additionally, Cox regression
multivariate analysis was performed to assess the possible
independent prognostic impact of SLAMF7 protein expression in
relation to the age, lymph node status, tumor grade and
histological type of the patients. In all tests, a value of
P<0.05 was considered to indicate a statistically significant
difference. SPSS® (IBM Corp.) software packages (PASW
Statistics for Windows, version 19) were used to perform all
statistical analyses.
In silico analysis of SLAMF7 mRNA
expression
To further validate the findings of the present
retrospective study that assessed the SLAMF protein expression in
BC, transcription data from The Cancer Genome Atlas (TCGA) were
analyzed using the freely available web application, The University
of ALabama at Birmingham CANcer data analysis Portal (UALCAN;
available at: http://ualcan.path.uab.edu/analysis.html) (46) and the online multi-omic exploration
tool of the University of California, Santa Cruz (UCSC Xena;
available at: http://xena.ucsc.edu/#overview) (47). Using the TCGA data repository,
which is a comprehensive, user-friendly and interactive web
resource allowing graphical and statistical analyses of cancer
OMICS data, SLAMF7 mRNA expression in BC was then analyzed
and compared with normal breast tissues as a control (available
from the same platform) using Student's t-test (P<0.05 was
considered to indicate a statistically significant difference).
Results
Expression profile of SLAMF7 in
primary BC samples
The results revealed that the cellular localization
of SLMAF7 protein expression was mainly cytoplasmic in the primary
BC samples, as well as in the lymph node-positive cases. It was
found that ~20% of the primary samples exhibited moderate/strong
(high) expression patterns, while the majority of the samples (80%)
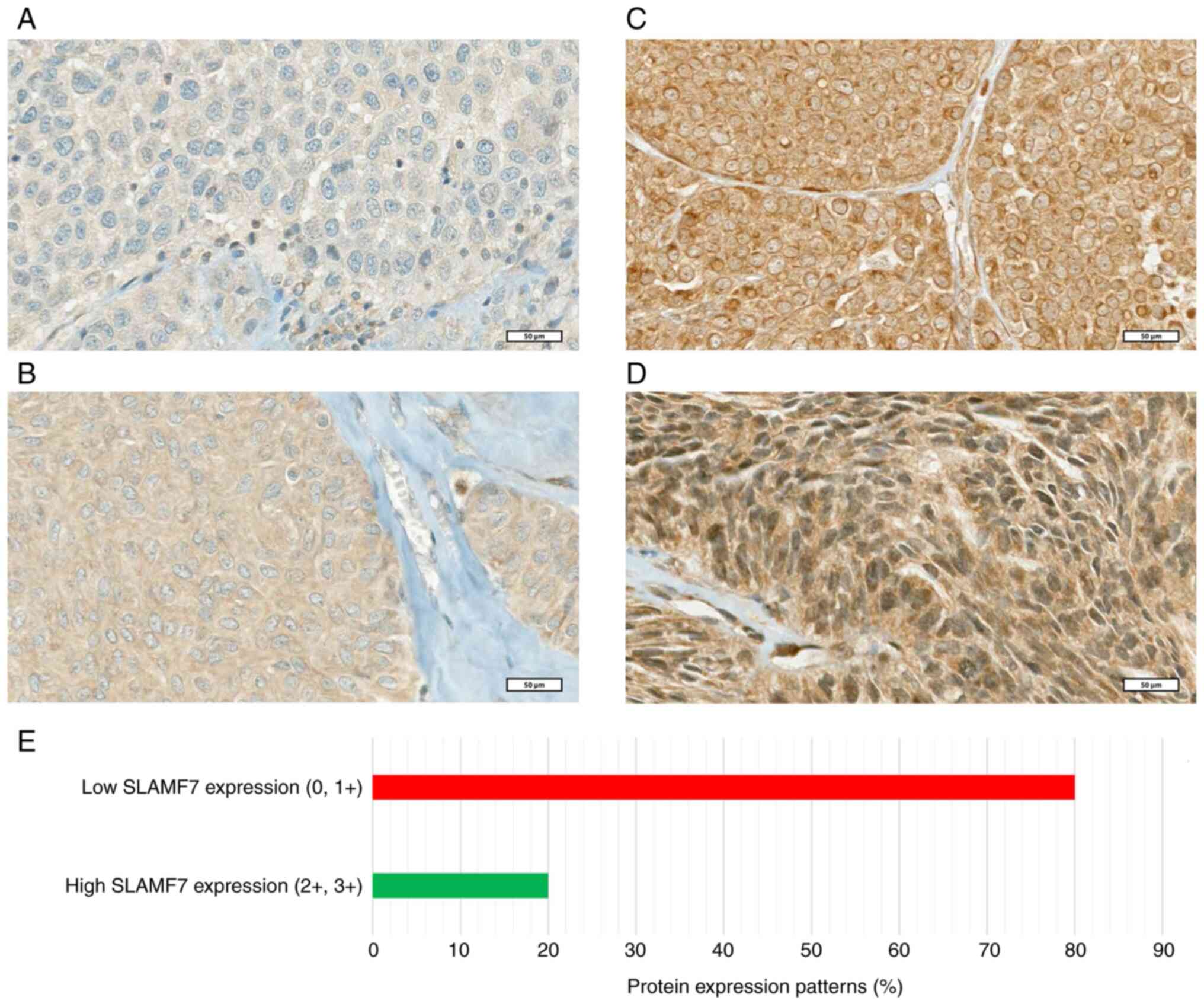
had either negative or weak (low) expression profiles (Fig. 1A-E). On the other hand, ~70% of the
cancerous tissues in the lymph node-positive samples exhibited a
high cytoplasmic expression (2+, 3+) while 30% of the samples
exhibited low cytoplasmic expression patterns (0, 1+) (data not
shown). The cytoplasmic expression patterns of 278 lymph
node-positive primary BC cases are illustrated in Fig. 1. Of note, a perinuclear-like
staining was observed. However, it was uncommon and difficult to
confirm.
