Introduction
As the most prevalent form of lung cancer, non-small
cell lung cancer (NSCLC) is reported to be one of the deadliest
types of cancer in the world, with 2,206,771 newly diagnosed cases
and 1,796,144 new deaths recorded in 2020 (1,2). In
patients with advanced NSCLC, platinum-based chemotherapy is the
first line treatment, but it is usually cytotoxic and has a short
progression-free survival (PFS) time of 3–5 months and an overall
survival (OS) time of ≤10 months (3). Targeted therapy has been developed to
prevent epidermal growth factor receptor (EGFR) activation
(3–6). EGFR is a transmembrane protein and a
potent transducer of altered signals in tumor cells. There are two
ways of blocking the EGFR: Either by blocking the ligand from
binding to the receptor extracellular domain with anti-EGFR
monoclonal antibodies (cetuximab) or by reversibly binding the
small molecule tyrosine kinase inhibitors (TKIs) to the receptor
intracellular tyrosine kinase domain (4,5).
Thus, the introduction of the first TKI generation drugs, gefitinib
and erlotinib, resulted in markedly higher treatment response rates
(73.7% for TKI compared with 30.7% for chemotherapy), and the
median PFS time increased to 10–13 months for patients with NSCLC
(6–8).
For accurate therapeutic decisions to be made in the
management of patients with NSCLC, it is essential to find
molecular markers that can identify patients who will respond most
effectively to treatment. Promising molecular identifiers include
mutations in the EGFR gene. In patients carrying exon 19
deletions and point mutations in exon 21, a significant clinical
benefit following treatment with TKIs was observed (9,10).
However, acquired resistance in connection with EGFR T790
mutations limited the efficacy of the EGFR-TKI (11,12).
The role of polymorphisms [including single nucleotide
polymorphisms (SNPs) and short tandem CA repeats] of the
EGFR gene as another potential molecular target that
improved clinical outcomes is well-established (13–16).
Recently, a meta-analysis elucidated that among rs712829
(−216G>T), rs11568315 (CA repeat), rs2293347 (D994D) and
rs4947492, −216G>T and variable CA repeat polymorphisms
significantly affected OS and PFS time in gefitinib- or
erlotinib-treated patients with NSCLC (17).
EGFR-TKI therapy is associated with side effects,
primarily in the form of skin or gastrointestinal toxicities (e.g.,
skin rash or diarrhea). Although skin toxicities are not lethal or
dose-limited, they frequently occur with EGFR-TKIs and affect
patient quality of life (18).
Among usual skin toxicities, such as xerosis, pruritus, paronychia,
mucositis and increased growth of eyelashes or facial hair, skin
rash is the most prevalent (19–22).
Notably, patients with NSCLC that develop skin rashes are better
responders to EGFR-TKI therapy and have a longer median overall
survival time (18–23). EGFR SNPs have been examined
in association with survival in NSCLC (17,18);
they may provide insight into therapy outcomes, particularly the
potential side effects associated with TKIs (23–28).
Literature analysis discovered notable inconsistency in previously
published reports. While some studies found associations with
EGFR genotypes and TKI toxicity (23–26),
others did not (27).
Additionally, previous meta-analyses investigated EGFR
mutations, but not EGFR polymorphisms and therapy side
effects in patients with NSCLC (29,30),
or toxicity in relation to radiotherapy (31). With regards to these discrepancies
and the role of EGFR SNPs as potential determinants of
treatment outcome, the aim of the present meta-analysis was to
determine whether the molecular mechanisms involving EGFR
SNPs were associated with EGFR-TKI therapy side effects.
Materials and methods
Search strategy and study
selection
The present study was performed according to the
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
guidelines (PRISMA) (32). The
systematic search for the relevant studies was performed using
electronic databases, PubMed, Scopus and ISI Web of Science.
Searches were performed considering EGFR polymorphisms and
side effects of TKI therapy in patients with NSCLC. The search had
the following retrieval strategy for the PubMed database:
[(‘receptor, epidermal growth factor’ (MeSH Terms) OR EGFR
(All Fields)) AND (gene(tiab) OR ‘polymorphism, genetic’(MeSH
Terms)) AND (‘carcinoma, non-small-cell lung’ (MeSH Terms) OR NSCLC
(All Fields)) AND (‘drug therapy’ (Subheading) OR treatment (All
Fields) OR ‘erlotinib hydrochloride’ (MeSH Terms) OR ‘gefitinib’
(MeSH Terms) OR TKI OR ‘TK inhibitors’ OR ‘tyrosine kinase
inhibitors’ OR ‘Tyrosine-kinase inhibitor’) AND response (All
Fields)) OR Prognosis (MeSH)) OR toxic (MeSH)) OR toxicity (MeSH))
OR side effect (MeSH)) AND (humans (MeSH))]. The Scopus and ISI Web
of Science databases were also searched with necessary
modifications to the PubMed search query. The full search string is
available from the corresponding author upon request. Finally,
additional studies were searched for in the bibliographies of the
selected eligible studies or reviews.
Selection criteria
All studies fulfilling the following inclusion
criteria were eligible: i) Studies published from January 1, 2009,
to February 13, 2019; ii) studies published in English; iii)
studies involving human subjects; iv) patients >18 years old
with histopathologically confirmed NSCLC who received EGFR-TKI
therapy and v) clinical trials or observational studies that
investigated associations between EGFR polymorphisms and any
side effects of TKI therapy. In the systematic review, studies were
excluded based on the following criteria: i) Meta-analyses,
editorials, letters, commentaries, systematic or narrative reviews;
ii) not in the English language; iii) duplicate publications or
studies involving animal or cell experimental models; iv) studies
investigating EGFR polymorphisms and TKI adverse effects but
not reporting their associations; v) single study reports of
EGFR polymorphisms associated with TKI toxicities (skin
toxicity or diarrhea), or other side effects (such as
hepatotoxicity) due to being unable to make comparisons due to the
lack of data from other studies and vi) randomized control trial
(RCT) studies that did not report genotype numbers data, even
though the odds ratio (OR) was reported.
Data extraction
Extracted studies from the electronic databases were
first merged and duplicates were removed. A total of 2 authors (JO
and JT) independently performed a manual search of titles and
abstracts of potentially eligible studies according to the
inclusion and exclusion criteria. Any discrepancies were resolved
by discussion or by consulting the third author (VJ). Finally, the
following data were extracted from the full texts based on the
prior determined datasheet: The first author, year of publication,
country, study type, study period, number of patients, median age,
sex and ethnicity of patients, percentage of smokers, clinical
stage, histology, median follow-up (in months), TKI treatment
dosage, additional therapy, toxicity assessment, adverse effects of
treatment, available EGFR genotype, variant location, SNP
database identifier and number of patients/genotype.
Quality assessment
The Newcastle-Ottawa Quality Assessment Scale (NOS)
(33) for cohort studies and the
Jadad Scale for RCTs (34) were
used to assess the methodological quality of the studies included.
For the NOS scale, the overall maximum quality score was 9 points;
for the Jadad Scale, the score was 5 points. The reviewers (JO and
JT) independently evaluated the quality of the studies with
discrepancies resolved by consensus.
Statistical analysis
When ≥2 studies had available EGFR
polymorphic genotypes associated with TKI therapy side effects,
meta-analysis was conducted. To examine heterogeneity between the
eligible studies, Cochran's Q statistics and I2
statistics were applied. I2 was interpreted as follows:
0, no heterogeneity; 25, low heterogeneity; 50, moderate
heterogeneity and 75%, high heterogeneity (35). The random effect model was used
when there was significant heterogeneity between studies
(P<0.05; I2>50%), otherwise, the fixed effect
model was applied (36).
Galbraith's plot was used to identify potential sources of
heterogeneity (37). If
heterogeneity was present, subgroup analyses of OR were conducted
according to the available EGFR SNPs. The dominant genetic
model (wild-type homozygote vs. heterozygote + mutant homozygote)
of all three EGFR SNPs (rs11568315, rs712829 and rs712830)
was used to calculate OR. The available adverse effects for the
analysis were skin toxicity (skin rash) and gastrointestinal
toxicity (diarrhea). For comparison, the adverse effects were
combined and used as any grade vs. the absence of adverse effects.
Sensitivity analysis was also performed to determine whether the
results would be affected by excluding the study with the smallest
sample size. The publication bias of the enrolled studies was
tested with Begg's and Egger's tests, as well as funnel plots
(38,39). P<0.05 was considered to indicate
a statistically significant difference. STATA software package v.15
(StataCorp LP) was used for all statistical analysis.
