Introduction
Osteosarcoma (OS) is the most common primary
malignant bone tumor in children and adolescents; ~1,000 new cases
of OS are diagnosed in the United States each year, approximately
half of which are in children and teenagers. The 5-year overall
survival rate is 60% (1).
Post-treatment factors contributing to poor survival include
incomplete surgical resection and poor response to chemotherapy
(2,3). Surgery, chemotherapy and radiotherapy
are currently the most common treatment options. Improvement in
treatment modalities has led to a better prognosis for patients
with OS but treatment efficacy remains insufficient (4,5).
Therefore, better understanding of the molecular mechanism
underlying OS development is key (6,7).
Protein serine kinase H1 (PSKH1) belongs to the
serine/arginine-rich domain protein family with a specific
localization in nuclear speckles (8,9). This
gene plays an important role in the trafficking of
serine/arginine-rich domains, which leads to pre-mRNA synthesis.
Previous studies have demonstrated that PSKH1 plays a role in
promoting cancer in humans (10,11).
For example, microRNA (miR)-566 directly targets PSKH1, regulating
colon cancer cell proliferation, migration and invasion (12). Various factors contribute to the
development of tumors via the p38/MAPK signaling pathway (13,14).
For example, downregulation of Non-SMC condensin I complex subunit
G (NCAPG) could suppress OC cell proliferation and invasion via
activating the p38 MAPK signaling pathway (13). The p38 pathway serves a crucial role
in facilitating muscle differentiation, which confers its
anti-tumor properties (15). The
activation of the p38/MAPK signaling pathway leads to the
reactivation of dormant disseminated tumor cells, resulting in the
development of metastatic prostate cancer (16). Other studies have shown that the
p38/MAPK signaling pathway promotes cancer by improving survival
and migration (17,18). In early stages of cancer, low p38
activity impairs tumor formation and growth, while a higher level
of activation of the p38/MAPK pathway contributes to survival and
proliferation of tumor cells for more advanced tumor stages
(19). In late stages of
tumorigenesis, p38 inhibits cancer cell migration to neighboring
tissue (20). Nuclear antigen Ki-67
is only found in proliferating cells (21) at the G1, S, and G2 phases of the
cell cycle and mitosis, but is absent from resting cells at G0
(22). In cancer cells, it is
widely used as a proliferation marker (23–25).
It has been found that Ki-67 is associated with tumor metastasis in
several studies (26,27). Zeng et al (28) reported that positive Ki-67
expression is associated with distant metastasis and overall
survival of OS. However, the biological role of PSKH1 in OS cells
and the association between PSKH1, p38/MAPK signaling pathway and
Ki-67 expression remain unclear.
The present study aimed to explore the role and
underlying molecular mechanism of PSKH1 in OS cells.
Materials and methods
Bioinformatics analysis
Gene expression profiles of 37 OS cases were
obtained from the Gene Expression Omnibus (GEO,
ncbi.nlm.nih.gov/geo/) database (accession no. GSE 39055).
Expression profiling was performed using an Illumina HumanHT-12
WG-DASL V4.0 R2 Expression BeadChip (Illumina, Inc.). All mRNA
expression datasets were retrieved from the GEO database. Analysis
of differentially expressed mRNAs of tumor tissue was performed
using the DESeq2 package in R (version 1.20.0) (29). Samples were stratified based on the
mean expression levels of PSKH1. Survival and ggplot2 packages in R
were used for survival analysis and plotting.
Gene set enrichment analysis
(GSEA)
GSEA was used to identify potential biological
pathways and processes associated with PSKH1. mRNA expression data
were obtained from ArrayExpress (https://www.ebi.ac.uk/biostudies/arrayexpress)
(accession no. E-MEXP-3628), including four normal bone and 14 OS
tissue samples. The dataset was extracted from the Molecular
Signatures Database on the GSEA website
(gsea-msigdb.org/gsea/index.jsp). Next, A weighted enrichment
analysis was carried out utilizing the GSEA 2.2.2 software, which
involved the selection of 1,000 random combinations (30).
