Introduction
Vestibular schwannoma (VS) is the most common tumor
in the cerebellopontine angle (1).
These tumors arise from the myelin-producing Schwann cells within
the vestibular branch of the eighth (VIII) cranial nerve (2). Although these tumors are
histologically benign, VSs most commonly present with hearing loss
and tinnitus and can also cause dizziness, facial paralysis, other
cranial neuropathies, and even death from brainstem compression.
Most of VS cases are sporadic and unilateral, while bilateral
tumors are associated with Neurofibromatosis type 2 (NF2).
The number of sporadic VS cases diagnosed has
increased dramatically over the last decade, mainly due to the
widespread adoption of, and easy access to magnetic resonance
imaging (MRI). The prevalence of sporadic VS is estimated to be 1
in 2,000 adults and 1 in every 500 persons aged 70 years or older
(3). The prevalence of NF2 patients
is estimated to be 1 in 56,161 (4).
Despite the increasing number of VS patients, the traditional
microsurgery treatment for VS via the retrosigmoid, middle cranial
fossa, or translabyrinthine approach has decreased (5). Serial imaging is now the most common
initial strategy for most patients with small- or medium-sized VS
tumors (6).
The NF2 gene located on chromosome 22 at
22q12.2, which encodes for the tumor suppressor protein Merlin, is
considered to be the most important gene for understanding VS
pathobiology in both sporadic VSs and familial (NF2 related) VSs
(7–9). Various pathways, including those
involving the NF2 gene, have been investigated for the
treatment of VSs (10). However,
NF2 alone cannot fully explain all tumor development and
growth. Therefore, it is essential to analyze genetic information
obtained from tumor tissue to gain novel insights into the
pathobiology of VSs and devise effective treatment strategies. In
this study, we focused on key tumor suppressor genes and
oncogenes.
To date, obtaining small tumor samples has become
increasingly difficult as nonsurgical management options have been
shown to produce good results in slow-growing and smaller VS
tumors, especially those with a total length of <2 cm (11). Tumor size does not correlate with
degree of hearing loss (12–14),
as small tumors limited to the auditory canal can still result in
hearing loss (15). Patients in the
small tumor (<1.0 cm) group presented more frequently with
tinnitus and sudden hearing loss than those in the large tumor
(>4.0 cm) group, suggesting differences in the biological
characteristics of large and small tumors (16).
Thus, it is important to investigate the genetic
profile of small tumors to understand the pathobiology of VS. To
elucidate the genetic landscape of small VSs, especially the genes
related to tumor genesis and tumor growth, this study involved a
comprehensive genomic analysis of all the exons from the key tumor
suppressor genes and oncogenes obtained from 10 sporadic, small VS
samples collected through surgical intervention.
Materials and methods
Patients
We enrolled a total of 15 patients who underwent
surgical resection for small unilateral VS (<2 cm total length)
between October 2012 and March 2017 at the Kindai University
Hospital. Ten out of 15 samples yielded genomic DNA of adequate
quality for library preparation. Five samples were excluded from
this study because of the inadequate quality of genomic DNA. These
tumors were classified using the Koos acoustic neuroma grading
system (17) and 10 of these
samples were then analyzed using next generation sequencing (NGS)
in this study. Our study was approved by the ethics committee at
the Kindai University Hospital (approval no. 29-015).
DNA extraction and library
preparation
Genomic DNA was extracted from the FFPE tumor
tissues using the QIAamp DNA FFPE Tissue Kit (Qiagen). This was
then used as the template in four independent multiplex polymerase
chain reactions (PCRs) which amplified 15,992 regions across 409
cancer-related genes (Table SI).
These amplifications were completed using an Ion AmpliSeq Library
Kit 2.0 with Comprehensive Cancer Panel (Thermo Fisher Scientific).
After multiplex PCR, Ion Xpress Barcode Adapters (Thermo Fisher
Scientific) were ligated to the PCR products, which were then
purified using Agencourt AMPure XP beads (Beckman Coulter).
Purified libraries were pooled and sequenced on an Ion Torrent
Proton instrument using an Ion Proton Hi-Q Sequencing Kit, and an
Ion PI v3 Chip (all from Thermo Fisher Scientific). DNA sequencing
data were then accessed using the Torrent Suite v.5.0 program
(Thermo Fisher Scientific).