Results
Study selection
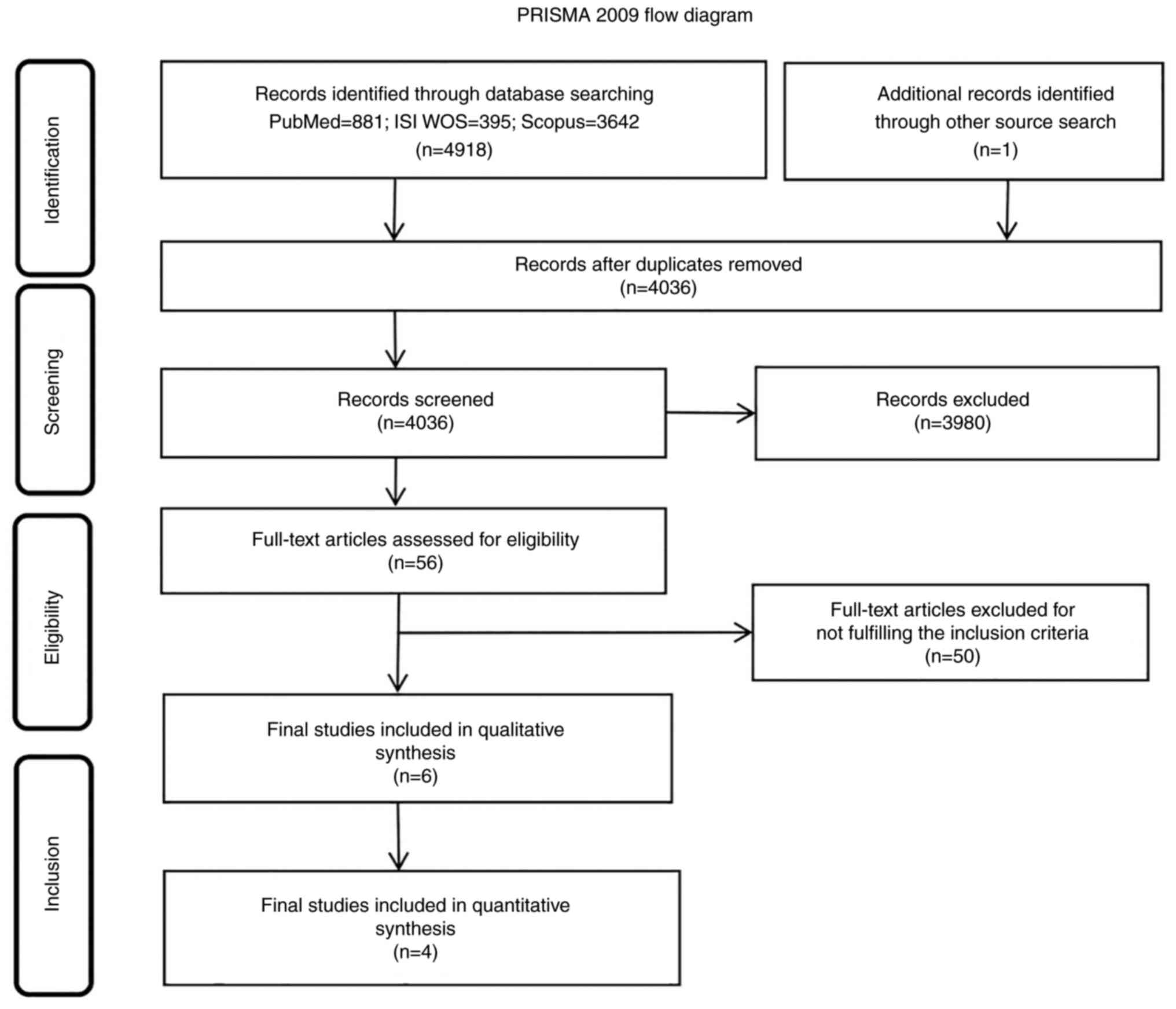
The initial search of databases identified 4,918
results (PubMed, 881; ISI Web of Science, 395; Scopus, 3,642;
Fig. 1). An additional study was
included after reading the bibliographies of the full-text
articles. After merging into the single datasheet and removing
duplicates, 4,036 studies remained. Of these, 3,980 were excluded
and 56 full-text articles were used to assess eligibility. Of these
56 articles, 50 were excluded due to not fulfilling the inclusion
criteria. Finally, 6 clinical trials were included in the
systematic review which contained 1,318 patients with NSCLC. A
total of 4 studies were included in the meta-analysis.
Characteristics of the studies
The six studies from the search included four cohort
studies (23,25,26,28)
containing 316 patients and 2 RCTs (24,27)
containing 1,002 patients. The studies were published from
2009–2017, with sample sizes ranging from 52–760 patients. A total
of two studies were from Asia (Taiwan and China), two from Europe
(Germany and Italy), one from Canada and one RCT was from a
consortium of counties (Canada, Italy, South Korea and Brazil). The
number of male patients in the studies was 33–67%. The percentage
of smokers was 12–76%, while the median age was 56–68 years. Most
of the patients had adenocarcinoma histology and were in clinical
stages IV, IIIB and IIIA (23–28).
Only one study reported a median follow-up of 12 months (23). The EGFR-TKI therapy type for
patients with NSCLC in all examined reports was gefitinib (250
mg/day) and erlotinib (150 mg/day), except in one study where
cetuximab (250 or 500 mg/m2) or panitumumab (6 mg/kg)
was prescribed (25).
Additionally, four studies reported patients that had been
previously treated with cisplatin (24–27).
Adverse effects were skin (rash) and gastrointestinal toxicity
(diarrhea and hepatotoxicity). Toxicity assessment was conducted
using the National Cancer Institute's Common Terminology Criteria
for Adverse Events (40). The
quality of the studies was rated acceptable using the NOS and the
Jadad scale (33,34). Adequacy of follow-up was the lowest
rated aspect. The characteristics and quality assessment of the
included studies are presented in Table I.
 | Table I.Characteristics of the included
studies. |
Table I.
Characteristics of the included
studies.
| First author,
year | Country | Study type | Study period | Patients, n | Median age, years
(range) | Males, % | Ethnicity, % | Smokers, % | Clinical stage
(%)a | Histology (%) | Median follow-up,
months | TKI treatment
(dose) | Additional
therapy | Toxicity
assessment | Adverse effect | Overall quality
score | (Refs.) |
|---|
| Huang et al,
2009 | Taiwan | Cohort | May 2005-September
1, 2006 | 52 | 66 (39–86) | 33 | NR | NR | IIIB (14), IV (87) | Adenoc-arcinoma
(96), Other (4) | 12 | Gefitinib (250
mg/day) | NR | NCI CTCA E
v3.0c | Skin | NOSh:
7 | (23) |
| Giovannetti et
al, 2010 | Italy | Cohort | NR | 96 | 64 (NR) | 57 | NR | 69 | IIIB (9), V (91) | Adenoc-arcinoma
(56), Other (44) | NR | Gefitinib (250
mg/day) | 85% of patients
were previously treated with chemot-herapyb | NCI CTC manual
versiond | Skin, diarrhea | NOS: 7 | (26) |
| Liu et al,
2012 | Canada | RCT Phase III | NR | 242 | 60 (NR) | 64 | East Asian
(6), Other (94) | 76 | IIIB (NR), IV
(NR) | Adenoc-arcinoma
(54), Other (46) | NR | Erlotinib (NR) | 93% patients were
previously treated with chemot-herapy | NCI
v2.0.e | Skin | Jadadi:
4 | (24) |
| Parmar et
al, 2013 | Germany | Cohort | September
2008-November 2010 | 109 | 68 (43–86) | 67 | NR | 12; 49.5
former | IIIA (2); IIIB (11), IV (83), Unknown (5) | NR | NR | Gefitinib (250
mg/day), erlotinib (150 mg/day), cetuximab (250 mg/m2,
with weekly or biweekly or | 31% patients were
previously treated with chemot-herapy panitumumab (6 mg/kg)
biweekly | NCI CTC
v3.0f | Skin | NOS: 6 | (25) |
| Kim et al,
2017 | Canada, Italy,
South Korea and Brazil | RCT Phase III | NR | 760 | 62 (56–66) | 66 | Caucasian (96),
East Asian (3), Other (1) | 26; 53 former | IIIB (11), IV (89) | Adenoc-arcinoma
(56), Other (44) | NR | Erlotinib (NR) | 50% of patients
were treated with erlotinib, followed by cisplatin and gemcita bine
at progression; 50% were treated with cisplatin and gemcita bine,
followed by erlotinib at progression | NR | Skin, diarrhea | Jadad: 3 | (27) |
| Ma et al,
2017 | China | Cohort | 2011-2014 | 59 | 56 (31–77) | 49 | NR | 61 | IIIB or IV
(64) | Adenoc-arcinoma
(90), Other (10) | NR | Gefitinib (250
mg/day) | NR | CTCA E
v4.0g | Skin, diarrhea,
hepat otoxicity | NOS: 7 | (28) |
A total of nine EGFR SNPs (rs11568315,
rs712829, rs712830, rs2227983, rs2075102, rs2293347, rs11977388,
rs4947492 and rs884225) relative to TKI toxicity was identified in
the literature search, which were provided by seven studies
(23–28,41).
