Forkhead box K2 (FOXK2), a central transcriptional
regulator of embryonic development and cell homeostasis, was
initially recognized as an essential member of the FOX family.
Since its identification, evidence available has shown that FOXK2
mediates a diverse range of biological processes, such as cancer
genetics and biology (1–4). However, the biological functions of
FOXK2, especially functional redundancy and non-functional
redundancy, remain largely unexplored. Functional redundancy is a
property of transcription factors (TFs) that allows one TF to
compensate for another due to their protein sequence homology, or
the shared molecular chaperone (5–7).
Non-functional redundancy of TFs serves a more important role in
the cell fate conversions. Thus, loss of FOXK2 function or the
absence of gain-of-function, directly and indirectly, affect
tumorigenesis. Over the past 30 years, the hallmarks of cancer are
defined as the collection of acquired biological capabilities
during the multistep development of human tumors (8–10).
It is well known that TFs are actively involved in the acquisition
of biological capabilities in human tumors. As, to date, there is
neither commercially available FOXK2 inhibitors/drugs nor
convincing clinical trials of its use as a therapeutic target, the
present review outlined the broad roles and possible mediating
mechanisms of FOXK2 in carcinogenesis. Finally, it highlighted that
the functional redundancy and non-functional redundancy of FOXK2
maps to tumor pathogenesis. This relationship may influence the
direction of future FOXK2 research in tumorigenesis.
FOXKs are members of an evolutionarily conserved TF
family that share a forkhead DNA-binding domain with their binding
partners. The binding occurs at a conserved core sequence
(TTGTTTAC) and mediates various chromatin events (11–14).
For example, FOXK2 recognizes and binds to a purine-rich motif in
the long terminal repeats of the human immunodeficiency virus and
is identified as an interleukin-enhancing factor-binding factor
(ILF). This behavior was demonstrated in a study of genes encoding
cytokines (15).
The role of alternative splicing in cancer is
multifaceted and the activity of tumor suppressors and oncogenes is
altered by alternative splicing (20,21).
These changes are preferentially found in cancer cells and often
manifest at the protein level as structural changes (22), removal of phosphorylation sites
(23), or changes in subcellular
localization (24). As with most
human genes, FOXK2 mRNA undergoes some degree of alternative
splicing (25) and three isomers
have been identified. The three isomers, termed ILF-1, ILF-2 and
ILF-3, encode proteins with lengths of 655, 609 and 323 amino
acids, respectively. The GenBank database (https://www.ncbi.nlm.nih.gov/genbank) labels them as
FOX protein K2 isoforms X1, X2 and X3.
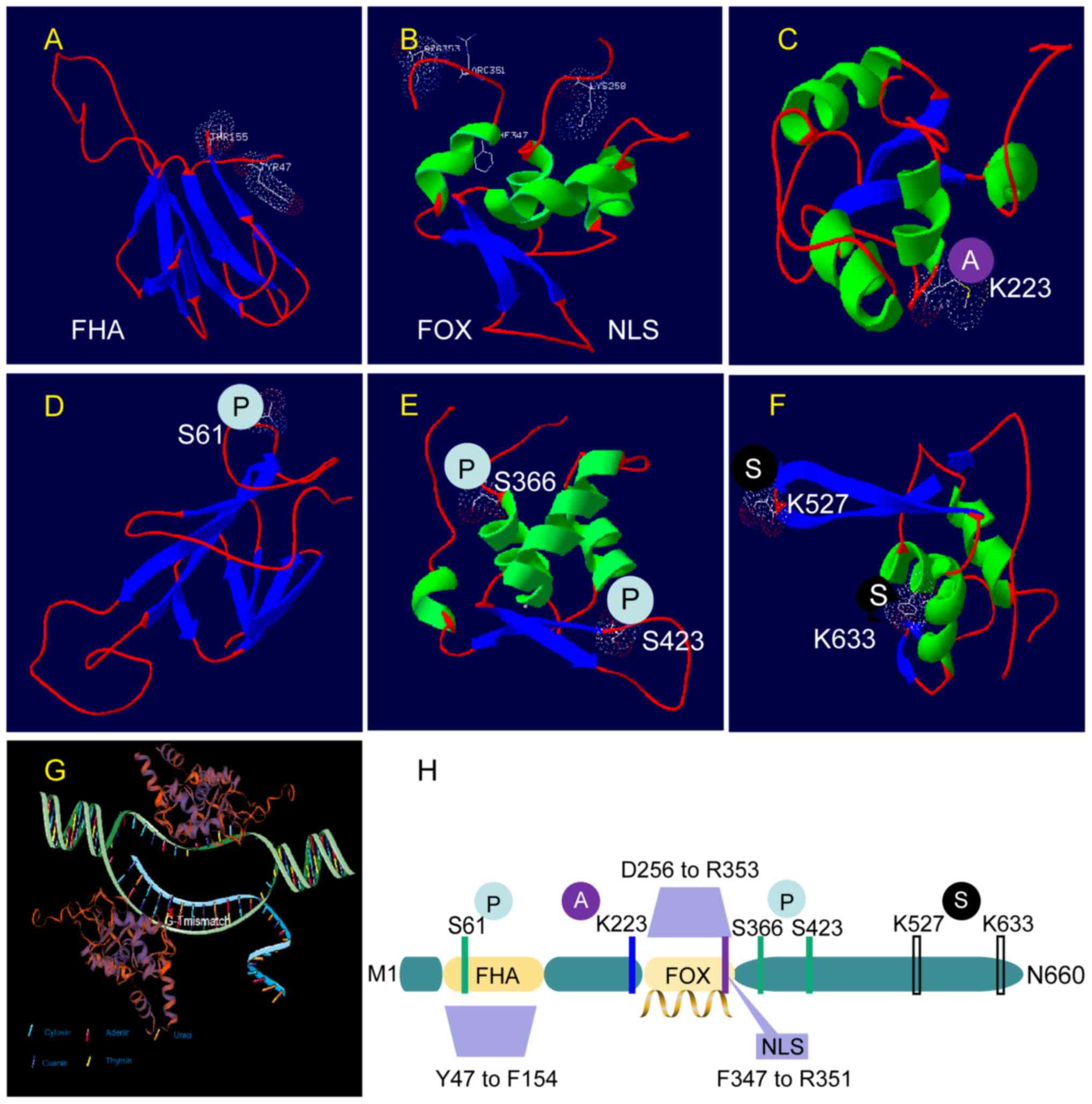
Structurally, all three proteins contain a signature
proline-rich FHA domain. However, in contrast to ILF-1 and ILF-2,
which contain a complete forkhead domain (FKH), ILF-3 contains a
partially missing FKH (NCBI; http://www.ncbi.nlm.nih.gov). Although the
significance of the complete FKH domain existence or absence is
unclear, ILF-3 does lose the majority of potential phosphorylation
sites in the COG5025 (GenBank, Ser180 to Gln577) region (26). There is evidence that alternative
splicing alters protein phosphorylation, thereby limiting the
effect of the kinase cascade signal (27–30).
However, there is still a lack of research data on FOXK2
alternative splicing. Chromatin immunoprecipitation followed by
sequencing (ChIP-seq) allows analysis of chromatin binding to TF
and this particular technique may help answer a number of open
questions about FOXK2 functions. The function of FOXK2 proteins is
also closely related to their dynamic allocation in different
subcellular structures. Therefore, understanding this new aspect
and studying the regulatory mechanism help to elucidate its dynamic
transcriptional role in mRNA expression of target genes. This
understanding is critical to evaluating how it promotes health and
disease (28,31).
The NLS is a motif that allows for active nuclear
import of large proteins. However, the nuclear translocation of
certain proteins does not appear to be dependent on the NLS of
FOXK2. For example, FOXK2 mediates Disheveled (DVL) nuclear
translocation according to its FHA and adjacent region (residue
Arg129-Pro171) (32). Thus, there
is no credible evidence to support the effect of NLS on FOXK2
functionality.
Over the past decade, the unexpected functional
redundancies and non-functional redundancies of FOXK2 have become
increasingly attractive prospects for researchers to explore. There
is growing evidence that FOXK2 serves a vital role in various
biological processes, especially in cancer cells, including in
proliferation, differentiation, cell cycle progression, apoptosis
and metabolic reprogramming.
The regulation of FOXK2 activity has been
extensively studied. In addition to regulation of mRNA expression,
post-translational modifications (PTMs; Fig. 1), non-coding RNA (ncRNAs) and
protein interactions also serve important roles in the loss or gain
of FOXKs functions (4,33) (Fig.
2, Fig. 3, Fig. 4). PTMs affect the stability of
transcriptionally active proteins. PTMs also control how these
proteins interact with other molecules and serve different roles in
various developmental processes in both internal and external
settings, whether favorable or unfavorable (34–39).
The most common PTMs are glycosylation modification,
phosphorylation, methylation, acetylation, ubiquitination,
sulphuration and reduction/oxidation (redox) modifications
(40). Notably, epigenetic
mechanisms including DNA cytosine modifications, histone
modifications and regulation by ncRNAs are prominent epigenetic
regulatory elements (41,42).
DNA methylation is an evolutionarily ancient
epigenetic modification that regulates FOXKs at the transcriptional
level (43,44). These epigenetic modifications are
closely associated with the aging process and regulate the
transcriptional profile of DNA fragments by packaging them
(43,44).
A considerable body of evidence suggests that ~1% of
the human genome is methylated and methylated markers of gene
promoter regions control gene expression (45–47).
In addition, DNA methylation has been implicated in mediating
transcriptional silencing, although the particular molecular
pathways are not fully understood (48). Transcriptional silencing serves a
vital role in critical biological processes such as replication,
division, development survival, aging, genomic imprinting and
embryonic development as facultative chromatin, especially in
cancer development (49–54).
Several studies have demonstrated a preference for
methylated markers for genomic site selection (55–57).
Methyl groups are attached to 5-methylcytosine (5mC) throughout the
genome, typically between cytosine and guanine (CpG) or within CpG
islands polymerized by CpGs (55).
This finding was further exemplified in a global causal analysis
involving firefighters exposed to various environmental hazards. As
expected, this controlled analysis revealed differential
methylation loci in FOXK2. Three CpG loci in FOXK2
were shown to be located in CpG islands and they exhibited reduced
methylation (56).
FOXK2 is an effector of DNA methylation. FOXK2
methylation is a meaningful indicator of fertility. High levels of
FOXK2 methylation are closely associated with male infertility
(58). Furthermore, FOXK2
hypomethylation induced by dioxin exposure can also negatively
affect male reproductive health (59).
The effect of FOXK2 methylation can also be observed
in the following examples of interaction with a range of
environmental factors. A recent study analyzed genome-wide DNA
methylation profiles of white blood cells. It found that FOXK2
hypermethylation levels were strongly associated with smoking
levels and also varied across racial/ethnic groups (60). Notably, hypermethylation can be
observed in patients with severe psychophysiological trauma
(61) and arsenic toxicity in
vivo (62). The potential
implication of this meaningful evidence is that FOXK2 methylation
levels are associated with physiological stresses caused by
environmental exposure. However, there is a lack of research on the
relationship between changes in FOXK2 methylation levels and
psychological stress and toxic transformation.
There is also considerable interest towards
understanding the effects of certain lifestyle factors on FOXK2
methylation modification. In CpG islands of adipose tissue,
methyltransferase nicotinamide n-methyltransferase (NNMT) levels
are influenced by diet and exercise. FOXK2 methylation levels are
inversely correlated with NNMT (57), further supporting the link between
environment and methylation levels.
Abnormal increases in methylation are associated
with the inactivation of tumor related genes (63,64).
A study examining genome-wide DNA methylation profiles of
fibromatoid-like fibroma tumors involving FOXK2 showed that
hypermethylation reduced FOXK2 mRNA expression (65).
Additionally, FOXK2 has also been identified as a
dynamic reader of DNA methylation, mediating the interaction of
methylated binding domain (MBD) deficient transcription factors
with methylated DNA (66,67). Several specific homologous
framework proteins and proteins with wing-like helix domains,
including FOXK2, can recognize methylated CpG (mCpG) (66,68,69).
FOXK2 has been shown to bind methylated DNA 5mC and the oxidative
derivative 5-formylcytosine to recruit relevant functional proteins
in mouse embryonic stem cells (66,67).
Although FOXK2 can be used for MBD screening and serves an
important role in coordinating transcriptional replication and DNA
repair, it is not clear how a number of MBD-deficient TFs might be
recruited by FOXK2 (70).
The dynamic regulation of phosphorylation is
undoubtedly the most common and well-studied PTM (71,72).
Phosphorylation with rapid and reversible properties profoundly
modulates a wide range of proteins at a relatively small dynamic
cost, controlling the balance between phosphorylation and
dephosphorylation events (73,74).
The unique FHA domain of FOXK2 makes it an
‘intelligent’ sensor in complex networks related to cell signal
cascades. It contributes to genomic stability, cell growth
maintenance, cell cycle regulation and signal transduction
(75). These functions have been
partially validated in yeast. The yeast FOX proteins Fkh1 and Fkh2
(homologs of human FOXK1 and FOXK2 proteins) are phosphorylated by
Cdc28p, the primary cycle-dependent kinase in yeast, in a cell
cycle-dependent manner (76,77).
Changes in Fkh2 activity can affect pseudomycelia growth through
transcriptional regulation of genes involved in M-phase transition
(76). The cyclin-dependent kinase
1 (CDK1) phosphorylates FOXK2 at Ser368 and Ser423 in the COG5025
domain. The phosphorylation fluctuates periodically and reaches its
peak in the M phase (26). In
human osteosarcoma cells, CDK1-mediated phosphorylation of FOXK2
(S368/S423) negatively regulates its stability and inhibitory
activity and thus inhibits tumor cell apoptosis (26).
Additionally, phosphorylation exerts a regulatory
effect by modulating the subcellular localization and translocation
of FOX family members and also regulates their interaction with
chaperone proteins (78–80), such as 14-3-3, a hub-protein of
complex network of protein-protein interaction that has several
hundred identified protein interaction partners and is therefore
involved in cellular processes and diseases (81). A recent study on autophagy showed
that ataxia-telangiectasia mutation (ATM) mediates phosphorylation
of checkpoint kinase 2 protein (CHK2) at Thr68 after DNA damage,
which is critical for binding to the FHA domain of FOXK. This
binding enables FOXK1 and FOXK2 to be phosphorylated by CHK2 at
Ser130 and Ser61, respectively. Then, the phosphorylated FOXK
protein is captured in the cytoplasm by 14-3-3γ and this affects
the transcription of autophagy-related (ATG) genes (1). Phosphorylation-induced subcellular
localization affecting cell metabolic reprogramming has also been
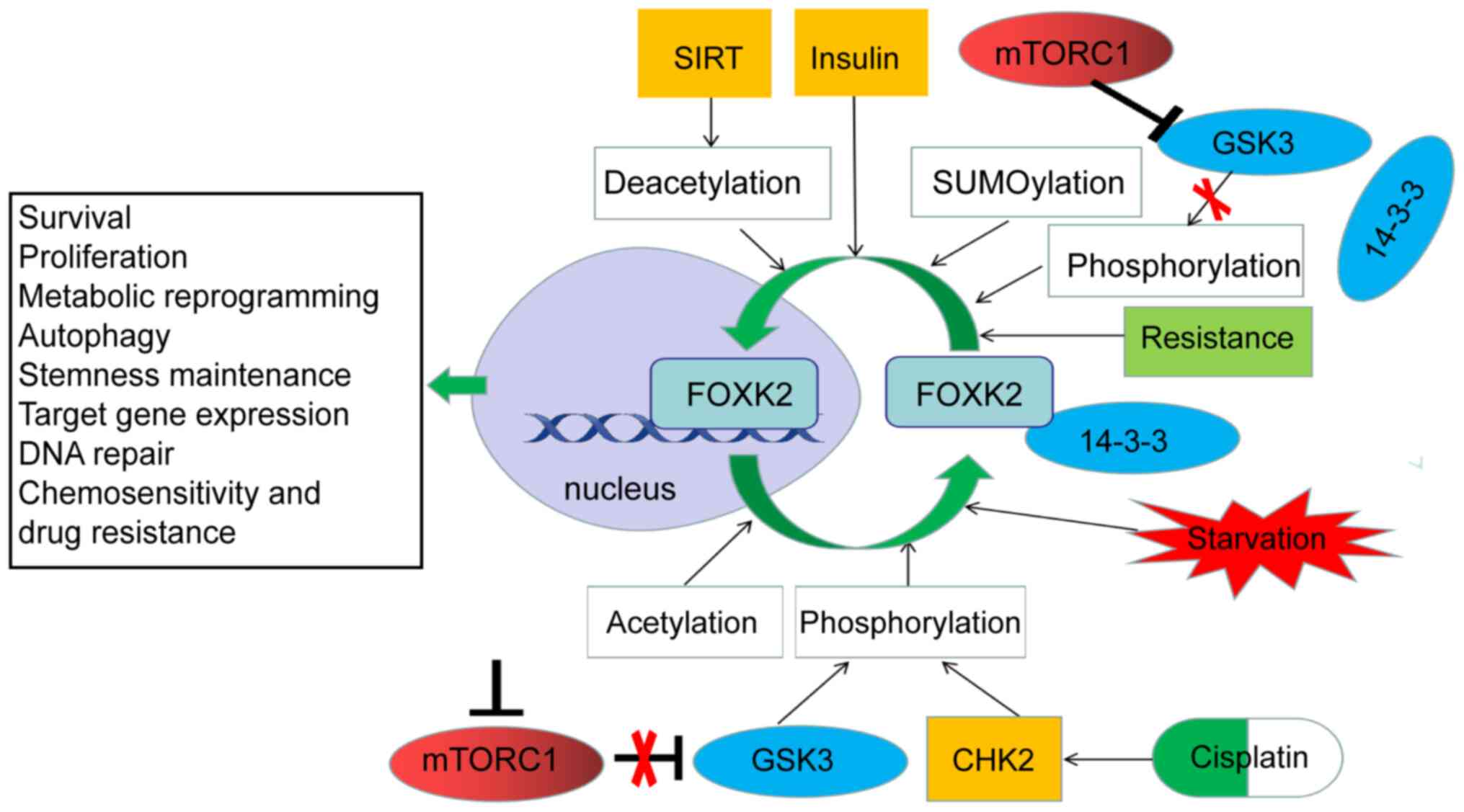
demonstrated. By blocking mTOR complex 1 (mTORC1) and then
eliminating its inhibitory effect on glycogen synthase kinase 3,
both FOXK1 phosphorylation and its interactions with 14-3-3 ε are
increased, resulting in reduced expression of multiple genes
involved in glycolysis related pathways (2). However, whether all 14-3-3 subtypes
or some of them indiscriminately trap FOXK2 remains unclear.
