Introduction
The inflammatory bowel diseases, ulcerative colitis
(UC) and Crohn's disease, are chronic inflammatory diseases of the
gastrointestinal tract, and have become global diseases that are
increasing in Westernized countries (1). UC is associated with an increased
risk of colitis-associated neoplasia, which increases with extended
and more active inflammation (2).
Ulcerative colitis-associated neoplasia (UCAN) was first reported
by Crohn and Rosenberg in 1925 (3), and it has been recognized as an
important complication of UC. Surveillance colonoscopy is widely
accepted as being important for the early detection and treatment
of UCAN, and UCAN surveillance is recommended in many countries
(4).
In this surveillance, clinicians need to distinguish
between sporadic neoplasms and UCAN based on endoscopic and
pathological findings. Generally, sporadic neoplasms and UCAN are
treated differently. Endoscopic resection is usually applied if the
lesion is diagnosed as a sporadic neoplasm; while total
proctocolectomy with ileal pouch-anal anastomosis or ileal
pouch-anal canal anastomosis is applied if the lesion is diagnosed
as UCAN (4). The American College
of Gastroenterology clinical guideline described subsequent
surveillance colonoscopy should initially be performed at shortened
intervals when dysplasia in the UC case in which discrete neoplasms
are completely removed endoscopically refusing the total
proctocolectomy (4). Therefore,
the differential diagnosis between UCAN and sporadic neoplasm is
important when determining the treatment strategy for neoplasms
arising in long-standing UC. However, it is often difficult to
distinguish between the two using endoscopic and histopathological
findings.
UCAN chemotherapy regimens are usually selected from
cytotoxic and molecular-targeted agents used for sporadic
colorectal cancer (CRC), based on genetic testing for markers such
as RAS, BRAF, and microsatellite instability (MSI) status
(5). The genomic landscape of
sporadic CRC has been fairly well studied using next-generation
sequencing (NGS) technology. Despite previous analyses showing that
UCAN harbors unique genetic alterations and mutational tendencies
(6–20), chemotherapy regimens for UCAN
continue to be extrapolated from those for sporadic CRC.
Clinical application of gene panel testing enables
us to practice precision medicine by tailoring treatment to
individual gene alterations, and we previously reported NGS-based
gene panel testing for management of solid tumors (21). We assumed that gene panel testing
would detect clinically important genetic alterations in UCAN, with
potential utility in UCAN diagnosis and treatment. In this
analysis, we aimed to identify genetic alterations of UCAN using
gene panel testing, and investigate the possibility of clinical
utility of gene panel testing in UCAN.
Materials and methods
Patients
We studied 15 patients with UCAN who had been
treated between 2009 and 2021 at Niigata University Medical and
Dental Hospital. We have previously reported on genetic alterations
in Japanese patients with sporadic CRC using gene panel testing
(22–27), but not in patients with UCAN. In
this analysis, we identified genetic alterations in the 15 patients
with UCAN, and compared them with those identified in Stage I–IV
203 patients with sporadic CRC according to the American Joint
Committee on Cancer guidelines, 8th edition (28), who had undergone primary tumor
resection between 2009 and 2015 at Niigata University Medical and
Dental Hospital or Niigata Cancer Centre Hospital. Endoscopic
diagnoses of UCAN were made by endoscopists specializing
inflammatory bowel disease (K. M. and J. Y.) according to the
SCIENIC international consensus statement (29). Histopathological diagnoses of UCAN
were made by a pathologist specializing in inflammatory bowel
disease (Y. A.) according to the classification proposed by the
Research Committee on Inflammatory Bowel Disease of the Japanese
Ministry of Health and Welfare (30) and the Riddell's classification
(31). p53 overexpression was
assessed as an aid to histopathological diagnosis of UCAN (32). We included 15 UCAN diagnosed as
UC-IV of the classification proposed by the Research Committee on
Inflammatory Bowel Disease of the Japanese Ministry of Health and
Welfare (30). UC-IV was defined
as carcinoma including intramucosal carcinoma. The diagnosis of
intramucosal carcinoma was to be made when there was a high grade
of cytological and structural atypia consistent with carcinoma
(30). This retrospective analysis
was performed in accordance with the Helsinki Declaration, and the
Ethics Committee of the School of Medicine, Niigata University
approved the study protocol (G2015-0816, G2020-0038). Written
informed consent was obtained from the patients.
NGS for detecting genetic
alterations
Formalin-fixed, paraffin-embedded biopsy or
endoscopically/surgically resected samples were used for evaluating
genetic alterations, as we have previously reported (22–27).
Briefly, hematoxylin and eosin-stained sections were used to assess
tumor content, ensuring >50% tumor content. Where applicable,
unstained sections were macro-dissected to enrich for tumor
content. DNA was extracted using a BioStic FFPE Tissue DNA
Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). All
sample preparations, NGS, and bioinformatics analyses were
performed in a Clinical Laboratory Improvement Amendments/College
of American Pathologists (CLIA/CAP)-accredited laboratory. First,
50–150 ng DNA fragment libraries were prepared and enriched for
CANCERPLEX version 3.0 (415-gene panel; KEW) or version 4.0
(435-gene panel; KEW). An average 500× sequencing depth was
achieved using the Illumina MiSeq and NextSeq platforms. A 10%
allelic fraction threshold for single nucleotide variants (SNVs)
and insertions/deletions was used, as well as thresholds of
>2.5-fold and 0.5-fold for gain and loss, respectively. MSI was
tested based on an extended loci panel: in addition to the Bethesda
panel, a collection of 950 regions consisting of tandem repeats of
1, 2 or 3 nucleotides with a minimum length of 10 bases was used.
Tumor mutational burden was calculated as the number of
non-synonymous mutations per megabase of sequence in the panel
(panel size=1.3 Mb). Mutational signatures were analyzed as
previously reported (27). Each
SNV was classified in a matrix of the 96 possible substitutions,
based on the sequence context comprising the nucleotides 5′ and 3′
to the position of the mutation. Mutational signatures were
extracted using non-negative matrix factorization analysis with the
SomaticSignatures R package (33)
(The R Foundation, Vienna, Austria) and plotted with the ggplots R
package (http://ggplot2.org/). We previously
confirmed reliability of the mutational signatures at the gene
panel level using data sets of gene panel testing and whole exome
sequencing data (27).
Statistical analysis
Statistical analyses were performed with IBM SPSS
Statistics 28 software (IBM Japan, Inc., Tokyo, Japan). The
frequency of genetic alterations between UCAN and sporadic CRC were
compared using Fisher's exact tests. The mutational signature ratio
between UCAN and sporadic CRC was also compared using a Fisher's
exact test. P-values of less than 0.05 were considered
significant.