Of these, four studies reported exact numbers of patients/genotype
of EGFR SNPs (rs11568315, rs712829 and rs712830) associated
with TKI-caused toxicity and were included in quantitative
synthesis (23,25,26,28).
Genotypes for all EGFR SNPs for the meta-analysis were
merged according to the dominant genetic model. The data for the
EGFR SNPs genotype and skin toxicity, diarrhea or
hepatotoxicity caused by EGFR TKI therapy are presented in
Table II. Certain studies or sets
of data were excluded from further analyses. The reasons are
outlined below.
 | Table II.EGFR genotypes and adverse
effects of tyrosine kinase inhibitor therapy. |
Table II.
EGFR genotypes and adverse
effects of tyrosine kinase inhibitor therapy.
| A, Patients with
skin toxicity (n=880, 78.36%) |
|---|
|
|---|
| dbSNP-ID | Variant type,
location, and/or consequence | First author,
year | Genotyping platform
used | Genotype | Total number of
patients, n | Patients with skin
toxicity, n | ORa | 95% CIa | za |
P-valuea | (Refs.) |
|---|
| rs11568315 | Intron
variant, | Huang et al,
2009 | PCR and | SS | 7 | 5 | Ref. |
| 2.13 | 0.032 | (23) |
|
|
g.55020560_5502 |
| direct | SL | 26 | 8 | 6.87 | 1.1734- | 70 | 6 |
|
|
| 0561AC[n] |
| sequencing | LL | 19 | 4 | 5 | 40.2797 |
|
|
|
|
|
| Parmar et
al, 2013 | Real-Time | SS | 22 | 19 | Ref. |
| 1.23 | 0.217 | (25) |
|
|
|
| PCR and | SL | 58 | 41 | 2.27 | 0.6156- | 30 | 7 |
|
|
|
|
| 16-capillary
electrophoresis | LL | 29 | 23 | 6 | 8.4150 |
|
|
|
|
|
|
| or KASPar |
|
|
|
|
|
|
|
|
|
|
| Giovannetti et
al, 2010 | TaqMan | SS | 27 | 18 | Ref. |
| 1.71 | 0.087 | (26) |
|
|
|
| PCR | SL-LL | 60 | 28 | 2.28 | 0.8863- | 00 | 2 |
|
|
|
|
|
|
|
|
| 57 | 5.8947 |
|
|
|
|
|
| Ma et al,
2017 | Sequenom | SS | 6 | 6 | NA | NA | NA | NA | (28) |
|
|
|
| Massarray | SL | 20 | 14 |
|
|
|
|
|
|
|
|
| system | LL | 16 | 10 |
|
|
|
|
|
|
|
| Kim et al,
2017 | Sanger | SS | 38 | 28 | Ref. |
| 0.15 | 0.874 | (27) |
|
|
|
| sequencing and | SL-LL | 96 | 72 | 0.93 | 0.3961- | 80 | 7 |
|
|
|
|
| Taqman PCR |
|
|
|
| 2.1994 |
|
|
|
|
|
| Liu et al,
2012 | PCR and direct
sequencing | LL-S- | NR | NR | 0.60 | 0.2–1.9 | NA | NA | (24) |
| rs712829 | 5′ UTR
variant, | Huang et al,
2009 | PCR and direct | GG | 45 | 14 | Ref. |
| 0.61 | 0.540 | (23) |
|
| g.5031G>T,
- |
| sequencing | GT | 5 | 1 | 0.60 | 0.1186- | 20 | 6 |
|
|
| 216G>T |
|
| TT | 2 | 2 | 22 | 3.0567 |
|
|
|
|
| | Parmar et
al, 2013 | Real-Time | GG | 49 | 33 | Ref. |
| 1.92 | 0.054 | (25) |
|
|
|
| PCR and | GT | 48 | 39 | 0.41 | 0.1670- | 00 | 9 |
|
|
|
|
| 16-capillary
electrophoresis | TT | 12 | 11 | 25 | 1.0188 |
|
|
|
|
|
|
| or KASPar |
|
|
|
|
|
|
|
|
|
|
| Giovannetti et
al, 2010 | TaqMan PCR | GG | 30 | 19 | Ref. |
| 1.41 | 0.158 | (26) |
|
|
|
|
| GT-TT | 57 | 27 | 1.91 | 0.7752- | 00 | 7 |
|
|
|
|
|
|
|
|
| 92 | 4.7513 |
|
|
|
|
|
| Kim et al,
2017 | Sanger sequencing
and | GG | 40 | 32 | Ref. |
| 0.91 | 0.358 | (27) |
|
|
|
| Taqman PCR | GT-TT | 83 | 60 | 1.53 | 0.62–3.82 | 90 | 2 |
|
|
|
| Liu et al,
2012 | PCR and direct
sequencing | GT-GG | NR | NR | 2.00 | 0.5–8.3 | NA | NA | (24) |
| rs712830 | 5′ UTR
variant, | Parmar et
al, 2013 | Real-Time | CC | 79 | 59 | Ref. |
| 0.58 | 0.561 | (25) |
|
| g.5056A>C,
- |
| PCR and | CA | 30 | 24 | 0.73 | 0.2637- | 00 | 7 |
|
|
| 191C>A |
| 16-capillary
electrophoresis or KASPar | AA | 0 | 0 | 75 | 2.0624 |
|
|
|
|
|
| Giovannetti et
al, 2010 | TaqMan | CC | 72 | 36 | Ref. |
| 1.16 | 0.245 | (26) |
|
|
|
| PCR | CA-AA | 15 | 10 | 0.5 | 0.1554- | 20 | 1 |
|
|
|
|
|
|
|
|
|
| 1.6089 |
|
|
|
|
|
| Kim et al,
2017 | Sanger sequencing
and | CC | 100 | 72 | Ref. |
| 1.44 | 0.147 | (27) |
|
|
|
| Taqman PCR | CA- AA | 23 | 20 | 0.38 | 0.1062- | 80 | 7 |
|
|
|
|
|
|
|
|
| 57 | 1.4007 |
|
|
|
|
|
| Liu et al,
2012 | PCR and direct
sequencing | CA-CC | NR | NR | 1.00 | 0.1–6.8 | NA | NA | (24) |
|
rs2227983b | Missense
variant, | Parmar et
al, 2013 | Real-Time PCR
and | GG | 57 | 49 | Ref. |
| 2.45 | 0.014 | (25) |
|
| (1562G>A, | | 16-capillary
electrophoresis | GA | 47 | 29 | 3.24 | 1.2657- | 10 | 2 |
|
|
| R497K) |
| or KASPar | AA | 5 | 5 | 26 | 8.3073 |
|
|
|
|
|
| Giovannetti et
al, 2010 | TaqMan | GG-GA | 75 | 38 | 0.58 | 0.1585- | 0.79 | 0.425 | (26) |
|
|
|
| PCR |
|
|
| 69 | 2.1734 | 80 | 0 |
|
|
|
|
|
| AA | 11 | 7 | Ref. |
|
|
|
|
|
|
| Huang et
al, | PCR and | GG | 12 | 4 | Ref. |
| 0.16 | 0.867 | (23) |
|
|
| 2009 | direct | GA | 28 | 8 | 1.12 | 0.2832- | 70 | 1 |
|
|
|
|
| sequencing | AA | 12 | 5 | 5 | 4.4695 |
|
|
|
|
| B, Patients with
diarrhea (n=233, 20.75%) |
|
|
|
|
|
|
|
|
Patients |
|
|
|
|
|
|
| Variant
type, |
|
|
| Total | with |
|
|
|
|
|
|
| location,
and/or |
|
|
| number
of |
diarrhea |
|
|
|
|
|
|
dbSNP-ID |
consequence | First author,
year | Genotyping
platform used |
Genotype | patients,
n | toxicity,
n | ORa | 95%
CIa | za |
P-valuea | (Refs.) |
|
| rs11568315 | Intron
variant, | Giovannetti et
al, 2010 | TaqMan assay | SS | 26 | 13 | Ref. |
| 0.76 | 0.446 | (26) |
|
| g.55020560_550 |
|
| SS-LL | 49 | 20 | 1.45 | 0.5569- | 10 | 6 |
|
|
| 20561AC[n] |
|
|
|
|
|
| 3.7751 |
|
|
|
|
|
| Ma et al,