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
Total SAOS2, HOS, MG63 and U2OS and osteoblastic
hFOB1.19 cells or transfected SAOS2, U2OS and MG63 cells RNA was
isolated using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). RNA was dissolved in 0.025 ml nuclease-free
water and stored at −80°C. cDNA synthesis was performed using the
SuperScript First-Strand Synthesis System (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Thermocycling was performed at 95°C for 10 min, followed by 40
cycles at 95°C for 15 sec and 60°C for 45 sec. qPCR was then
performed using SYBR Green PCR Master Mix (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. PCR
amplification was performed in triplicate. Primers were as follows:
PSKH1: Forward, 5′-GCTGTGGGACAAGCAAGG-3′ and reverse,
5′-TGGTGGTTCTGAGGGAGG-3′ and β-actin: Forward,
5′-AATGAGCGGTTCCGTTGC-3′ and reverse, 5′-TCTTCATGGTGCTGGGAG-3′.
β-actin was used as an internal reference to calculate the relative
expression of PSKH1. Relative gene expression was analyzed using
2-ΔΔCq method (31).
Western blotting
The SAOS2, HOS, MG63 and U2OS and osteoblastic
hFOB1.19 cells or transfected SAOS2, U2OS and MG63 cells lysate was
prepared using ice-cold RIPA lysis buffer (Beyotime Institute of
Biotechnology). BCA Protein Assay kit was used to determine the
concentration of protein. Whole cell lysate (25 µg/lane) was
electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gel
and semi-dry transferred onto polyvinylidene difluoride membranes.
Membranes were blocked with 5% skimmed milk in Tris-buffered saline
with 0.5% Tween-20 (TBST) for 1 h at room temperature, then
incubated overnight at 4°C with primary antibodies in TBST
containing 5% skimmed milk. Primary antibodies were as follows:
PSKH1 (1:500, cat. no. Sc-514401, Santa Cruz Biotechnology, Inc.),
p38 (1:2,000, cat. no. Ab170099, Abcam), phosphorylated (p)-p38
(1:1,000; cat. no. Ab47363, Abcam) and β-actin (1:5,000; cat. no.
66009-1-lg, Proteintech Group, Inc.). Membranes were washed and
incubated at room temperature for 30 min with horseradish
peroxidase-conjugated with goat anti-rabbit (1:1,000; cat. no.
A0208, Shanghai Biyuntian Bio-Technology Co. Ltd.) and anti-mouse
secondary antibodies (1:1,000; cat. no. A0216, Shanghai Biyuntian
Bio-Technology Co. Ltd.). Membranes were washed three times with
TBST and bound proteins were visualized using Immobilon
chemiluminescent HRP substrate (Millipore, Sigma) and detected
using BioImaging Systems (Tanon Science and Technology Co., Ltd.).
β-actin was used as an internal control to verify basal protein
expression levels using Image Lab software (version 6.0; Bio-Rad
Laboratories, Inc.).
Cell culture
Human OS SAOS2, HOS, MG63 and U2OS and osteoblastic
hFOB1.19 cell lines were purchased from the American Type Culture
Collection (ATCC). Standard protocols for cell culture were
followed (32). SAOS2, HOS and MG63
cells were cultured in a Minimum Essential Medium (Hyclone)
supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1%
penicillin/streptomycin (Beijing Solarbio Science & Technology
Co., Ltd.). U2OS and hFOB1.19 cells were cultured in RPMI-1640
(Hyclone) supplemented with 10% FBS and 1% penicillin/streptomycin
(Beijing Solarbio Science & Technology Co., Ltd.). Cells were
incubated at 37°C in a humidified incubator with 5% CO2.
In rescue experiments, cells were pretreated with p38/MAPK
inhibitor SB203580 (cat. no. ab120162; Abcam) or DMSO for 1 h at
37°C.