Sequence analysis
DNA sequencing data were accessed using the Torrent
Suite ver. 5.2 software program and the reads were aligned against
the hg19 human reference genome. Variants were called using the
variant caller ver. 5.2. and the VCF files were annotated using CLC
Genomics Workbench ver. 9.0.1 (Qiagen). Variants were filtered
based on quality (quality score >100, read depth >50 and
allele frequency >5%) and were manually checked using the
integrative genomics viewer (IGV; Broad Institute). Germline
mutations were excluded using the use of the Human Genetic
Variation Database (http://www.genome.med.kyoto-u.ac.jp/SnpDB) and the
Exome Aggregation Consortium (ExAC). The pathogenicity of the
mutations was predicted using SIFT (18), PolyPhen2 (19), FATHMM (20), and CScape (21).
Results
Clinical characteristics of the
patients
The clinical characteristics of the 10 patients
profiled in this study are summarized in Table I. This cohort was comprised of five
men and five women, with a mean age of 61.4 years (median 62.5,
range 40–73). Tumor size ranged from those limited to the internal
auditory canal to a maximum diameter of 15 mm. A total of 6
patients presented with Koos grade I (intracanalicular) tumors
while the other 4 had Koos grade II (small tumor with protrusion
into the CPA; no contact with the brainstem) tumors. The middle
cranial fossa approach was used in five patients, and
translabyrinthine approach was used in the other five patients.
Complete resection of the tumor was reported for nine patients, and
a 99% resection was performed in one patient.
 | Table I.Clinical characteristics of ten
patients with sporadic vestibular schwannoma. |
Table I.
Clinical characteristics of ten
patients with sporadic vestibular schwannoma.
| No. | Sex | Age, years | Side | Tumor size, mm | Koos grade | HL affected side,
dB HL | HL intact side, dB
HL | Surgery | Resection mode |
|---|
| 1 | M | 58 | L | 15 | II | 63 | 16 | MCF | Total |
| 2 | M | 65 | L | IAC
(6.0)a | I | 48 | 25 |
TL | Total |
| 3 | M | 60 | L | 6.8 | II | 101 | 10 |
TL | Total |
| 4 | F | 72 | L | 7.7 | II | 89 | 48 |
TL | Total |
| 5 | F | 48 | L | 7.2 | II | 38 | 26 | MCF | Total |
| 6 | F | 40 | R | 6.2 | II | 6 | 9 | MCF | Total |
| 7 | F | 69 | L | 8.3 | II | 56 | 30 | MCF | Total |
| 8 | M | 73 | R | IAC
(6.6)a | I | 36 | 36 | MCF | 99% |
| 9 | F | 58 | R | IAC
(6.0)a | I | s.o | 14 |
TL | Total |
| 10 | M | 71 | R | IAC
(5.9)a | I | 86 | 13 |
TL | Total |
Genetic landscape of our small
vestibular schwannoma samples
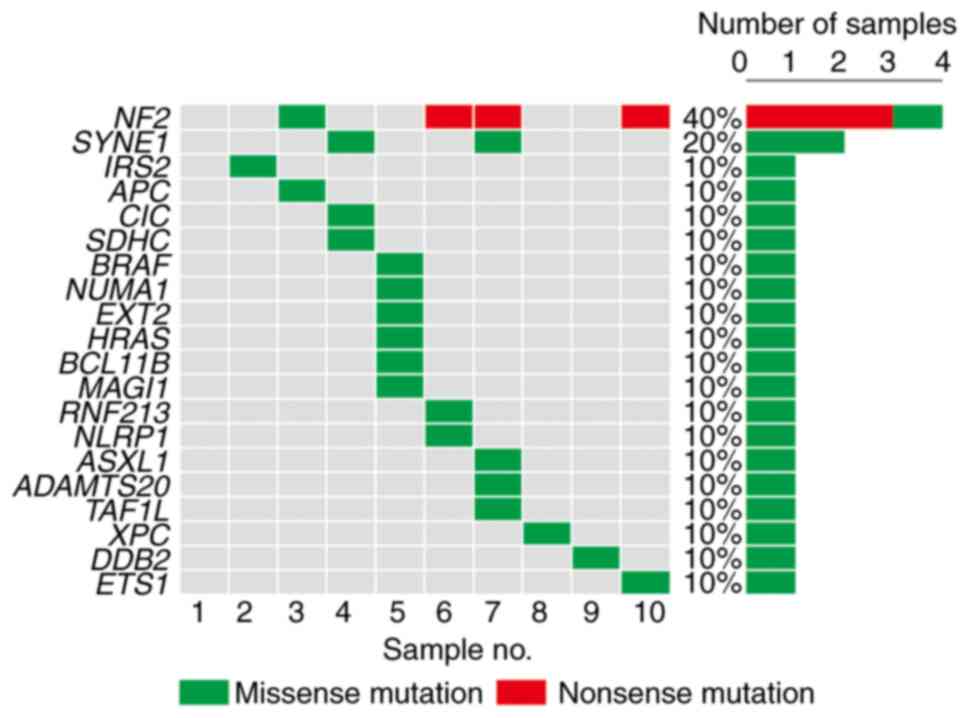
NF2 (40%) was the most frequently mutated
cancer-related gene in these VS samples (Fig. 1) and was followed by SYNE1
(20%), IRS2 (10%), APC (10%), CIC (10%),
SDHC (10%), BRAF (10%), NUMA1 (10%),