Association of SLAMF7 protein
expression patterns with clinicopathological features
The association of cytoplasmic SLAMF7 protein
expression in lymph node-positive BC with the patient
clinicopathological characteristics using different cut-off values
revealed that the cut-off value for low (0, 1+) SLAMF7 protein
expression compared to high (2+, 3+) SLAMF7 protein expression (low
expression vs. high expression) was the strongest discriminator.
Based on the aforementioned discriminator, the results revealed
that there were significant associations between the cytoplasmic
SLAMF7 protein expression profile and the age of the patients at
diagnosis, in that the proportion of primary BC tissues with higher
SLAMF7 protein expression was greater in younger patients (<50
years; 27%) compared with that in older patients (>50 years;
13%) (P<0.007; Table I). In
addition, a significant association was found between the SLAMF7
expression profile and tumor invasion. BC tissues with a high
invasive characteristic had a lower SLAMF7 protein expression than
less invasive tumors (P<0.008; Table I).
The same tendency was observed between the SLAMF7
protein expression patterns and vascular invasion. Indeed, tumors
with highly vascular invasive cells exhibited a lower SLAMF7
expression pattern than those with low vascular invasion (P=0.05).
However, the other clinicopathological features did not exhibit any
significant associations with the SLAMF7 protein expression
profiles, including the hormonal and HER2 protein status (P=0.9 and
P=0.2, respectively), as well as tumor grade (P=0.8), tumor margin
(P=0.7) and tumor size (P=0.2) (Table
I).
Association of SLAMF7 protein
expression patterns with survival outcomes
In Kaplan-Meier analysis, the tier two cut-off [no
expression (0) vs. expression (1+, 2+ and 3+)] was the strongest
discriminator. Using this cut-off, survival analysis revealed that
patients with BC with positive SLAMF7 protein expression patterns
in their lymph node positive tissues (1+, 2+ and 3+) had a lower
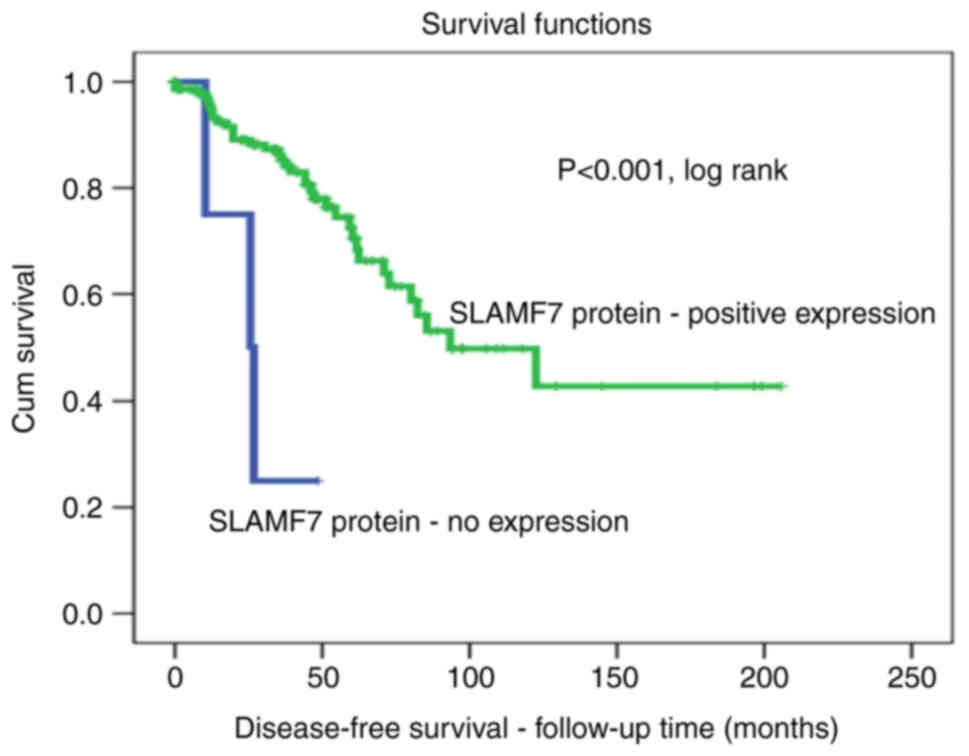
relapse rate (DFS) than those without SLAMF7 expression profiles.
For example, after 5 years of follow-up, all patients with lymph
node-positive BC without SLAMF7 expression (100%) exhibited disease
recurrence, compared with a recurrence rate of only 40% in the
lymph nodes of with BC with a positive SLAMF7 protein expression
(P<0.001, log-rank; Fig.
2).
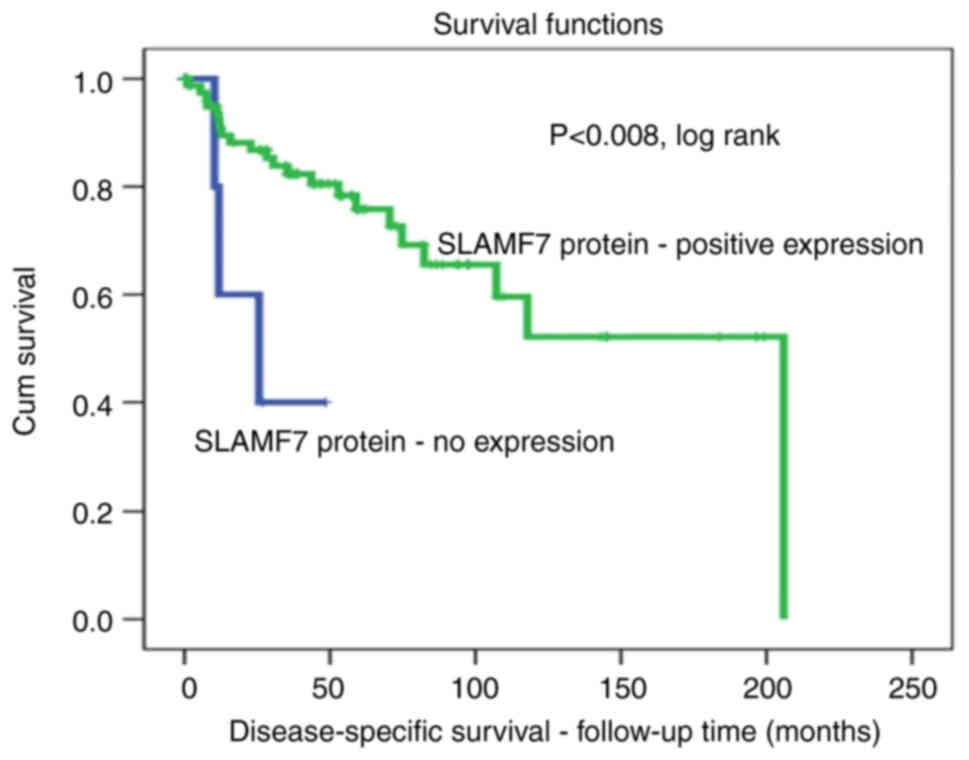
The assessment of DSS using the same cut-off points
[negative (0) vs. positive (1+, 2+ and 3+)] also revealed a
significant association. In fact, patients with lymph node-positive
BC positive for SLAMF7 protein expression survived for a longer
period of time. After 5 years of follow-up, ~30% of patients with
BC with lymph node-positive tumors with a positive SLAMF7
expression died compared to a 100% death rate in those without a
SLAMF7 expression pattern (P<0.008, log-rank; Fig. 3).