Nutrition-related signals such as insulin and amino acids activate
mTORC1 to induce protein phosphatase 2A (PP2A)-mediated
dephosphorylation of the FOXK1 (82). This effect reduces the production
of insulin-induced C-C chemokine ligand 2 (CCL2) (83). FOXK1 and FOXK2, like their FOXO
subfamily partners, are downstream targets of insulin action
(82). However, unlike FOXO1,
insulin stimulation directs the translocation of FOXK proteins from
the cytoplasm to the nucleus in an AKT-mTOR pathway-dependent
manner (84). This translocation
controls the expression of genes involved in regulating
mitochondrial β-oxidation and biogenesis in the nucleus (85). Descriptions of FOXK2 translocation
between nucleus and cytoplasm are helpful in understanding the
functions of FOXK2 in signal transduction and gene expression
regulation, but studies on this shuttle mechanism are scarce.
It is important to note that although some studies
only investigated FOXK1, the results have practical reference
significance for FOXK2 due to their high degree of similarities in
the domain and protein sequence, with amino acid homology
approaching 50% (86,87). Furthermore, studies on the
clustering of FOXK1 and FOXK2 samples support the hypothesis of
functional overlap of FOXKs. For example, one study found that
single and double knockdowns of FOXK1/FOXK2 upregulated the
expression of apoptosis-related genes and downregulated the
expression of genes related to cell cycle and lipid metabolism
(84). Additionally, FOXK1 and
FOXK2 have a significant positive effect on the regulation of
glycolysis, as they share a common regulatory substrate preference
(glucose and fatty acid) and can upregulate the expression of
enzymes required in glycolysis, which in turn regulate lactic acid
production (88).
In addition to the examples mentioned above, in
FOXK1 knock-out (KO) cells, several genes that participate in
hormone biosynthesis, monoamine transport regulation, hematopoietic
stem cell differentiation and integrin activation are
downregulated. This gene regulatory profile is similar to that of
FOXK2 KO cells, suggesting functional similarities between FOXK1
and FOXK2 in regulatory targets (89). However, the extent to which they
cause physiological or pathological overlap, as well as
non-redundant functions, remain to be elucidated.
Several studies have shown that FOX-protein
stability, DNA binding activity and interactions with chaperones
are also regulated by SUMOylation (90–94).
These regulatory activities are well represented in breast cancer
cells, where FOXK2 SUMOylation serves a key role in mediating
chemical sensitivity and resistance to paclitaxel. Paclitaxel
treatment of breast cancer cells requires the SUMOylation of FOXK2
at the K527 and K633 sites for their cytotoxic function. The
SUMOylation of FOXK2 significantly increased its binding to the
FOXO3 promoter, leading to upregulation of FOXO3 mRNA and
protein levels. Conversely, FOXK2 accumulates in the nucleus of
paclitaxel-resistant breast cancer cells, but recruitment to
endogenous FOXO3 promotors is impaired (95). FOXK2 does this by dynamically
regulating subcellular localization and binding to target genes
such as tumor suppressor FOXO3. However, the more detailed
regulatory mechanisms remain to be elucidated.
The mechanisms of ubiquitination have been
extensively studied. Ubiquitination primarily consists of
mono-ubiquitination and polyubiquitination and it regulates a
diverse range of physiological and pathological activities
(96,97). Ubiquitination and deubiquitination
events of substrate proteins have significant effects on several
aspects of cell life, such as cell cycle regulation, apoptosis,
receptor downregulation and gene transcription (98–100). The ubiquitin-proteasome system
include ubiquitin ligases (E1, E2 and E3), proteasomes and
deubiquitination enzymes (DUBs) (96,97).
The unique roles that these proteasomes and enzymes serve in tumor
inhibition and tumor inhibition pathways are well documented, such
as in tumor metabolism regulation, immune tumor microenvironment
(TME) regulation and cancer stem cell maintenance (101).
The PcG-repressive (PR)-DUB complex catalyzes the
deubiquitination of H2A at lysine 119 (102). Although several models for the
PR-DUB complex's inhibitory function have been reported, how it
mediates gene inhibition is still not fully understood (103–106). FOXK1 and FOXK2 are considered to
be indispensable components of three mammalian PR-DUB complexes,
including breast cancer type 1 susceptibility protein (BRCA1)
associated protein-1 (BAP1, homolog of human Calypso), host cell
factor C1 (HCFC1) and additional sex combs-like proteins (103). Notably, BAP1 DUB has been
reported to function as a FOXK2 chaperone in human cells in a FOXK2
FHA-dependent manner (107).
Furthermore, BAP1 functions as an important tumor suppressor
protein in several different tumor types and can deubiquitinate
histone H2A to regulate transcription (108–110). In the absence of BAP1, FOXK2
fails to recruit BAP1, losing the ability to inhibit oncogenesis by
directing BAP1 to its target gene (111). The relationships between the
various components of the PR-DUB complex have been extensively
studied. However, the link between FOXK2 and enzymes responsible
for regulating protein O-GlcNAc modification, including OGT and
glycoside enzyme (O-Glcnase, OGA), has not been adequately
studied.
Acetylation is involved in almost all cellular
biological processes, including cancer. FOXK2 can affect the
acetylation of proteins of interest and the transcription of target
genes. A study of the FOXK proteins in hunger-induced atrophy and
initiation of autophagy found that FOXK1 and FOXK2 restrict the
acetylation of the target genomic protein H4 and the expression of
essential autophagy genes. FOXK1 and FOXK2 achieve this restriction
by recruiting the suppressor-interacting 3A (Sin3a) histone
deacetylation enzyme (HDAC) complex (85). There is also evidence that FOXK2,
as a transcription inhibitor, can interact with proteins in the
Sin3a HDAC co-inhibitory complex in human cells (105). Despite growing evidence of
non-histone acetylation affecting a range of cellular processes
(112,113), the regulatory role of lysine
acetylation in cancer cells remains to be elucidated. Lysine
residue in FOXK2 can also be modified by acetylation. The
acetylation levels at the K223 site in FOXK2 are directly related
to the sensitivity of tumor cells to cisplatin. In cancer cells,
cisplatin can enhance the acetylation of FOXK2 K223, reduce the
nuclear distribution of FOXK2, significantly downregulate the
expression of cell-cycle-related genes and significantly upregulate
the expression of apoptosis-related genes. FOXK2 K223
hyperacetylation can even promote mitotic catastrophe (114). However, in cisplatin-treated
cancer cells, the silencing of information regulator 2-related
enzyme 1 reduces the effect of deacetylation of FOXK2 K233 on cell
apoptosis (114). This finding
has far-reaching implications for understanding chemical
sensitivity and drug resistance in cancer and warrants further
in-depth studies in the future.
Thanks to rapid advances in sequencing technologies,
several unique ncRNA sequences have been identified. MicroRNAs
(miRNAs/miRs), circular RNAs (circRNAs) and long ncRNAs (lncRNAs)
control numerous molecular targets, mediate cellular processes and
determine cell fate (115,116).
ncRNAs are RNA molecules lacking protein-coding
regions responsible for regulating gene expression at the
transcriptional or post-transcriptional level (117–120). These functional regulators link
relevant genes into regulatory networks, with some ncRNAs, such as
miRNAs and lncRNAs, possibly regulating the mRNAs of several target
genes. Moreover, the mRNA of a specific gene can be regulated by
multiple miRNAs (121,122). Notably, some ncRNA crosstalk
imparts robustness to biological processes, supporting their role
as crucial regulators. The noise of these complex interactions can
profoundly impact cell fate, especially in cancer (121,123).
miRNAs are endogenous and abundant. They are RNA
sequences that are ~22 nucleotides long and can be associated with
the corresponding miRNA response elements (124,125). These miRNAs, composed of 18–25
nucleotides, bind with other proteins to form RNA-induced silencing
complexes that target the 3′-untranslated region (3′-UTR) of mRNA.
This function regulates the translation of mRNAs involved in
biological processes such as cell proliferation, apoptosis,
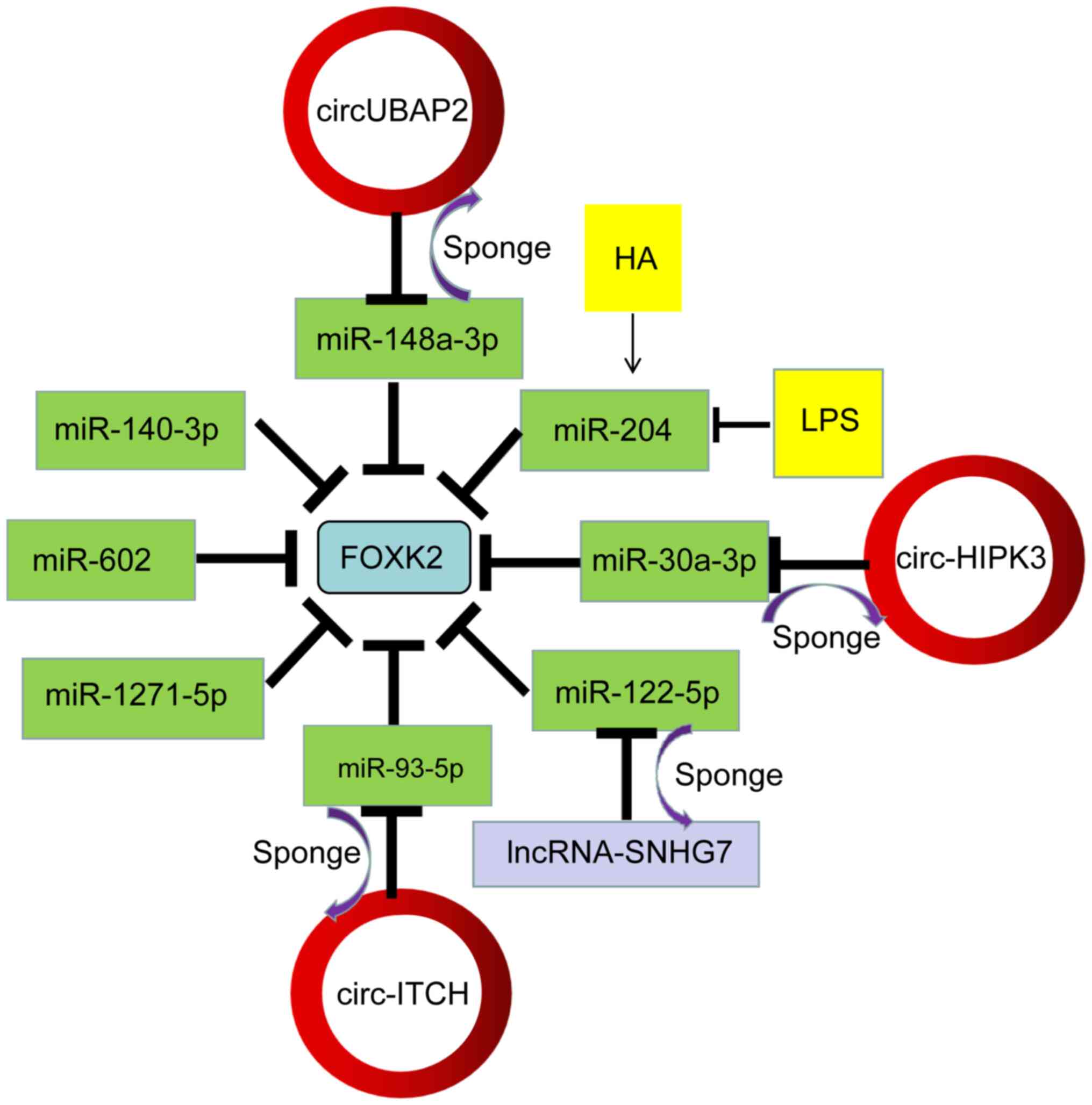
differentiation and transformation (126–129). In a study involving granulosa
cells (GC), miR-204, a downstream regulator of the phospho-inositol
3-kinase (PI3K)/AKT/mTOR signaling pathway, directly targets FOXK2
and results in promoting GC proliferation and inhibiting apoptosis
(130). In hepatocellular
carcinoma (HCC), FOXK2 is a direct target of miR-1271, which
negatively regulates FOXK2 at the mRNA and protein levels (131). A study assessing
epithelial-mesenchymal transition and proliferation in non-small
cell lung cancer (NSCLC) confirmed that the FOXK2 3′-UTR site
(position 40–47) (GUGCCAA) is directly targeted and negatively
regulated by miR-1271 (132).
Notably, miRNAs that regulate FOXK2 are affected by
epigenetic and environmental changes. In various esophageal
squamous cell carcinoma (ESCC) studies (133,134), hypomethylation of the miR-602
promoter induced expression and negatively regulated the target
gene FOXK2. It regulated the cell cycle by promoting the
proliferation and metastasis of ESCC in vitro and in
vivo. Notably, reduced FOXK2 expression significantly
accelerated the biological pathway mediated by miR-602
overexpression (134).
Some metabolic substrates and drugs also induce
miRNA expression. Under high glucose conditions, the expression of
miR-140-3p in endothelial cells (ECs) was significantly decreased.
The low level of miR-140-3p lost the inhibitory effect on the
expression of FOXK2, thereby enhancing the angiogenic function of
ECs (135). In hirsutanol A
(HA)-pretreated A549 cells, upregulation of miR-204 directly
targets FOXK2, promoting cell viability by reducing apoptosis and
inhibiting the release of inflammatory factors by attenuating NF-κB
activation (136).
circRNAs possess a continuous loop of at least a few
hundred nucleotides and a covalently closed loop, resulting in a
higher degree of stability compared with most linear RNAs (137,138). Ashwal-Fluss et al showed
that circRNAs are produced through co-transcription and competition
with conventional splicing (139). Several circRNAs are closely
associated with tumor development and progression; however, the
details of their regulatory mechanisms remain inconclusive
(140–142). Intriguingly, two studies
conducted in 2013 showed that two circRNAs, CDR1-as (also known as
CIRS-7) and sex-determining region Y (Sry), act as sponges for
miRNAs that regulate transcription of specific miRNAs (143,144). That circRNAs act as sponges for
miRNAs to influence the transcription of target genes is now widely
accepted (145,146).
Circ-ITCH has been reported to significantly affect
several biological characteristics of tumors by acting as a tumor
suppressor (147,148). Knockdown of circ-ITCH expression
in human cervical cancer (CC) tissues and cell lines attenuated the
inhibitory effects of circ-ITCH on the malignancy of CC cells
(149). The presence of a
circ-ITCH/miR-93-5p/FOXK2 axis was further explored in that study;
miR-93-5p has been shown to function as a tumor promoter in several
types of cancer (150–152) and it is significantly upregulated
in CC tissues and cell lines (153). The researchers confirmed that
circ-ITCH could directly bind to miR-93-5p using bioinformatics
tools and this was confirmed using a luciferase reporter assay.
FOXK2 expression was significantly downregulated in CC tissues. The
study also confirmed that FOXK2 was a target of miR-93-5p using
TargetScan and this was verified using a luciferase reporter assay.
miR-93-5p mimics significantly inhibit FOXK2 expression in HeLa
cells and FOXK2 knockdown significantly reduced FOXK2 expression in
HeLa cells transfected with a miR-93-5p inhibitor (153). In summary, circ-ITCH achieves its
tumor-suppressive activity by sponging miR-93-5p to regulate FOXK2
expression and its role as a tumor suppressor in several types of
cancer is well established and reviewed elsewhere (154).
Another example of a circRNA acting as a sponge can
be found in clear cell kidney cells (ccRCC). The novel circRNA
UBAP2 acts as a miRNA sponge to regulate miR-148a-3p, which itself
affects FOXK2 mRNA and protein levels and influences ccRCC cell
proliferation, migration and invasion (155). In addition, a study on pulmonary
fibrosis showed that circHIPK3 enhances FOXK2 expression by
sponging miR-30A-3p, thereby promoting fibroblast activation
proliferation and glycolysis (156).
lncRNAs are transcripts that do not encode proteins
and are often >200 nucleotides (157,158). lncRNAs are presently hypothesized
to serve vital roles in several cellular processes, including cell
cycle regulation (159),
differentiation (160–162), metabolism (163) and various diseases (164,165). One study has shown that certain
lncRNAs are involved in cancer progression through the adsorption
of miRNAs via sponging (166).
lncRNAs can also regulate FOXK2 expression. Emerging evidence
suggests that lncRNA tumor protein 53 target gene 1
(TP53TG1), enriched in CC, sponges the FOXK2-targeting
miR-33a-5p and thus increases FOXK2 expression. This increase in
FOXK2 expression promotes related protein activity, activates the
PI3K/AKT/mTOR signaling pathway and increases tumor biological
activity (167). It has been
reported that TP53TG1 functions as an oncogene in several
types of cancer (168,169) and miR-33A-5P can function as a
tumor suppressor gene in several other types of cancer (170–172). Similarly, lncRNA small nucleolar
RNA host gene 7 (SNHG7) also functions as an oncogene to
promote HCC tumor growth in vivo via a miR-122-5p/FOXK2
axis. In addition, lncRNA SNHG7 abrogated the negative regulation
on FOXK2 through the sponging of miR-122-5p and promoted the
occurrence and development of liver cancer (173). The mechanism of FOXK2 as a
repressor and activator of gene transcription remains to be further
studied.