2017 | Sequenom | SS | 6 | 2 | Ref. |
| 0.50 | 0.612 | (28) |
|
|
|
| Massarray | SL | 20 | 9 | 0.62 | 0.1012- | 60 | 8 |
|
|
|
|
| system | LL | 16 | 7 | 5 | 3.8585 |
|
|
|
|
|
| Kim et al,
2017 | Sanger sequencing
and | SS | 38 | 15 | Ref. |
| 0.12 | 0.902 | (27) |
|
|
|
| Taqman PCR | SL-LL | 96 | 39 | 0.95 | 0.4424- | 20 | 5 |
|
|
|
|
|
|
|
|
|
| 2.0535 |
|
|
|
| rs712829 | 5′ UTR
variant, | Kim et al,
2017 | Sanger sequencing
and | GG | 40 | 18 | Ref. |
| 0.94 | 0.346 | (27) |
|
| g.5031G>T,
- |
| Taqman PCR | GT-TT | 83 | 30 | 1.44 | 0.6711- | 10 | 6 |
|
|
| 216G>T |
|
|
|
|
| 55 | 3.1131 |
|
|
|
| rs712830 | 5′ UTR
variant, | Kim et al,
2017 | Sanger sequencing
and | CC | 100 | 37 | Ref. |
| 0.95 | 0.339 | (27) |
|
| g.5056A>C,
- |
| Taqman PCR | CA-AA | 23 | 11 | 0.64 | 0.2570- | 50 | 4 |
|
|
| 191C>A |
| |
|
|
|
| 1.597 |
|
|
|
|
rs2227983b | Missense
variant, | Giovannetti et
al, 2010 | TaqMan PCR | GG-GA | 74 | 26 | 2.76 | 0.7163- | 1.47 | 0.139 | (26) |
|
| (1562G>A, |
|
|
|
|
| 92 | 10.7057 | 60 | 8 |
|
|
| R497K) |
|
| AA | 10 | 6 | Ref. |
|
|
|
|
|
| C, Patients with
hepatotoxicity (n=10, 0.89%) |
|
|
|
|
|
|
|
|
Patients |
|
|
|
|
|
|
| Variant
type, |
|
|
| Total | with |
|
|
|
|
|
|
| location,
and/or |
|
Genotyping |
| number
of |
hepatoto |
|
|
|
|
|
|
dbSNP-ID |
consequence | Author,
year | platform
used |
Genotype | patients,
n | xicity,
n | ORa | 95%
CIa | za |
P-valuea | (Refs.) |
|
| rs1156 | Intron
variant, | Ma et al,
2017 | Sequenom
Massarray | SS | 6 | 1 | Ref. |
| 0.40 | 0.682 | (28) |
| 8315 | g.55020560_550 |
| system | SL | 21 | 6 | 0.62 | 0.0640- | 90 | 7 |
|
|
| 20561AC[n] |
|
| LL | 16 | 3 | 22 | 6.0507 |
|
|
|
Due to lack of data from other studies for
comparison, one study was excluded from further analysis, although
the study did identify rs884225, a 3′-untranslated region variant
c.*774T>C associated with EGFR TKI toxicity (41). Similarly, data for the
hepatotoxicity, as well as for four EGFR SNPs were not
included (rs2075102, rs2293347, rs11977388 and rs4947492) (27). An RCT study reported pre-calculated
ORs for three examined EGFR SNPs, but without precise
numbers of patients per genotype (24), and was therefore included in the
qualitative, but not the quantitative, analysis. Consequently,
another RCT study was excluded from quantitative analysis (27) to avoid comparison between
observational and RCT studies, and prevent potential heterogeneity.
If zeros present in patient genotype numbers for rs2227983 and
rs11568315 interfered with computation or if there were
insufficient data for analysis, studies were excluded from
quantitative synthesis (23–25,28).
In the literature search, three other studies explored EGFR
TKI toxicity, as well as EGFR SNPs, but failed to find any
associations between them (42–44).
Side effects of EGFR-TKIs
There was notable inconsistency in the scientific
reports describing the association between EGFR SNPs and TKI
toxicity in patients with NSCLC. While some articles reported
evidence of association with skin toxicity (23–26)
or severe diarrhea (26), one
article found no association with skin or gastrointestinal
toxicities (27).
In patients treated with gefitinib, there was a
significant association between SS genotype in CA repeat
polymorphism and early G2/3 skin rash (P=0.031), meaning these
patients were more likely to develop early G2/3 rash (23). Despite this, the EGFR
polymorphisms −216G>T and R521K were not associated with early
G2/3 rash (P=0.104 and P=0.720, respectively) (23). Another study on patients treated
with erlotinib found a similar result for three EGFR
polymorphisms and skin rash: −191C>A, −216G>T and CA repeats
(P=1.00, P=0.13 and P=0.34, respectively) (24). Only the EGFR −216/-191GC
haplotype was associated with the appearance of skin rash (P=0.029)
(25). Nevertheless, the absence
of association with skin rash was evidenced for the single
EGFR SNPs −191C>A (P=0.62), −216G>T (P=0.147) and CA
repeats (P=0.36) (25). Diarrhea
was a less frequent toxicity and no significant association between
any of the EGFR SNPs or haplotypes with diarrhea was
observed (25). However, in
another study, severe diarrhea occurred in patients with NSCLC
treated with gefitinib, most frequently in carriers of −191C>A,
−191A>A (P<0.0001) and −216G>G genotypes (P<0.01)
(26). There was no significant
association between EGFR CA repeat polymorphisms and skin or
gastrointestinal toxicity, nor any association between EGFR
polymorphism and skin toxicity (26).
Toxicity
The most common adverse effects associated with TKIs
in treating advanced NSCLC were skin toxicity (78.36%) and diarrhea
(20.75%; Table II). One study
reported hepatotoxicity (0.89%) (27), but the study was excluded since
there were no data from other studies for comparison. Among the
studies available for the meta-analysis, gefitinib (250 mg/day) or
erlotinib (150 mg/day) were predominant. For data available for
genotypes relative to skin toxicity, the OR and 95% confidence
interval (CI) were calculated and their effect was summarized in
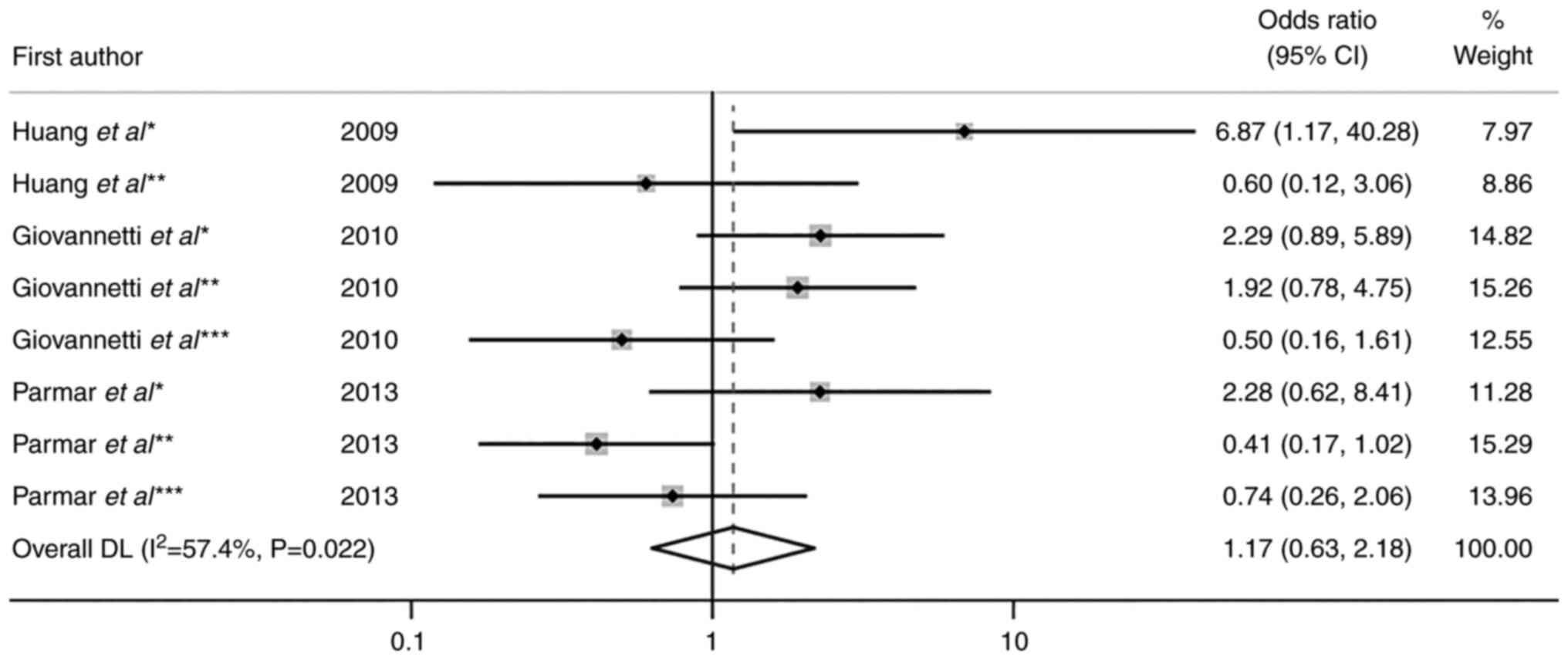
the quantitative synthesis (Fig.