Multivariate Cox regression analysis revealed that
the SLAMF7 expression profile (low, 0 and 1+; vs. high, 2+ and 3+)
was not an independent factor for a poor DFS and DSS in relation to
patient age, lymph node status, tumor grade and vascular invasion
(Table II).
 | Table II.Cox regression analysis of the
prognostic values of cytoplasmic SLAMF7 protein expression, age at
diagnosis, lymph node status, grade and tumor vascular invasion in
association with the survival of patients with breast cancer. |
Table II.
Cox regression analysis of the
prognostic values of cytoplasmic SLAMF7 protein expression, age at
diagnosis, lymph node status, grade and tumor vascular invasion in
association with the survival of patients with breast cancer.
| Parameter | P-value | SE value | Relative risk | 95% CI |
|---|
| SLAMF7 | 0.58 | 0.431 | 1.270 | 0.338-1.830 |
| Age at
diagnosis | 0.12 | 0.425 | 0.512 | 0.848-4.490 |
| Lymph node
status | 0.10 | 0.453 | 0.478 | 0.861-5.077 |
| Tumor grade | 0.42 | 0.307 | 1.282 | 0.428-1.423 |
| Tumor vascular
invasion | 0.060 | 0.489 | 2.50 | 0.154-1.043 |
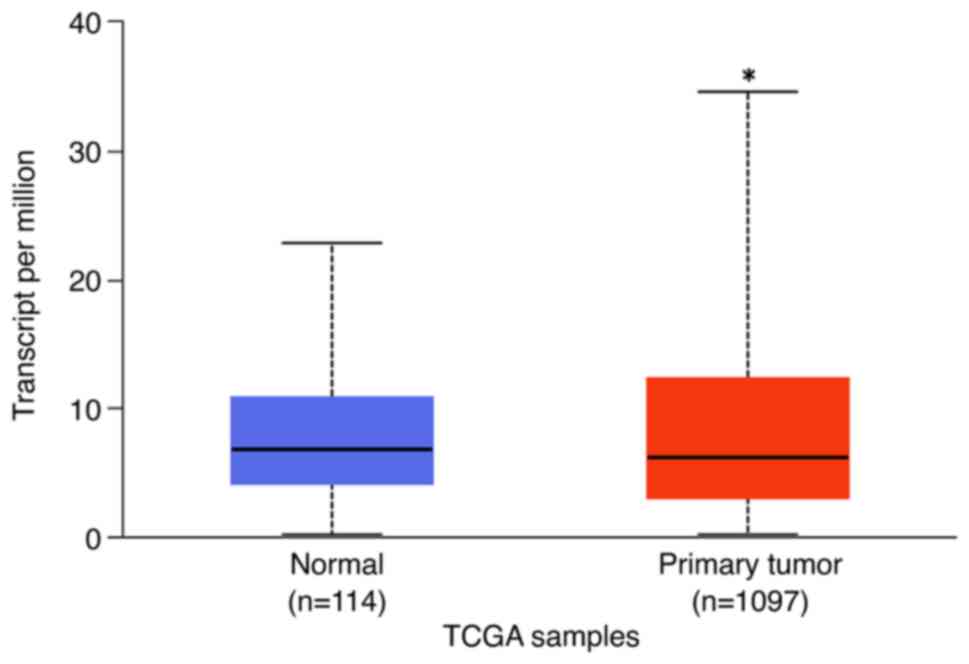
SLAMF7 mRNA expression
The in silico analysis of SLAMF7 mRNA
expression available in the freely available UCSC Xena or UALCAN
transcriptomic databases confirmed that the SLAMF7
transcript was expressed in the breast invasive carcinoma cohort.
Using the Student's t-test, the expression of this mRNA was shown
to be higher than that in normal breast tissues according to the
UALCAN database (P<0.001; Fig.
4).