A number of molecules have been shown to interact
with FOXK2, which interacts with other transcription factors and
active proteins as a key to carrying out its regulatory functions.
Protein kinases are one of the most common partners that interact
with FOXK2. Their interactions are involved in a variety of
cellular functions, including metabolism (84,88),
autophagy (1), cell cycle
regulation (26), cell
proliferation and survival (174)
and changes in subcellular localization (2). FOXK2 binds to oncoproteins; Qian
et al (175) reported that
the sex-determining region Y box 9 (SOX9) oncoprotein directly
binds to the FOXK2 promoter, significantly upregulating its mRNA
expression levels. FOXK2 also interacts with activating and
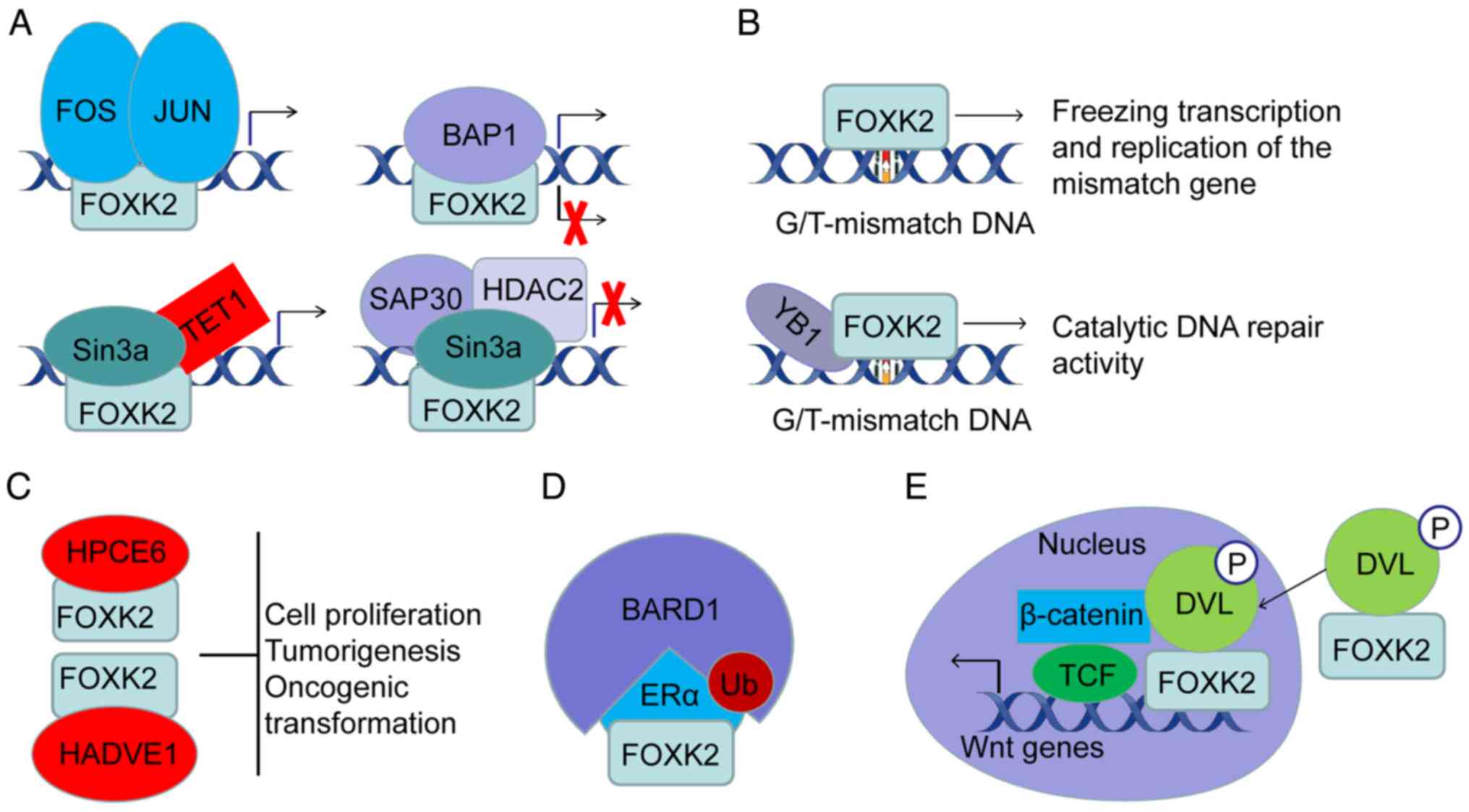
inhibiting proteins. For example, FOXK2 efficiently recruits
activating protein-1 (AP-1) transcription factors to chromatin and
binds to them, contributing to AP-1-dependent gene expression
changes (176). In addition,
FOXK1 and FOXK2 can recruit the Sin3a HDAC complex to inhibit the
expression of essential autophagy genes (87). Notably, FOXK2 appears to recruit
Sin3a HDAC complex and BAP1 impartially. The mechanism of this
differential recruitment is unclear and further studies are
necessary to elucidate the molecular mechanisms underlying the
differing epigenetic modification preferences of local chromatin.
FOXK2 functions well with proteins exhibiting similar functions.
For example, methyl-CpG binding domain proteins (MBD6) and FOXK2
are prime candidates for MBD proteins and DNA methyl-dissociation
reading. However, MBD6 is recruited to laser-induced DNA damage
sites in a manner independent of its MBD domain and interacts with
PR-DUB (69,70,177,178).
An excellent example of FOX interfamily interactions
is the dynamic occupancy model of FOXK2 and FOXO3a for shared
binding modes. The two genes dynamically correlate and isolate
rather than directly competing to control their FOXO-dependent gene
expression functions (179).
One study demonstrated that FOXK1 and FOXK2 interact
with the c-terminal region of the adenovirus (HAdV) protein E1A,
inhibiting HAdV E1A-induced proliferation and transformation in
cells (180). DVL2, an adaptor
protein of Wnt/β-catenin signaling, serves an important role in the
development of colitis-associated colorectal cancer (CRC) by
linking the inflammatory NF-κB signaling pathway to the
Wnt/β-catenin signaling pathway (181). FOXK2 associates with DVL2 and
migrates to the nucleus to positively regulate the Wnt/β-catenin
signaling pathway (181). The PDZ
domain of DVL2 and a four-amino acid motif named IVLT are necessary
when binding to the FHA domain on FOXK2 and its adjacent region
(residue ~129-171) (32).
Scaffold proteins are high-order complexes that bind
at least two protein partners together, specifically recruiting
signaling proteins, within the delicate tissue framework to achieve
temporal and spatial control of specific pathways (182–185). For example, a breast cancer study
showed that FOXK2 interacts with BRCA1 as a scaffold protein for
BReast-CAncer susceptibility gene 1 (BRCA1)/BRCA1-associated RING
domain protein 1 (BARD1) and estrogen receptor α (ERα), resulting
in enhanced degradation of ERα and ultimately reduced
transcriptional activity (186).
Proteins typically do not function as single modules
in the biological processes of living cells. Instead, they function
with other proteins in dynamic networks, interacting with numerous
biologically active substances. For example, in a recent study of
tumor-derived morphological mutations, BAP1 was isolated from
wild-type ASXL1 mutants whose C-terminus was truncated and whose
regulation of target genes was lost through the ASXL1-BAP1-FOXK1/K2
axis (89). This example
demonstrates that numerous proteins can interact amongst themselves
in tandem within intricate complexes. Furthermore, their
interactions occasionally span multiple complexes, giving
fascinating and elaborate protein-protein relationships.
In conclusion, FOXK2 regulates target genes through
a combination of multiple transcription factors. FOXK2 and
chaperone proteins form various complexes and the specific
interactions between FOXK2 and each component of the complex lead
to the diversity of its regulatory functions. However, the
mechanisms by which FOXK2 interacts with other transcription
factors and active factors are not well understood. The current
studies neither reveal how FOXK2 selects for preferred interacting
partner nor address the biological significance or evolutionary
advantages of this selection.
In cancer, FOXK2 functions as a gatekeeper of DNA
repair and mutation prevention. Studies have shown that genes
mediating the DNA repair process are inextricably linked to
potential mutations in cancer (187–189). Furthermore, FOX proteins regulate
several aspects of cell biology by inducing the transcription of
target genes (190,191). The ability of FOXK2 to regulate
fundamental biological processes is evident in cell proliferation
(132,174), DNA repair (192), apoptosis (193) and regulation of cell metabolism
(84,88) (Fig.
5). Indeed, there is growing evidence that FOXK2 is closely
related to the development of tumors, but it may also serve
opposing roles based on the specific type of tumor. Several studies
have reported that FOXK2 expression is low in breast cancer
(186,194), NSCLC (132), glioma (195) and ccRCC (193). Its role as a tumor suppressor
gene is not evident. Conversely, the increase in FOXK2 expression
is closely related to the occurrence and development of tumors.
Other studies have found that FOXK2 expression is upregulated in
papillary thyroid carcinoma (PTC) (196), anaplastic thyroid cancer (ATC)
(197), CRC (198) and HCC tissues (131). These conflicting findings suggest
that tumor-specificity may determine the role of FOXK2 and dictate
its function as an oncogene or tumor suppressor gene in
tumorigenesis and progression. However, general rules cannot be
extrapolated from current data and these results are far from
helping the understanding of the general regulatory pattern of
FOXK2.
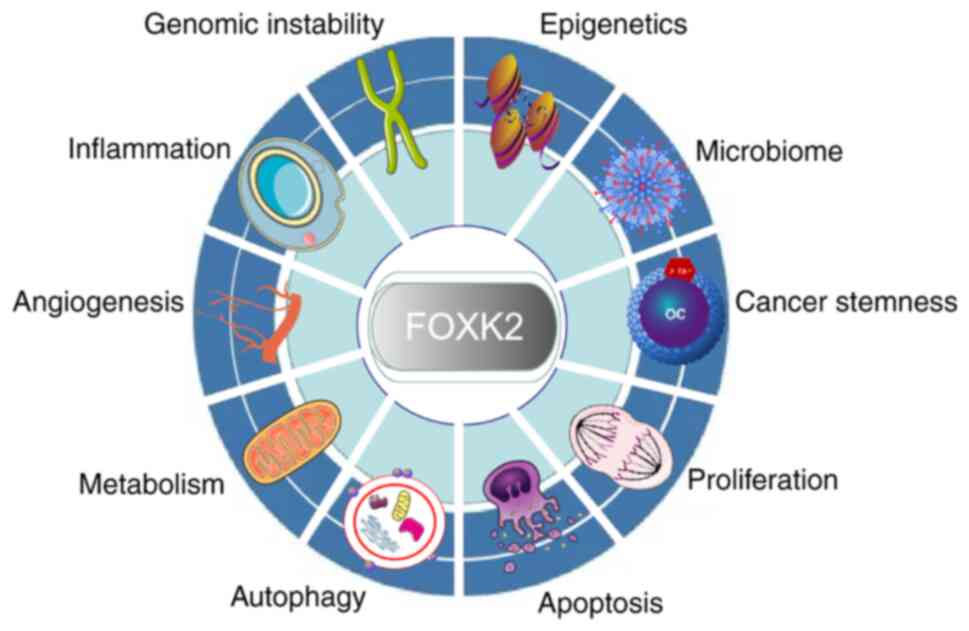
Enabling hallmarks are the precise molecular and
cellular mechanisms that allow for the evolution of
tumor-initiating cells to develop and acquire core signature
capabilities, typically during tumor development and malignant
progression. In addition, enabling hallmarks assist in the linkage
of cancer marker phenotypes to the TME and explains various aspects
relevant to cancer (9,10). These enabling features are present
across all stages of cancer progression.
Genomic instability in cancer cells has been
considered the primary hallmark, resulting in random mutations and
chromosomal rearrangement (9,10,199,200). It has been reported that a
patient with West syndrome, a severe intellectual disability and
malformation, was identified as partial tetrasomy 17q25.3 and the
breakpoint of chromosome 17q25.3 rearrangement was located in the
FOXK2 (3). Certain mutated
genotypes confer advantages in subclonal selection and growth in
the local tissue environment (201). A roster of alterations in
conditions of genomic instability have been suggested for DNA
damage prevention, DNA repair system activation, DNA repair
defects, centrosomes and telomerase (202).
Genomic stability is a prerequisite for high
fidelity DNA and thus DNA is subject to precise and complex control
mechanisms (9,10). Throughout the cell cycle of a
normal cell, the integrity of the genome is protected by
checkpoints (9,10). An abnormal number of chromosomes
during cancer development indicates the failure of one or more cell
cycle checkpoints (203). It is
well established that CHK2 functions as an effector kinase of the
ATM-CHk2-p53 pathway in DNA damage repair and its phosphorylation
activity is critical for the DNA double-stranded break (DSB)
response (204,205). A transcriptional control study of
autophagy showed that Ser61 within FOXK2 is phosphorylated by CHK2,
which inhibits apoptosis of cancer cells via DNA damage (1).
DNA mismatch repair (MMR) is vital in ensuring
replication fidelity and maintaining genomic stability and is
critical in the prevention of mutations (206). MMR defects that lead to a high
mutational burden were exemplified in a study of breast cancer
(207). Researchers have modeled
the initiation of MMR (208).
FOXK2, as a novel G/T mismatch-specific binding protein, may sense
G/T mismatches and recruit BAP1 to trigger the DNA repair mechanism
(192,209). The phosphorylation of BAP1 and
its catalytic activity are necessary prerequisites for its repair
function (209). Mechanistically,
FOXK2 may act as a DNA-binding protein that binds to the distorted
conformation of DNA resulting from mismatches, facilitating the
recruitment of other repair proteins (210). In response to laser
micro-irradiation, MBD6 was recruited to laser-induced DNA damage
sites independently of PR-DUB. It was also found that FOXK2/PR-DUB
and MBD6 share a genome target gene subset (69). A study of yeast FOX proteins showed
that lexa-FHA fusion proteins bind to chromatin, induced by DSBs,
and subsequently recruit donors in an FHA-domain-dependent manner
(211). Importantly, the presence
or absence of the N-terminal coding region (139–459 bp) of the FHA
domain determines whether FOXK2 binds specifically to the G/T
mismatch. This specific binding either recruits DNA repair proteins
such as YB-1 to form complexes that initiate DNA mismatch repair,
or freezes transcription and replication of mismatched genes
leading to cell death (192).
Together, these findings suggest that FOXK2 serves an important
role in the regulatory mechanisms of DNA repair.
Loss of telomere protection can lead to a telomere
crisis, a widespread state of genomic instability that can amplify
or drive aging-related cancer development (212,213). Conversely, telomerase activation
provides an opportunity to eliminate the telomere crisis, leading
to the formation of cancer clones with genomic rearrangements
(212,213). Although the relationship between
FOXK2 and telomeres or telomerase has not been studied in-depth,
studies have implicated FOXK1 in cellular telomere fusion (33,214).
Epigenetic effects serve a significant role in
regulating the interactions between genomes and the environment and
this has attracted considerable research interest (215). These epigenetic effects may also
influence gene expression patterns and drive cancer development
(64,216,217). Non-mutational epigenetic
regulation of gene expression was initially interpreted, a decade
ago, as a powerful mechanism that mediates development and
differentiation (218,219). This concept has received
increased attention in recent years regarding its significance in
cancer biology (10,220,221). It has been shown that crucial
regulatory elements constitute a set of epigenetic regulations
(44). The emerging field of
epitranscriptomics, involving modifications of mRNAs and lncRNAs as
well as newly identified DNA cytosine modifications, is a key
mechanism of epigenetic regulation promoting cancer progression
(222). As described earlier,
epigenetic modification of FOXK2 mediates several chromatin events
critical in the multi-step process of cancer development. The
present review highlighted a possible link between environmental
exposure to cancer risk and epigenetic modifications of DNA. One
study focusing on differences in DNA methylation amongst ethnic
firefighters showed that FOXK2 hypomethylation partly explains the
differences in epigenetic susceptibility to cancer risk associated
with toxic exposure between ethnic groups (56). Chemical exposure, including but not
limited to polycyclic aromatic hydrocarbons, may lead to
differential methylation of FOXK2 CpG sites across ethnic groups
(56). Other suitable examples are
the hypermethylation of leukocytes caused by smoking, which is
positively correlated with smoking level (60). Certain lifestyles, such as exercise
and bariatric surgery, reduce NNMT expression and lead to increased
levels of FOXK2 methylation (57).
However, the relationship between more environmental factors,
lifestyle and psychological stress and FOXK2 methylation has not
been well explored. In addition, few studies have been published on
the regulatory mechanism of FOXK2 methylation and its effect on
downstream target genes.