Construction of lentivirus and cell
transfection
For the knockdown of PSKH1, human short hairpin
(sh)RNA sequences (shRNA-1,
5′-CCGGTCCCAGCAGCAAGTCAGTATCTCGAGATACTGACTTGCTGCTGGTGTTTTG-3′,
shRNA-2,
5′-CCGGTGCTCTTTGACCGCATCATTCTCGAGAATGATGCGGTCAAAGAGCTTTTTG-3′ and
shRNA-3,
5′-CCGGTCCTGAGAATCTGCTCTACTCTCGAGAGTAGAGCAGATTCTCAGGTTTTTG-3′) and
negative control (NC) shRNA
(5′-CCGGTGTTCAGTGCCTACACGATTCTCGAGAATCGTGTAGGCACTGAACTTTTTG-3′)
were synthesized by Genewiz, Inc. at the final concentration of 1
µg/µl and cloned into the PLKO.1 plasmid (Addgene, Inc.) to
generate PLKO.1-shPSKH1. The transfection of 293T cells (at 90%
confluence) with a combination of PLKO.1-shPSKH1 (1,000 ng), psPAX2
(900 ng), and pMD2G (100 ng; both Addgene, Inc.) was performed
utilizing the 2nd generation system and Lipofectamine 2000 (by
Invitrogen, Thermo Fisher Scientific, Inc.) following the
manufacturer's protocols.293T cells were obtained from ATCC.
Transfection was performed for 4 h at 37°C, then DMEM (Gibco) was
replaced with complete medium for 72 h at 37°C. The supernatant of
293T cells was collected, centrifuged at 4,000 × g for 10 min at
4°C to remove cell debris, filtered, centrifuged at 7,000 × g for 5
min at 4°C and resuspended in ice-cold PBS to detect the titer. The
infectious titer was determined by hole-by-dilution titer assay.
The quantity of lentiviral plasmid used for transfection was 5 µg.
Cells were divided into PSKH1 knockdown (LvshPSKH1) and NC groups.
The SAOS2 and U2OS cells at a density of 5×104
cells/well in a 6-well culture plate were infected with LvshPSKH1
or shNC with a multiplicity of infection of 20. After 48 h
transfection at 37°C, the selection of stable cell lines was
performed with puromycin (Sigma-Aldrich, Merck KGaA) at 4 µg/ml for
2 weeks. The concentration of puromycin used for maintenance was 1
µg/ml. SAOS2 and U2OS cells were used in further experiments.
In addition, human PSKH1 mRNA sequences were
synthesized by Genewiz, Inc. and cloned into the pLVX-Puro vector
(Clontech; Takara Bio USA) to generate pLVX-Puro-PSKH1 Plasmids.
All constructs were verified by DNA sequencing. A 2nd generation
system was used to package the lentivirus. A total of 1,000 ng
constructed plasmids together with packaging (100 ng psPAX2) and
envelope plasmid (900 ng pMD2G; both Addgene, Inc.) was transfected
for 4 h at room temperature using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) in 293T cells (90% confluency)
according to the manufacturer's instructions. DMEM was replaced
with full culture medium for 48 h at 37°C. The supernatant of 293T
cells was collected, centrifuged at 4,000 × g for 10 min at 4°C to
remove cell debris, filtered, centrifuged at 7,000 × g for 5 min at
4°C and resuspended in ice-cold PBS to detect the titer. The
infectious titer was determined by hole-by-dilution titer assay.
The quantity of lentiviral plasmid used for transfection was 5 µg.
Cells were divided into PSKH1 overexpression (oePSKH1) and vector
groups. MG63 cells at a density of 5×104 cells/well in a
6-well culture plate were infected with oePSKH1 with a multiplicity
of infection of 5. After 48 h transfection at 37°C,stable cell
lines were selected by puromycin (Sigma-Aldrich; Merck KGaA) at 4
µg/ml for 2 weeks. The concentration of puromycin used for
maintenance was 1 µg/ml. The cells were used for downstream assay
or transplantation.
Cell proliferation assay
Cell Counting Kit-8 (SAB Biotech; cat. no. CP002)
was used to measure cell proliferation. A total of
~3×104 transfected SAOS2, U2OS and MG63 cells/well was
seeded in 96-well plates and cultured overnight at 37°C. A total of
~10 ml CCK-8 solution was mixed with 100 ml medium and added to
each well. Samples were incubated for 1 h at 37°C. The optical
density was measured at 450 nm using a microplate reader (Perlong
Medical Equipment Co., Ltd.).