EXT2 (105), HRAS (10%), BCL11B (10%),
MAGI1 (10%), RNF123 (10%), NLRP1 (10%),
ASXL1 (10%), ADAMTS20 (10%), TAF1L (10%),
XPC (10%), DDB2 (10%), and ETS1 (10%). Most of
these mutations were predicted to be pathogenic (Table II). Closer evaluation of the cases
harboring mutations in the NF2 revealed that one case
harbored a missense mutation in exon 4 (case no. 3) and its
pathogenicity was predicted as ‘damaging’ by SIFT, ‘probably
damaging’ by Polyphen2, ‘pathogenic’ by FATHMM-XF, and ‘oncogenic
with high confidence’ by CScape. Two cases harbored a nonsense
mutation in exon 2 (case no. 7 and 10) and its pathogenicity was
predicted as ‘benign’ by FATHMM-XF, and ‘oncogenic with high
confidence’ by CScape. One case harbored a nonsense mutation in
exon 6 (case no. 6) and its pathogenicity was predicted as
‘pathogenic’ by FATHMM-XF, and ‘oncogenic with high confidence’ by
CScape.
 | Table II.The pathogenicity of the somatic
mutations in sporadic vestibular schwannoma using SIFT, PolyPhen2,
FATHMM and CScape. |
Table II.
The pathogenicity of the somatic
mutations in sporadic vestibular schwannoma using SIFT, PolyPhen2,
FATHMM and CScape.
| Gene symbol | Effect | Sample |
Genomepositiona | cDNA change | Amino acid
change | VAF, % | SIFT | Polyphen2 | FATHMM-XF | CScape |
|---|
| NF2 | Missense | 3 | chr22:30038274 | c.447G>T | p.Lys149Asn | 33.2 | D | Prob-D | P | O (HC) |
| NF2 | Non-sense | 6 | chr22:30070927 | c.1443C>G | p.Tyr481* | 36.3 | NA | NA | P | O (HC) |
| NF2 | Non-sense | 7,10 | chr22:30032794 | c.169C>T | p.Arg57* | 46.5/61.2 | NA | NA | B | O (HC) |
| SYNE1 | Missense | 4 | chr6:152774819 | c.2929G>A | p.Ala977Thr | 45.5 | T | B | B | O |
| SYNE1 | Missense | 7 |
chr6:152658141-152658142 |
c.12362_12363delAGinsGT | p.Lys4121Ser | 47.7 | NA | NA | B | O |
| IRS2 | Missense | 2 |
chr13:110434998 | c.3403G>A | p.Val1135Ile | 46.1 | D | Prob-D | P | B |
| APC | Missense | 3 | chr5:112177052 | c.5761G>A | p.Gly1921Ser | 44.4 | T | B | B | B |
| CIC | Missense | 4 | chr19:42796489 | c.5773C>T | p.Pro1925Ser | 50.5 | T | Prob-D | B | O |
| SDHC | Missense | 4 | chr1:161298236 | c.128A>G | p.Asn43Ser | 50.2 | T | B | B | B |
| BRAF | Missense | 5 | chr7:140500192 | c.950C>T | p.Ser317Phe | 52.4 | T | B | B | O |
| NUMA1 | Missense | 5 | chr11:71726642 | c.1907C>T | p.Thr636Ile | 51.3 | T | B | B | B |
| EXT2 | Missense | 5 | chr11:44129270 | c.107C>T | p.Ala36Val | 48.5 | NA | B | P | O (HC) |
| HRAS | Missense | 5 | chr11:532688 | c.518C>T | p.Pro173Leu | 48.6 | D | B | P | B |
| BCL11B | Missense | 5 | chr14:99697747 | c.575C>T | p.Ser192Leu | 47.1 | T | Poss-D | P | O (HC) |
| MAGI1 | Missense | 5 | chr3:65464432 | c.592A>G | p.Ser198Gly | 48.0 | T | B | B | O |