The link between chronic inflammation and the
development and progression of cancer has been long established
(223,224). Inflammatory mediators and cell
effectors promote tumor development by changing the local TME. The
altered microenvironment disrupts the immune response and
contributes to the proliferation and survival of malignant cells
(224). Inflammation in the TME
is mainly characterized by the accumulation of innate and adaptive
immune cells (223,225), both of which promote tumor
progression (226,227). However, the association of these
immune cells with FOXK2 has not been fully studied. Encouragingly,
a study of early immune networks suggested that FOXK2 is involved
in immune regulation in early life (the 1st, 2nd, 3rd trimester of
gestations, birth, newborn and infant periods) (228). Other studies have also shown that
FOXK2 affects the activation of T lymphocytes and serves a role in
the development of immune networks (229,230). NF-κB is a TF involved in the
inflammation and immune response cellular pathways (231) and it has been shown to be
involved in tumorigenesis (232,233). FOXK2 expression is positively
regulated by NF-κB. For example, epidermal growth factor (EGF)
promotes FOXK2 expression through the NF-κB pathway (198). Alterations in TME caused by
cellular inflammation can also induce DNA damage, which in turn
assists tumor cells to acquire a variety of biological abilities
(227,234,235). As previously described, FOXK2
functions as a G/T mismatch-specific binding protein that initiates
DNA damage repair or freezes transcription and replication of
mismatched genes. However, this function in cancer is still poorly
characterized. Together, these studies suggest that the immune
networks are closely related to FOXK2 and serves a crucial role in
oncogenesis. Studies have shown that the adaptive immune system can
promote tumor suppressor gene inactivation (224,236); however, whether FOXK2 is involved
in this regulation is unknown.
The relationship between the microbiome and human
cancer is complex and contested and the relationships are well
described elsewhere (237,238).
Recently, polymorphic variations in an organism's microbial
community have been suggested as constituting a uniquely
advantageous trait for acquiring signature abilities (10). This view is becoming increasingly
compelling. Since human HAdVs were first isolated from an adenoid
tissue nearly 70 years ago (239), the ability of specific categories
of viruses to induce tumor growth has been demonstrated in
different mammalian models (240,241). The possible role of HAdVs in
malignant diseases in humans has been continuously explored, but
their role in human cancer remains unclear (242). One study found that the binding
and interaction of the c-terminal region of the viral E1A protein
with FOXK2 is essential for suppressing HAdV-mediated tumor
formation in vivo and in vitro (180). Furthermore, HAdV E1A protein
interaction with FOXK1 and FOXK2 is dependent on the levels of
phosphorylated E1A.
Human papillomavirus (HPV) is another interesting
virus in relation to FOXK2. The E6 protein of HPV 21/14 exerts its
antitumor effects by targeting FOXK1 and FOXK2 in tandem with a
conserved Thr-Ser motif (180).
In addition, the interaction of the E6 protein with FOXK1 and FOXK2
in epithelial cells may drive viral infection replication and
differentiation rather than transformation (180). However, the relationship between
FOXK2 and more viruses has not received sufficient attention.
The most fundamental characteristic of cancer cells
is their ability to maintain chronic proliferation. The degree of
proliferation is directly related to the development and
progression of cancer (9). During
development, growth factor signaling pathways induce proliferation,
migration, differentiation and death in select populations of cells
to ensure adherence to programmed organ sizes and functions. The
expression of cycle-related proteins and signaling pathways in
cancer cells is often altered, resulting in oncogenic activation of
growth factor signals or inhibition of cell death, leading to
pathological proliferation and tumor growth. These shared signaling
pathways primarily include hypoxia-inducible factor-1 (HIF-1),
CDKs, NF-κB, PI3K/AKT, insulin-like growth factor receptor (IGF-1R)
and estrogen receptor signaling (201).
Different studies have assessed the effects of
FOXK2 on the signaling pathways aforementioned. FOXK2 knockdown
induces non-neoplastic immortal cell death, proliferation and
survival as FOXK2 deletion leads to increased expression of p53
up-regulated modulator of apoptosis (PUMA) and NOXA (174).
The transcription factor HIF-1 structurally acts as
a heterodimer and regulates inducible genes that respond to changes
in oxygen levels (243,244). Nuclear localization of this
molecule in conditions of low oxygen concentrations induces
transcription of several genes responsible for tumor invasiveness
(245). The network crosstalk
between FOXK2 and HIF-1 is complex. FOXK2 interacts with ASXL1, a
vital component of PR-DUB, to regulate HIF-1α and STAT3 signaling
pathways (89).
PI3Ks are a family of lipid kinases initially
hypothesized to be involved in the transformational ability of
viral oncoproteins. Subsequent studies found that PI3Ks were
involved in regulating various cellular processes, including cell
proliferation and differentiation (131,250). The effects of FOXK2 and PI3Ks are
multi-dimensional. In certain clinical samples, such as patients
who only received surgery without preoperative chemotherapy or
radiotherapy, FOXK2 is negatively regulated by miR-1271-5p and
exerts carcinogenic activity by activating the PI3K/AKT signaling
pathway in HCC cells (131).
TP53TG1 promotes the occurrence and development of CC by regulating
miR-33A-5P targeting FOXK2 (167). FOXK2, as a downstream regulator
of the PI3K/AKT/mTOR signaling pathway, promotes GC proliferation
and inhibits apoptosis (130).
CDKs are serine/threonine kinases that rely on a
cyclin regulatory subunit to initiate cell division and
transcription, particularly in cancer progression (251). FOX TFs, including FOXKs, control
cellular processes during physiological development and in the
development and progression of cancer (190,252,253). Furthermore, in prokaryotes and
metazoans, a fundamental process controlled by FOX TFs is cell
cycle progression (76,254–257). In addition to regulating the
transcription of target genes in the cell cycle, FOX TFs are
regulated by cyclically regulated kinases. There are extensive and
complex links between cell cycle-regulated kinases and the FOX
transcription factor family (257). Studies involving the regulatory
function of FKH2 (a homolog of human FOXKs) on the cell cycle
support the link between cycle-regulated kinase and the FOX protein
(76,77).
Furthermore, another study identified FOXK2 as a
target of the CDK-cyclin complex in human cells and found that
FOXK2 levels are cell cycle-dependent, reaching a maximum
concentration during the M-phase (26). This study also found that FOXK2
mRNA levels did not change significantly during the cell cycle,
suggesting that FOXK2 is regulated via PTMs. Notably, endogenous
FOXK2 stably translocates to the nucleus of most asynchronously
growing U2OS cells (26), while
the subcellular localization of other FOX TFs varies with the stage
of the cell cycle (257,258). FOXK2 relocalization away from the
DNA during mitosis (26) also
differs from the persistent association of FOXK1 with DNA (259). However, the significance of this
small change in nuclear localization has not been thoroughly
studied.
Cancer stem cells are the source of tumor cells,
granting them the ability to achieve a state of cell immortality.
Recent research has shown that FOXK2, a highly expressed stem
cell-specific TF in ovarian cancer, binds to an intron regulatory
element of the sensor ERN1. This binding triggers an unfolded
protein response that directly upregulates inositol-requiring
enzyme 1α (IRE1α, ERN1 gene) expression. In addition, it
results in the X-box-binding selective active splicing of protein 1
(XBP1) and activation of stemness-related pathways (264). However, there is still a lack of
broader studies on the effect of FOXK2 on cancer stem cell stemness
and little is known about the regulatory mechanism of FOXK2
upstream regulatory signals in the maintenance of stemness.
High-throughput sequencing has proved to be an
invaluable tool in cancer research. The ability to avoid
anti-growth signals and the loss of tumor suppressor factors leads
cancer cells to exhibit disorderly and uncontrolled growth, which
is widely accepted as a hallmark of cancer (9,265).
Several tumor suppressor genes function together to determine cell
fate (265). In addition to BAP1,
BRCA1 and DVL as aforementioned, the relationship between FOXKs and
other tumor-related factors is discussed in the present review.
Phosphatase and TENsin Homolog (PTEN) is a phosphatase that
dephosphorylates phosphatidylinositol-triphosphate (PIP3) to PIP2
(266–268). PTEN is a well-known tumor
suppressor gene involved in several types of cancer, negatively
regulating the PI3K/AKT signaling pathway (266). However, loss-of-function
mutations of PTEN are often found in tumors (269). Through ChIP and dual luciferin
reporter assays, FOXK1 was shown to directly bind to the miR-32
promoter. It was also shown to positively regulate the expression
of miR-32 and transmembrane protein 245 gene (TMEM245). In CRC,
FOXK1 was shown to inhibit the expression of PTEN through
transcriptional regulation of TMEM245/miR-32, thereby enhancing the
proliferation, migration and invasion of CRC cells and reducing the
apoptosis of CRC cells (270).
Another study further demonstrated the existence of a core promoter
region in the −320 to −1 bp range of the 5′ flanks of the
TMEM245/miR-32 gene and inhibitory regulatory elements in the-606
to −320 bp range (271).
FOXK2 also interacts with other tumor suppressor
proteins. For example, after S-phase DNA damage, FOXK1-53BP1
interaction is dependent on ATM/CHK2, which reduces the correlation
between 53BP1 and its downstream factors RIF1 and PTIP (214). In addition, FOXK2 interacts with
the transcriptional co-suppressor complex NCoR/SMRT Sin3a NuRD and
REST/CoREST to exert its anti-tumor role. As a result, FOXK2
inhibits genes such as HIF-1β and EZH2 and modulates several
signaling pathways, including the hypoxia response (194).
Maintenance of the balance of pro-apoptotic and
anti-apoptotic proteins is crucial in determining cell apoptosis
development. The struggle between apoptosis and anti-apoptosis is
present in all stages of cancer, from precancerous lesions to tumor
formation. The deregulation of apoptosis is associated with the
development and progression of cancer through unmoderated cell
proliferation and cancer resistance to drug therapy (272). Therefore, the ability to evade
apoptosis is considered a defining cancer hallmark (9). Among the several anti-apoptotic
pathways, the overexpression of anti-apoptotic proteins is the
primary strategy cancer cells use to avoid apoptosis (201). The role of the Bcl-2 family
members in apoptosis is well established. The Bcl-2 homologous (BH)
domain is the structural basis for interaction among its members
and drives pro-apoptotic or anti-apoptotic functions (273,274).
One study found that knockdown of FOXK2 led to
reduced proliferation and increased apoptosis in mouse NIH3T3
fibroblasts and mouse breast cancer NMuMG cells (174). After FOXK2 gene KO, expression of
the pro-apoptotic proteins PUMA and NOXA was significantly
upregulated (174) and PUMA and
NOXA are members of the pro-apoptotic Bcl-2 family (275). A positive association between
FOXK2 and increased phospho-AKT levels has been shown (131). Additional studies have
demonstrated that AKT is phosphorylated by PDK1 on one or two
specific sites, which is necessary for its full catalytic activity.
Activated AKT inactivates the expression of pro-apoptotic proteins
such as Bcl-2 and FOXO TFs, positively affecting cell survival
(276,277). Although there is evidence
suggesting that FOXK2 exerts an anti-apoptotic role, research on
the effects of FOXK2 on cell proliferation and survival is limited.
Thus, further studies are required to understand the function and
mechanism underlying the anti-apoptotic effects of FOXK2 are
required.
Angiogenesis, defined as the growth of new
capillaries from existing blood vessels, involves endothelial cell
migration, invasion and duct formation. Angiogenesis is essential
for dividing cells and this is especially true for tumor cells, as
the new vessels provide oxygen and nutrients to maintain cell
division (278,279). The activation of angiogenesis can
result from the imbalance between pro-angiogenic and
anti-angiogenic molecules (280,281). Of all the angiogenic factors, the
most influential are VEGF, EGF and platelet-derived growth factor
(PDGF) (282). VEGFA exerts an
angiogenic effect by binding to VEGFR-1 and VEGFR-2 and its
co-receptors neuropilin-1 and neuropil-2 (NRP-1 and NRP-2)
(283,284). In addition, VEGFA regulates
endothelial cell survival and enhances the mobilization of bone
marrow-derived endothelial precursor cells (285,286). VEGFA/VEGFR-2 signaling is widely
considered the most important angiogenic mechanism.
It has previously been reported that VEGFA
expression is increased in ATC following apatinib treatment
(287). Furthermore, the ChIP
dual-luciferase reporter system and functional assays confirm that
FOXK2 promotes ATC angiogenesis by inducing VEGFA transcription
(197). When VEGFR-2 is blocked,
VEGFA then binds to VEGFR-1, promoting angiogenesis by activating
ERK, PI3K/AKT and P38/MAPK signaling in human umbilical vein
endothelial cells, which compensates for VEGFR2 blockage (197,288). Notably, VEGFA binding to VEGFR-1
can promote FOXK2-mediated VEGFA transcription and angiogenesis
through a positive feedback loop (197).
In a diabetes mellitus mouse model and in human
ECs, miR-140-3p transcription inhibits FOXK2 signaling, promoting
key angiogenic steps, including EC proliferation, cell migration
and endovascular formation (135). Conversely, FOXK1 inhibits
angiogenesis by inhibiting VEGFA transcription (289). However, the controversial works
aforementioned also leave a number of unanswered questions. The key
to solve these problems is to further study the regulatory
mechanism of FOXK2 and angiogenesis related metabolic
remodeling.
Tumor cells are especially adept at adapting to the
environment and extracting energy. Their ability to increase
glucose uptake and lactic acid production (the Warburg effect) is
an excellent example of the evolution of substrate metabolic fate
(290). This well-evolved
flexibility is necessary to ensure enhanced biomass synthesis while
maintaining redox equilibrium and cellular homeostasis (291,292). These properties reflect a balance
between the availability of growth-signaling chemical nutrients,
the subsequent adaptive metabolic remodeling and the everyday needs
of the cell (293).
A study showed that FOXK1 and FOXK2, experimentally
associated with nutritional stress, may function as regulators to
induce aerobic glycolysis reprogramming (88). Mechanistically, FOXK1 and FOXK2
induce aerobic glycolysis by upregulating hexokinase 2 (HK2),
phosphofructokinase, pyruvate kinase (PK) and lactate dehydrogenase
(LDH). These enzymes are associated with glucose metabolism and
regulate the flow of glycolysis (88,294). Further studies have shown that an
increase in pyruvate dehydrogenase (PDH) kinases 1 and 4 activity
prevents the conversion of pyruvate to acetyl-CoA in the
mitochondria and pyruvate is instead reduced to lactic acid
(88,290,294).
Thioredoxin interacting protein (TXNIP) is an
α-inhibitory protein. TXNIP modulates glucose homeostasis through
strong negative regulation of glucose uptake and aerobic glycolysis
(295,296). FOXK1/FOXK2 can directly bind to
the TXNIP promoter to exert an influence on aerobic glycolysis
(89). It is also found that
structurally and functionally deficient PR-DUB complexes, including
the absence of FOXK1/FOXK2, significantly reduce TXNIP protein
levels, resulting in increased glucose uptake and increased
intracellular lactic acid and ATP levels (89).
The FOXK transcription factor regulates glucose
consumption by altering its own subcellular localization and
affecting HIF-1α gene expression through a process regulated by
mTOR (2,87). PDH can be hyper-phosphorylated by
pyruvate dehydrogenase kinases (PDKs), which are upregulated by
HIF-1α, resulting in its inactivation. This inactivation inhibits
the conversion of pyruvate to acetyl-CoA and increases lactic acid
production (290). Additionally,
HIF1α regulates several major glycolytic proteins, including the
glycolytic enzymes HK2, phosphoglycerate kinase 1, LDHA and PKM2
(297,298).
The present review examined the most influential
roles of FOXK2 in cancer development. It highlighted the complexity
of the function of FOXK2 and gave an outlook on what has to be
investigated in future work. In addition, it described the current
understanding of FOXK2 and its global capabilities, providing
context for explaining how FOXK2 functions in cancer, both
individually and as a part of numerous complex systems. The
extensive expression of FOXK2 and inherent structural
characteristics distinguish it from ~1,600 other human TFs
(304).
FOXK2 functions in several different contexts by
cooperating with other active molecules. The suggestion that TFs
work together to achieve their function is widely accepted
(305,306). The properties of FOXK2 and other
members of the FOX family determine its precise function in
biology. For example, there are also binding differences between
FOXK2 and other members of the FOX family among functionally
specific target genes partly influenced by the flanking region
(179,307). Theoretical and practical
observations show that synergistic binding and co-regulation are
the primary synergistic modes of TFs, which help bioactive
molecules bind to DNA, influencing chromatin accessibility and
downstream gene transcription (308). However, specific modes of action
of FOXK2 expression in different time and space under physiological
and pathological conditions have not been clearly demonstrated and
general principles cannot be inferred from the current study.
FOXK2 mediates several functionally significant
chromatin events. This suggests that DNA-mediated cooperative
binding is crucial for the function of TFs (309). TFs that can bind to target sites
on nucleosome DNA are known as pioneer factors (310,311). These pioneer TFs are responsible
for opening chromatin or changing the conformation of the
nucleosome by initiating nucleosome displacement (312–314). The above are necessary
prerequisites for recruiting other bioactive substances and other
TFs (315). These pioneer TFs
control cell fate by locally opening chromatin to initiate
transcription (316,317). Its stability is partly influenced
by steric hindrance (318) and
nucleosome affinity for active chromatin remodeling (319). Members of the FOX family have
been shown to function as pioneer factors as they can bind to
nucleosome DNA and open chromatin, thereby exposing DNA binding
modes allowing it to bind to other TFs and subsequently regulate
gene expression (320,321). Based on this evidence and the
regulation of chromatin events by FOXK2 described above, FOXK2 may
be considered a pioneer factor. Unraveling local chromatin regions
without the help of ATP-dependent chromatin remodeling factors
(322,323) is a valuable characteristic for
consideration of FOXK2 as a pioneer candidate.