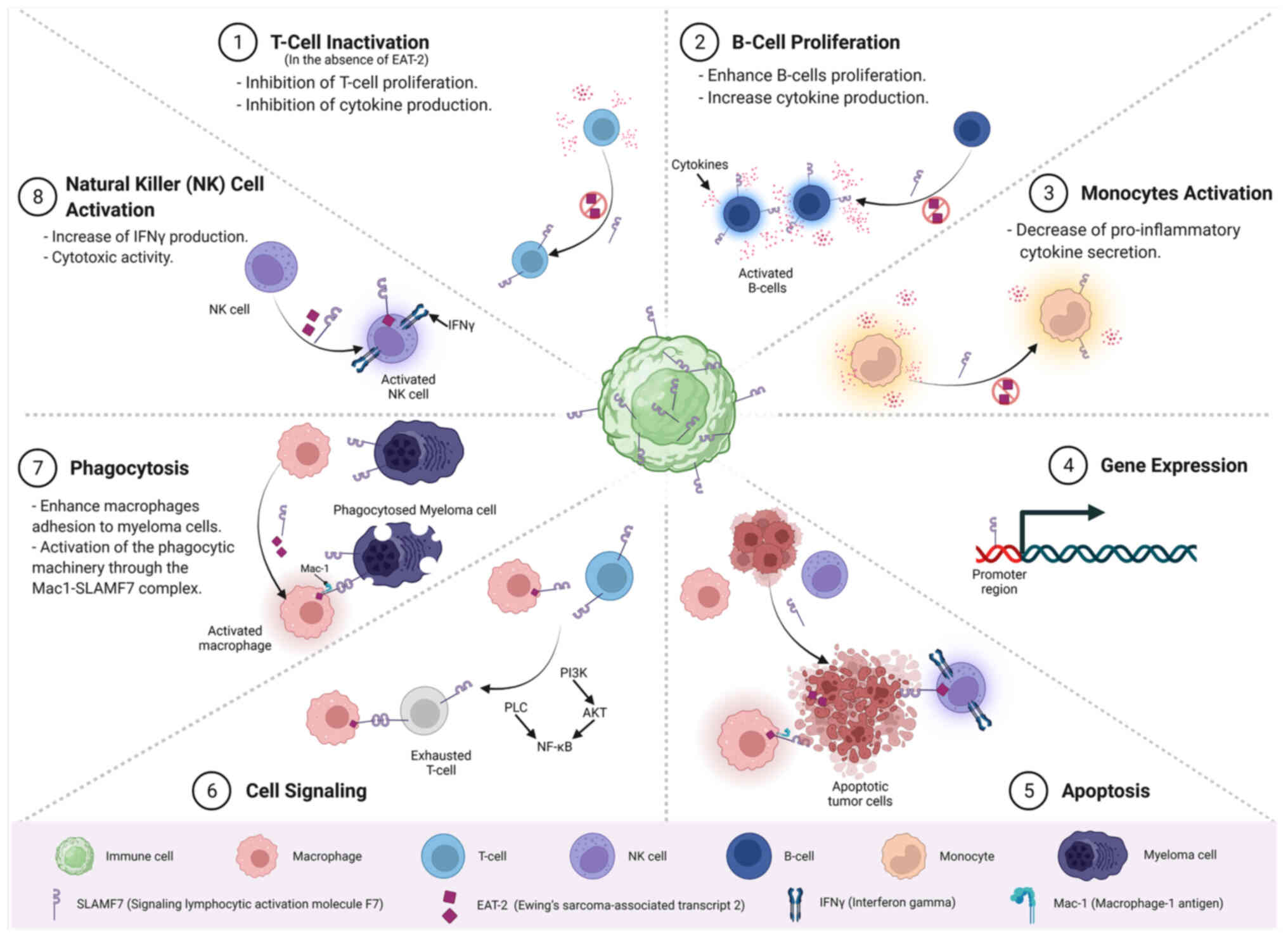
An overview of the main molecular, cellular and
signaling functions of SLAMF7 as regards its role in the immune
system, as well as in other tissues is presented in Fig. 5.
Discussion
BC remains a major health concern and a very common
cause of cancer-related mortality among women worldwide, with an
inherent complexity and molecular heterogeneity (48,49).
Moreover, the current treatment modalities for BC are based on
histopathological features, such as age, stage, grade, tumor size
and receptor status (50).
However, patients with BC with similar conditions and diagnoses may
have different prognoses, responses to treatment and disease
courses when treated with the same therapeutic regimen. This
inherent complexity is due to different morphological,
pathophysiological, clinical and environmental characteristics
(51). In the post-genomic era,
great efforts have been made to overcome this heterogeneity of BC
and to further elucidate this complexity. In addition to ER, PR and
HER2 receptors, other promising biomarkers have been proposed to
further elucidate the molecular heterogeneity of BC (52). Since the majority of tumors are no
longer considered as a single disease with predefined molecular
features, the identification of more relevant clinical and
molecular features of BC is urgently required in order to identify
therapeutic targets and define pathways of disease progression
leading to an earlier diagnosis, better prognosis and more precise
therapeutics (53).
Multiomics approaches have led to substantial
progress being made in the accurate molecular stratification of BC
into different subtypes to identify more appropriate/precise
therapeutic options (54–56). However, much still remains to be
done before precision theranostics for BC can be achieved, and thus
more molecular biomarkers are required for this aggressive
disease.
In this context, SLAMF7 is expressed in selected
immune cells and functions as an inhibitor in monocytes to modulate
pro-inflammatory immune responses (57). Chen et al (30) recently discovered that SLAMF7 is
required for the phagocytosis of hematopoietic malignancy cells,
which is crucial for cancer treatment. Notably, elotuzumab is the
targeted drug that specifically targets SLAMF7 in patients with
myeloma via ADCC (26,33,34).
Although SLAMF7 has been reported to be rarely
expressed in normal tissues, it is expressed in certain types of
cancer, such as colorectal cancer and multiple myeloma (26,58).
In addition, analysis of data from TCGA has demonstrated that
SLAMF7 is also expressed in certain solid tumors at either the RNA
and/or protein level (59–61); however, no specific study has yet
been conducted using BC tissue, at least to the best of our
knowledge. In addition, it is considered that the availability of
an approved anti-SLAMF7 monoclonal antibody (elotuzumab) would be
an advantage that would not only increase the value of studies on
patients with BC, but would also be beneficial for oncologists,
pathologists and cancer researchers.
To the best of our knowledge, this is the first
study to investigate the expression patterns of SLAMF7 in BC tissue
and to demonstrate its potential prognostic value in BC. The
results of the present study confirmed the presence of SLAMF7
protein in BC tissues. Indeed, SLAMF7 was strongly expressed in the
cytoplasm of BC tissue. Of note, it was more overexpressed in the
samples from patients with BC with a positive lymph node status
(advanced stages) than in those with only primary BC. Moreover, the
cytoplasmic expression of SLAMF7 was significantly associated with
several clinicopathological characteristics in the present cohort,
including age (P<0.007), tumor invasion (P<0.008) and
vascular invasion (P=0.05). The results of the present study
clearly demonstrated that a low expression of SLAMF7 protein was
associated with more aggressive and invasive BC cases. In fact, the
low expression of SLAMF7 was found in 81 and 86% of patients with
BC with positive tumor invasion and positive vascular invasion,
respectively (Table I).