| RNF213 | Missense | 6 | chr17:78343382 | c.12236A>C | p.Lys4079Thr | 51.5 | D | NA | B | B |
| NLRP1 | Missense | 6 | chr17:5462461 | c.1555G>A | p.Val519Met | 42.8 | D | B | B (HC) | B |
| ASXL1 | Missense | 7 | chr20:31025035 | c.4520C>T | p.Ala1507Val | 55.7 | T | B | B | B |
|
ADAMTS20 | Missense | 7 | chr12:43826171 | c.3032G>A | p.Arg1011Gln | 30.1 | T | B | B | O |
| TAF1L | Missense | 7 | chr9:32634817 | c.761G>T | p.Arg254Leu | 46.7 | D | Poss-D | B | O |
| XPC | Missense | 8 | chr3:14190071 | c.2411C>G | p.Ser804Cys | 48.0 | T | B | B | O |
| DDB2 | Missense | 9 | chr11:47256837 | c.897T>C | p.Met240Thr | 49.0 | NA | NA | B | B |
| ETS1 | Missense | 10 |
chr11:128332425 | c.1289A>T | p.Tyr430Phe | 37.7 | D | Prob-D | P | O (HC) |
Discussion
We investigated the genomic landscape of the small
VSs using a comprehensive genomic analysis of all the exons from
key tumor suppressor genes and oncogenes in 10 small sporadic VS
tumors. These evaluations identified NF2, SYNE1, IRS2, APC, CIC,
SDHC, BRAF, NUMA1, EXT2, HRAS, BCL11B, MAGI1, RNF123, NLRP1, ASXL1,
ADAMTS20, TAF1L, XPC, DDB2, and ETS1 as mutated genes.
Five mutations were identified in a single tumor sample (sample no.
5), but did not indicate high tumor mutation burden (TMB) (22).
To date, the genetic profile of sporadic VS is not
completely understood. The only consistent genetic alteration in
these tumors appears to be the inactivation of the NF2.
Merlin, a cytoskeletal protein encoded by the NF2 gene,
suppresses tumorigenesis by interacting with integrins and receptor
tyrosine kinases. Mutational analysis of the NF2 gene in
sporadic VSs has been reported to be identified in 49 to 100% of
analyzed samples (23–35). In this study, we identified
mutations in NF2 in four of ten cases (40%), and revealed that even
in small sporadic VSs, genetic mutations were most frequently found
in the NF2.
The second most frequent mutation was found in
SYNE1, and appeared in 20% of the samples. Synaptic nuclear
envelope protein 1 (SYNE1) encodes a series of spectrin structural
proteins that play important roles in cytoskeletal, nuclear, and
vesicular anchoring (36), and
mutations in this gene are associated with a form of cerebellar
ataxia (37). Recently, it has also
been suggested that altered expression, somatic mutations, and
single nucleotide polymorphisms in the SYNE1 are associated
with the development and progression of lung cancer (38), oral cancer (39), and hepatocellular carcinoma
(40). There have been no reports
of these mutations in schwannomas. The amino acid changes
‘p.Ala977Thr’ and ‘p.Lys4121Ser’ in the SYNE1 gene
identified in this study have not been reported in any of the
germline mutation databases. Therefore, we considered these
mutations to be somatic.