Protein-protein interactions are regulatory
mechanisms for TFs that are well understood. Studies using
single-molecule imaging confirm that when multiple TFs bind with
DNA at consistently spaced intervals with a consistent orientation,
the binding sites are occupied for longer periods of time,
conferring additional stability (324,325). FOXK2 has been shown to form
complexes with several proteins to perform different functions.
According to its nature, eukaryotic gene expression regulation can
be divided into instantaneous (reversible) (326,327) and development (irreversible)
regulation (328,329). Instantaneous regulation
determines the fate of metabolic substrates and hormonal
fluctuations in enzyme activity. It also dictates the substrate or
hormonal concentrations at different stages of the cell cycle.
Development regulation influences overall eukaryotic cell
differentiation, growth and development processes. The present
review provides an overview of the contribution of FOXK2 to
transient and developmental regulation and highlights the role of
epigenetic modifications in controlling chromatin accessibility and
protein interactions.
The present review also discussed the multifaceted
role of FOXK2 in cancer. FOXK2 is involved in the pathogenesis of
numerous types of cancer. Whether FOXK2 functions as an oncoprotein
or tumor suppressor appears to be closely related to its partners
and its spatio-temporal properties and is thus tumor-specific. A
human cancer genome survey elucidated several salient features of
oncogenes involving the types of sequence alterations identified,
oncogenic mutations in cancer classes and protein domains encoded
by cancer genes (330). Indeed,
proteins encoded by cancer genes typically regulate cell
proliferation, differentiation and death. A functional review of
FOXK2 also supports the hypothesis of FOXK2 as an oncogene.
However, the genes that have been reported with precise causal
associations with tumorigenesis have been identified and initially
reported based on sufficient genetic evidence. Mutated genes that
provide cancer cells a growth advantage are highly suitable
candidates for oncogenes. Genes with translocations or copy number
alterations supported by convincing genetic data are another group
of candidates. Genes whose expression levels are altered only in
cancer cells are not suitable oncogene candidates, lacking any
mutations in DNA that cannot be conclusively linked to
tumorigenesis. However, FOXK2 mutations have not been reported
previously to the best of the authors' knowledge. Based on the
evidence, the biological regulatory functions of FOXK2, such as the
regulation of glucose metabolism and autophagy, may be used as
hijacking tools by tumor cells to enable unlimited proliferation
and survival of tumor cells. Of course, these assumptions are
contested and unproven. In the absence of more extensive research,
one should be cautious about making conclusive claims. However,
what is certain is that FOXK2 is vital in the development and
progression of cancer. In the foreseeable future, in-depth studies
targeting the regulatory features of FOXK2 may reveal its role in
tumorigenesis.
Although a similar review was published in 2019
regarding FOXK2 and its roles in cancer (4), the present review has updated the
recent findings about FOXK2 by a number publications since then.
First, it detailed the nomenclature and structural differences of
the three isoforms of FOXK2 and suggested that alternative splicing
of FOXK2 may be related to the role of kinase cascade signaling.
Second, for the regulatory mechanism of FOXK2, it considered both
its roles as a regulator and being regulated, with systematically
and clearly description in terms of gene level, post-translational
modification and protein interaction. Third, it summarized the
roles of FOXK2 as pioneer factor, G/T mismatch DNA-binding protein,
virus-binding protein, scaffold protein and transfer vector and
proposed that FOXK2 can be a candidate as a pioneer factor. Fourth,
it used the widely accepted concept of cancer hallmarks to describe
the broad role of FOXK2 in tumorigenesis in detail. Fifth, it took
a cautious attitude toward the definition of FOXK2 as an oncogene
or a tumor suppressor. FOXK2 may act as a hijacked molecular to
achieve its spatiotemporal and tumor-specific functions. Finally,
it objectively noted the shortcomings of current studies and the
directions for future research on FOXK2 in the hope that the
present review will provide useful information for researchers
working in this field.
Not applicable.
The present study was supported by the Shandong Provincial
Natural Science Foundation of China (grant nos. ZR2020KC016 and
ZR2020QH096); and the Weifang Science and Technology Bureau (grant
no. 2020YQFK013).
Not applicable.
ZhaoW and XL developed the idea, and wrote and
revised the manuscript. ZhanW and ZH supervised the study and
contributed to critical reading and revising of the manuscript. All
the authors have read and approved the final manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Chen Y, Wu J, Liang G, Geng G, Zhao F, Yin
P, Nowsheen S, Wu C, Li Y, Li L, et al: CHK2-FOXK axis promotes
transcriptional control of autophagy programs. Sci Adv.
6:eaax58192020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He L, Gomes AP, Wang X, Yoon SO, Lee G,
Nagiec MJ, Cho S, Chavez A, Islam T, Yu Y, et al: mTORC1 promotes
metabolic reprogramming by the suppression of GSK3-dependent Foxk1
phosphorylation. Mol Cell. 70:949–960.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hackmann K, Stadler A, Schallner J, Franke
K, Gerlach EM, Schrock E, Rump A, Fauth C, Tinschert S and Oexle K:
Severe intellectual disability, west syndrome, Dandy-Walker
malformation, and syndactyly in a patient with partial tetrasomy
17q25.3. Am J Med Genet A. 161A:3144–3149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nestal de Moraes G, Carneiro LD, Maia RC,
Lam EW and Sharrocks AD: FOXK2 transcription factor and its
emerging roles in cancer. Cancers (Basel). 11:3932019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gitter A, Siegfried Z, Klutstein M, Fornes
O, Oliva B, Simon I and Bar-Joseph Z: Backup in gene regulatory
networks explains differences between binding and knockout results.
Mol Syst Biol. 5:2762009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dai Z, Dai X, Xiang Q and Feng J:
Robustness of transcriptional regulatory program influences gene
expression variability. BMC Genomics. 10:5732009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu WS and Lai FJ: Functional redundancy of
transcription factors explains why most binding targets of a
transcription factor are not affected when the transcription factor
is knocked out. BMC Syst Biol. 9 (Suppl 6):S22015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaestner KH, Knochel W and Martinez DE:
Unified nomenclature for the winged helix/forkhead transcription
factors. Genes Dev. 14:142–146. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lam EW, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: Tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y, Ao X, Ding W, Ponnusamy M, Wu W,
Hao X, Yu W, Wang Y, Li P and Wang J: Critical role of FOXO3a in
carcinogenesis. Mol Cancer. 17:1042018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakagawa S, Gisselbrecht SS, Rogers JM,
Hartl DL and Bulyk ML: DNA-binding specificity changes in the
evolution of forkhead transcription factors. Proc Natl Acad Sci
USA. 110:12349–12354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li C, Lai CF, Sigman DS and Gaynor RB:
Cloning of a cellular factor, interleukin binding factor, that
binds to NFAT-like motifs in the human immunodeficiency virus long
terminal repeat. Proc Natl Acad Sci USA. 88:7739–7743. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang JT and Lee V: Identification and
characterization of a novel human FOXK1 gene in silico. Int
J Oncol. 25:751–757. 2004.PubMed/NCBI
|
|
17
|
Mahajan A, Yuan C, Lee H, Chen ES, Wu PY
and Tsai MD: Structure and function of the
phosphothreonine-specific FHA domain. Sci Signal. 1:re122008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Durocher D and Jackson SP: The FHA domain.
FEBS Lett. 513:58–66. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reinhardt HC and Yaffe MB:
Phospho-Ser/Thr-binding domains: Navigating the cell cycle and DNA
damage response. Nat Rev Mol Cell Biol. 14:563–580. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kalnina Z, Zayakin P, Silina K and Linē A:
Alterations of pre-mRNA splicing in cancer. Genes Chromosomes
Cancer. 42:342–357. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roy M, Xu Q and Lee C: Evidence that
public database records for many cancer-associated genes reflect a
splice form found in tumors and lack normal splice forms. Nucleic
Acids Res. 33:5026–5033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bates DO, Cui TG, Doughty JM, Winkler M,
Sugiono M, Shields JD, Peat D, Gillatt D and Harper SJ: VEGF165b,
an inhibitory splice variant of vascular endothelial growth factor,
is down-regulated in renal cell carcinoma. Cancer Res.
62:4123–4131. 2002.PubMed/NCBI
|
|
23
|
Hu Y, Fang C and Xu Y: The effect of
isoforms of the cell polarity protein, human ASIP, on the cell
cycle and Fas/FasL-mediated apoptosis in human hepatoma cells. Cell
Mol Life Sci. 62:1974–1983. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Duke L, Zhang PS, Arlinghaus RB,
Symmans WF, Sahin A, Mendez R and Dai JL: Alternative splicing
disrupts a nuclear localization signal in spleen tyrosine kinase
that is required for invasion suppression in breast cancer. Cancer
Res. 63:4724–4730. 2003.PubMed/NCBI
|
|
25
|
Nirula A, Moore DJ and Gaynor RB:
Constitutive binding of the transcription factor interleukin-2
(IL-2) enhancer binding factor to the IL-2 promoter. J Biol Chem.
272:7736–7745. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Marais A, Ji Z, Child ES, Krause E, Mann
DJ and Sharrocks AD: Cell cycle-dependent regulation of the
forkhead transcription factor FOXK2 by CDK·cyclin complexes. J Biol
Chem. 285:35728–35739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pan Q, Shai O, Lee LJ, Frey BJ and
Blencowe BJ: Deep surveying of alternative splicing complexity in
the human transcriptome by high-throughput sequencing. Nat Genet.
40:1413–1415. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li C, Lusis AJ, Sparkes R, Nirula A and
Gaynor R: Characterization and chromosomal mapping of the gene
encoding the cellular DNA binding protein ILF. Genomics.
13:665–671. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang ET, Sandberg R, Luo S, Khrebtukova I,
Zhang L, Mayr C, Kingsmore SF, Schroth GP and Burge CB: Alternative
isoform regulation in human tissue transcriptomes. Nature.
456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Merkin J, Russell C, Chen P and Burge CB:
Evolutionary dynamics of gene and isoform regulation in mammalian
tissues. Science. 338:1593–1599. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Climente-González H, Porta-Pardo E, Godzik
A and Eyras E: The functional impact of alternative splicing in
cancer. Cell Rep. 20:2215–2226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang W, Li X, Lee M, Jun S, Aziz KE, Feng
L, Tran MK, Li N, McCrea PD, Park JI and Chen J: FOXKs promote
Wnt/β-catenin signaling by translocating DVL into the nucleus. Dev
Cell. 32:707–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu Y, Ding W, Ge H, Ponnusamy M, Wang Q,
Hao X, Wu W, Zhang Y, Yu W, Ao X and Wang J: FOXK transcription
factors: Regulation and critical role in cancer. Cancer Lett.
458:1–12. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Giardina B, Messana I, Scatena R and
Castagnola M: The multiple functions of hemoglobin. Crit Rev
Biochem Mol Biol. 30:165–196. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Arbez N, Ratovitski T, Roby E, Chighladze
E, Stewart JC, Ren M, Wang X, Lavery DJ and Ross CA:
Post-translational modifications clustering within proteolytic
domains decrease mutant huntingtin toxicity. J Biol Chem.
292:19238–19249. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Snider NT and Omary MB: Post-translational
modifications of intermediate filament proteins: Mechanisms and
functions. Nat Rev Mol Cell Biol. 15:163–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Richard SA, Jiang Y, Xiang LH, Zhou S,
Wang J, Su Z and Xu H: Post-translational modifications of high
mobility group box 1 and cancer. Am J Transl Res. 9:5181–5196.
2017.PubMed/NCBI
|
|
38
|
Corujo D and Buschbeck M:
Post-translational modifications of H2A histone variants and their
role in cancer. Cancers (Basel). 10:592018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Iavarone F, Desiderio C, Vitali A, Messana
I, Martelli C, Castagnola M and Cabras T: Cryptides: Latent
peptides everywhere. Crit Rev Biochem Mol Biol. 53:246–263. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang H, Arighi CN, Ross KE, Ren J, Li G,
Chen SC, Wang Q, Cowart J, Vijay-Shanker K and Wu CH: iPTMnet: An
integrated resource for protein post-translational modification
network discovery. Nucleic Acids Res. 46:D542–D550. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yao B, Christian KM, He C, Jin P, Ming GL
and Song H: Epigenetic mechanisms in neurogenesis. Nat Rev
Neurosci. 17:537–549. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu MY, DeNizio JE, Schutsky EK and Kohli
RM: The expanding scope and impact of epigenetic cytosine
modifications. Curr Opin Chem Biol. 33:67–73. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jones MJ, Goodman SJ and Kobor MS: DNA
methylation and healthy human aging. Aging Cell. 14:924–932. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tsuchida T, Mano T, Koshi-Mano K, Bannai
T, Matsubara T, Yamashita S, Ushijima T, Nagata K, Murayama S, Toda
T, et al: Methylation changes and aberrant expression of FGFR3 in
Lewy body disease neurons. Brain Res. 1697:59–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pan XY, Yang Y, Meng HW, Li HD, Chen X,
Huang HM, Bu FT, Yu HX, Wang Q, Huang C, et al: DNA methylation of
PTGIS enhances hepatic stellate cells activation and liver
fibrogenesis. Front Pharmacol. 9:5532018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hopp L, Löffler-Wirth H, Galle J and
Binder H: Combined SOM-portrayal of gene expression and DNA
methylation landscapes disentangles modes of epigenetic regulation
in glioblastoma. Epigenomics. 10:745–764. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lopez-Serra P and Esteller M: DNA
methylation-associated silencing of tumor-suppressor microRNAs in
cancer. Oncogene. 31:1609–1622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Le TN, Schumann U, Smith NA, Tiwari S, Au
PC, Zhu QH, Taylor JM, Kazan K, Llewellyn DJ, Zhang R, et al: DNA
demethylases target promoter transposable elements to positively
regulate stress responsive genes in Arabidopsis. Genome Biol.
15:4582014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jung M and Pfeifer GP: Aging and DNA
methylation. BMC Biol. 13:72015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bormann F, Rodríguez-Paredes M, Lasitschka
F, Edelmann D, Musch T, Benner A, Bergman Y, Dieter SM, Ball CR,
Glimm H, et al: Cell-of-Origin DNA methylation signatures are
maintained during colorectal carcinogenesis. Cell Rep.
23:3407–3418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: How the genome integrates intrinsic
and environmental signals. Nat Genet. 33 (Suppl):S245–S254. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Egger G, Liang G, Aparicio A and Jones PA:
Epigenetics in human disease and prospects for epigenetic therapy.
Nature. 429:457–463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Robertson KD: DNA methylation and human
disease. Nat Rev Genet. 6:597–610. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bird A, Taggart M, Frommer M, Miller OJ
and Macleod D: A fraction of the mouse genome that is derived from
islands of nonmethylated, CpG-rich DNA. Cell. 40:91–99. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Goodrich JM, Furlong MA, Caban-Martinez
AJ, Jung AM, Batai K, Jenkins T, Beitel S, Littau S, Gulotta J,
Wallentine D, et al: Differential DNA methylation by hispanic
ethnicity among firefighters in the United States. Epigenet
Insights. Mar 26–2021.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Crujeiras AB, Pissios P, Moreno-Navarrete
JM, Diaz-Lagares A, Sandoval J, Gomez A, Ricart W, Esteller M,
Casanueva FF and Fernandez-Real JM: An epigenetic signature in
adipose tissue is linked to nicotinamide N-methyltransferase gene
expression. Mol Nutr Food Res. Apr 24–2018.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Camprubí C, Salas-Huetos A, Aiese-Cigliano
R, Godo A, Pons MC, Castellano G, Grossmann M, Sanseverino W,
Martin-Subero JI, Garrido N and Blanco J: Spermatozoa from
infertile patients exhibit differences of DNA methylation
associated with spermatogenesis-related processes: An array-based
analysis. Reprod Biomed Online. 33:709–719. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nwanaji-Enwerem JC, Jenkins TG, Colicino
E, Cardenas A, Baccarelli AA and Boyer EW: Serum dioxin levels and
sperm DNA methylation age: Findings in Vietnam war veterans exposed
to agent orange. Reprod Toxicol. 96:27–35. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Park SL, Patel YM, Loo LW, Mullen DJ,
Offringa IA, Maunakea A, Stram DO, Siegmund K, Murphy SE,
Tiirikainen M and Le Marchand L: Association of internal smoking
dose with blood DNA methylation in three racial/ethnic populations.
Clin Epigenetics. 10:1102018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yehuda R, Daskalakis NP, Bierer LM, Bader
HN, Klengel T, Holsboer F and Binder EB: Holocaust exposure induced
intergenerational effects on FKBP5 methylation. Biol Psychiatry.
80:372–380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hughes MF: Arsenic toxicity and potential
mechanisms of action. Toxicol Lett. 133:1–16. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
64
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Timbergen MJM, Boers R, Vriends ALM, Boers
J, van IJcken WFJ, Lavrijsen M, Grünhagen DJ, Verhoef C, Sleijfer
S, Smits R, et al: Differentially methylated regions in
desmoid-type fibromatosis: A comparison between CTNNB1 S45F and
T41A tumors. Front Oncol. 10:5650312020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Spruijt CG, Gnerlich F, Smits AH,
Pfaffeneder T, Jansen PW, Bauer C, Münzel M, Wagner M, Müller M,
Khan F, et al: Dynamic readers for 5-(hydroxy)methylcytosine and
its oxidized derivatives. Cell. 152:1146–1159. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Iurlaro M, Ficz G, Oxley D, Raiber EA,
Bachman M, Booth MJ, Andrews S, Balasubramanian S and Reik W: A
screen for hydroxymethylcytosine and formylcytosine binding
proteins suggests functions in transcription and chromatin
regulation. Genome Biol. 14:R1192013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen
HN, Shin J, Cox E, Rho HS, Woodard C, et al: DNA methylation
presents distinct binding sites for human transcription factors.