Cell cycle analysis
Stable transfected SAOS2, U2OS and MG63 cells were
washed twice with ice-cold PBS and fixed with ice-cold 70% ethanol
overnight at 4°C, cells were harvested by centrifugation at 1,000 ×
g for 5 min at room temperature and resuspended in PBS. Cells were
stained with 0.5 ml staining buffer, 25 µl propidium iodide and 10
µl RNase A at 37°C for 30 min in the dark. Cell cycle were detected
using FACS Caliber flow cytometer (Beckman Coulter, Inc.) The
results were analyzed using FlowJo software (version 10.6.2; BD
Biosciences) to determine cell cycle phase.
Wound healing assay
A wound assay was employed to evaluate cell
migration, as previously described (33), Stable transfected SAOS2, U2OS and
MG63 cells (5×105/well) were seeded in 12-well plates
and grown to nearly 100% confluence in 10% FBS-containing medium,
before being washed with PBS and transferred to serum-free medium
overnight at 37°C. A 100-microliter pipette tip was used to create
an artificial wound through the monolayer. After washing three
times with PBS, the wound size was measured after 0, 12 and 24 h.
Wound areas were photographed under an inverted fluorescence
microscope (XDS-500C, Cai Kang Optical Instrument Co., Ltd.,
Shanghai, China) with a magnification of ×40. The area of wound
closure was quantified using ImageJ software (v1.8.0; National
Institutes of Health).
Transwell assay
Matrigel (BD Biosciences) and Transwell chambers
with 8-µm pores (Corning, Inc.) were used for Transwell invasion
assay. Defrosted Matrigel glue at 4°C was diluted in MEM (Hyclone)
at a 1:2 ratio. In the upper chamber, 80 µl diluted Matrigel glue
was used for the invasion assay at 37°C for 30 min. Transwell
chambers were placed in 24-well plates. The lower chamber was
filled with 800 µl MEM containing 10% FBS. The upper chamber was
filled with 200 µl serum-free medium and 2×104
transfected SAOS2, U2OS and MG63 cells per well were seeded for 24
h at 37°C. A total of 1 ml 0.5% crystal violet solution was added
to each well. After 30 min at room temperature, each well was
washed three times with 1X PBS. Cells in five randomly selected
areas under the light microscope (20× magnification) were counted
(CX41RF, Olympus).
Colony formation assay
Stable transfected SAOS2, U2OS and MG63 cells were
seeded in 96-well-plates at 1,000 cells/well and cultivated at 37°C
for 21 days until clones (>10 cells) could be seen with the
naked eye. Cells were fixed with 100% methanol and incubated for 15
min at room temperature. 0.5% Crystal violet staining was used for
30 min at room temperature to visualize the cells. Each sample was
tested in triplicate. The cell clones were counted with the naked
eye. The clone formation rate was calculated as follows: (Number of
clones/number of inoculated cells) ×100.
Xenografts in athymic nude mice
A total of 40 nude female BALB/c mice (age, 4–6
weeks; weight, 18–20 g) were obtained from Silaike Experimental
Animal Limited Liability Company (Shanghai, China). Mice were
maintained in a pathogen-free facility at Tongji Hospital with each
cage containing 4–5 mice. Mice were maintained under standard
12/12-h light/dark conditions with food and water ad
libitum. They were kept in a controlled environment with
regulated temperature (22–24°C) and humidity (65–70%). The present
study was approved by the Ethics Committee for Animal Experiments
of Tongji Hospital. Animals were handled according to The Guide for
the Care and Use of Laboratory Animals (34). All mice were euthanized at the end
of the experiment. The total duration of the experiment was 5
weeks. No mice died prematurely during the experiment. Mice were
subjected to 4% isoflurane for inhalant anesthesia induction and
1.5% for maintenance. The mice were subcutaneously injected with
5×106 U2OS cells in the armpits of nude mice. A total of
16 mice were randomly distributed into shNC (mice injected
subcutaneously with U2OS cells transduced with shNC) and shPSKH1
groups (mice injected subcutaneously with U2OS cells transduced
with shPSKH1) (n=8/group). Measurements of tumor length and width
were taken weekly. Tumor volume was calculated as follows: Tumor
volume=(Length × width2)/2. After 33 days, mice were
anesthetized with 4% isoflurane, sacrificed by cervical dislocation
and death was confirmed by breathing cessation. Tumors were
dissected, weighed and fixed in 10% formalin overnight at 4°C. No
animals reached the humane endpoints, as follows: Total tumor
volume of 1,500 mm3, 15% loss of body weight, vomiting
or inability to stand to reach food and water.