The genes whose mutations were predicted as
‘damaging’, ‘pathogenic’, or ‘oncogenic’ by at least two
pathogenicity prediction models were NF2, IRS2, CIC, EXT2, HRAS,
BCL11B, TAF1L, and ETS1 (Table II). IRS2 encodes the insulin
receptor substrate 2, a cytoplasmic signaling molecule regulating
the effects of insulin, insulin-like growth factor 1, and other
cytokines (41). Alterations in
IRS2 have been reported in several cancers such as
colorectal cancer, lung cancer, and breast cancer (42). Capicua transcriptional repressor
(CIC), a part of the high mobility group (HMG)-box family,
encodes a transcription repressor protein.
CIC mutations occur most frequently in
oligodendroglioma (43).
EXT2 (exostosin glycosyltransferase 2) encodes
glycosyltransferases responsible for heparan sulfate biosynthesis.
Mutations in EXT2 cause the type II form of hereditary
multiple exostoses (44).
HRAS (Harvey rat sarcoma viral oncogene homolog) encodes the
GTPase HRas protein, one of the three human RAS proteins.
HRAS mutations occur in various cancers such as head and
neck squamous cell carcinoma and bladder urothelial carcinoma
(45). BCL11B encodes B-cell
leukemia/lymphoma 11B, a C2H2-type zinc finger protein that
functions as a transcriptional repressor. BCL11B mutations
occur in T-cell acute lymphoblastic leukemia (T-ALL) (46). TATA-Box Binding Protein Associated
Factor 1 (TAF1) possesses intrinsic protein kinase, histone
acetyl-transferase and ubiquitin-conjugating activities.
TAF1L encodes TAF1 Like (TAF1L), a TAF1 homolog that shows
95% amino acid identity with TAF1. TAF1L mutations occur in
oral squamous cell carcinoma (47),
and gastric and colorectal cancers (48). ETS proto-oncogene 1 (ETS1)
encodes a transcription factor and is mutated in cancers such as
colon carcinoma (42).
Here, we focused on previously resected tumors, but
our current strategy for evaluating and treating small VSs is to
follow up with serial MRI scans and audiological tests. Serial
imaging is the most common initial strategy for slow-growing,
especially small-sized VS with a length of less than 2 cm (6,11).
Previous reports describing tumor size in the genetically analyzed
sporadic VS samples reported that the mean size of their surgically
removed tumors ranged from 24.8 to 31.0 mm in length (33–35,49)
(Table III). In this study, the
average size of the sporadic VSs was 8.3 mm. In the present study,
we did not identify any gene mutations that were specific to small
tumors. However, we did find that one gene mutation in
smaller-sized Koos grade I tumors (cases 2, 8, and 9). Only two
mutations, those in NF2 and ETS1, were found in case
10. The mean number of gene mutations detected in the four Koos
grade I tumors was 1.25. In the six Koos grade II tumors, the mean
number of gene mutations detected was 3.0. It is possible that
tumors with larger sizes may harbor a greater number of mutated
genes, but this will require further examination in a future study
featuring a larger number of cases.
 | Table III.Summary of studies describing tumor
size in the genetically analyzed sporadic VS samples. |
Table III.
Summary of studies describing tumor
size in the genetically analyzed sporadic VS samples.
| First author,
year | Sample size range,
mm (average) | Number of VS
samples | (Refs.) |
|---|
| This study | 6.2–15 (8.3) | 10 | - |
| Aaron et al,
2020 | 13–35 (24.8) | 12 | (49) |
| Havik et al,
2018 | 16–56 (31.0) | 46 | (33) |
| Carlson et
al, 2018 | 6–49 (24.8) | 23 | (35) |
| Chen et al,
2017 | 8–70 (26.8) | 281 | (34) |
This study has several limitations. First, the small
sample size limited the accuracy of the conclusions. Second, the
samples lacked reference materials such as blood samples, so we
need to rely on databases to determine whether the mutations were
somatic or germline. Third, the pathogenicity of the mutant genes
in schwannomas needs to be further investigated, as do the effects
of these potentially pathogenic genes and their underlying
mechanisms. The current study could not draw any new conclusions
about the relationship between VS-related hearing loss and gene
mutations. However, this study revealed that NF2 was the
most frequently mutated gene in small sporadic VSs. Moreover, the
novel mutations in SYNE1 were identified by comprehensively
analyzing the genomic data obtained from small tumors. In the
future, establishing diagnostic and prognostic biomarkers using
blood samples may be a critical strategy to predict tumor growth
and hearing loss, since tumor specimens are difficult to obtain
from small VSs.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by JSPS KAKENHI (grant nos. JP17849003
and 19147146).