Elife. 2:e007262013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Baymaz HI, Fournier A, Laget S, Ji Z,
Jansen PW, Smits AH, Ferry L, Mensinga A, Poser I, Sharrocks A, et
al: MBD5 and MBD6 interact with the human PR-DUB complex through
their methyl-CpG-binding domain. Proteomics. 14:2179–2189. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Du Q, Luu PL, Stirzaker C and Clark SJ:
Methyl-CpG-binding domain proteins: Readers of the epigenome.
Epigenomics. 7:1051–1073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li X, Wilmanns M, Thornton J and Köhn M:
Elucidating human phosphatase-substrate networks. Sci Signal.
6:rs102013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sacco F, Perfetto L, Castagnoli L and
Cesareni G: The human phosphatase interactome: An intricate family
portrait. FEBS Lett. 586:2732–2739. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fukami Y and Lipmann F: Reversal of Rous
sarcoma-specific immunoglobulin phosphorylation on tyrosine (ADP as
phosphate acceptor) catalyzed by the src gene kinase. Proc Natl
Acad Sci USA. 80:1872–1876. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kole HK, Abdel-Ghany M and Racker E:
Specific dephosphorylation of phosphoproteins by protein-serine and
-tyrosine kinases. Proc Natl Acad Sci USA. 85:5849–5853. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Almawi AW, Matthews LA and Guarné A: FHA
domains: Phosphopeptide binding and beyond. Prog Biophys Mol Biol.
127:105–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhu G, Spellman PT, Volpe T, Brown PO,
Botstein D, Davis TN and Futcher B: Two yeast forkhead genes
regulate the cell cycle and pseudohyphal growth. Nature. 406:90–94.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Pic-Taylor A, Darieva Z, Morgan BA and
Sharrocks AD: Regulation of cell cycle-specific gene expression
through cyclin-dependent kinase-mediated phosphorylation of the
forkhead transcription factor Fkh2p. Mol Cell Biol. 24:10036–10046.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ma RY, Tong TH, Cheung AM, Tsang AC, Leung
WY and Yao KM: Raf/MEK/MAPK signaling stimulates the nuclear
translocation and transactivating activity of FOXM1c. J Cell Sci.
118:795–806. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Li A, Wang J, Wu M, Zhang X and Zhang H:
The inhibition of activated hepatic stellate cells proliferation by
arctigenin through G0/G1 phase cell cycle arrest: Persistent
p27(Kip1) induction by interfering with PI3K/Akt/FOXO3a signaling
pathway. Eur J Pharmacol. 747:71–87. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Aitken A: 14-3-3 proteins: A historic
overview. Semin Cancer Biol. 16:162–172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Nakatsumi H, Oka T, Higa T, Shirane M and
Nakayama KI: Nuclear-cytoplasmic shuttling protein PP2AB56
contributes to mTORC1-dependent dephosphorylation of FOXK1. Genes
Cells. 23:599–605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Nakatsumi H, Matsumoto M and Nakayama KI:
Noncanonical pathway for regulation of CCL2 expression by an
mTORC1-FOXK1 axis promotes recruitment of tumor-associated
macrophages. Cell Rep. 21:2471–2486. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Sakaguchi M, Cai W, Wang CH, Cederquist
CT, Damasio M, Homan EP, Batista T, Ramirez AK, Gupta MK, Steger M,
et al: FoxK1 and FoxK2 in insulin regulation of cellular and
mitochondrial metabolism. Nat Commun. 10:15822019. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Amaya MJ, Oliveira AG, Guimarães ES,
Casteluber MC, Carvalho SM, Andrade LM, Pinto MC, Mennone A,
Oliveira CA, Resende RR, et al: The insulin receptor translocates
to the nucleus to regulate cell proliferation in liver. Hepatology.
59:274–283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Katoh M and Katoh M: Identification and
characterization of human FOXK1 gene in silico. Int J Mol
Med. 14:127–132. 2004.PubMed/NCBI
|
|
87
|
Bowman CJ, Ayer DE and Dynlacht BD: Foxk
proteins repress the initiation of starvation-induced atrophy and
autophagy programs. Nat Cell Biol. 16:1202–1214. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sukonina V, Ma H, Zhang W, Bartesaghi S,
Subhash S, Heglind M, Foyn H, Betz MJ, Nilsson D, Lidell ME, et al:
FOXK1 and FOXK2 regulate aerobic glycolysis. Nature. 566:279–283.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Xia YK, Zeng YR, Zhang ML, Liu P, Liu F,
Zhang H, He CX, Sun YP, Zhang JY, Zhang C, et al: Tumor-derived
neomorphic mutations in ASXL1 impairs the BAP1-ASXL1-FOXK1/K2
transcription network. Protein Cell. 12:557–577. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Danciu TE, Chupreta S, Cruz O, Fox JE,
Whitman M and Iñiguez-Lluhí JA: Small ubiquitin-like modifier
(SUMO) modification mediates function of the inhibitory domains of
developmental regulators FOXC1 and FOXC2. J Biol Chem.
287:18318–18329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Sutinen P, Rahkama V, Rytinki M and
Palvimo JJ: Nuclear mobility and activity of FOXA1 with androgen
receptor are regulated by SUMOylation. Mol Endocrinol.
28:1719–1728. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Song JG, Xie HH, Li N, Wu K, Qiu JG, Shen
DM and Huang CJ: SUMO-specific protease 6 promotes gastric cancer
cell growth via deSUMOylation of FoxM1. Tumour Biol. 36:9865–9871.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Meredith LJ, Wang CM, Nascimento L, Liu R,
Wang L and Yang WH: The key regulator for language and speech
development, FOXP2, is a novel substrate for SUMOylation. J Cell
Biochem. 117:426–438. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Rocca DL, Wilkinson KA and Henley JM:
SUMOylation of FOXP1 regulates transcriptional repression via CtBP1
to drive dendritic morphogenesis. Sci Rep. 7:8772017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Nestal de Moraes G, Ji Z, Fan LY, Yao S,
Zona S, Sharrocks AD and Lam EW: SUMOylation modulates
FOXK2-mediated paclitaxel sensitivity in breast cancer cells.
Oncogenesis. 7:292018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Shmueli A and Oren M: Life, death and
ubiquitin: Taming the mule. Cell. 121:963–965. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
López-Otín C and Hunter T: The regulatory
crosstalk between kinases and proteases in cancer. Nat Rev Cancer.
10:278–292. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ikeda F and Dikic I: Atypical ubiquitin
chains: New molecular signals. ‘Protein modifications: Beyond the
usual suspects’ review series. EMBO Rep. 9:536–542. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Suryadinata R, Roesley SN, Yang G and
Sarčević B: Mechanisms of generating polyubiquitin chains of
different topology. Cells. 3:674–689. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Rajalingam K and Dikic I: SnapShot:
Expanding the ubiquitin code. Cell. 164:1074–1074.e1. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Deng L, Meng T, Chen L, Wei W and Wang P:
The role of ubiquitination in tumorigenesis and targeted drug
discovery. Signal Transduct Target Ther. 5:112020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Scheuermann JC, de Ayala Alonso AG, Oktaba
K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW and Müller
J: Histone H2A deubiquitinase activity of the polycomb repressive
complex PR-DUB. Nature. 465:243–247. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Abdel-Wahab O, Gao J, Adli M, Dey A,
Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, Lafave
LM, et al: Deletion of Asxl1 results in myelodysplasia and severe
developmental defects in vivo. J Exp Med. 210:2641–2659. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
LaFave LM, Béguelin W, Koche R, Teater M,
Spitzer B, Chramiec A, Papalexi E, Keller MD, Hricik T,
Konstantinoff K, et al: Loss of BAP1 function leads to
EZH2-dependent transformation. Nat Med. 21:1344–1349. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Micol JB and Abdel-Wahab O: The role of
additional sex combs-like proteins in cancer. Cold Spring Harb
Perspect Med. 6:a0265262016. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Campagne A, Lee MK, Zielinski D, Michaud
A, Le Corre S, Dingli F, Chen H, Shahidian LZ, Vassilev I, Servant
N, et al: BAP1 complex promotes transcription by opposing
PRC1-mediated H2A ubiquitylation. Nat Commun. 10:3482019.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ji Z, Mohammed H, Webber A, Ridsdale J,
Han N, Carroll JS and Sharrocks AD: The forkhead transcription
factor FOXK2 acts as a chromatin targeting factor for the
BAP1-containing histone deubiquitinase complex. Nucleic Acids Res.
42:6232–6242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Abdel-Wahab O and Dey A: The ASXL-BAP1
axis: New factors in myelopoiesis, cancer and epigenetics.
Leukemia. 27:10–15. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Carbone M, Yang H, Pass HI, Krausz T,
Testa JR and Gaudino G: BAP1 and cancer. Nat Rev Cancer.
13:153–159. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Chittock EC, Latwiel S, Miller TC and
Müller CW: Molecular architecture of polycomb repressive complexes.
Biochem Soc Trans. 45:193–205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Okino Y, Machida Y, Frankland-Searby S and
Machida YJ: BRCA1-associated protein 1 (BAP1) deubiquitinase
antagonizes the ubiquitin-mediated activation of FoxK2 target
genes. J Biol Chem. 290:1580–1591. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ivanov GS, Ivanova T, Kurash J, Ivanov A,
Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F III,
Reinberg D and Barlev NA: Methylation-acetylation interplay
activates p53 in response to DNA damage. Mol Cell Biol.
27:6756–6769. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Li G, Margueron R, Hu G, Stokes D, Wang YH
and Reinberg D: Highly compacted chromatin formed in vitro reflects
the dynamics of transcription activation in vivo. Mol Cell.
38:41–53. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wang XW, Guo QQ, Yu Y, Zhou TT, Zhang SY,
Wang Z, Liu JW, Tang J, Jiang XY, Wang SS, et al: The deacetylation
of Foxk2 by Sirt1 reduces chemosensitivity to cisplatin. J Cell Mol
Med. 26:491–506. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Bejerano G, Pheasant M, Makunin I, Stephen
S, Kent WJ, Mattick JS and Haussler D: Ultraconserved elements in
the human genome. Science. 304:1321–1325. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Johnsson P, Lipovich L, Grandér D and
Morris KV: Evolutionary conservation of long non-coding RNAs;
sequence, structure, function. Biochim Biophys Acta.
1840:1063–1071. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Kentwell J, Gundara JS and Sidhu SB:
Noncoding RNAs in endocrine malignancy. Oncologist. 19:483–491.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Lieberman J: Tapping the RNA world for
therapeutics. Nat Struct Mol Biol. 25:357–364. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Gomes CPC, Schroen B, Kuster GM, Robinson
EL, Ford K, Squire IB, Heymans S, Martelli F, Emanueli C and Devaux
Y; EU-CardioRNA COST Action (CA17129), : Regulatory RNAs in heart
failure. Circulation. 141:313–328. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yamamura S, Imai-Sumida M, Tanaka Y and
Dahiya R: Interaction and cross-talk between non-coding RNAs. Cell
Mol Life Sci. 75:467–484. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Fabian MR, Mathonnet G, Sundermeier T,
Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel
J, Doudna JA, et al: Mammalian miRNA RISC recruits CAF1 and PABP to
affect PABP-dependent deadenylation. Mol Cell. 35:868–880. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Min KW, Jo MH, Shin S, Davila S, Zealy RW,
Kang SI, Lloyd LT, Hohng S and Yoon JH: AUF1 facilitates
microRNA-mediated gene silencing. Nucleic Acids Res. 45:6064–6073.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Sun M, Ding J, Li D, Yang G, Cheng Z and
Zhu Q: NUDT21 regulates 3′-UTR length and microRNA-mediated gene
silencing in hepatocellular carcinoma. Cancer Lett. 410:158–168.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Chen D, Wang H, Chen J, Li Z, Li S, Hu Z,
Huang S, Zhao Y and He X: MicroRNA-129-5p regulates glycolysis and
cell proliferation by targeting the glucose transporter SLC2A3 in
gastric cancer cells. Front Pharmacol. 9:5022018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Cui Z, Liu L, Kwame Amevor F, Zhu Q, Wang
Y, Li D, Shu G, Tian Y and Zhao X: High expression of miR-204 in
chicken atrophic ovaries promotes granulosa cell apoptosis and
inhibits autophagy. Front Cell Dev Biol. 8:5800722020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Lin MF, Yang YF, Peng ZP, Zhang MF, Liang
JY, Chen W, Liu XH and Zheng YL: FOXK2, regulted by miR-1271-5p,
promotes cell growth and indicates unfavorable prognosis in
hepatocellular carcinoma. Int J Biochem Cell Biol. 88:155–161.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Chen S, Jiang S, Hu F, Xu Y, Wang T and
Mei Q: Foxk2 inhibits non-small cell lung cancer
epithelial-mesenchymal transition and proliferation through the
repression of different key target genes. Oncol Rep. 37:2335–2347.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Harada K, Baba Y, Ishimoto T, Shigaki H,
Kosumi K, Yoshida N, Watanabe M and Baba H: The role of microRNA in
esophageal squamous cell carcinoma. J Gastroenterol. 51:520–530.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Liu M, Yu J, Wang D, Niu Y, Chen S, Gao P,
Yang Z, Wang H, Zhang J, Zhang C, et al: Epigenetically upregulated
MicroRNA-602 is involved in a negative feedback loop with FOXK2 in
esophageal squamous cell carcinoma. Mol Ther. 27:1796–1809. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Wang D, Wang H, Liu C, Mu X and Cheng S:
Hyperglycemia inhibition of endothelial miR-140-3p mediates
angiogenic dysfunction in diabetes mellitus. J Diabetes
Complications. 33:374–382. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Li S, Zhao L, Li X, Shang G, Gao L, Song Z
and Li T: Mir-204 regulates LPS-induced A549 cell damage by
targeting FOXK2. J Healthc Eng. 2021:74046712021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Patop IL and Kadener S: circRNAs in
cancer. Curr Opin Genet Dev. 48:121–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Zhang M and Xin Y: Circular RNAs: A new
frontier for cancer diagnosis and therapy. J Hematol Oncol.
11:212018. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Hu W, Bi ZY, Chen ZL, Liu C, Li LL, Zhang
F, Zhou Q, Zhu W, Song YY, Zhan BT, et al: Emerging landscape of
circular RNAs in lung cancer. Cancer Lett. 427:18–27. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Hua Q, Chen Y, Liu Y, Li M, Diao Q, Xue H,
Zeng H, Huang L and Jiang Y: Circular RNA 0039411 is involved in
neodymium oxide-induced inflammation and antiproliferation in a
human bronchial epithelial cell line via sponging miR-93-5p.
Toxicol Sci. 170:69–81. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Han D, Wang Y, Wang Y, Dai X, Zhou T, Chen
J, Tao B, Zhang J and Cao F: The tumor-suppressive human circular
RNA CircITCH sponges miR-330-5p to ameliorate doxorubicin-induced
cardiotoxicity through upregulating SIRT6, survivin and SERCA2a.
Circ Res. 127:e108–e125. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Li J, Guo R, Liu Q, Sun J and Wang H:
Circular RNA Circ-ITCH inhibits the malignant behaviors of cervical
cancer by microRNA-93-5p/FOXK2 axis. Reprod Sci. 27:860–868. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Shi X, Liu TT, Yu XN, Balakrishnan A, Zhu
HR, Guo HY, Zhang GC, Bilegsaikhan E, Sun JL, Song GQ, et al:
microRNA-93-5p promotes hepatocellular carcinoma progression via a
microRNA-93-5p/MAP3K2/c-Jun positive feedback circuit. Oncogene.
39:5768–5781. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Ma DH, Li BS, Liu JJ, Xiao YF, Yong X,
Wang SM, Wu YY, Zhu HB, Wang DX and Yang SM: miR-93-5p/IFNAR1 axis
promotes gastric cancer metastasis through activating the STAT3
signaling pathway. Cancer Lett. 408:23–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Chen X, Chen S, Xiu YL, Sun KX, Zong ZH
and Zhao Y: RhoC is a major target of microRNA-93-5P in epithelial
ovarian carcinoma tumorigenesis and progression. Mol Cancer.
14:312015. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Li J, Chu ZP, Han H, Zhang Y, Tian F,
Zhang JQ and Huang XH: Suppression of miR-93-5p inhibits high-risk
HPV-positive cervical cancer progression via targeting of BTG3. Hum
Cell. 32:160–171. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Li Y, Ge YZ, Xu L and Jia R: Circular RNA
ITCH: A novel tumor suppressor in multiple cancers. Life Sci.