Results
Clinicopathological characteristics of
UCAN
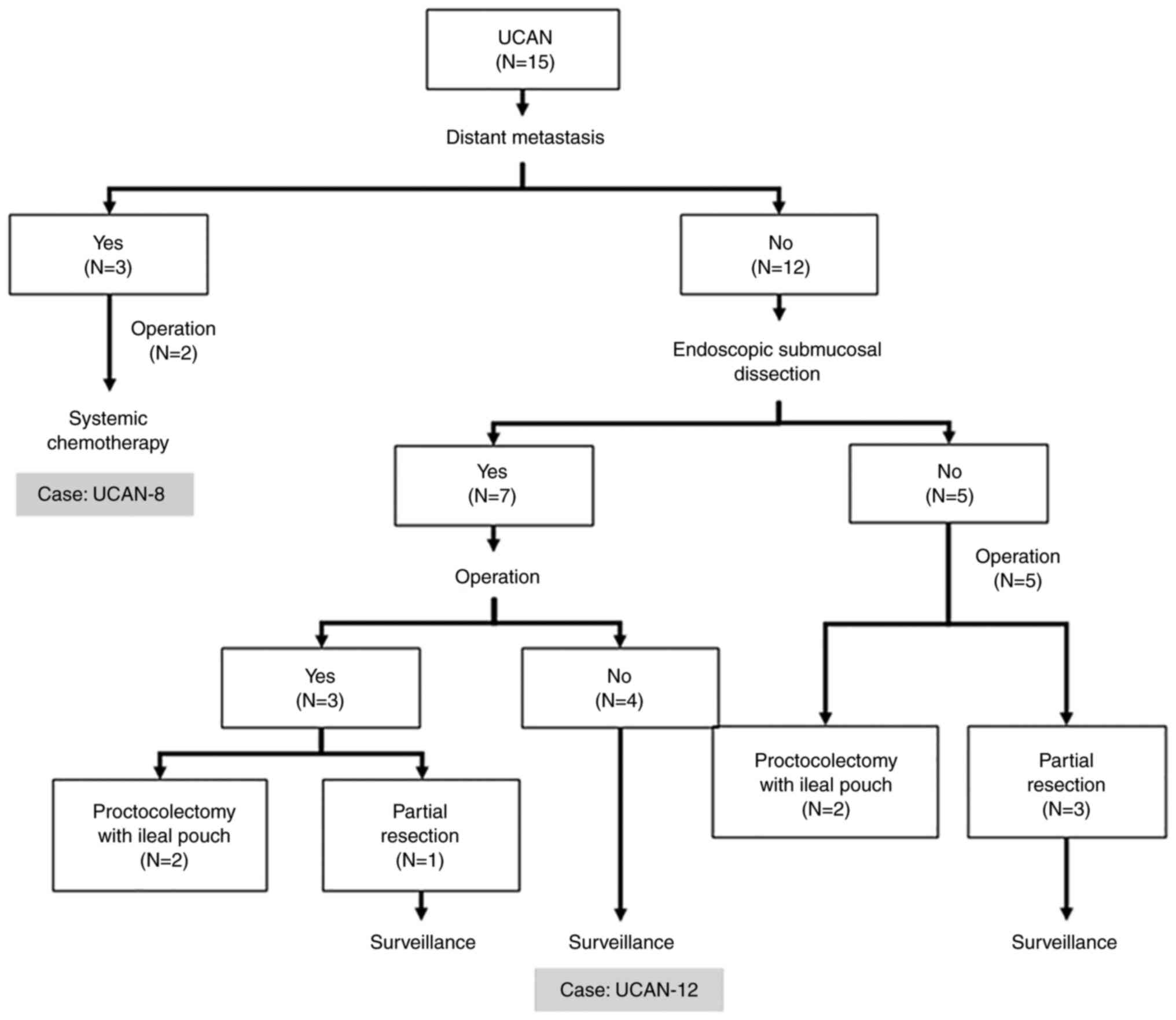
In this analysis of 15 patients with UCAN, UCAN was
significantly associated with a lower T stage, N stage, and M stage
compared with sporadic CRC (Table
SI). The median disease duration of UC was 25 years (range,
9–45 years). Fourteen patients were diagnosed with UCAN caused by
chronic inflammation of the large intestine, while one patient was
diagnosed with UCAN caused by chronic pouchitis. Eight of 15
patients (53%) had UCAN in the rectum. Six patients had carcinoma
in situ, while nine patients had a tumor invading to the submucosal
layer or deeper. Detailed information for each patient is shown in
Fig. 1 and Table I. Three patients had distant
metastasis, and received systemic chemotherapy, as indicated for
sporadic CRC. Four patients without distant metastasis underwent
proctocolectomy with ileal pouch, while four patients without
distant metastasis underwent partial resection of the large
intestine because of the patients' request and have been
followed-up using annual surveillance colonoscopy under informed
consent. Seven patients who had a lesion localized in the mucosal
layer or slightly invading into the submucosal layer underwent
endoscopic submucosal dissection (ESD) for accurate diagnosis of
UCAN. Four of the seven patients who underwent ESD have been
followed-up using annual surveillance colonoscopy.
 | Table I.Clinicopathological characteristics
of 15 patients with UCAN. |
Table I.
Clinicopathological characteristics
of 15 patients with UCAN.
| ID | Age, years | Sex | UC disease
duration, years | Primary tumor
location | TNM
stagea | Histological
classificationb | p53 IHC
overexpression | TMB (/Mb) | MSI status | APC | TP53 | KRAS | RNF43 | TP53
mutation alone | Treatment |
|---|
| UCAN-1 | 67 | M | 45 | Ascending | 0 | UC-IV + UC-III | Absent | 26.94 | MSI-H |
|
|
| p.G659fs |
| Partial
resection |
| UCAN-2 | 66 | F | 35 | Rectum | IIIB | UC-IV | NA | 26.17 | MSI-H |
| p.I195T |
|
| Yes | Partial
resection |
| UCAN-3 | 67 | M | 40 | Rectum | IVB | UC-IV | NA | 17.7 | MSS | p.S1198X | p.R273H, p.R248Q,
loss |
|
|
| Systemic
chemotherapy |
| UCAN-4 | 45 | M | 23 | Ascending | I | UC-IV + UC-III | Absent | 16.9 | MSS | loss | p.C135F |
|
|
| Partial
resection |
| UCAN-5 | 62 | F | 15 | Transverse | I | UC-IV + UC-III | Present | 14.6 | MSS |