Availability of data and materials
The datasets generated and analyzed during the
current study are not publicly available due to the research
proposal approved by the ethics committee of our institution but
are available from the corresponding author on reasonable
request.
Authors' contributions
TF and KSak designed the study. KSak and KN
conducted the genetic analyses. TF and KSak confirm the
authenticity of all raw data. TF and NU substantially contributed
to the manuscript drafting. NU, HK, YH, AM, MS, YO, KSai and KD
significantly contributed to data analysis and interpretation. All
authors critically reviewed and revised the manuscript draft, and
read and approved the final manuscript.
Ethics approval and consent to
participate
The study complied with the standards of the
Declaration of Helsinki and was approved (approval no. 29-015) by
the Institutional Review Board of Kindai University Hospital
(Osaka, Japan). Opt-out informed consent from patients was obtained
by exhibiting the research information on the official website of
our hospital (Kindai University Hospital, Osaka, Japan). The
authors guarantee the opportunity for refusal by document, call or
e-mail whenever possible. Patients who rejected participation in
this study were excluded.
Patient consent for publication
Opt-out informed consent from patients was obtained
by exhibiting the research information on the official website of
our hospital (Kindai University Hospital, Osaka, Japan). The
authors guarantee the opportunity for refusal by document, call or
e-mail whenever possible. Patients who rejected publication of
their information in this study were excluded.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moffat DA and Ballagh RH: Rare tumours of
the cerebellopontine angle. Clin Oncol (R Coll Radiol). 7:28–41.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Neff BA, Welling DB, Akhmametyeva E and
Chang LS: The molecular biology of vestibular schwannomas:
Dissecting the pathogenic process at the molecular level. Otol
Neurotol. 27:197–208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marinelli JP, Grossardt BR, Lohse CM and
Carlson ML: Prevalence of sporadic vestibular schwannoma:
Reconciling temporal bone, radiologic, and population-based
studies. Otol Neurotol. 40:384–390. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Evans DG, Howard E, Giblin C, Clancy T,
Spencer H, Huson SM and Lalloo F: Birth incidence and prevalence of
tumor-prone syndromes: Estimates from a UK family genetic register
service. Am J Med Genet A. 152A:327–332. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan SA, Marinelli JP, Hahs-Vaughn DL, Nye
C, Link MJ and Carlson ML: Evolution in management trends of
sporadic vestibular schwannoma in the United States over the last
half-century. Otol Neurotol. 42:300–305. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Macielak RJ, Driscoll CLW, Link MJ, Haynes
DS, Lohse CM and Carlson ML: Vestibular schwannoma practice
patterns: An International cross-specialty survey. Otol Neurotol.
41:e1304–e1313. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Trofatter JA, MacCollin MM, Rutter JL,
Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG,
Pulaski K, et al: A novel moesin-, ezrin-, radixin-like gene is a
candidate for the neurofibromatosis 2 tumor suppressor. Cell.
75:8261993.PubMed/NCBI
|
|
8
|
Rouleau GA, Merel P, Lutchman M, Sanson M,
Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C,
Plougastel B, et al: Alteration in a new gene encoding a putative
membrane-organizing protein causes neuro-fibromatosis type 2.
Nature. 363:515–521. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Vries M, van der Mey AG and Hogendoorn
PC: Tumor biology of vestibular schwannoma: A review of
experimental data on the determinants of tumor genesis and growth
characteristics. Otol Neurotol. 36:1128–1136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tamura R and Toda M: A critical overview
of targeted therapies for vestibular schwannoma. Int J Mol Sci.
23:54622022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goshtasbi K, Abouzari M, Moshtaghi O,
Sahyouni R, Sajjadi A, Lin HW and Djalilian HR: The changing
landscape of vestibular schwannoma diagnosis and management: A
cross-sectional study. Laryngoscope. 130:482–486. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nadol JB Jr, Diamond PF and Thornton AR:
Correlation of hearing loss and radiologic dimensions of vestibular
schwannomas (acoustic Neuromas). Am J Otol. 17:312–316.