254:1171762020. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Sun J, Yin A, Zhang W, Lv J, Liang Y, Li
H, Li Y and Li X: CircUBAP2 inhibits proliferation and metastasis
of clear cell renal cell carcinoma via targeting miR-148a-3p/FOXK2
pathway. Cell Transplant. 29:9636897209257512020. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Xu Q, Cheng D, Li G, Liu Y, Li P, Sun W,
Ma D and Ni C: CircHIPK3 regulates pulmonary fibrosis by
facilitating glycolysis in miR-30a-3p/FOXK2-dependent manner. Int J
Biol Sci. 17:2294–2307. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
St Laurent G, Wahlestedt C and Kapranov P:
The landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Kitagawa M, Kitagawa K, Kotake Y, Niida H
and Ohhata T: Cell cycle regulation by long non-coding RNAs. Cell
Mol Life Sci. 70:4785–4794. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Ballarino M, Morlando M, Fatica A and
Bozzoni I: Non-coding RNAs in muscle differentiation and
musculoskeletal disease. J Clin Invest. 126:2021–2030. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Brazão TF, Johnson JS, Müller J, Heger A,
Ponting CP and Tybulewicz VL: Long noncoding RNAs in B-cell
development and activation. Blood. 128:e10–e19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Delás MJ, Sabin LR, Dolzhenko E, Knott SR,
Munera Maravilla E, Jackson BT, Wild SA, Kovacevic T, Stork EM,
Zhou M, et al: lncRNA requirements for mouse acute myeloid leukemia
and normal differentiation. Elife. 6:e256072017. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Sirey TM, Roberts K, Haerty W,
Bedoya-Reina O, Rogatti-Granados S, Tan JY, Li N, Heather LC,
Carter RN, Cooper S, et al: The long non-coding RNA Cerox1 is a
post transcriptional regulator of mitochondrial complex I catalytic
activity. Elife. 8:e450512019. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Liao D, Liu X, Yuan X, Feng P, Ouyang Z,
Liu Y and Li C: Long non-coding RNA tumor protein 53 target gene 1
promotes cervical cancer development via regulating microRNA-33a-5p
to target forkhead box K2. Cell Cycle. 21:572–584. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Diaz-Lagares A, Crujeiras AB, Lopez-Serra
P, Soler M, Setien F, Goyal A, Sandoval J, Hashimoto Y,
Martinez-Cardús A, Gomez A, et al: Epigenetic inactivation of the
p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc
Natl Acad Sci USA. 113:E7535–E7544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Chen B, Lan J, Xiao Y, Liu P, Guo D, Gu Y,
Song Y, Zhong Q, Ma D, Lei P and Liu Q: Long noncoding RNA TP53TG1
suppresses the growth and metastasis of hepatocellular carcinoma by
regulating the PRDX4/β-catenin pathway. Cancer Lett. 513:75–89.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Pan J, Fang S, Tian H, Zhou C, Zhao X,
Tian H, He J, Shen W, Meng X, Jin X and Gong Z: lncRNA
JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis
of lung cancer by activating Wnt/β-catenin signaling. Mol Cancer.
19:92020. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Lin C, Xiang Y, Sheng J, Liu S, Cui M and
Zhang X: Long non-coding RNA CRNDE promotes malignant progression
of hepatocellular carcinoma through the miR-33a-5p/CDK6 axis. J
Physiol Biochem. 76:469–481. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Sasaki M, Ishikawa T, Ishiguro M, Okazaki
S, Yamauchi S, Kikuchi A, Matsuyama T, Kawada K, Tokunaga M, Uetake
H and Kinugasa Y: The effectiveness of plasma miR-33a-5p as a
predictive biomarker for the efficacy of colorectal cancer
chemotherapy. Oncol Lett. 21:4892021. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Zhao Z, Gao J and Huang S: LncRNA SNHG7
promotes the HCC progression through miR-122-5p/FOXK2 axis. Dig Dis
Sci. 67:925–935. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
van der Heide LP, Wijchers PJ, von Oerthel
L, Burbach JP, Hoekman MF and Smidt MP: FoxK2 is required for
cellular proliferation and survival. J Cell Physiol. 230:1013–1023.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Qian Y, Xia S and Feng Z: Sox9 mediated
transcriptional activation of FOXK2 is critical for colorectal
cancer cells proliferation. Biochem Biophys Res Commun.
483:475–481. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Ji Z, Donaldson IJ, Liu J, Hayes A, Zeef
LA and Sharrocks AD: The forkhead transcription factor FOXK2
promotes AP-1-mediated transcriptional regulation. Mol Cell Biol.
32:385–398. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Meehan RR, Lewis JD, McKay S, Kleiner EL
and Bird AP: Identification of a mammalian protein that binds
specifically to DNA containing methylated CpGs. Cell. 58:499–507.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Hendrich B and Bird A: Identification and
characterization of a family of mammalian methyl-CpG binding
proteins. Mol Cell Biol. 18:6538–6547. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Chen X, Ji Z, Webber A and Sharrocks AD:
Genome-wide binding studies reveal DNA binding specificity
mechanisms and functional interplay amongst forkhead transcription
factors. Nucleic Acids Res. 44:1566–1578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Komorek J, Kuppuswamy M, Subramanian T,
Vijayalingam S, Lomonosova E, Zhao LJ, Mymryk JS, Schmitt K and
Chinnadurai G: Adenovirus type 5 E1A and E6 proteins of low-risk
cutaneous beta-human papillomaviruses suppress cell transformation
through interaction with FOXK1/K2 transcription factors. J Virol.
84:2719–2731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Tang F, Cao F, Lu C, He X, Weng L and Sun
L: Dvl2 facilitates the coordination of NF-κB and Wnt signaling to
promote colitis-associated colorectal progression. Cancer Sci.
113:565–575. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Good MC, Zalatan JG and Lim WA: Scaffold
proteins: Hubs for controlling the flow of cellular information.
Science. 332:680–686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Pan CQ, Sudol M, Sheetz M and Low BC:
Modularity and functional plasticity of scaffold proteins as
p(l)acemakers in cell signaling. Cell Signal. 24:2143–2165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Kagan JC, Magupalli VG and Wu H: SMOCs:
Supramolecular organizing centres that control innate immunity. Nat
Rev Immunol. 14:821–826. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Langeberg LK and Scott JD: Signalling
scaffolds and local organization of cellular behaviour. Nat Rev Mol
Cell Biol. 16:232–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Liu Y, Ao X, Jia Z, Bai XY, Xu Z, Hu G,
Jiang X, Chen M and Wu H: FOXK2 transcription factor suppresses
ERα-positive breast cancer cell growth through down-regulating the
stability of ERα via mechanism involving BRCA1/BARD1. Sci Rep.
5:87962015. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Parsons R, Li GM, Longley MJ, Fang WH,
Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B
and Modrich P: Hypermutability and mismatch repair deficiency in
RER+ tumor cells. Cell. 75:1227–1236. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Fishel R, Lescoe MK, Rao MR, Copeland NG,
Jenkins NA, Garber J, Kane M and Kolodner R: The human mutator gene
homolog MSH2 and its association with hereditary nonpolyposis colon
cancer. Cell. 75:1027–1038. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Leach FS, Nicolaides NC, Papadopoulos N,
Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen LA,
Nyström-Lahti M, et al: Mutations of a mutS homolog in hereditary
nonpolyposis colorectal cancer. Cell. 75:1215–1225. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Katoh M, Igarashi M, Fukuda H, Nakagama H
and Katoh M: Cancer genetics and genomics of human FOX family
genes. Cancer Lett. 328:198–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Michailidou K, Lindström S, Dennis J,
Beesley J, Hui S, Kar S, Lemaçon A, Soucy P, Glubb D, Rostamianfar
A, et al: Association analysis identifies 65 new breast cancer risk
loci. Nature. 551:92–94. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Fujii Y and Nakamura M: FOXK2
transcription factor is a novel G/T-mismatch DNA binding protein. J
Biochem. 147:705–709. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Zhang F, Ma X, Li H, Zhang Y, Li X, Chen
L, Guo G, Gao Y, Gu L, Xie Y, et al: FOXK2 suppresses the malignant
phenotype and induces apoptosis through inhibition of EGFR in
clear-cell renal cell carcinoma. Int J Cancer. 142:2543–2557. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Shan L, Zhou X, Liu X, Wang Y, Su D, Hou
Y, Yu N, Yang C, Liu B, Gao J, et al: FOXK2 elicits massive
transcription repression and suppresses the hypoxic response and
breast cancer carcinogenesis. Cancer Cell. 30:708–722. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Wang B, Zhang X, Wang W, Zhu Z, Tang F,
Wang D, Liu X, Zhuang H and Yan X: Forkhead box K2 inhibits the
proliferation, migration, and invasion of human glioma cells and
predicts a favorable prognosis. Onco Targets Ther. 11:1067–1075.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Li S, Wang P, Ju H, Zhu T, Shi J and Huang
Y: FOXK2 promotes the proliferation of papillary thyroid cancer
cell by down-regulating autophagy. J Cancer. 13:858–868. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Feng H, Jin Z, Liang J, Zhao Q, Zhan L,
Yang Z, Yan J, Kuang J, Cheng X and Qiu W: FOXK2 transcriptionally
activating VEGFA induces apatinib resistance in anaplastic thyroid
cancer through VEGFA/VEGFR1 pathway. Oncogene. 40:6115–6129. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Du F, Qiao C, Li X, Chen Z, Liu H, Wu S,
Hu S, Qiu Z, Qian M, Tian D, et al: Forkhead box K2 promotes human
colorectal cancer metastasis by upregulating ZEB1 and EGFR.
Theranostics. 9:3879–3902. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Baylin SB and Jones PA: Epigenetic
determinants of cancer. Cold Spring Harb Perspect Biol.
8:a0195052016. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Jones PA, Issa JP and Baylin S: Targeting
the cancer epigenome for therapy. Nat Rev Genet. 17:630–641. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Block KI, Gyllenhaal C, Lowe L, Amedei A,
Amin AR, Amin A, Aquilano K, Arbiser J, Arreola A, Arzumanyan A, et
al: Designing a broad-spectrum integrative approach for cancer
prevention and treatment. Semin Cancer Biol. 35 (Suppl
1):S276–S304. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Duijf PHG, Nanayakkara D, Nones K, Srihari
S, Kalimutho M and Khanna KK: Mechanisms of genomic instability in
breast cancer. Trends Mol Med. 25:595–611. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Rusin M, Zajkowicz A and Butkiewicz D:
Resveratrol induces senescence-like growth inhibition of U-2 OS
cells associated with the instability of telomeric DNA and
upregulation of BRCA1. Mech Ageing Dev. 130:528–537. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Falck J, Mailand N, Syljuåsen RG, Bartek J
and Lukas J: The ATM-Chk2-Cdc25A checkpoint pathway guards against
radioresistant DNA synthesis. Nature. 410:842–847. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Matsuoka S, Huang M and Elledge SJ:
Linkage of ATM to cell cycle regulation by the Chk2 protein kinase.
Science. 282:1893–1897. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Mas-Ponte D and Supek F: DNA mismatch
repair promotes APOBEC3-mediated diffuse hypermutation in human
cancers. Nat Genet. 52:958–968. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Barroso-Sousa R, Jain E, Cohen O, Kim D,
Buendia-Buendia J, Winer E, Lin N, Tolaney SM and Wagle N:
Prevalence and mutational determinants of high tumor mutation
burden in breast cancer. Ann Oncol. 31:387–394. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
LeBlanc SJ, Gauer JW, Hao P, Case BC,
Hingorani MM, Weninger KR and Erie DA: Coordinated protein and DNA
conformational changes govern mismatch repair initiation by MutS.
Nucleic Acids Res. 46:10782–10795. 2018.PubMed/NCBI
|
|
209
|
Yu H, Pak H, Hammond-Martel I, Ghram M,
Rodrigue A, Daou S, Barbour H, Corbeil L, Hébert J, Drobetsky E, et
al: Tumor suppressor and deubiquitinase BAP1 promotes DNA
double-strand break repair. Proc Natl Acad Sci USA. 111:285–290.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Kundert K and Fraser JS: DNA-binding
proteins meet their mismatch. Nature. 587:199–200. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Li J, Coïc E, Lee K, Lee CS, Kim JA, Wu Q
and Haber JE: Regulation of budding yeast mating-type switching
donor preference by the FHA domain of Fkh1. PLoS Genet.
8:e10026302012. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Maciejowski J and de Lange T: Telomeres in
cancer: Tumour suppression and genome instability. Nat Rev Mol Cell
Biol. 18:175–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Chakravarti D, LaBella KA and DePinho RA:
Telomeres: History, health and hallmarks of aging. Cell.
184:306–322. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Tang M, Feng X, Pei G, Srivastava M, Wang
C, Chen Z, Li S, Zhang H, Zhao Z, Li X and Chen J: FOXK1
participates in DNA damage response by controlling 53BP1 function.
Cell Rep. 32:1080182020. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Lichtenstein P, Holm NV, Verkasalo PK,
Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A and Hemminki
K: Environmental and heritable factors in the causation of
cancer-analyses of cohorts of twins from Sweden, Denmark, and
Finland. N Engl J Med. 343:78–85. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Berdasco M and Esteller M: Aberrant
epigenetic landscape in cancer: How cellular identity goes awry.
Dev Cell. 19:698–711. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Bitman-Lotan E and Orian A: Nuclear
organization and regulation of the differentiated state. Cell Mol
Life Sci. 78:3141–3158. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Goldberg AD, Allis CD and Bernstein E:
Epigenetics: A landscape takes shape. Cell. 128:635–638. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Nam AS, Chaligne R and Landau DA:
Integrating genetic and non-genetic determinants of cancer
evolution by single-cell multi-omics. Nat Rev Genet. 22:3–18. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Feng Y, Liu X and Pauklin S: 3D chromatin
architecture and epigenetic regulation in cancer stem cells.
Protein Cell. 12:440–454. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Toh TB, Lim JJ and Chow EK: Epigenetics in
cancer stem cells. Mol Cancer. 16:292017. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Dvorak HF: Tumors: Wounds that do not
heal. Similarities between tumor stroma generation and wound
healing. N Engl J Med. 315:1650–1659. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Pagès F, Galon J, Dieu-Nosjean MC, Tartour
E, Sautès-Fridman C and Fridman WH: Immune infiltration in human
tumors: A prognostic factor that should not be ignored. Oncogene.
29:1093–1102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Qian BZ and Pollard JW: Macrophage
diversity enhances tumor progression and metastasis. Cell.
141:39–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
van Bilsen JHM, Dulos R, van Stee MF,
Meima MY, Rouhani Rankouhi T, Neergaard Jacobsen L, Staudt
Kvistgaard A, Garthoff JA, Knippels LMJ, Knipping K, et al: Seeking
windows of opportunity to shape lifelong immune health: A
network-based strategy to predict and prioritize markers of early
life immune modulation. Front Immunol. 11:6442020. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Oh H and Ghosh S: NF-κB: Roles and
regulation in different CD4(+) T-cell subsets. Immunol Rev.
252:41–51. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Blanchett S, Boal-Carvalho I, Layzell S
and Seddon B: NF-κB and extrinsic cell death pathways-entwined
do-or-die decisions for T cells. Trends Immunol. 42:76–88. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Gilmore TD: Introduction to NF-kappaB:
Players, pathways, perspectives. Oncogene. 25:6680–6684. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Karin M and Greten FR: NF-kappaB: Linking
inflammation and immunity to cancer development and progression.
Nat Rev Immunol. 5:749–759. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Li Q, Withoff S and Verma IM:
Inflammation-associated cancer: NF-kappaB is the lynchpin. Trends
Immunol. 26:318–325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
DeNardo DG, Andreu P and Coussens LM:
Interactions between lymphocytes and myeloid cells regulate
pro-versus anti-tumor immunity. Cancer Metastasis Rev. 29:309–316.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Ohnishi S, Ma N, Thanan R, Pinlaor S,
Hammam O, Murata M and Kawanishi S: DNA damage in
inflammation-related carcinogenesis and cancer stem cells. Oxid Med
Cell Longev. 2013:3870142013. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Martin TD, Patel RS, Cook DR, Choi MY,
Patil A, Liang AC, Li MZ, Haigis KM and Elledge SJ: The adaptive
immune system is a major driver of selection for tumor suppressor
gene inactivation. Science. 373:1327–1335. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Sepich-Poore GD, Zitvogel L, Straussman R,
Hasty J, Wargo JA and Knight R: The microbiome and human cancer.
Science. 371:eabc45522021. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Gopalakrishnan V, Helmink BA, Spencer CN,
Reuben A and Wargo JA: The influence of the gut microbiome on
cancer, immunity, and cancer immunotherapy. Cancer Cell.
33:570–580. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Rowe WP, Huebner RJ, Gilmore LK, Parrott
RH and Ward TG: Isolation of a cytopathogenic agent from human
adenoids undergoing spontaneous degeneration in tissue culture.
Proc Soc Exp Biol Med. 84:570–573. 1953. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Trentin JJ, Yabe Y and Taylor G: The quest
for human cancer viruses. Science. 137:835–841. 1962. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Javier RT: Adenovirus type 9 E4 open
reading frame 1 encodes a transforming protein required for the
production of mammary tumors in rats. J Virol. 68:3917–3924. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Sanchez-Prieto R, de Alava E, Palomino T,
Guinea J, Fernandez V, Cebrian S, LLeonart M, Cabello P, Martin P,
San Roman C, et al: An association between viral genes and human
oncogenic alterations: The adenovirus E1A induces the Ewing tumor
fusion transcript EWS-FLI1. Nat Med. 5:1076–1079. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Wang GL, Jiang BH, Rue EA and Semenza GL:
Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS
heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci
USA. 92:5510–5514. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Semenza GL: Hypoxia-inducible factor 1:
Master regulator of O2 homeostasis. Curr Opin Genet Dev. 8:588–594.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Vaupel P and Mayer A: Hypoxia in cancer:
Significance and impact on clinical outcome. Cancer Metastasis Rev.
26:225–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Sui H, Fan S, Liu W, Li Y, Zhang X, Du Y
and Bao H: LINC00028 regulates the development of TGFβ1-treated
human tenon capsule fibroblasts by targeting miR-204-5p. Biochem
Biophys Res Commun. Feb 19–2020.(Epub ahead of print). View Article : Google Scholar
|
|
247
|
Wittstatt J, Weider M, Wegner M and
Reiprich S: MicroRNA miR-204 regulates proliferation and
differentiation of oligodendroglia in culture. Glia. 68:2015–2027.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Zhang J, Su M and Yin Z: Construction of
inflammatory directed polymer micelles and its application in acute
lung injury. AAPS PharmSciTech. 21:2172020. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Wang S, Liu Z, Wang L and Zhang X:
NF-kappaB signaling pathway, inflammation and colorectal cancer.