2). This involved three EGFR SNPs (rs11568315, rs712829
and rs712830) obtained from three studies for skin toxicity
(23,25,26).
Of these, two examined rs11568315 and diarrhea (26,28).
The pooled OR for skin toxicity and rs11568315, rs712829 and
rs712830 was 1.17 (95% CI, 0.63–2.18; P=0.616) with moderate
heterogeneity (I2=57.4%; P=0.022; Fig. 2).
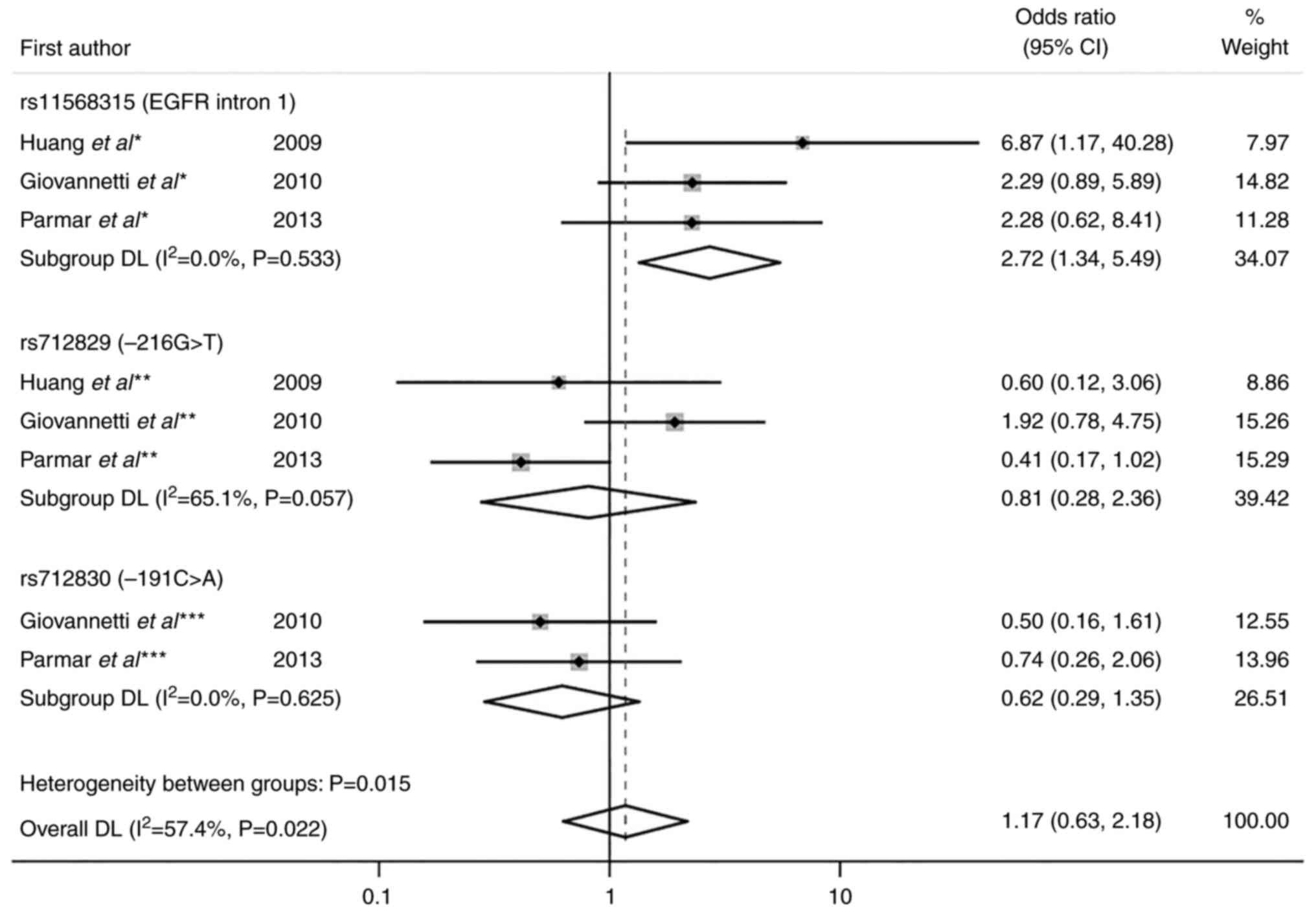
To test heterogeneity, random effect model and
subgroup analyses were performed. Subgroup analysis for skin
toxicity showed that the OR for rs11568315 was 2.72 (95% CI,
1.34–5.49; P=0.005) without heterogeneity (I2=0.0%;
P=0.533). A statistically significant result for skin toxicity
(z=2.785 and P=0.005) were obtained under the dominant genetic
model for rs11568315 (SS vs. SL + LL). OR for rs712829 was 0.81
(95% CI, 0.28–2.36; P=0.700) with moderate heterogeneity
(I2=65.1%; P=0.057) and OR for rs712830 was 0.62 (95%
CI, 0.29–1.35; P=0.229) with no heterogeneity (I2=0.0%,
P=0.625; Fig. 3). Data for
diarrhea was only available for rs11568315 (data not shown). It was
tested in two studies using the fixed effects model (OR, 1.21; 95%
CI, 0.52–2.82), with no evidence of heterogeneity
(I2=0.0%; P=0.422) and without statistically significant
association (P=0.661) (26,28).
Publication bias and sensitivity
analysis
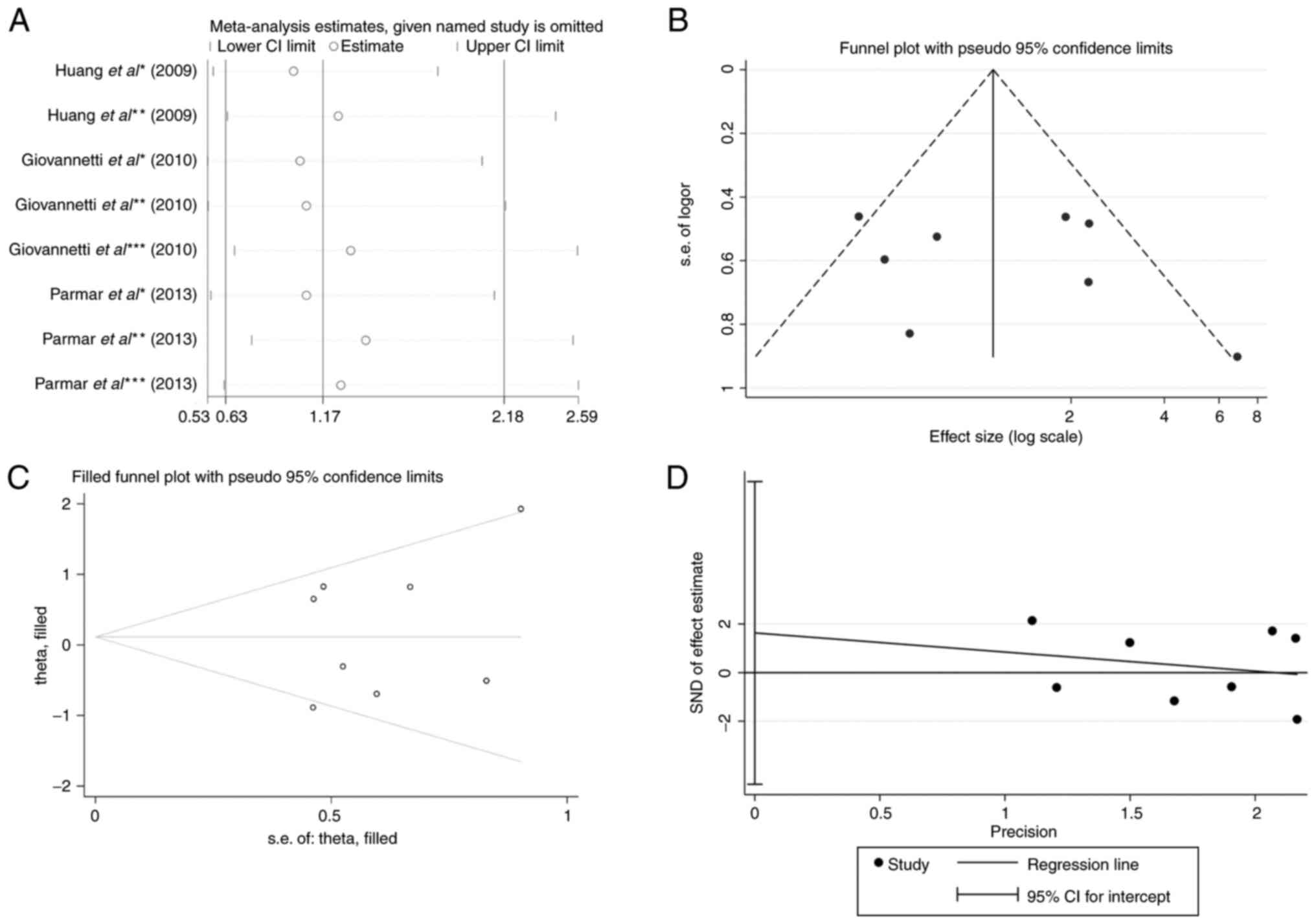
The results of the sensitivity analysis regarding
toxicity were relatively stable. The overall effective size was not
affected by exclusion of each of the studies, even by a study with
a smaller sample size (OR, 6.87; 95% CI, 1.17–2.28; Fig. 4A) (23). The funnel plot for EGFR SNPs
and TKI skin toxicity in patients with NSCLC was roughly symmetric
(Fig. 4B). Begg's funnel plot and
Egger's regression test (P=0.545) were used to test the publication
bias, but no significantly different results were obtained
(Fig. 4C and D). Similarly, the
funnel plot revealed no potential bias of rs11568315 (CA repeat)
and TKI-caused diarrhea (data not shown). Galbraith's plot
identified no source of heterogeneity relative to skin toxicity
(data not shown).