1996.PubMed/NCBI
|
|
13
|
Roosli C, Linthicum FH Jr, Cureoglu S and
Merchant SN: Dysfunction of the cochlea contributing to hearing
loss in acoustic neuromas: An underappreciated entity. Otol
Neurotol. 33:473–480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lassaletta L, Calvino M, Morales-Puebla
JM, Lapunzina P, Rodriguez-de la Rosa L, Varela-Nieto I and
Martinez-Glez V: Biomarkers in vestibular schwannoma-associated
hearing loss. Front Neurol. 10:9782019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fujita T, Saito K, Kashiwagi N, Sato M,
Seo T and Doi K: The prevalence of vestibular schwannoma among
patients treated as sudden sensorineural hearing loss. Auris Nasus
Larynx. 46:78–82. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kiyofuji S, Neff BA, Carlson ML, Driscoll
CLW and Link MJ: Large and small vestibular schwannomas: Same, yet
different tumors. Acta Neurochir (Wien). 163:2199–2207. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Koos WT, Day JD, Matula C and Levy DI:
Neurotopographic considerations in the microsurgical treatment of
small acoustic neurinomas. J Neurosurg. 88:506–512. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kumar P, Henikoff S and Ng PC: Predicting
the effects of coding non-synonymous variants on protein function
using the SIFT algorithm. Nat Protoc. 4:1073–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rogers MF, Shihab HA, Mort M, Cooper DN,
Gaunt TR and Campbell C: FATHMM-XF: Accurate prediction of
pathogenic point mutations via extended features. Bioinformatics.
34:511–513. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rogers MF, Shihab HA, Gaunt TR and
Campbell C: CScape: A tool for predicting oncogenic single-point
mutations in the cancer genome. Sci Rep. 7:115972017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Galuppini F, Dal Pozzo CA, Deckert J,
Loupakis F, Fassan M and Baffa R: Tumor mutation burden: From
comprehensive mutational screening to the clinic. Cancer Cell Int.
19:2092019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Antinheimo J, Sallinen SL, Sallinen P,
Haapasalo H, Helin H, Horelli-Kuitunen N, Wessman M, Sainio M,
Jääskeläinen J and Carpén O: Genetic aberrations in sporadic and
neurofibromatosis 2 (NF2)-associated schwannomas studied by
comparative genomic hybridization (CGH). Acta Neurochir (Wien).
142:1099–1104; discussion 1104-5. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Welling DB, Lasak JM, Akhmametyeva E,
Ghaheri B and Chang LS: cDNA microarray analysis of vestibular
schwannomas. Otol Neurotol. 23:736–748. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ikeda T, Hashimoto S, Fukushige S, Ohmori
H and Horii A: Comparative genomic hybridization and mutation
analyses of sporadic schwannomas. J Neurooncol. 72:225–230. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bian LG, Tirakotai W, Sun QF, Zhao WG,
Shen JK and Luo QZ: Molecular genetics alterations and tumor
behavior of sporadic vestibular schwannoma from the People's
Republic of China. J Neurooncol. 73:253–260. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hadfield KD, Smith MJ, Urquhart JE,
Wallace AJ, Bowers NL, King AT, Trump D, Newman WG and Evans DG:
Rates of loss of heterozygosity and mitotic recombination in NF2
schwannomas, sporadic vestibular schwannomas and schwannomatosis
schwannomas. Oncogene. 29:6216–6221. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aarhus M, Bruland O, Saetran HA, Mork SJ,
Lund-Johansen M and Knappskog PM: Global gene expression profiling
and tissue microarray reveal novel candidate genes and
down-regulation of the tumor suppressor gene CAV1 in sporadic
vestibular schwannomas. Neurosurgery. 67:998–1019. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee JD, Kwon TJ, Kim UK and Lee WS:
Genetic and epigenetic alterations of the NF2 gene in sporadic
vestibular schwannomas. PLoS One. 7:e304182012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lassaletta L, Torres-Martin M,
Pena-Granero C, Roda JM, Santa-Cruz-Ruiz S, Castresana JS, Gavilan
J and Rey JA: NF2 genetic alterations in sporadic vestibular
schwannomas: Clinical implications. Otol Neurotol. 34:1355–1361.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Torres-Martin M, Lassaletta L,
San-Roman-Montero J, De Campos JM, Isla A, Gavilan J, Melendez B,
Pinto GR, Burbano RR, Castresana JS and Rey JA: Microarray analysis
of gene expression in vestibular schwannomas reveals SPP1/MET
signaling pathway and androgen receptor deregulation. Int J Oncol.