Cell Mol Immunol. 6:327–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Malumbres M: Cyclin-dependent kinases.
Genome Biol. 15:1222014. View
Article : Google Scholar : PubMed/NCBI
|
|
252
|
Greer EL and Brunet A: FOXO transcription
factors at the interface between longevity and tumor suppression.
Oncogene. 24:7410–7425. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
254
|
Koranda M, Schleiffer A, Endler L and
Ammerer G: Forkhead-like transcription factors recruit Ndd1 to the
chromatin of G2/M-specific promoters. Nature. 406:94–98. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Pic A, Lim FL, Ross SJ, Veal EA, Johnson
AL, Sultan MR, West AG, Johnston LH, Sharrocks AD and Morgan BA:
The forkhead protein Fkh2 is a component of the yeast cell cycle
transcription factor SFF. EMBO J. 19:3750–3761. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Kumar R, Reynolds DM, Shevchenko A,
Shevchenko A, Goldstone SD and Dalton S: Forkhead transcription
factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control
transcription required for M-phase. Curr Biol. 10:896–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Ho KK, Myatt SS and Lam EW: A number of
forks in the path: Cycling with FoxO. Oncogene. 27:2300–2311. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Laoukili J, Stahl M and Medema RH: FoxM1:
At the crossroads of ageing and cancer. Biochim Biophys Acta.
1775:92–102. 2007.PubMed/NCBI
|
|
259
|
Yan J, Xu L, Crawford G, Wang Z and
Burgess SM: The forkhead transcription factor FoxI1 remains bound
to condensed mitotic chromosomes and stably remodels chromatin
structure. Mol Cell Biol. 26:155–168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
260
|
Liang J and Shang Y: Estrogen and cancer.
Annu Rev Physiol. 75:225–240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
261
|
Douglas CC, Johnson SA and Arjmandi BH:
Soy and its isoflavones: The truth behind the science in breast
cancer. Anticancer Agents Med Chem. 13:1178–1187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
262
|
Eroles P, Bosch A, Pérez-Fidalgo JA and
Lluch A: Molecular biology in breast cancer: Intrinsic subtypes and
signaling pathways. Cancer Treat Rev. 38:698–707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
263
|
Nestal de Moraes G, Khongkow P, Gong C,
Yao S, Gomes AR, Ji Z, Kandola N, Delbue D, Man EP, Khoo US, et al:
Forkhead box K2 modulates epirubicin and paclitaxel sensitivity
through FOXO3a in breast cancer. Oncogenesis. 4:e1672015.
View Article : Google Scholar : PubMed/NCBI
|
|
264
|
Zhang Y, Wang Y, Zhao G, Tanner EJ, Adli M
and Matei D: FOXK2 promotes ovarian cancer stemness by regulating
the unfolded protein response pathway. J Clin Invest.
132:e1515912022. View Article : Google Scholar : PubMed/NCBI
|
|
265
|
Amin ARMR, Karpowicz PA, Carey TE, Arbiser
J, Nahta R, Chen ZG, Dong JT, Kucuk O, Khan GN, Huang GS, et al:
Evasion of anti-growth signaling: A key step in tumorigenesis and
potential target for treatment and prophylaxis by natural
compounds. Semin Cancer Biol. 35 (Suppl 1):S55–S77. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
266
|
Milella M, Falcone I, Conciatori F, Cesta
Incani U, Del Curatolo A, Inzerilli N, Nuzzo CM, Vaccaro V, Vari S,
Cognetti F and Ciuffreda L: PTEN: Multiple functions in human
malignant tumors. Front Oncol. 5:242015. View Article : Google Scholar : PubMed/NCBI
|
|
267
|
Trinquand A, Tanguy-Schmidt A, Ben
Abdelali R, Lambert J, Beldjord K, Lengliné E, De Gunzburg N,
Payet-Bornet D, Lhermitte L, Mossafa H, et al: Toward a
NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of
adult T-cell acute lymphoblastic leukemia: A group for research in
adult acute lymphoblastic leukemia study. J Clin Oncol.
31:4333–4342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
268
|
Tesio M, Trinquand A, Macintyre E and
Asnafi V: Oncogenic PTEN functions and models in T-cell
malignancies. Oncogene. 35:3887–3896. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
269
|
Liu Y, Easton J, Shao Y, Maciaszek J, Wang
Z, Wilkinson MR, McCastlain K, Edmonson M, Pounds SB, Shi L, et al:
The genomic landscape of pediatric and young adult T-lineage acute
lymphoblastic leukemia. Nat Genet. 49:1211–1218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
270
|
Wu W, Chen Y, Ye S, Yang H, Yang J and
Quan J: Transcription factor forkhead box K1 regulates miR-32
expression and enhances cell proliferation in colorectal cancer.
Oncol Lett. 21:4072021. View Article : Google Scholar : PubMed/NCBI
|
|
271
|
Wu W, Tan W, Ye S, Zhou Y and Quan J:
Analysis of the promoter region of the human miR-32 gene in
colorectal cancer. Oncol Lett. 17:3743–3750. 2019.PubMed/NCBI
|
|
272
|
Opel D, Schnaiter A, Dodier D, Jovanovic
M, Gerhardinger A, Idler I, Mertens D, Bullinger L, Stilgenbauer S
and Fulda S: Targeting inhibitor of apoptosis proteins by Smac
mimetic elicits cell death in poor prognostic subgroups of chronic
lymphocytic leukemia. Int J Cancer. 137:2959–2970. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
273
|
Mergny JL, Lacroix L, Teulade-Fichou MP,
Hounsou C, Guittat L, Hoarau M, Arimondo PB, Vigneron JP, Lehn JM,
Riou JF, et al: Telomerase inhibitors based on quadruplex ligands
selected by a fluorescence assay. Proc Natl Acad Sci USA.
98:3062–3067. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
274
|
Yin XM, Oltvai ZN and Korsmeyer SJ: BH1
and BH2 domains of Bcl-2 are required for inhibition of apoptosis
and heterodimerization with Bax. Nature. 369:321–323. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
275
|
Youle RJ and Strasser A: The BCL-2 protein
family: Opposing activities that mediate cell death. Nat Rev Mol
Cell Biol. 9:47–59. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
276
|
Asnaghi L, Calastretti A, Bevilacqua A,
D'Agnano I, Gatti G, Canti G, Delia D, Capaccioli S and Nicolin A:
Bcl-2 phosphorylation and apoptosis activated by damaged
microtubules require mTOR and are regulated by Akt. Oncogene.
23:5781–5791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
277
|
Van Der Heide LP, Hoekman MF and Smidt MP:
The ins and outs of FoxO shuttling: Mechanisms of FoxO
translocation and transcriptional regulation. Biochem J.
380:297–309. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
278
|
Folkman J: Angiogenesis in cancer,
vascular, rheumatoid and other disease. Nat Med. 1:27–31. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
279
|
Folkman J: What is the evidence that
tumors are angiogenesis dependent? J Natl Cancer Inst. 82:4–6.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
280
|
Baeriswyl V and Christofori G: The
angiogenic switch in carcinogenesis. Semin Cancer Biol. 19:329–337.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
281
|
Cao Y: Antiangiogenic cancer therapy.
Semin Cancer Biol. 14:139–145. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
282
|
Bergers G and Benjamin LE: Tumorigenesis
and the angiogenic switch. Nat Rev Cancer. 3:401–410. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
283
|
Song Y, Zeng S, Zheng G, Chen D, Li P,
Yang M, Luo K, Yin J, Gu Y, Zhang Z, et al: FOXO3a-driven miRNA
signatures suppresses VEGF-A/NRP1 signaling and breast cancer
metastasis. Oncogene. 40:777–790. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
284
|
Karaman S, Leppänen VM and Alitalo K:
Vascular endothelial growth factor signaling in development and
disease. Development. 145:dev1510192018. View Article : Google Scholar : PubMed/NCBI
|
|
285
|
Ellis LM and Hicklin DJ: VEGF-targeted
therapy: Mechanisms of anti-tumour activity. Nat Rev Cancer.
8:579–591. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
286
|
El Atat O, Fakih A and El-Sibai M: RHOG
activates RAC1 through CDC42 leading to tube formation in vascular
endothelial cells. Cells. 8:1712019. View Article : Google Scholar : PubMed/NCBI
|
|
287
|
Jin Z, Cheng X, Feng H, Kuang J, Yang W,
Peng C, Shen B and Qiu W: Apatinib inhibits angiogenesis via
suppressing Akt/GSK3β/ANG signaling pathway in anaplastic thyroid
cancer. Cell Physiol Biochem. 44:1471–1484. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
288
|
Wang S, Xiao Z, Hong Z, Jiao H, Zhu S,
Zhao Y, Bi J, Qiu J, Zhang D, Yan J, et al: FOXF1 promotes
angiogenesis and accelerates bevacizumab resistance in colorectal
cancer by transcriptionally activating VEGFA. Cancer Lett.
439:78–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
289
|
Sun T, Wang H, Li Q, Qian Z and Shen C:
Forkhead box protein k1 recruits TET1 to act as a tumor suppressor
and is associated with MRI detection. Jpn J Clin Oncol. 46:209–221.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
290
|
Bensinger SJ and Christofk HR: New aspects
of the Warburg effect in cancer cell biology. Semin Cell Dev Biol.
23:352–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
291
|
Palm W and Thompson CB: Nutrient
acquisition strategies of mammalian cells. Nature. 546:234–242.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
292
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
293
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
294
|
Tamada M, Suematsu M and Saya H: Pyruvate
kinase M2: Multiple faces for conferring benefits on cancer cells.
Clin Cancer Res. 18:5554–5561. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
295
|
Waldhart AN, Dykstra H, Peck AS,
Boguslawski EA, Madaj ZB, Wen J, Veldkamp K, Hollowell M, Zheng B,
Cantley LC, et al: Phosphorylation of TXNIP by AKT mediates acute
influx of glucose in response to insulin. Cell Rep. 19:2005–2013.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
296
|
Sheth SS, Castellani LW, Chari S, Wagg C,
Thipphavong CK, Bodnar JS, Tontonoz P, Attie AD, Lopaschuk GD and
Lusis AJ: Thioredoxin-interacting protein deficiency disrupts the
fasting-feeding metabolic transition. J Lipid Res. 46:123–134.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
297
|
Luo W, Hu H, Chang R, Zhong J, Knabel M,
O'Meally R, Cole RN, Pandey A and Semenza GL: Pyruvate kinase M2 is
a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell.
145:732–744. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
298
|
Denko NC: Hypoxia, HIF1 and glucose
metabolism in the solid tumour. Nat Rev Cancer. 8:705–713. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
299
|
Takamura A, Komatsu M, Hara T, Sakamoto A,
Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K and Mizushima N:
Autophagy-deficient mice develop multiple liver tumors. Genes Dev.
25:795–800. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
300
|
Sun T, Li X, Zhang P, Chen WD, Zhang HL,
Li DD, Deng R, Qian XJ, Jiao L, Ji J, et al: Acetylation of beclin
1 inhibits autophagosome maturation and promotes tumour growth. Nat
Commun. 6:72152015. View Article : Google Scholar : PubMed/NCBI
|
|
301
|
Kimmelman AC and White E: Autophagy and
tumor metabolism. Cell Metab. 25:1037–1043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
302
|
Nakatogawa H, Suzuki K, Kamada Y and
Ohsumi Y: Dynamics and diversity in autophagy mechanisms: Lessons
from yeast. Nat Rev Mol Cell Biol. 10:458–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
303
|
Kim J, Kim YC, Fang C, Russell RC, Kim JH,
Fan W, Liu R, Zhong Q and Guan KL: Differential regulation of
distinct Vps34 complexes by AMPK in nutrient stress and autophagy.
Cell. 152:290–303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
304
|
Lambert SA, Jolma A, Campitelli LF, Das
PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT:
The human transcription factors. Cell. 172:650–665. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
305
|
Reiter F, Wienerroither S and Stark A:
Combinatorial function of transcription factors and cofactors. Curr
Opin Genet Dev. 43:73–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
306
|
Wunderlich Z and Mirny LA: Different gene
regulation strategies revealed by analysis of binding motifs.
Trends Genet. 25:434–440. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
307
|
Kuroyanagi H: Fox-1 family of RNA-binding
proteins. Cell Mol Life Sci. 66:3895–3907. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
308
|
Morgunova E and Taipale J: Structural
perspective of cooperative transcription factor binding. Curr Opin
Struct Biol. 47:1–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
309
|
Klemm SL, Shipony Z and Greenleaf WJ:
Chromatin accessibility and the regulatory epigenome. Nat Rev
Genet. 20:207–220. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
310
|
Iwafuchi-Doi M and Zaret KS: Pioneer
transcription factors in cell reprogramming. Genes Dev.
28:2679–2692. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
311
|
Soufi A, Garcia MF, Jaroszewicz A, Osman
N, Pellegrini M and Zaret KS: Pioneer transcription factors target
partial DNA motifs on nucleosomes to initiate reprogramming. Cell.
161:555–568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
312
|
Swinstead EE, Miranda TB, Paakinaho V,
Baek S, Goldstein I, Hawkins M, Karpova TS, Ball D, Mazza D, Lavis
LD, et al: Steroid receptors reprogram FoxA1 occupancy through
dynamic chromatin transitions. Cell. 165:593–605. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
313
|
Hughes AL, Jin Y, Rando OJ and Struhl K: A
functional evolutionary approach to identify determinants of
nucleosome positioning: A unifying model for establishing the
genome-wide pattern. Mol Cell. 48:5–15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
314
|
Struhl K and Segal E: Determinants of
nucleosome positioning. Nat Struct Mol Biol. 20:267–273. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
315
|
Swinstead EE, Paakinaho V, Presman DM and
Hager GL: Pioneer factors and ATP-dependent chromatin remodeling
factors interact dynamically: A new perspective: Multiple
transcription factors can effect chromatin pioneer functions
through dynamic interactions with ATP-dependent chromatin
remodeling factors. Bioessays. 38:1150–1157. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
316
|
Zhu F, Farnung L, Kaasinen E, Sahu B, Yin
Y, Wei B, Dodonova SO, Nitta KR, Morgunova E, Taipale M, et al: The
interaction landscape between transcription factors and the
nucleosome. Nature. 562:76–81. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
317
|
Iwafuchi-Doi M and Zaret KS: Cell fate
control by pioneer transcription factors. Development.
143:1833–1837. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
318
|
Allis CD and Jenuwein T: The molecular
hallmarks of epigenetic control. Nat Rev Genet. 17:487–500. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
319
|
Dann GP, Liszczak GP, Bagert JD, Müller
MM, Nguyen UTT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R, et
al: ISWI chromatin remodellers sense nucleosome modifications to
determine substrate preference. Nature. 548:607–611. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
320
|
Iwafuchi-Doi M, Donahue G, Kakumanu A,
Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH and Zaret KS: The
pioneer transcription factor FoxA maintains an accessible
nucleosome configuration at enhancers for tissue-specific gene
activation. Mol Cell. 62:79–91. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
321
|
Iwafuchi M, Cuesta I, Donahue G, Takenaka
N, Osipovich AB, Magnuson MA, Roder H, Seeholzer SH, Santisteban P
and Zaret KS: Gene network transitions in embryos depend upon
interactions between a pioneer transcription factor and core
histones. Nat Genet. 52:418–427. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
322
|
Cirillo LA, Lin FR, Cuesta I, Friedman D,
Jarnik M and Zaret KS: Opening of compacted chromatin by early
developmental transcription factors HNF3 (FoxA) and GATA-4. Mol
Cell. 9:279–289. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
323
|
Shim EY, Woodcock C and Zaret KS:
Nucleosome positioning by the winged helix transcription factor
HNF3. Genes Dev. 12:5–10. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
324
|
Chen J, Zhang Z, Li L, Chen BC, Revyakin
A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al:
Single-molecule dynamics of enhanceosome assembly in embryonic stem
cells. Cell. 156:1274–1285. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
325
|
Gebhardt JC, Suter DM, Roy R, Zhao ZW,
Chapman AR, Basu S, Maniatis T and Xie XS: Single-molecule imaging
of transcription factor binding to DNA in live mammalian cells. Nat
Methods. 10:421–426. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
326
|
Mazza D, Abernathy A, Golob N, Morisaki T
and McNally JG: A benchmark for chromatin binding measurements in
live cells. Nucleic Acids Res. 40:e1192012. View Article : Google Scholar : PubMed/NCBI
|
|
327
|
Morisaki T, Müller WG, Golob N, Mazza D
and McNally JG: Single-molecule analysis of transcription factor
binding at transcription sites in live cells. Nat Commun.
5:44562014. View Article : Google Scholar : PubMed/NCBI
|
|
328
|
Marchive C, Roudier F, Castaings L,
Bréhaut V, Blondet E, Colot V, Meyer C and Krapp A: Nuclear
retention of the transcription factor NLP7 orchestrates the early
response to nitrate in plants. Nat Commun. 4:17132013. View Article : Google Scholar : PubMed/NCBI
|
|
329
|
Rey G, Cesbron F, Rougemont J, Reinke H,
Brunner M and Naef F: Genome-wide and phase-specific DNA-binding
rhythms of BMAL1 control circadian output functions in mouse liver.
PLoS Biol. 9:e10005952011. View Article : Google Scholar : PubMed/NCBI
|
|
330
|
Futreal PA, Coin L, Marshall M, Down T,
Hubbard T, Wooster R, Rahman N and Stratton MR: A census of human
cancer genes. Nat Rev Cancer. 4:177–183. 2004. View Article : Google Scholar : PubMed/NCBI
|