Discussion
The present systematic review involved the analysis
of two RCTs and four cohort studies to test the association of
EGFR polymorphisms with the potential toxicity of TKI
therapy regimens in patients with NSCLC. A total of 1,123 patients
per genotype were observed with any TKI-associated toxicity. A
total of four studies provided data for the meta-analysis (23,25,26,28),
while six were involved in quality analysis (23–28).
In the literature search, nine EGFR SNPs relative to TKI
toxicity were identified: rs11568315, rs712829, rs712830,
rs2227983, rs2075102, rs2293347, rs11977388, rs4947492 and rs884225
(23–28,41).
Of these, enough data was available for three (rs11568315, rs712829
and rs712830) to be included in the meta-analysis.
Our recent meta-analysis showed that CA repeat
polymorphism and −216G>T significantly affected survival in
patients with NSCLC treated with TKI (17). In light of the inconsistency of
previous reports (23–31), the present meta-analysis was
performed to extend our previous findings and to analyze the effect
of the EGFR polymorphisms and TKIs on NSCLC.
A number of studies in the present review founded an
association between some EGFR polymorphisms and TKI-related
skin toxicity (23,25,26)
or diarrhea (26,27). Contradictory results were also
detected in previous studies published before 2009 (45–47).
The most common TKI adverse effects in the present meta-analysis
were skin toxicity and diarrhea, which were 78.36% and 20.75%,
respectively (concerning any grade of toxicity vs. no toxicity).
They were separately analyzed in the meta-analysis. The pooled OR
for three EGFR SNPs (rs11568315, rs712829 and rs712830) was
1.17 (95% CI, 0.63–2.18) without a statistically significant
overall effect on skin toxicity (P=0.616). In further analysis, a
moderate overall heterogeneity (I2=57.4%; P=0.022) was
observed. To explore the heterogeneity further, a subgroup analysis
was performed and the random effect model was applied. The subgroup
analysis involved three EGFR SNPs (rs11568315, rs712829 and
rs712830) concerning skin toxicity (23,25,26).
The source of heterogeneity (I2=65.1%; P=0.057) was
likely due to the −216G>T (rs712829) polymorphism (26). The CIs were overlapping the line of
no effect for all three studies, suggesting the result was not
statistically significant. A total of two studies favored the GG
genotype for −216G>T (rs712829) and skin toxicity (23,25),
which contrasts the GT+TT genotype favored by Giovannetti et
al (26). Most importantly,
there was no heterogeneity for the other two SNPs examined
(rs11568315, I2=0.0%; P=0.533; rs712830,
I2=0.0%; P=0.625).
Chemotherapy is the first-line treatment for
patients with NSCLC, but notable improvements in the response rate
have been observed following the application of the TKIs gefitinib
and erlotinib (6–8,48).
However, resistance, as well as adverse effects, is common in this
therapy regimen. Typical side effects of the drugs used in NSCLC
treatment (for both monoclonal antibodies and small molecule TKIs)
are skin rash and diarrhea (49,50).
Since the EGFR is commonly affected by somatic mutations in altered
neoplastic cells and the EGFR gene is highly polymorphic,
the potential cause of those toxic manifestations of drugs may be
EGFR genetic variability (11,13–16).
SNPs or microsatellite tandem repeats are typically found in the
EGFR promoter region and intron 1. These notably affect
EGFR gene expression and may mediate response to TKI
therapy. A CA single sequence repeat polymorphism (rs11568315) is
located in EGFR intron 1 and it usually comprises 14–21
variable short tandem repeats. The shorter allele is associated
with increased EGFR expression and carriers of this
polymorphism are better responders to TKI therapy and have
prolonged overall survival time (13,14,47,51–54).
Among side effects of TKI therapy, typical skin rash
manifestations were in the form of papules and pustules on the
scalp, face, neck and upper trunk. To the best of our knowledge,
the mechanism of skin rash development has not yet been elucidated.
One hypothesis is that there is a genetic susceptibility for rash
development, where altered EGFR expression alters the TKI
response (11,13–16,28).
Another is that poor vascularization of the tumor tissue and drug
concentrations at a level that does not inhibit tumor growth may
cause a skin rash by over-saturation of EGFR (18,55).
There is evidence of a significant association between skin rash
and an improved outcome in patients with NSCLC (18,56).
Skin rash has been reported to be a predictor of tumor response
(25) and EGFR CA repeat is
a valuable predictor of early G2/3 rash (23). Previous studies have reported that
lower number CA repeat carriers develop skin toxicity when treated
with gefitinib (13,23,57),
while other studies did not (24–28,45,47,53).
In another study where patients with NSCLC were treated with
erlotinib, SL allele length was associated with a higher risk of
diarrhea (46). In the present
meta-analysis, the pooled OR values for CA repeats (rs11568315) and
skin toxicity were 2.72 (95% CI, 1.34–5.49). A significant
association with skin toxicity was evident under the dominant
genetic model. Namely, heterozygote and long alleles (SL + LL) or
prevalently long CA repeat carriers were more likely to develop
TKI-related skin toxicity (P=0.005). However, it is probable that
short CA carriers would be less likely develop skin rash. There was
no association between CA repeats and diarrhea (P=0.661).
The other well-examined SNPs, −191C>A (rs712830)
and −216G>T (rs712829) polymorphisms, are located in the
EGFR promoter region and are associated with enhanced
EGFR mRNA expression (14,53,58).
A previous meta-analysis revealed that any genotype with T allele
for −216G>T showed an association with higher response and
disease control rates and longer PFS and OS times than GG
homozygote carriers (59). Another
meta-analysis elucidated that the −216G>T polymorphism
significantly affected OS and PFS times in patients with NSCLC
treated with gefitinib or erlotinib (17). Both of the aforementioned
polymorphisms are reported to be in linkage disequilibrium (D'=1.0)
(25). Considering their
association with toxicities, a study reported the haplotypes
showing association with the appearance of skin rash (25). Other studies reported that the T
allele of −216G>T was significantly associated with high-risk of
TKI-induced skin rash (24) or
diarrhea (14). An association
between −216G>T and −191C>A with grade >1 diarrhea has
also been reported (26). Contrary
to these findings, the present meta-analysis observed no
significant association for EGFR SNPs −216G>T and
−191C>A with skin toxicity (P=0.700 and P=0.229, respectively),
which agreed with the findings from previous studies (23,24,27,45).
The advantage of the present meta-analysis over
previous meta-analyses is the examination of commonly used TKIs
(such as gefitinib and erlotinib) and their toxicity, while other
studies involved a single therapeutic agent (60,61).