| p.P152L, loss | p.G12V |
|
| Proctocolectomy
with ileal pouch |
| UCAN-6 | 37 | M | 18 | Ascending | 0 | UC-IV + UC-III | Absent | 13.9 | MSS |
| loss |
| p.E742X, |
| Proctocolectomy
with ileal pouch |
|
|
|
|
|
|
|
|
|
|
|
|
|
| p.R132X |
|
|
| UCAN-7 | 61 | M | 25 | Rectum | I | UC-IV + UC-III | Present | 12.32 | MSS |
| p.R248Q |
|
| Yes | ESD followed by
partial resection |
| UCAN-8 | 43 | F | 26 | Ileal pouch | IVA | UC-IV + UC-III | NA | 11.55 | MSS |
| p.G199V | p.G12D |
|
| Resection of ileal
pouch followed by systemic chemotherapy |
| UCAN-9 | 42 | M | 17 | Rectum | IVC | UC-IV | NA | 8.5 | MSS |
| p.Y234C |
|
| Yes | Partial resection
followed by systemic chemotherapy |
| UCAN-10 | 53 | M | 15 | Rectum | 0 | UC-IV + UC-III | Present | 6.9 | MSS |
| p.R248W | p.G13D |
|
| ESD |
| UCAN-11 | 67 | M | 27 | Rectum | 0 | UC-IV + UC-III | Present | 6.2 | MSS |
| p.V173M |
|
|
| ESD followed by
proctocolectomy with ileal pouch |
| UCAN-12 | 65 | F | 30 | Sigmoid | 0 | UC-IV + UC-III | Present | 6.2 | MSS |
| Splice variant |
|
| Yes | ESD |
| UCAN-13 | 53 | F | 9 | Ascending | 0 | UC-IV | Absent | 2.3 | MSS |
|
|
| p.G659fs |
| ESD |
| UCAN-14 | 49 | F | 15 | Rectum | I | UC-IV + UC-III | NA | 1.5 | MSS |
| p.I254T | p.G13D |
|
| ESD followed by
proctocolectomy with ileal pouch |
| UCAN-15 | 66 | F | 31 | Rectum | I | UC-IV + UC-III | Present | 0.8 | MSS |
| p.R196X |
| p.D95fs |
| ESD |
Gene panel testing of UCAN and
sporadic CRC
Characteristic mutational tendencies in the WNT
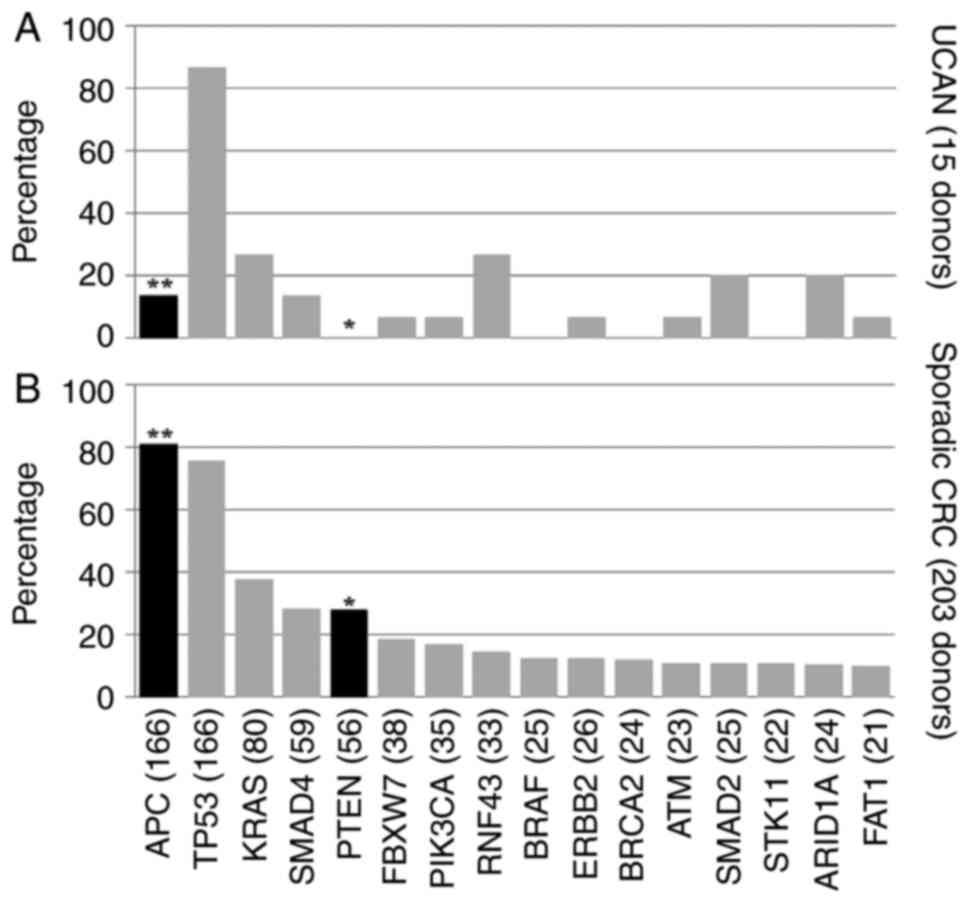
signaling pathway, including APC and RNF43, were
identified in patients with UCAN. APC mutations were
significantly less frequent in patients with UCAN compared with
sporadic CRC (P<0.001), being identified in 2/15 patients (13%)
with UCAN and 164/203 patients (81%) with sporadic CRC (Fig. 2). RNF43 frameshift or
nonsense mutations were significantly more frequent in patients
with UCAN compared with sporadic CRC (P=0.025), being identified in
4/15 patients (27%) with UCAN and 14/203 patients (7%) with
sporadic CRC (Figs. S1 and
2). PTEN mutations were
significantly less frequent in patients with UCAN compared with
sporadic CRC (P=0.014), being completely absent in patients with
UCAN (Fig. 2). TP53
mutations were identified in 13/15 patients (87%) with UCAN, with