Several studies among Saudi women have reported that
the majority were diagnosed with BC at <50 years of age
(62–65). These findings were also confirmed
in the present study cohort, in which >51% of the patients with
BC were <50 years of age when they were diagnosed with BC.
However, in the USA, the SEER cancer statistics review reported
that the median age of American women at the time of diagnosis of
BC was 62 years (66). These
results indicate that the onset of BC is delayed by >10 years in
the USA as compared to Saudi women. This significant early-onset of
BC among Saudi women may be due to a number of factors, including
population ageing, economic and social disparities, lifestyle
choices and environmental factors (62,67).
Of note, the results of the present study demonstrated that the
expression of SLAMF7 was higher in younger patients with BC than in
their older counterparts, which may be attributed to, at least in
part, the lower activity of immune cells in the elderly (68).
Using Kaplan-Meier analysis, significant
associations were found between SLAMF7 expression in BC and both
DFS and DSS (P<0.001 and P<0.008; log-rank test,
respectively). The results revealed that patients with BC who
overexpressed SLAMF7 had better survival outcomes with longer
survival and lower recurrence rates. In fact, all (100%) the
patients with BC without SLAMF7 expression had disease recurrence
after 5 years of follow-up, whereas only 35% of patients with a
positive expression of SLAMF7 relapsed during the same period
(Fig. 2). Similarly, as regards
DSS, 100% of the patients without SLAMF7 expression succumbed to
the disease during the 5-year follow-up period compared with only
25% of those with expressed SLAMF7 protein (Fig. 3). The survival results of the
patients with BC in the present study are consistent with those in
patients with MM, in whom a higher SLAMF7 mRNA expression
was a significant prognosticator of a longer survival and has been
proposed as a useful tool for classifying hematologic malignancies
into molecular subgroups for therapeutic purposes (69). In addition, elotuzumab (a
monoclonal antibody against SLAMF7 approved by the FDA in November,
2015), in combination with other targeted antimyeloma therapies
that stimulate host immunity, has been shown to exert an effective
immunotherapeutic effect (70),
improving DFS and thus reducing relapse [as reviewed by Boudreault
et al (71)]. Although
immunotherapy is an established adjunct therapeutic strategy that
improves the survival and treatment of cancer patients (72), SLAMF7 remains poorly studied in
solid tumors. In addition to the present study, which documented
the clear cytoplasmic protein expression in BC, SLAMF7 has also
been shown to be expressed in colorectal cancer (37), and at the transcript level in both
ovarian and cervical cancers (38,39);
however, those studies did not evaluate its prognostic/predictive
value. Consistent with the findings presented herein, data from
TCGA have revealed that a low SLAMF7 gene expression is
strongly associated with poor survival outcomes in ovarian cancer
(61). Furthermore, the findings
obtained herein were compared with those from other freely
available transcriptomic data of SLAMF7 in cancer genomics
databases, mainly the UCSC Xena or UALCAN databases. Using the UCSC
Xena portal, the SLAMF7 transcript was expressed in the majority of
the TCGA breast invasive carcinoma cohort (47), and this expression was
significantly higher than that in normal breast tissues according
to the UALCAN database (P<0.001) (73).
SLAMF7 is a transmembrane marker and a promising
molecular marker that plays multiple immune-molecular and signaling
roles in modulating the cellular immune response (Fig. 5). SLAMF7 is involved in the
inhibition of T-cell proliferation, the overexpression of
growth-promoting cytokines in B-cells and cytokine production
following antigen stimulation (74). In NK cells, SLAMF7 mediates
activating signals via Ewing's sarcoma-associated transcript 2
involving phospholipase Cγ1 and the ERK1/2 signaling pathways and
calcium influx, leading to cell-mediated cytotoxicity (28,75).
It also plays a critical role in mediating cellular adhesion in
macrophages through SLAM family receptors (76).
Since SLAMF7 has an FDA-approved monoclonal antibody
(elotuzumab), the results of the present study suggest that once
validated, it may have immense potential for monoclonal antibody
therapy, which could improve the prognosis and therapeutic outcomes
of patients with BC. It can also serve as an additional molecular
classifier and make a noticeable contribution as a prognosticator
and therapeutic target, particularly for patients with TNBC where
outcomes are still challenging. The present study revealed that
only 10% of TNBC cases expressed SLAMF7, whereas, on the other
hand, 78% of TPBC cases had no expression of SLAMF7. Further
studies are warranted to further investigate the association
between TNBC cases and SLAMF7 using larger BC cohorts. This
research approach is essential in order to identify a promising
therapeutic option for the aggressive TNBC molecular subtype, known
by the limited treatment modalities compared with its TPBC
counterpart.
It is important to highlight that the lack of
SLAMF7 mRNA expression in the studied tumor tissues is a
limitation of the present study, since it would be a valuable
validation of the protein expression findings. Moreover, and as
aforementioned, the molecular effects of SLAMF7 are affected not
only by its level of expression, but also by its soluble fraction
(sSLAMF7) shown to be involved in lymphocyte proliferation. The
authors aim to perform additional studies in the future to
investigate the mRNA expression patterns, roles and molecular
mechanisms underlying the action of SLAMF7/sSLAMF7 in solid tumors
in general and BC, in particular, in order to validate the IHC
findings and optimize its use towards precision BC therapies.
In conclusion, to the best of our knowledge, the
present study was the first to report the cytoplasmic expression of
SLAMF7 in BC. The lack of or a low expression of SLAMF7 protein was
associated with both tumor and vascular invasion. It was also a
strong prognosticator of poor survival outcomes, including a higher
number of BC recurrences and mortality. Therefore, elotuzumab may
prove to be an additional targeted therapy for patients with
BC.