Other analyses investigated EGFR mutations, not EGFR
polymorphisms (29,30) or their association with toxicity
(59), or they only investigated
toxicity in relation to radiotherapy (31). The present meta-analysis has
certain limitations. Firstly, some studies included in the analysis
had small sample sizes so consistent conclusions could not be
obtained, as with the RCTs that have larger sample sizes. A total
of two RCTs were excluded from the meta-analysis (24,27),
since one study alone did not provide enough data to be tested. In
particular, exact numbers of patients with NSCLC with each
EGFR SNP genotype were not reported and the study presented
only pre-calculated data for OR (24). The aforementioned RCTs obtained low
NOS scores in the quality analysis, whereas the other studies
included in the present meta-analysis had relatively good scores.
Also, the results of the present meta-analysis were not adjusted
for other factors (i.e., demographic factors), although the
majority of the studies did not report ethnicity for the examined
subjects. Potential bias in the results may be due to the absence
of a consensus in the literature of an exact number of CA repeats
when reporting short vs. long alleles. The linkage disequilibrium
between examined SNPs was not taken into account and selection bias
may be present.
In conclusion, the results of the present
meta-analysis revealed that out of nine EGFR SNPs related to
TKI side effects, rs11568315, rs712829 and rs712830 were associated
with skin toxicity. NSCLC carriers of long CA repeats (rs11568315,
SL + LL) were more likely to develop TKI-associated skin toxicity
than short CA repeats (rs11568315, SS). To establish clear
inter-individual benefits of TKI therapy, future RCTs that include
a broader genetic panel are required to determine genetic
susceptibility to TKI-induced toxicity in patients with NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Ministry of Education,
Science and Technological Development, Serbia (no. OI 175056 under
agreement number 451-03-68/2022-14/200378).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
Conceptualization and supervision of the study were
conducted by VJ. The selection of papers, formal analysis,
investigation and writing were conducted by JO. The acquisition of
data was conducted by JT. All authors have read and approved the
final manuscript. JO, JT and VJ confirm the authenticity of all the
raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics. CA Cancer J Clin. 71:7–33. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang ZY, Liu L, Mao C, Wu XY, Huang YF, Hu
XF and Tang JL: Chemotherapy with cetuximab versus chemotherapy
alone for chemotherapy-naive advanced non-small cell lung cancer.
Cochrane Database Syst Rev. 17:CD0099482014.PubMed/NCBI
|
|
4
|
Hirsch FR, Herbst RS, Olsen C, Chansky K,
Crowley J, Kelly K, Franklin WA, Bunn PA Jr, Varella-Garcia M and
Gandara DR: Increased EGFR gene copy number detected by fluorescent
in situ hybridization predicts outcome in non-small-cell lung
cancer patients treated with cetuximab and chemotherapy. J Clin
Oncol. 26:3351–3357. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pirker R, Pereira JR, von Pawel J,
Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WEE,
Paz-Ares L, Störkel S, et al: EGFR expression as a predictor of
survival for first-line chemotherapy plus cetuximab in patients
with advanced non-small-cell lung cancer: Analysis of data from the
phase 3 FLEX study. Lancet Oncol. 13:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee CK, Davies L, Wu YL, Mitsudomi T,
Inoue A, Rosell R, Zhou C, Nakagawa K, Thongprasert S, Fukuoka M,
et al: Gefitinib or erlotinib vs chemotherapy for EGFR
mutation-positive lung cancer: Individual patient data
meta-analysis of overall survival. J Natl Cancer Inst.
109:djw2792017. View Article : Google Scholar
|
|
9
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yun CH, Mengwasser KE, Toms AV, Woo MS,
Greulich H, Wong KK, Meyerson M and Eck MJ: The T790M mutation in
EGFR kinase causes drug resistance by increasing the affinity for
ATP. Proc Natl Acad Sci U S A. 105:2070–2075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goldberg SB, Oxnard GR, Digumarthy S,
Muzikansky A, Jackman DM, Lennes IT and Sequist LV: Chemotherapy
with Erlotinib or chemotherapy alone in advanced non-small cell
lung cancer with acquired resistance to EGFR tyrosine kinase
inhibitors. Oncologist. 18:1214–1220. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Amador ML, Oppenheimer D, Perea S, Maitra
A, Cusatis G, Iacobuzio-Donahue C, Baker SD, Ashfaq R, Takimoto C,
Forastiere A and Hidalgo M: An epidermal growth factor receptor
intron 1 polymorphism mediates response to epidermal growth factor
receptor inhibitors. Cancer Res. 64:9139–9143. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu G, Gurubhagavatula S, Zhou W, Wang Z,
Yeap BY, Asomaning K, Su L, Heist R, Lynch TJ and Christiani DC:
Epidermal growth factor receptor polymorphisms and clinical
outcomes in non-small-cell lung cancer patients treated with
gefitinib. Pharmacogenomics J. 8:129–138. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jung M, Cho BC, Lee CH, Park HS, Kang YS,
Kim SK, Chang J, Kim DJ, Rha SY, Kim JH and Lee JH: EGFR
polymorphism as a predictor of clinical outcome in advanced lung
cancer patients treated with EGFR-TKI. Yonsei Med J. 53:1128–1135.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Winther-Larsen A, Ebert EB, Meldgaard P
and Sorensen BS: EGFR gene polymorphism predicts improved outcome
in patients with EGFR mutation-positive non-small cell lung cancer
treated with erlotinib. Clin Lung Cancer. 20:161–166.e161. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jurisic V, Vukovic V, Obradovic J,
Gulyaeva LF, Kushlinskii NE and Djordjević N: EGFR polymorphism and
survival of NSCLC patients treated with TKIs: A systematic review
and meta-analysis. J Oncol. 2020:19732412020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peréz-Soler R and Saltz L: Cutaneous
adverse effects with HER1/EGFR-targeted agents: Is there a silver
lining? J Clin Oncol. 23:5235–5246. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Segaert S and Van Cutsem E: Clinical
signs, pathophysiology and management of skin toxicity during
therapy with epidermal growth factor receptor inhibitors. Ann
Oncol. 16:1425–1433. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agero AL, Dusza SW, Benvenuto-Andrade C,
Busam KJ, Myskowski P and Halpern AC: Dermatologic side effects
associated with the epidermal growth factor receptor inhibitors. J
Am Acad Dermatol. 55:657–670. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu JC, Sadeghi P, Pinter-Brown LC, Yashar
S and Chiu MW: Cutaneous side effects of epidermal growth factor
receptor inhibitors: Clinical presentation, pathogenesis, and
management. J Am Acad Dermatol. 56:317–326. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bianchini D, Jayanth A, Chua YJ and
Cunningham D: Epidermal growth factor receptor inhibitor-related
skin toxicity: Mechanisms, treatment, and its potential role as a
predictive marker. Clin Colorectal Cancer. 7:33–43. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang CL, Yang CH, Yeh KH, Hu FC, Chen KY,
Shih JY, Lin ZZ, Yu CJ, Cheng AL and Yang PC: EGFR intron 1
dinucleotide repeat polymorphism is associated with the occurrence
of skin rash with gefitinib treatment. Lung Cancer. 64:346–351.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu G, Cheng D, Ding K, Maitre AL, Liu N,
Patel D, Chen Z, Seymour L, Shepherd FA and Tsao MS:
Pharmacogenetic analysis of BR.21, a placebo-controlled randomized
phase III clinical trial of erlotinib in advanced non-small cell
lung cancer. J Thorac Oncol. 7:316–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parmar S, Schumann C, Rüdiger S, Boeck S,
Heinemann V, Kächele V, Seeringer A, Paul T, Seufferlein T and
Stingl JC: Pharmacogenetic predictors for EGFR-inhibitor-associated
skin toxicity. Pharmacogenomics J. 13:181–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Giovannetti E, Zucali PA, Peters GJ,
Cortesi F, D'Incecco A, Smit EF, Falcone A, Burgers JA, Santoro A,
Danesi R, et al: Association of polymorphisms in AKT1 and EGFR with
clinical outcome and toxicity in non-small cell lung cancer
patients treated with gefitinib. Mol Cancer Ther. 9:581–593. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim L, Saieg M, Di Maio M, Gallo C, Butts
C, Ciardiello F, Feld R, Cheng D, Gebbia V, Burgio MA, et al:
Biomarker analysis of the phase 3 TORCH trial for first line
erlotinib versus chemotherapy in advanced non-small cell lung
cancer patients. Oncotarget. 8:57528–57536. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Xin S, Huang M, Yang Y, Zhu C, Zhao
H, Zhang Y, Chen L, Zhao Y, Li J, et al: Determinants of Gefitinib
toxicity in advanced non-small cell lung cancer (NSCLC): A
pharmacogenomic study of metabolic enzymes and transporters.