most consisting of SNV, identified in the DNA-binding domain
(Fig. S3). Interestingly, 4/15
patients (27%) with UCAN had no genetic alterations other than a
TP53 mutation, while this occurred in 1/203 patients (0.5%)
with sporadic CRC (P<0.001) (Fig.
S4).
MSI-H was identified in 2/15 patients (13%) with
UCAN and 13/203 patients (6%) with sporadic CRC. Despite the
reported clinical utility of immune checkpoint inhibitors (ICIs) in
some patients with MSI-H CRC, no patients received ICIs in this
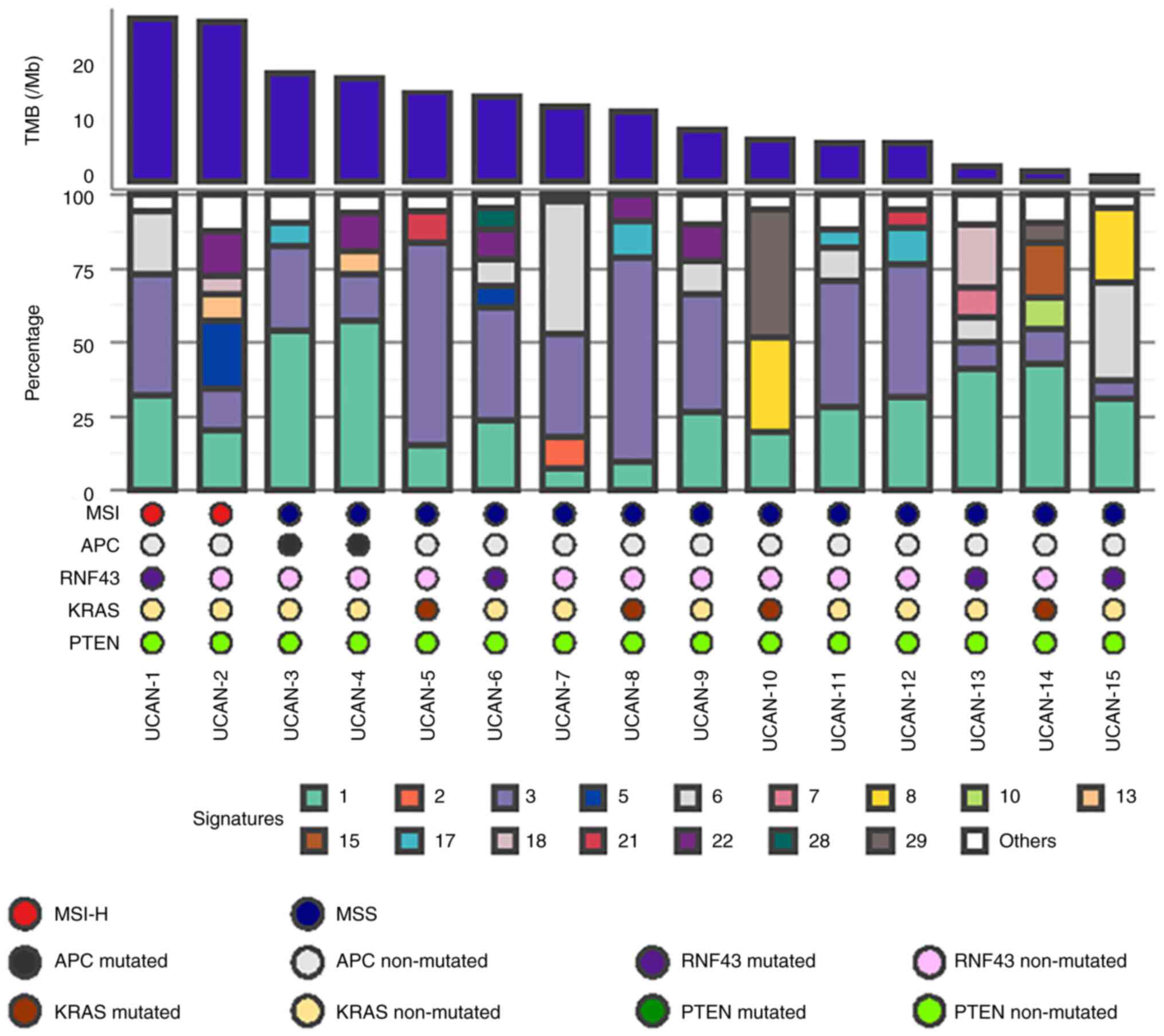
cohort. Mutational signature 3, which is associated with failure of
DNA double-strand break repair by homologous recombination
deficiency (HRD), was detected in 14/15 patients (93%) with UCAN
(Fig. 3), and enriched in UCAN
compared with sporadic CRC (P=0.030) (Fig. S5). Among all the 15 patients with
MSI-H in this cohort (two UCAN, 13 sporadic CRC), the two UCAN
patients exhibited mutational signature 3 (Fig. S6). An oxaliplatin-based regimen,
which is considered effective for tumors with failed DNA
double-strand break repair, was used in the two patients with
mutational signature 3, who had stage IV disease.
Possibility of clinical utility of
gene panel testing for diagnosis in UCAN
A 65-year-old-woman with a 30-year history of
extensive UC was found to have a flat mucosal lesion in her sigmoid
colon by surveillance colonoscopy (Fig. 4A). ESD was performed to diagnose
UCAN (Fig. 4B), and the lesion was
diagnosed as a well-differentiated adenocarcinoma (Fig. 4C). Gene panel testing revealed that
the tumor had a TP53 splice site variant, with no other
mutations detected (Fig. 4D). The
genome profile was considered to be characteristic of UCAN, and
served an auxiliary role for diagnosis of UCAN. Although the
patient was recommended proctocolectomy with an ileal pouch, she
declined surgery and instead chose follow-up by annual surveillance
colonoscopy. She has been treated with mesalazine, and her
condition has been well controlled. No new lesion has been detected
five years after ESD (Fig.
4A).
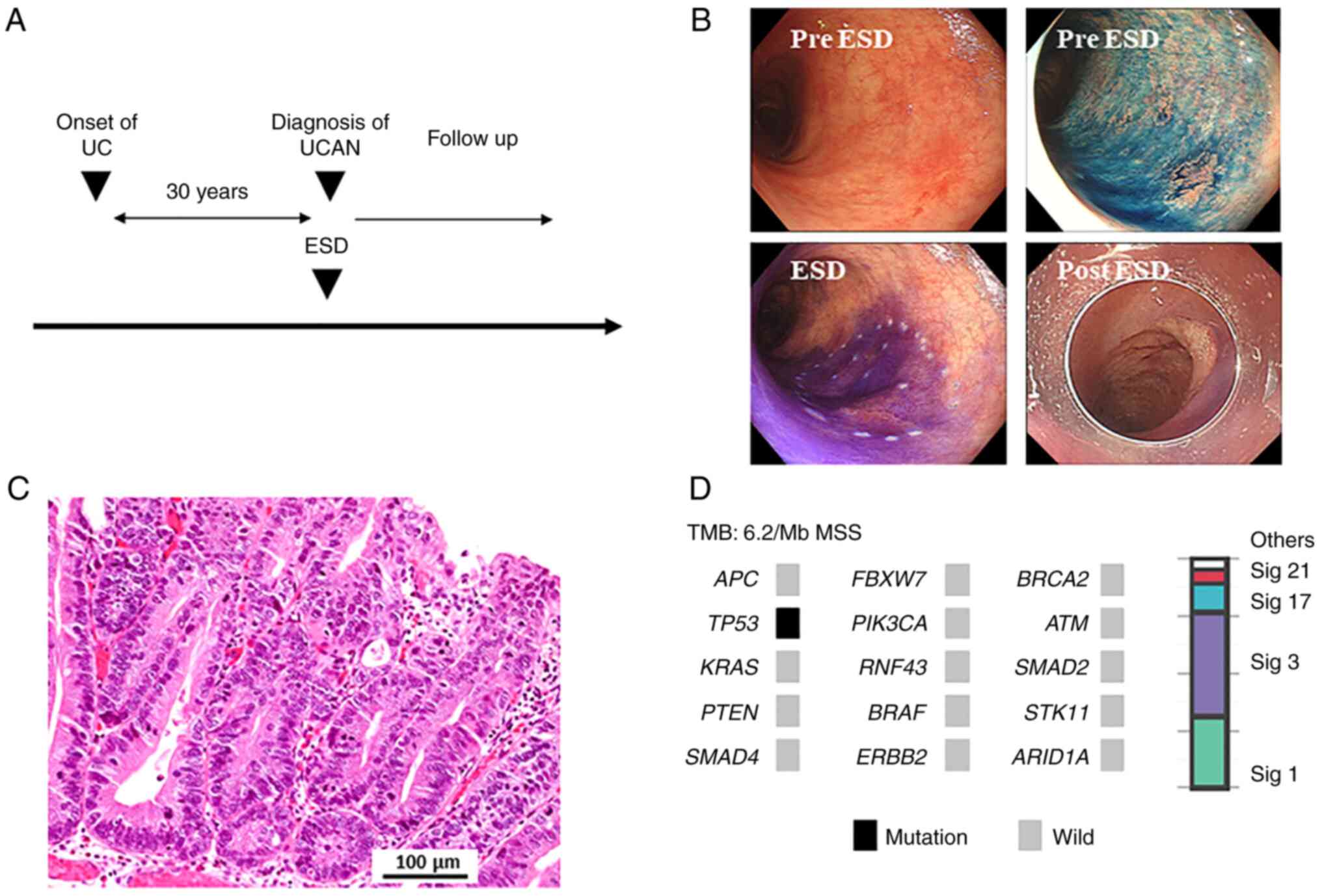 | Figure 4.A patient (UCAN-12) with UCAN that
underwent ESD followed by surveillance without proctocolectomy. (A)
Clinical course of the patient. (B) ESD for UCAN in sigmoid colon.
(C) Histopathological assessment, hematoxylin and eosin staining.