Acknowledgements
The author would like to thank Professor Abdelbaset
Buhmeida, Center of Excellence in Genomic Medicine Research, King
Abdulaziz University, for validating the immunohistochemistry
scoring, as well as Ms. F. Yahia from the Department of
Biochemistry, King Abdulaziz University, for improving and
exporting Fig. 5 using
Biorender.com.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Author's contributions
MA is the sole contributor to this work. MA
conceived and designed the study, analyzed the data and wrote the
whole manuscript. MA confirms the authenticity of all the raw data.
The author has read and approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval for the present study was obtained
from the Research Ethics Committee of The Center of Excellence in
Genomic Medicine Research (IRB no. 08-CEGMR-02-ETH). The study was
in line with the declaration of Helsinki. All participants provided
signed informed consent before inclusion in this study.
Patient consent for publication
Not applicable.
Competing interests
The author declares that he has no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Parkin DM, Piñeros M, Znaor A and Bray F: Cancer statistics for the
year 2020: An overview. Int J Cancer. 149:778–789. 2021. View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pfeiffer RM, Webb-Vargas Y, Wheeler W and
Gail MH: Proportion of US trends in breast cancer incidence
attributable to long-term changes in risk factor distributions.
Cancer Epidemiol Biomarkers Prev. 27:1214–1222. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Globocan, . Saudi Arabia Fact Sheets.
International Agency for Research on Cancer (IARC); Lyon, France:
https://gco.iarc.fr/today/data/factsheets/populations/682-saudi-arabia-fact-sheets.pdfApril
7–2022
|
|
5
|
Albeshan SM and Alashban YI: Incidence
trends of breast cancer in Saudi Arabia: A Joinpoint regression
analysis (2004–2016). J King Saud Univ Sci. 33:1015782021.
View Article : Google Scholar
|
|
6
|
Jazieh AR, Da'ar OB, Alkaiyat M, Zaatreh
YA, Saad AA, Bustami R, Alrujaib M and Alkattan K: Cancer incidence
trends from 1999 to 2015 and contributions of various cancer types
to the overall burden: Projections to 2030 and extrapolation of
economic burden in Saudi Arabia. Cancer Manag Res. 11:9665–9674.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Albeshan SM, Mackey MG, Hossain SZ,
Alfuraih AA and Brennan PC: Breast cancer epidemiology in gulf
cooperation council countries: A regional and international
comparison. Clin Breast Cancer. 18:e381–e392. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Assidi M, Jafri MA, Abu-Elmagd M, N
Pushparaj P, Saddick S, Messaoudi S, Alkhatabi H, Al-Maghrabi J,
Anfinan N, Sait M, et al: Prognostic value of E-Cadherin and its
tumor suppressor role in Saudi women with advanced epithelial
ovarian cancer. Libyan J Med. 16:19947412021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lumachi F, Luisetto G, MM Basso S, Basso
U, Brunello A and Camozzi V: Endocrine therapy of breast cancer.
Current Med Chem. 18:513–522. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang CY, Ju DT, Chang CF, Reddy PM and
Velmurugan BK: A review on the effects of current chemotherapy
drugs and natural agents in treating non-small cell lung cancer.
Biomedicine (Taipei). 7:232017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Allison KH, Hammond MEH, Dowsett M,
McKernin SE, Carey LA, Fitzgibbons PL, Hayes DF, Lakhani SR,
Chavez-MacGregor M, Perlmutter J, et al: Estrogen and progesterone
receptor testing in breast cancer: ASCO/CAP guideline update. J
Clin Oncol. 38:1346–1366. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Purdie CA, Quinlan P, Jordan LB, Ashfield
A, Ogston S, Dewar JA and Thompson AM: Progesterone receptor
expression is an independent prognostic variable in early breast
cancer: A population-based study. Br J Cancer. 110:565–572. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Petroni S, Caldarola L, Scamarcio R,
Giotta F, Latorre A, Mangia A and Simone G: FISH testing of HER2
immunohistochemistry 1+ invasive breast cancer with unfavorable
characteristics. Oncol Lett. 12:3115–3122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brown JR, DiGiovanna MP, Killelea B,
Lannin DR and Rimm DL: Quantitative assessment Ki-67 score for
prediction of response to neoadjuvant chemotherapy in breast
cancer. Lab Invest. 94:98–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lampelj M, Arko D, Cas-Sikosek N, Kavalar
R, Ravnik M, Jezersek-Novakovic B, Dobnik S, Dovnik NF and Takac I:
Urokinase plasminogen activator (uPA) and plasminogen activator
inhibitor type-1 (PAI-1) in breast cancer-correlation with
traditional prognostic factors. Radiol Oncol. 49:357–364. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Parrales A and Iwakuma T: Targeting
oncogenic mutant p53 for cancer therapy. Front Oncol. 5:2882015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bi H, Li S, Qu X, Wang M, Bai X, Xu Z, Ao
X, Jia Z, Jiang X, Yang Y and Wu H: DEC1 regulates breast cancer
cell proliferation by stabilizing cyclin E protein and delays the
progression of cell cycle S phase. Cell Death Dis. 6:e1891. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Al-Keilani MS, Elstaty RI, Alqudah MA and
Alkhateeb AM: Immunohistochemical expression of substance P in
breast cancer and its association with prognostic parameters and
Ki-67 index. PLoS One. 16:e02526162021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tray N, Weber JS and Adams S: Predictive
biomarkers for checkpoint immunotherapy: Current status and
challenges for clinical application. Cancer Immunol Res.
6:1122–1128. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Emens LA, Cruz C, Eder JP, Braiteh F,
Chung C, Tolaney SM, Kuter I, Nanda R, Cassier PA, Delord JP, et
al: Long-term clinical outcomes and biomarker analyses of
Atezolizumab therapy for patients with metastatic Triple-negative
breast cancer: A phase 1 study. JAMA Oncol. 5:74–82. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Waldman AD, Fritz JM and Lenardo MJ: A
guide to cancer immunotherapy: From T cell basic science to
clinical practice. Nat Rev Immunol. 20:651–668. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Keefe DMK and Bateman EH: Potential
successes and challenges of targeted cancer therapies. J Natl
Cancer Inst Monogr. 2019:lgz0082019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Akbulut H, Babahan C, Abgarmi SA, Ocal M
and Besler M: Recent advances in cancer stem cell targeted therapy.