Pharmacogenomics J. 17:325–330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Petrelli F, Borgonovo K, Cabiddu M, Lonati
V and Barni S: Relationship between skin rash and outcome in
non-small-cell lung cancer patients treated with anti-EGFR tyrosine
kinase inhibitors: A literature-based meta-analysis of 24 trials.
Lung Cancer. 78:8–15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wo H, He J, Zhao Y, Yu H, Chen F and Yi H:
The efficacy and toxicity of gefitinib in treating non-small cell
lung cancer: A meta-analysis of 19 randomized clinical trials. J
Cancer. 9:1455–1465. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Xu Y, Tang W and Liu L: Efficacy
and safety of radiotherapy plus EGFR-TKIs in NSCLC patients with
brain metastases: A meta-analysis of published data. Transl Oncol.
11:1119–1127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Moher D, Liberati A, Tetzlaff J and Altman
DG: Preferred reporting items for systematic reviews and
meta-analyses: The PRISMA statement. BMJ. 339:b25352009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ottawa Hospital Research Institute, . NOS
Manual. Available from:. http://www.ohri.ca/programs/clinical_epidemiology/nos_manual.pdfMarch
8–2021
|
|
34
|
Jadad AR, Moore RA, Carroll D, Jenkinson
C, Reynolds DJ, Gavaghan DJ and McQuay HJ: Assessing the quality of
reports of randomized clinical trials: Is blinding necessary?
Control Clin Trials. 17:1–12. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Higgins JPT, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials revisited. Contemp Clin Trials. 45:139–145.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Galbraith RF: A note on graphical
presentation of estimated odds ratios from several clinical trials.
Stat Med. 7:889–894. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Egger M, Smith GD, Schneider M and Minder
C: Bias in meta-analysis detected by a simple, graphical test. BMJ.
315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
National Cancer Institue, . Common
Terminology Criteria for Adverse Events (CTCAE) Version 4.
Available at. https://stacks.stanford.edu/file/druid:nw036fx4646/CTCAE_4.03_2010-06-14_QuickReference_5×7.pdfFebruary
12–2021
|
|
41
|
Ruan Y, Jiang J, Guo L, Li Y, Huang H,
Shen L, Luan M, Li M, Du H, Ma C, et al: Genetic association of
curative and adverse reactions to tyrosine kinase inhibitors in
chinese advanced non-small cell lung cancer patients. Sci Rep.
6:233682016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nie Q, Yang XN, An SJ, Zhang XC, Yang JJ,
Zhong WJ, Liao RQ, Chen ZH, Su J, Xie Z and Wu YL: CYP1A1*2A
polymorphism as a predictor of clinical outcome in advanced lung
cancer patients treated with EGFR-TKI and its combined effects with
EGFR intron 1 (CA)n polymorphism. Eur J Cancer. 47:1962–1970. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tiseo M, Rossi G, Capelletti M, Sartori G,
Spiritelli E, Marchioni A, Bozzetti C, De Palma G, Lagrasta C,
Campanini N, et al: Predictors of gefitinib outcomes in advanced
non-small cell lung cancer (NSCLC): Study of a comprehensive panel
of molecular markers. Lung Cancer. 67:355–360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chilingirova N, Hammoudeh Z, Balabanski L,
Ivanov S, Vazharova R, Nikolova D, Kurteva G, Toncheva D and
Chilingirov P: TruSight cancer sequencing panel reveals
pharmacogenetic variants associated with sensitivity to
chemotherapy in lung cancer. Memo-Magazine of European Medical
Oncology. 9:30–38. 2016. View Article : Google Scholar
|
|
45
|
Cusatis G, Gregorc V, Li J, Spreafico A,
Ingersoll RG, Verweij J, Ludovini V, Villa E, Hidalgo M, Sparreboom
A and Baker SD: Pharmacogenetics of ABCG2 and adverse reactions to
gefitinib. J Natl Cancer Inst. 98:1739–1742. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rudin CM, Liu W, Desai A, Karrison T,
Jiang X, Janisch L, Das S, Ramirez J, Poonkuzhali B, Schuetz E, et
al: Pharmacogenomic and pharmacokinetic determinants of erlotinib
toxicity. J Clin Oncol. 26:1119–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tiseo M, Capelletti M, De Palma G,
Franciosi V, Cavazzoni A, Mozzoni P, Alfieri RR, Goldoni M, Galetti
M, Bortesi B, et al: Epidermal growth factor receptor intron-1
polymorphism predicts gefitinib outcome in advanced non-small cell
lung cancer. J Thorac Oncol. 3:1104–1111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cappuzzo F, Ligorio C, Jänne PA, Toschi L,
Rossi E, Trisolini R, Paioli D, Holmes AJ, Magrini E, Finocchiaro
G, et al: Prospective study of gefitinib in epidermal growth factor
receptor fluorescence in situ
hybridization-positive/phospho-Akt-positive or never smoker
patients with advanced non-small-cell lung cancer: The ONCOBELL
trial. J Clin Oncol. 25:2248–2255. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
O'Byrne KJ, Bondarenko I, Barrios C,
Eschbach C, Martens U, Kortsik YH, Celik I, Stroh C and Pirker R:
Molecular and clinical predictors of outcome for cetuximab in
non-small cell lung cancer (NSCLC): Data from the FLEX study. J
Clin Oncol. 27:8007. 2009. View Article : Google Scholar
|
|
50
|
Su X, Lacouture ME, Jia Y and Wu S: Risk
of high-grade skin rash in cancer patients treated with
cetuximab-an antibody against epidermal growth factor receptor:
Systemic review and meta-analysis. Oncology. 77:124–133. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dubey S, Stephenson P, Levy DE, Miller JA,
Keller SM, Schiller JH, Johnson DH and Kolesar JM; Eastern
Cooperative Oncology Group, : EGFR dinucleotide repeat polymorphism
as a prognostic indicator in non-small cell lung cancer. J Thorac
Oncol. 1:406–412. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gebhardt F, Bürger H and Brandt B:
Modulation of EGFR gene transcription by secondary structures, a
polymorphic repetitive sequence and mutations-a link between
genetics and epigenetics. Histol Histopathol. 15:929–936.
2000.PubMed/NCBI
|
|
53
|
Han SW, Jeon YK, Lee KH, Keam B, Hwang PG,
Oh DY, Lee SH, Kim DW, Im SA, Chung DH, et al: Intron 1 CA
dinucleotide repeat polymorphism and mutations of epidermal growth
factor receptor and gefitinib responsiveness in non-small-cell lung
cancer. Pharmacogenet Genomics. 17:313–319. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ichihara S, Toyooka S, Fujiwara Y, Hotta
K, Shigematsu H, Tokumo M, Soh J, Asano H, Ichimura K, Aoe K, et
al: The impact of epidermal growth factor receptor gene status on
gefitinib-treated Japanese patients with non-small-cell lung
cancer. Int J Cancer. 120:1239–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dienstmann R, Braña I, Rodon J and
Tabernero J: Toxicity as a biomarker of efficacy of molecular
targeted therapies: Focus on EGFR and VEGF inhibiting anticancer
drugs. Oncologist. 16:1729–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wacker B, Nagrani T, Weinberg J, Witt K,
Clark G and Cagnoni PJ: Correlation between development of rash and
efficacy in patients treated with the epidermal growth factor
receptor tyrosine kinase inhibitor erlotinib in two large phase III
studies. Clin Cancer Res. 13:3913–3921. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Perea S, Oppenheimer D, Amador M, Cusati
G, Baker S, Takimoto C, Maitra A, Iocobuzio-Donahue C and Hidalgo
M: Genotypic bases of EGFR inhibitors pharmacological actions. J
Clin Oncol. 22:3005. 2004. View Article : Google Scholar
|
|
58
|
Liu W, Innocenti F, Wu MH, Desai AA, Dolan
EM, Cook EH Jr and Ratain MJ: A functional common polymorphism in a
Sp1 recognition site of the epidermal growth factor receptor gene
promoter. Cancer Res. 65:46–53. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang HX, Tang Y, Wang L, Wei SX, Liu QX,
Li F and Yuan XL: EGFR-216 G/T polymorphism as a predictor of
clinical outcomes in advanced non-small cell lung cancer patients
treated with EGFR-TKIs: A meta-analysis. Int J Clin Exper Med.
9:10273–10280. 2016.
|
|
60
|
Biaoxue R, Hua L, Wenlong G and Shuanying
Y: Efficacy and safety of icotinib in treating non-small cell lung
cancer: A systematic evaluation and meta-analysis based on 15
studies. Oncotarget. 7:86902–86913. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yi L, Fan J, Qian R and Luo P: Efficacy
and safety of osimertinib in treating EGFR-mutated advanced NSCLC:
A meta-analysis. Int J Cancer. 145:284–294. 2019. View Article : Google Scholar : PubMed/NCBI
|