Scale bar, 100 µm. (D) Genome profile and mutational signature of
the patient. APC, adenomatous polyposis coli; ARID1A, AT-rich
interaction domain 1A; ATM, ataxia telangiectasia mutated; ERBB2,
erb-b2 receptor tyrosine kinase 2; ESD, endoscopic submucosal
dissection; FBXW7, F-box and WD repeat domain containing 7; MSS,
microsatellite stable; PIK3CA,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α;
RNF43, ring finger protein 43; Sig, signature; STK11,
serine/threonine kinase 11; TMB, tumor mutational burden; UC,
ulcerative colitis; UCAN, ulcerative colitis-associated
neoplasia. |
Possibility of clinical utility of
gene panel testing for treatment in UCAN
A 43-year-old-woman received proctocolectomy with an
ileal pouch for severe UC 26 years ago. She had a 13-year history
of pouchitis, and had received various medical treatments for
pouchitis (Fig. 5A). Pouchoscopy
revealed an irregular ulcerative mass lesion in the ileal pouch
near the anastomotic site, suggesting that the lesion might have
developed at the remnant rectal tissue. Histopathological diagnosis
of the biopsy specimen was poorly-differentiated adenocarcinoma.
Taken together, she was diagnosed with UCAN arising from the ileal
pouch (Fig. 5B). Abdominal
computed tomography and magnetic resonance imaging revealed a
solitary liver metastasis in S8 (Fig.
5C). She underwent resection of the ileal pouch, and gene panel
testing of the primary tumor found TP53, PIK3CA and
KRAS mutations. Moreover, mutational signature 3 was also
identified (Fig. 5D). She received
neoadjuvant chemotherapy including oxaliplatin (one course of
CapeOx and four courses of mFOLFOX6), which seems to be effective
for tumors with failure of DNA double-strand break repair by HRD.
After the chemotherapy, the liver metastasis was shrunk. Then,
partial resection of the liver (S8) was performed, and no viable
cancer cells were detected by histopathological assessment
(Fig. 5C).
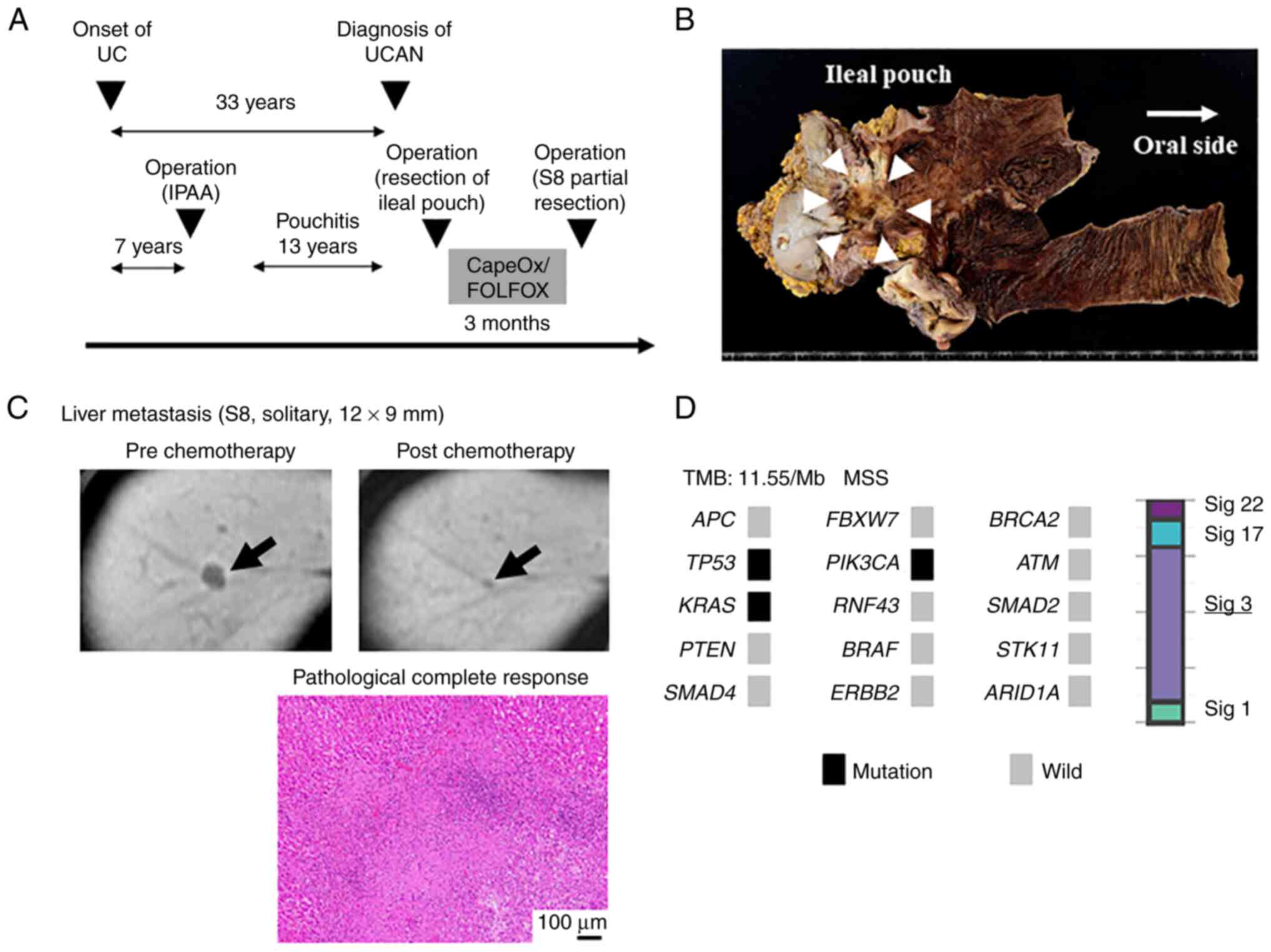 | Figure 5.A patient (UCAN-8) with UCAN
diagnosed as carcinoma arising from the ileal pouch with a solitary
liver metastasis. (A) Clinical course of the patient. (B) Surgical
specimen of the ileal pouch. (C) Radiological and histopathological
assessment of liver metastasis. Scale bar, 100 µm. (D) Genome
profile and mutational signature of the patient. Arrows indicate
lesion in each state of pre and post chemotherapy. Hematoxylin and
eosin staining demonstrates pathological complete response after
chemotherapy. APC, adenomatous polyposis coli; ARID1A, AT-rich
interaction domain 1A; ATM, ataxia telangiectasia mutated; ERBB2,
erb-b2 receptor tyrosine kinase 2; FBXW7, F-box and WD repeat
domain containing 7; FOLFOX, fluorouracil, leucovorin, oxaliplatin;
IPAA, ileal pouch-anal anastomosis; PIK3CA,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α;
RNF43, ring finger protein 43; Sig, signature; STK11,
serine/threonine kinase 11; UC, ulcerative colitis; UCAN,
ulcerative colitis-associated neoplasia. |
Discussion
This analysis of genetic alterations in UCAN using
gene panel testing generated two main findings. First, we
demonstrated UCAN has a distinct genomic profile compared with
sporadic CRC. Second, we identified genetic alterations and
mutational signature characteristics that may be associated with
diagnosis and treatment in UCAN. These results indicate gene panel
testing can be useful for differential diagnosis of UCAN and
sporadic CRC, and for tailoring treatment to individual genome
profiles in UCAN.