Crit Rev Oncog. 24:1–20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tai YT, Dillon M, Song W, Leiba M, Li XF,
Burger P, Lee AI, Podar K, Hideshima T, Rice AG, et al: Anti-CS1
humanized monoclonal antibody HuLuc63 inhibits myeloma cell
adhesion and induces antibody-dependent cellular cytotoxicity in
the bone marrow milieu. Blood. 112:1329–1337. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boles KS and Mathew PA: Molecular cloning
of CS1, a novel human natural killer cell receptor belonging to the
CD2 subset of the immunoglobulin superfamily. Immunogenetics.
52:302–307. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hsi ED, Steinle R, Balasa B, Szmania S,
Draksharapu A, Shum BP, Huseni M, Powers D, Nanisetti A, Zhang Y,
et al: CS1, a potential new therapeutic antibody target for the
treatment of multiple myeloma. Clin Cancer Res. 14:2775–2784. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lonial S, Dimopoulos M, Palumbo A, White
D, Grosicki S, Spicka I, Walter-Croneck A, Moreau P, Mateos MV,
Magen H, et al: Elotuzumab therapy for relapsed or refractory
multiple myeloma. N Engl J Med. 373:621–631. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bouchon A, Cella M, Grierson HL, Cohen JI
and Colonna M: Cutting edge: Activation of NK cell-mediated
cytotoxicity by a SAP-independent receptor of the CD2 family. J
Immunol. 167:5517–5521. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kumaresan PR, Lai WC, Chuang SS, Bennett M
and Mathew PA: CS1, a novel member of the CD2 family, is homophilic
and regulates NK cell function. Mol Immunol. 39:1–8. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Zhong MC, Guo H, Davidson D,
Mishel S, Lu Y, Rhee I, Pérez-Quintero LA, Zhang S, Cruz-Munoz ME,
et al: SLAMF7 is critical for phagocytosis of haematopoietic tumour
cells via Mac-1 integrin. Nature. 544:493–497. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pazina T, James AM, MacFarlane AW IV,
Bezman NA, Henning KA, Bee C, Graziano RF, Robbins MD, Cohen AD and
Campbell KS: The anti-SLAMF7 antibody elotuzumab mediates NK cell
activation through both CD16-dependent and-independent mechanisms.
Oncoimmunology. 6:e13398532017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Simmons DP, Nguyen HN, Gomez-Rivas E,
Jeong Y, Jonsson AH, Chen AF, Lange JK, Dyer GS, Blazar P, Earp BE,
et al: SLAMF7 engagement superactivates macrophages in acute and
chronic inflammation. Sci Immunol. 7:eabf28462022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Campbell KS, Cohen AD and Pazina T:
Mechanisms of NK cell activation and clinical activity of the
therapeutic SLAMF7 antibody, elotuzumab in multiple myeloma. Front
Immunol. 9:25512018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lonial S, Dimopoulos M, Palumbo A, White
D, Grosicki S, Spicka I, Walter-Croneck A, Moreau P, Mateos MV,
Magen H, et al: Elotuzumab therapy for relapsed or refractory
multiple myeloma. N Engl J Med. 373:621–631. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee JL, Roh SA, Kim CW, Kwon YH, Ha YJ,
Kim SK, Kim SY, Cho DH, Kim YS and Kim JC: Clinical assessment and
identification of immuno-oncology markers concerning the 19-gene
based risk classifier in stage IV colorectal cancer. World J
Gastroenterol. 25:1341–1354. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li X, Zhou H, Huang W, Wang X, Meng M, Hou
Z, Liao L, Tang W, Xie Y, Wang R, et al: Retrospective analysis of
the efficacy of adjuvant cytokine-induced killer cell immunotherapy
combined with chemotherapy in colorectal cancer patients after
surgery. Clin Transl Immunol. 11:e13682022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roh SA, Kwon YH, Lee JL, Kim SK and Kim
JC: SLAMF7 and TREM1 mediate immunogenic cell death in colorectal
cancer cells: Focus on microsatellite stability. Anticancer Res.
41:5431–5444. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Su R, Jin C, Zhou L, Gao Y, Kuang M, Li L
and Xiang J: Construction of a ceRNA network of hub genes affecting
immune infiltration in ovarian cancer identified by WGCNA. BMC
Cancer. 21:9702021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen H, Wang T, Huang S and Zeng P: New
novel non-MHC genes were identified for cervical cancer with an
integrative analysis approach of transcriptome-wide association
study. J Cancer. 12:840–848. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun H, Kim E, Ryu J, Lee H, Shin EA, Lee
M, Lee H, Lee HJ, Yoon JH, Song DG, et al: TM4SF5-mediated liver
malignancy involves NK cell exhaustion-like phenotypes. Cell Mol
Life Sci. 79:492021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
O'Connell P, Blake MK, Pepelyayeva Y,
Hyslop S, Godbehere S, Angarita AM, Pereira-Hicks C, Amalfitano A
and Aldhamen YA: Adenoviral delivery of an immunomodulatory protein
to the tumor microenvironment controls tumor growth. Mol Ther
Oncolytics. 24:180–193. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Al-Maghrabi J, Emam E, Gomaa W, Saggaf M,
Buhmeida A, Al-Qahtani M and Al-Ahwal M: c-MET immunostaining in
colorectal carcinoma is associated with local disease recurrence.