42:848–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Agnihotri S, Jalali S, Wilson MR, Danesh
A, Li M, Klironomos G, Krieger JR, Mansouri A, Khan O, Mamatjan Y,
et al: The genomic landscape of schwannoma. Nat Genet.
48:1339–1348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Havik AL, Bruland O, Myrseth E, Miletic H,
Aarhus M, Knappskog PM and Lund-Johansen M: Genetic landscape of
sporadic vestibular schwannoma. J Neurosurg. 128:911–922. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen H, Xue L, Wang H, Wang Z and Wu H:
Differential NF2 gene status in sporadic vestibular schwannomas and
its prognostic impact on tumour growth patterns. Sci Rep.
7:54702017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Carlson ML, Smadbeck JB, Link MJ, Klee EW,
Vasmatzis G and Schimmenti LA: Next generation sequencing of
sporadic vestibular schwannoma: Necessity of Biallelic NF2
inactivation and implications of accessory Non-NF2 Variants. Otol
Neurotol. 39:e860–e871. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Puckelwartz MJ, Kessler E, Zhang Y, Hodzic
D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn
SK, et al: Disruption of nesprin-1 produces an Emery Dreifuss
muscular dystrophy-like phenotype in mice. Hum Mol Genet.
18:607–620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gama MTD, Braga-Neto P, Dutra LA, Alessi
H, Maria LA, Gadelha AA, Ortiz BB, Kunii I, Correia-Silva SR, Dias
da Silva MR, et al: Cognitive and psychiatric evaluation in SYNE1
Ataxia. Cerebellum. 18:731–737. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Xiao X, Bosse Y, Gorlova O, Gorlov
I, Han Y, Byun J, Leighl N, Johansen JS, Barnett M, et al: Genetic
interaction analysis among oncogenesis-related genes revealed novel
genes and networks in lung cancer development. Oncotarget.
10:1760–1774. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shah K, Patel S, Modi B, Shah F and Rawal
R: Uncovering the potential of CD44v/SYNE1/miR34a axis in salivary
fluids of oral cancer patients. J Oral Pathol Med. 47:345–352.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Faraj Shaglouf LH, Ranjpour M, Wajid S and
Jain SK: Elevated expression of cellular SYNE1, MMP10, and GTPase1
and their regulatory role in hepatocellular carcinoma progression.
Protoplasma. 257:157–167. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shaw LM: The insulin receptor substrate
(IRS) proteins: At the intersection of metabolism and cancer. Cell
Cycle. 10:1750–1756. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
AACR Project GENIE Consortium, . AACR
Project GENIE: Powering precision medicine through an International
consortium. Cancer Discov. 7:818–831. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lee Y: Regulation and function of capicua
in mammals. Exp Mol Med. 52:531–537. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pacifici M: Hereditary multiple exostoses:
New Insights into pathogenesis, clinical complications, and
potential treatments. Curr Osteoporos Rep. 15:142–152. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hobbs GA, Der CJ and Rossman KL: RAS
isoforms and mutations in cancer at a glance. J Cell Sci.
129:1287–1292. 2016.PubMed/NCBI
|
|
46
|
Lennon MJ, Jones SP, Lovelace MD,
Guillemin GJ and Brew BJ: Bcl11b-A critical neurodevelopmental
transcription factor-roles in health and disease. Front Cell
Neurosci. 11:892017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nakagaki T, Tamura M, Kobashi K, Koyama R,
Fukushima H, Ohashi T, Idogawa M, Ogi K, Hiratsuka H, Tokino T and
Sasaki Y: Profiling cancer-related gene mutations in oral squamous
cell carcinoma from Japanese patients by targeted amplicon
sequencing. Oncotarget. 8:59113–59122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Oh HR, An CH, Yoo NJ and Lee SH:
Frameshift mutations in the mononucleotide repeats of TAF1 and
TAF1L genes in gastric and colorectal cancers with regional
heterogeneity. Pathol Oncol Res. 23:125–130. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Aaron KA, Manojlovic Z, Tu N, Xu Y, Jin Y,
Chang S, Kwok E, Webb M, Hurth K and Friedman RA: What genes can
tell: A closer look at vestibular schwannoma. Otol Neurotol.
41:522–529. 2020. View Article : Google Scholar : PubMed/NCBI
|