Genetic alterations in the WNT pathway, including
APC and RNF43, are different in UCAN compared with
sporadic CRC. APC is a negative regulator that controls
beta-catenin concentrations and interacts with E-cadherin, which
are involved in cell adhesion (34). Almost all sporadic CRC have genetic
alterations in APC, where it plays an important role in
tumorigenesis (35). Conversely,
previous reports have shown UCAN has fewer genetic alterations in
APC compared with sporadic CRC (6–20),
which is consistent with our findings (Table II). RNF43 is thought to negatively
regulate the WNT pathway, and loss of RNF43 plays an important role
in sporadic CRC through the enhancement of WNT signaling (36). We previously reported on the
clinical significance of RNF43 mutations in sporadic CRC,
which were associated with aggressive tumor biology along with
BRAF mutations in right-sided CRC (26). Fujita et al (9) reported that somatic mutation of
RNF43 is the driver genetic alteration that links chronic
inflammation and cancer development in about 10% of patients with
UCAN. In this analysis, we found RNF43 frameshift or
nonsense mutations, thought to be associated with functional loss
of RNF43, were significantly more frequent in UCAN compared with
sporadic CRC. These differences in WNT pathway genetic alterations
may be due to differences in developmental mechanisms between
sporadic CRC and UCAN, and are some of the most important genetic
differences for the differential diagnosis of sporadic CRC and
UCAN.
 | Table II.Previously published genomic analyses
of colitis-associated cancer. |
Table II.
Previously published genomic analyses
of colitis-associated cancer.
| First author/s,
year | Number of
cases | Sequencing
method | APC mutant,
% | TP53 mutant,
% | Hyper-mutated,
% | MSI-H, % | Other findings | (Refs.) |
|---|
| Robles et
al, 2016 | 31 (UC, 15; CD, 14;
indeterminate, 2) | Whole-exome | 13 | 65 | 6 | 3 | High frequency of
SOX9, EP300, NRG1 and IL16 mutations in CAC | (6) |
| Yaeger et
al, 2016 | 47 (UC, 29; CD,
18) | Targeted | 21 | 89 | NA | NA | High frequency of
IHD1 R132 mutations in CD, and high frequency of MYC
amplification in CAC | (8) |
| Fujita et
al, 2017 | 90 (UC, 58; CD,
32) | Targeted | 16 | 66 | NA | NA | High frequency of
RNF43 mutations, and RNF43-mutated CACs had elevated
expression of c-MYC | (9) |
| Tanaka et
al, 2017 | 12 (UC, 12) | Targeted | 17 | 58 | NA | NA | CDH1 and
FGFR2 became mutated at an early stage in colitic
carcinogenesis | (10) |
| Din et al,
2018 | 31 (UC, 16; CD,
15) | Whole-exome | 29 | 65 | 29 | 21 | Hypermutated-CAC
had increased numbers of predicted neo-peptides | (11) |
| Yan et al,
2019 | 9 (UC, 9) | Whole-exome | 22 | 33 | NA | NA | High frequency of
KMT2D and NCOA6 mutations | (12) |
| Baker et al,
2019 | 12 (UC, 9; CD,
3) | Exome | 40a | 80a | 17 | 17 | Precancerous clones
bearing SNAs and CNAs at dysplastic and non-dysplastic mucosa | (13) |
| Alpert et
al, 2019 | 35 (UC, 35; CD, 18;
indeterminate colitis, 2) | Targeted | 15 | 69 | NA | NA | Potentially
targetable IDH1 R132 mutation was present in 7% of CAC
cases | (14) |
| Wanders et
al, 2020 | 25 (UC, 15; CD,
10) | Targeted | 16 | 48 | NA | NA | FBXW7
mutation was more frequent in IBD-associated dysplastic lesions
than in sporadic adenomas. | (15) |
| Hirsch et
al, 2021 | 23 (UC, 23) | Targeted | 22 | 87 | NA | 0 | TP53
mutation and chromosomal aneuploidies including gains of chromosome
arm 5p | (16) |
| Matsumoto et
al, 2021 | 36 (UC, 36) | Targeted | 47 | 44 | NA | NA | KRAS and
TP53 mutations were mutually exclusive in CAC | (17) |
| Mäki-Nevala et
al, 2021 | 27 (UC, 27) | Targeted | 11 | 52 | 37 | 4 | Hypermutated was
divided into two distinct subgroups: Hypermutated
microsatellite-stable and hypermutated microsatellite-unstable | (18) |
| Rajamäki et
al, 2021 | 31 (UC, 27; CD, 2;
unclassified IBD, 2) | Whole-genome | 22b | 63b | NA | 10 | AXIN2 and
RNF43 were strongly downregulated in CAC | (19) |
| Present study | 15 (UC, 15) | Targeted | 13 | 87 | 13 | 13 | TP53
mutation alone was a characteristic gene profile in CAC, and
signature 3 was enriched in CAC | - |
Previous reports suggested that genetic alteration
in TP53 is a late event in sporadic CRC, but an early event
in UCAN (37). Most TP53
mutations in UCAN occurred in the DNA-binding domain (8,9,18),
and these mutations were considered to be oncogenic and, for some
of the missense mutations, potentially gain-of-function (8). Our analysis found a characteristic
genome profile for UCAN, where 4/15 patients (27%) had a
TP53 mutation alone, whereas only 1/203 patients (0.5%) with
sporadic CRC had a TP53 mutation alone. This provides both
mechanistic insight into UCAN tumorigenesis, as well as the
diagnostic potential of gene panel testing.
In this analysis, UCAN diagnoses were made by a
pathologist specializing in inflammatory bowel disease, and p53
overexpression was assessed as an aid to UCAN diagnosis. We
consider that p53 overexpression is not essential for the diagnosis
of UCAN; hence, we included four cases which has no p53
overexpression (Table I). We
speculate that the results of p53 immunohistochemical staining are
unlikely to have resulted in selection bias and influenced the
profile of genetic alterations in UCAN.
The clinical utility of ICIs has been observed in a
subset of patients with MSI-H CRC. Clinical studies have
demonstrated MSI status as an accepted response biomarker for ICIs
with progression-free survival rates of up to 78% in MSI-H CRC
compared with 11% in microsatellite stable (MSS) CRC (38). The rate and timing of MSI-H are
similar in UCAN and sporadic CRC, as is the prevalence of
MLH1 hypermethylation and silencing (6). Schulmann et al (39) reported that 18/107 lesions (17%)
showed MSI-H in UCAN, and the profiles of coding microsatellite
mutations differed between MSI-H UCAN and MSI-H sporadic CRC. In
this analysis, MSI-H was identified in 2/15 patients (13%) with
UCAN. Although ICIs were not used in either patient, they might be
potential candidates for ICIs. However, knowledge regarding the
clinical and molecular events underlying UCAN with MSI-H are
limited, and it is unclear whether ICIs have the same effect on
MSI-H sporadic CRC and MSI-H UCAN.