BMC Cancer. 15:6762015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nedjadi T, Al-Maghrabi J, Assidi M, Dallol
A, Al-Kattabi H, Chaudhary A, Al-Sayyad A, Al-Ammari A, Abuzenadah
A, Buhmeida A and Al-Qahtani M: Prognostic value of HER2 status in
bladder transitional cell carcinoma revealed by both IHC and BDISH
techniques. BMC Cancer. 16:6532016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Assidi M, Yahya FM, Al-Zahrani MH,
Elkhatib R, Zari A, Elaimi A, Al-Maghrabi J, Dallol A, Buhmeida A
and Abu-Elmagd M: Leptin protein expression and promoter
methylation in ovarian cancer: A strong prognostic value with
theranostic promises. Int J Mol Sci. 22:128722021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Buhmeida A, Dallol A, Merdad A,
Al-Maghrabi J, Gari MA, Abu-Elmagd MM, Chaudhary AG, Abuzenadah AM,
Nedjadi T, Ermiah E, et al: High fibroblast growth factor 19
(FGF19) expression predicts worse prognosis in invasive ductal
carcinoma of breast. Tumour Biol. 35:2817–2824. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cronin KA, Lake AJ, Scott S, Sherman RL,
Noone AM, Howlader N, Henley SJ, Anderson RN, Firth AU, Ma J, et
al: Annual report to the nation on the status of cancer, part I:
National cancer statistics. Cancer. 124:2785–2800. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiang M, Yang JS, Li K, Liu J, Jing XG and
Tang MQ: Insights into the theranostic value of precision medicine
on advanced radiotherapy to breast cancer. Int J Med Sci.
18:626–638. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Society AC: Cancer facts & figures
2012. Atlanta: American Cancer Society; pp. 9–11. 2012
|
|
51
|
Lai FM, Chen P, Ku HC, Lee MS, Chang SC,
Chang TM and Liou SH: A case-control study of parity, age at first
full-term pregnancy, breast feeding and breast cancer in Taiwanese
women. Proc Natl Sci Counc Repub China B. 20:71–77. 1996.PubMed/NCBI
|
|
52
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Reis-Filho JS and Pusztai L: Gene
expression profiling in breast cancer: Classification,
prognostication, and prediction. Lancet. 378:1812–1823. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Garrido-Castro AC, Lin NU and Polyak K:
Insights into molecular classifications of triple-negative breast
cancer: Improving patient selection for treatment. Cancer Discov.
9:176–198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dai X, Xiang L, Li T and Bai Z: Cancer
hallmarks, biomarkers and breast cancer molecular subtypes. J
Cancer. 7:1281–1294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Goldhirsch A, Wood WC, Coates AS, Gelber
RD, Thürlimann B and Senn HJ: Strategies for subtypes-dealing with
the diversity of breast cancer: Highlights of the St. Gallen
International Expert Consensus on the Primary Therapy of Early
Breast Cancer 2011. Ann Oncol. 22:1736–1747. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kim JR, Horton NC, Mathew SO and Mathew
PA: CS1 (SLAMF7) inhibits production of proinflammatory cytokines
by activated monocytes. Inflamm Res. 62:765–772. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li X, Zhou H, Huang W, Wang X, Meng M, Hou
Z, Liao L, Tang W, Xie Y, Wang R, et al: Retrospective analysis of
the efficacy of adjuvant cytokine-induced killer cell immunotherapy
combined with chemotherapy in colorectal cancer patients after
surgery. Clin Transl Immunol. 11:e13682022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Thul PJ and Lindskog C: The human protein
atlas: A spatial map of the human proteome. Protein Sci.
27:233–244. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Tissue-based map of the human
proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357:eaan25072017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zekri J, Saadeddin A and Alharbi H:
Frequency and clinical characteristics of HER2 over-expressed
breast cancer in Saudi Arabia: A retrospective study. BMC Womens
Health. 21:102021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Weigel MT and Dowsett M: Current and
emerging biomarkers in breast cancer: Prognosis and prediction.
Endocrine-related Cancer. 17:R245–R262. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Saggu S, Rehman H, Abbas ZK and Ansari AA:
Recent incidence and descriptive epidemiological survey of breast
cancer in Saudi Arabia. Saudi Med J. 36:1176–1180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Albasri A, Hussainy AS, Sundkji I and
Alhujaily A: Histopathological features of breast cancer in
Al-Madinah region of Saudi Arabia. Saudi Med J. 35:1489–1493.
2014.PubMed/NCBI
|
|
66
|
Howlader N, Noone A, Krapcho M, Miller D,
Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, et al:
SEER cancer statistics review, 1975–2016, National Cancer
Institute. Bethesda, MD: https://seer.cancer.gov/csr/1975_2016/based on
November 2018 SEER data submission, posted to the SEER web site.
April. 2019
|
|
67
|
Najjar H and Easson A: Age at diagnosis of
breast cancer in Arab nations. Int J Surg. 8:448–452. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Müller L and Pawelec G: Aging and
immunity-impact of behavioral intervention. Brain Behav Immunity.
39:8–22. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Valcárcel LV, Amundarain A, Kulis M,
Charalampopoulou S, Melnick A, San Miguel J, Martín-Subero JI,
Planes FJ, Agirre X and Prosper F: Gene expression derived from
alternative promoters improves prognostic stratification in
multiple myeloma. Leukemia. 35:3012–3016. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Malaer JD and Mathew PA: CS1 (SLAMF7,
CD319) is an effective immunotherapeutic target for multiple
myeloma. Am J Cancer Res. 7:1637–1641. 2017.PubMed/NCBI
|
|
71
|
Boudreault JS, Touzeau C and Moreau P: The
role of SLAMF7 in multiple myeloma: Impact on therapy. Expert Rev
Clin Immunol. 13:67–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Pham T, Roth S, Kong J, Guerra G,
Narasimhan V, Pereira L, Desai J, Heriot A and Ramsay R: An update
on immunotherapy for solid tumors: A review. Ann Surg Oncol.
25:3404–3412. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chandrashekar DS, Karthikeyan SK, Korla
PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne
U, et al: UALCAN: An update to the integrated cancer data analysis
platform. Neoplasia. 25:18–27. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Detre C, Keszei M, Romero X, Tsokos GC and
Terhorst C: SLAM family receptors and the SLAM-associated protein
(SAP) modulate T cell functions. Semin Immunopathol. 32:157–171.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wu N and Veillette A: SLAM family
receptors in normal immunity and immune pathologies. Curr Opin
Immunol. 38:45–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Claus M, Urlaub D, Fasbender F and Watzl
C: SLAM family receptors in natural killer cells-mediators of
adhesion, activation and inhibition via cis and Trans interactions.
Clin Immunol. 204:37–42. 2019. View Article : Google Scholar : PubMed/NCBI
|