Mutational signature 3, which is associated with
failed DNA double-strand break repair, is one of the genomic
features of HRD, in addition to loss of heterozygosity, telomeric
allelic imbalance, and large-scale state transitions (40). Tumors with HRD show high
sensitivity to platinum compounds and poly(ADP-ribose) polymerase
inhibitors in several malignancies, such as breast (41), ovarian, prostate, and pancreatic
cancers. However, only few data are available regarding the role of
HRD alterations in CRC (42), so
it is unclear whether CRC patients with HRD show high sensitivity
to platinum compounds and poly(ADP-ribose) polymerase inhibitors.
The TRIBE2 study reported that patients with MSS and HRD tumors
showed longer overall survival than patients with MSS and
homologous recombination proficient tumors (40.2 vs. 23.8 months;
P=0.04) (43). The TRIBE2 study
was designed to assign 679 patients with unresectable, previously
untreated metastatic CRC to receive two first-line
oxaliplatin-based regimens: FOLFOX plus bevacizumab or FOLFOXIRI
plus bevacizumab (43). We
consider these results might imply HRD tumors have high sensitivity
to an oxaliplatin-based regimen compared with homologous
recombination proficient tumors in CRC. In this analysis, we
demonstrated that 14/15 patients (93%) with UCAN had signature 3,
and that signature 3 was enriched in UCAN compared with sporadic
CRC. Moreover, we presented a rare case of UCAN arising from the
ileal pouch, which showed a remarkable response to
oxaliplatin-based chemotherapy. Taken together, we think that UCAN
might respond well to an oxaliplatin-based regimen.
This study has several limitations. First, this
study included a small number of patients, with only 15 patients
with UCAN. Second, we did not compare UCAN with sporadic CRC in
patients with UC because its number was limited as well as UCAN.
Third, there was selection bias of sporadic CRC, which included
more patients with distant metastasis compared with UCAN. Forth, we
did not treat any patients based on the results of gene panel
testing. However, to the best of our knowledge, this is the first
report that focused on the clinical utility of gene panel testing
for UCAN. We have shown a potential role for gene panel testing in
the diagnosis and treatment of UCAN, and the clinicians might be
able to develop more effective strategy in UCAN based on message
from gene panel testing.
In conclusion, gene panel testing can detect
important genetic alterations that can be useful for diagnosis and
treatment in UCAN, and may provide clinicians with important
information for tailored treatment strategies for UCAN.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This project was partly supported by JSPS KAKENHI (grant numbers
20K09003, 17K10624, and 21K08750) and by Denka Co., Ltd. (Tokyo,
Japan).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to the informed
consent obtained not including unrestricted disclosure of
sequencing data but are available from the corresponding author on
reasonable request.
Authors' contributions
YS, STe, YA and TW made substantial contributions to
the design and interpretation of data, and drafting of the article.
MaeN, KIM, JY, AM, KT, HO, MasN, YH, HI, JS, HK, YT and MS made
substantial contributions to acquisition of clinical data and
interpretation of data. YL, STa and SO made substantial
contributions to statistical analysis of the data and creation of
the figures. SO and YL confirm the authenticity of all the raw
data. TW critically revised the work and provided final approval of
article. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This retrospective analysis was performed in
accordance with the Helsinki Declaration, and the Ethics Committee
of the School of Medicine, Niigata University (Niigata, Japan)
approved the study protocol (G2015-0816, G2020-0038). Written
informed consent was obtained from the patients.
Patient consent for publication
Not applicable.
Competing interests
SO received research funding from Denka Co., Ltd. TW
received remuneration and research funding from Denka Co., Ltd. All
other authors declare that they have no competing interests.
References
|
1
|
Ng SC, Shi HY, Hamidi N, Underwood FE,
Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et
al: Worldwide incidence and prevalence of inflammatory bowel
disease in the 21st century: A systematic review of
population-based studies. Lancet. 390:2769–2778. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beaugerie L and Itzkowitz SH: Cancers
complicating inflammatory bowel disease. N Engl J Med.
372:1441–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crohn BB and Rosenberg H: The
sigmoidoscopic picture of chronic ulcerative colitis. Am J Med Sci.
170:220–228. 1925. View Article : Google Scholar
|
|
4
|
Rubin DT, Ananthakrishnan AN, Siegel CA,
Sauer BG and Long MD: ACG clinical guideline: Ulcerative colitis in
adults. Am J Gastroenterol. 114:384–413. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
National Comprehensive Cancer Network, .
NCCN clinical practice guidelines in oncology-colon cancer (version
1. 2022). https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf28–July.
2022
|
|
6
|
Robles AI, Traverso G, Zhang M, Roberts
NJ, Khan MA, Joseph C, Lauwers GY, Selaru FM, Popoli M, Pittman ME,
et al: Whole-exome sequencing analyses of inflammatory bowel
disease-associated colorectal cancers. Gastroenterology.
150:931–943. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grivennikov SI and Cominelli F:
Colitis-associated and sporadic colon cancers: Different diseases,
different mutations? Gastroenterology. 150:808–810. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yaeger R, Shah MA, Miller VA, Kelsen JR,
Wang K, Heins ZJ, Ross JS, He Y, Sanford E, Yantiss RK, et al:
Genomic alterations observed in colitis-associated cancers are
distinct from those found in sporadic colorectal cancers and vary
by type of inflammatory bowel disease. Gastroenterology.
151:278–287.e6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fujita M, Matsubara N, Matsuda I, Maejima
K, Oosawa A, Yamano T, Fujimoto A, Furuta M, Nakano K, Oku-Sasaki
A, et al: Genomic landscape of colitis-associated cancer indicates
the impact of chronic inflammation and its stratification by
mutations in the Wnt signaling. Oncotarget. 9:969–981. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tanaka T, Kobunai T, Yamamoto Y, Emoto S,
Murono K, Kaneko M, Sasaki K, Otani K, Nishikawa T, Kawai K, et al:
Colitic cancer develops through mutational alteration distinct from
that in sporadic colorectal cancer: A comparative analysis of
mutational rates at each step. Cancer Genomics Proteomics.
14:341–348. 2017.PubMed/NCBI
|
|
11
|
Din S, Wong K, Mueller MF, Oniscu A,
Hewinson J, Black CJ, Miller ML, Jiménez-Sánchez A, Rabbie R,
Rashid M, et al: Mutational analysis identifies therapeutic
biomarkers in inflammatory bowel disease-associated colorectal
cancers. Clin Cancer Res. 24:5133–5142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan P, Wang Y, Meng X, Yang H, Liu Z, Qian
J, Zhou W and Li J: Whole exome sequencing of ulcerative
colitis-associated colorectal cancer based on novel somatic
mutations identified in Chinese patients. Inflamm Bowel Dis.
25:1293–1301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baker AM, Cross W, Curtius K, Al Bakir I,
Choi CR, Davis HL, Temko D, Biswas S, Martinez P, Williams MJ, et
al: Evolutionary history of human colitis-associated colorectal
cancer. Gut. 68:985–995. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alpert L, Yassan L, Poon R, Kadri S, Niu
N, Patil SA, Mujacic I, Montes D, Galbo F, Wurst MN, et al:
Targeted mutational analysis of inflammatory bowel
disease-associated colorectal cancers. Hum Pathol. 89:44–50. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wanders LK, Cordes M, Voorham Q, Sie D, de
Vries SD, d'Haens GRAM, de Boer NKH, Ylstra B, van Grieken NCT,
Meijer GA, et al: IBD-associated dysplastic lesions show more
chromosomal instability than sporadic adenomas. Inflamm Bowel Dis.
26:167–180. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hirsch D, Hardt J, Sauer C,
Heselmeyer-Hadded K, Witt SH, Kienle P, Ried T and Gaiser T:
Molecular characterization of ulcerative colitis-associated
colorectal carcinomas. Mod Pathol. 34:1153–1166. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Matsumoto K, Urabe Y, Oka S, Inagaki K,
Tanaka H, Yuge R, Hayashi R, Kitadai Y, Arihiro K, Shimamoto F, et
al: Genomic landscape of early-stage colorectal neoplasia
developing from the ulcerative colitis mucosa in the Japanese
population. Inflamm Bowel Dis. 27:686–696. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mäki-Nevala S, Ukwattage S, Olkinuora A,
Almusa H, Ahtiainen M, Ristimäki A, Seppälä T, Lepistö A, Mecklin
JP and Peltomäki P: Somatic mutation profiles as molecular
classifiers of ulcerative colitis-associated colorectal cancer. Int
J Cancer. 148:2997–3007. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rajamäki K, Taira A, Katainen R, Välimäki
N, Kuosmanen A, Plaketti RM, Seppälä TT, Ahtiainen M, Wirta EV,
Vartiainen E, et al: Genetic and epigenetic characteristics of
inflammatory bowel disease-associated colorectal cancer.
Gastroenterology. 161:592–607. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kameyama H, Nagahashi M, Shimada Y, Tajima
Y, Ichikawa H, Nakano M, Sakata J, Kobayashi T, Narayanan S, Takabe
K and Wakai T: Genomic characterization of colitis-associated
colorectal cancer. World J Surg Oncol. 16:1212018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nagahashi M, Shimada Y, Ichikawa H,
Kameyama H, Takabe K, Okuda S and Wakai T: Next generation
sequencing-based gene panel tests for the management of solid
tumors. Cancer Sci. 110:6–15. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nagahashi M, Wakai T, Shimada Y, Ichikawa
H, Kameyama H, Kobayashi T, Sakata J, Yagi R, Sato N, Kitagawa Y,
et al: Genomic landscape of colorectal cancer in Japan: Clinical
implications of comprehensive genomic sequencing for precision
medicine. Genome Med. 8:1362016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shimada Y, Yagi R, Kameyama H, Nagahashi
M, Ichikawa H, Tajima Y, Okamura T, Nakano M, Nakano M, Sato Y, et
al: Utility of comprehensive genomic sequencing for detecting
HER2-positive colorectal cancer. Hum Pathol. 66:1–9. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oyanagi H, Shimada Y, Nagahashi M,
Ichikawa H, Tajima Y, Abe K, Nakano M, Kameyama H, Takii Y,
Kawasaki T, et al: SMAD4 alteration associates with invasive-front
pathological markers and poor prognosis in colorectal cancer.
Histopathology. 74:873–882. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shimada Y, Muneoka Y, Nagahashi M,
Ichikawa H, Tajima Y, Hirose Y, Ando T, Nakano M, Sakata J,
Kameyama H, et al: BRAF V600E and SRC mutations as molecular
markers for predicting prognosis and conversion surgery in Stage IV
colorectal cancer. Sci Rep. 9:24662019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matsumoto A, Shimada Y, Nakano M, Oyanagi
H, Tajima Y, Nakano M, Kameyama H, Hirose Y, Ichikawa H, Nagahashi
M, et al: RNF43 mutation is associated with aggressive tumor
biology along with BRAF V600E mutation in right-sided colorectal
cancer. Oncol Rep. 43:1853–1862. 2020.PubMed/NCBI
|
|
27
|
Shimada Y, Okuda S, Watanabe Y, Tajima Y,
Nagahashi M, Ichikawa H, Nakano M, Sakata J, Takii Y, Kawasaki T,
et al: Histopathological characteristics and artificial
intelligence for predicting tumor mutational burden-high colorectal
cancer. J Gastroenterol. 56:547–559. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. 8th edition.
Springer International; New York, NY: 2017, View Article : Google Scholar
|
|
29
|
Laine L, Kaltenbach T, Barkun A, McQuaid
KR, Subramanian V and Soetikno R; SCENIC guideline development
panel, : SCENIC international consensus statement on surveillance
and management of dysplasia in inflammatory bowel disease.
Gastroenterology. 148:639–651.e28. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Konishi F, Wakasa H, Kino I, Watanabe H,
Nagura H and Muto T: Histological classification of the neoplastic
changes arising in ulcerative colitis: A new proposal in Japan. J
Gastroenterol. 30 (Suppl 8):20–24. 1995.PubMed/NCBI
|
|
31
|
Riddell RH, Goldman H, Ransohoff DF,
Appelman HD, Fenoglio CM, Haggitt RC, Ahren C, Correa P, Hamilton
SR and Morson BC: Dysplasia in inflammatory bowel disease:
Standardized classification with provisional clinical applications.
Hum Pathol. 14:931–968. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Matsuda K, Watanabe H, Ajioka Y, Kobayashi
M, Saito H, Sasaki M, Yasuda K, Kuwabara A, Nishikura K and Muto T:
Ulcerative colitis with overexpression of p53 preceding overt
histological abnormalities of the epithelium. J Gastroenterol.
31:860–867. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gehring JS, Fischer B, Lawrence M and
Huber W: SomaticSignatures: Inferring mutational signatures from
single-nucleotide variants. Bioinformatics. 31:3673–3675.
2015.PubMed/NCBI
|
|
34
|
Markowitz SD and Bertagnolli MM: Molecular
origins of cancer: Molecular basis of colorectal cancer. N Engl J
Med. 361:2449–2460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vogelstein B, Fearon ER, Hamilton SR, Kern
SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM and Bos
JL: Genetic alterations during colorectal-tumor development. N Engl
J Med. 319:525–532. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Serra S and Chetty R: Rnf43. J Clin
Pathol. 71:1–6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brentnall TA, Crispin DA, Rabinovitch PS,
Haggitt RC, Rubin CE, Stevens AC and Burmer GC: Mutations in the
p53 gene: An early marker of neoplastic progression in ulcerative
colitis. Gastroenterology. 107:369–378. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schulmann K, Mori Y, Croog V, Yin J, Olaru
A, Sterian A, Sato F, Wang S, Xu Y, Deacu E, et al: Molecular
phenotype of inflammatory bowel disease-associated neoplasms with
microsatellite instability. Gastroenterology. 129:74–85. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Børresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Robson M, Im SA, Senkus E, Xu B, Domchek
SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, et al:
Olaparib for metastatic breast cancer in patients with a germline
BRCA mutation. N Engl J Med. 377:523–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Arena S, Corti G, Durinikova E, Montone M,
Reilly NM, Russo M, Lorenzato A, Arcella P, Lazzari L, Rospo G, et
al: A subset of colorectal cancers with cross-sensitivity to
olaparib and oxaliplatin. Clin Cancer Res. 26:1372–1384. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Moretto R, Elliott A, Zhang J, Arai H,
Germani MM, Conca V, Xiu J, Stafford P, Oberley M, Abraham J, et
al: Homologous recombination deficiency alterations in colorectal
cancer: Clinical, molecular, and prognostic implications. J Natl
Cancer Inst. 114:271–279. 2022. View Article : Google Scholar : PubMed/NCBI
|