Hematoxylin and eosin (HE)
staining
HE staining was performed according to standard
protocols (35). The tumor tissue
was excised and fixed in 10% formalin for 24 h at 4°C. After
alcohol gradient dehydration, ~6-µm-thick coronal sections were
embedded in transparent paraffin, and dewaxing was performed with
xylene. Each tissue section was incubated at 62°C for 30 min. A
series of descending ethanol (100, 90, 80 and 70%) was used to
dehydrate samples for 5–10 min and samples were rinsed with
distilled water for 10 min. The tissue section was subjected to
hematoxylin staining and differentiated using 1% hydrochloric acid
alcohol to reduce nonspecific background staining and enhance
visual contrast. The tissue was rinsed with water, followed by
counterstaining with 5% eosin. The slide was subsequently
dehydrated using sequential washes with 80, 95, and 100% alcohol
and finally cleared with xylene. The sections were sealed with
neutral resin and placed in an oven at 65°C for 15 min. These
slides were observed optical microscope (Nikon Corporation) at ×200
magnification.
Immunofluorescence (IF) staining
The tumor tissue was excised and fixed in 10%
formalin for 24 h at 4°C. Paraffin-embedded tissue samples were cut
into 5-µm-thick sections for IHC. Sections were deparaffinized in
xylene and rehydrated through graded alcohol series to water.
Sections were rinsed for 10 min in distilled water, and antigen
retrieval was performed by autoclaving the sections in 0.01 M
citrate buffer (pH 6.0) for 15 min at 121°C. After washing with
PBS, non-specific protein binding was prevented by incubating the
sample with 5% skimmed milk at room temperature for 30 min. The
primary antibody for E Cadherin (cat. no. ab231303; Abcam), Ki-67
(cat. no. ab16667; Abcam) was diluted at 1:100 in a humidified
container and incubated overnight at 4°C. Following three washes in
PBS, slides were incubated with Alexa Fluor 488-conjugated goat
anti-mouse IgG secondary antibodies (dilution1:100, cat. no. A0428)
and anti-rabbit IgG secondary antibodies (dilution1:100, cat.no.
A0423,both Beyotime Institute of Technology) for 1 h at room
temperature. The nuclei were counterstained with
4′,6-diamidino-2-phenylindole (DAPI). All sections were
photographed on a Nikon Eclipse fluorescence microscope (Nikon
Corporation) at ×400 magnification.
Experimental metastasis model
BALB/c nude mice were injected in the tail vein with
1×107 U2OS cells. Animals were randomly distributed into
shNC and shPSKH1 groups (n=12/group). All animals were euthanized
after 8 weeks. Metastatic nodules in the lungs were counted
manually and photographed.
Statistical analysis
All data are expressed as the mean ± SD. For
comparisons between multiple groups, one-way ANOVA followed by
Tukey's post hoc test was conducted using GraphPad Prism (GraphPad
Prism 8; GraphPad Software, Inc.). Additionally, log-rank test was
used to compare Kaplan-Meier survival curves. P<0.05 was
considered to indicate a statistically significant difference.
Results
High PSKH1 expression is associated
with poor survival in patients with OS
To determine the promotive or suppressive effect of
PSKH1 on overall survival of patients, PSKH1 expression was
evaluated in OS and normal tissue based on the ArrayExpress
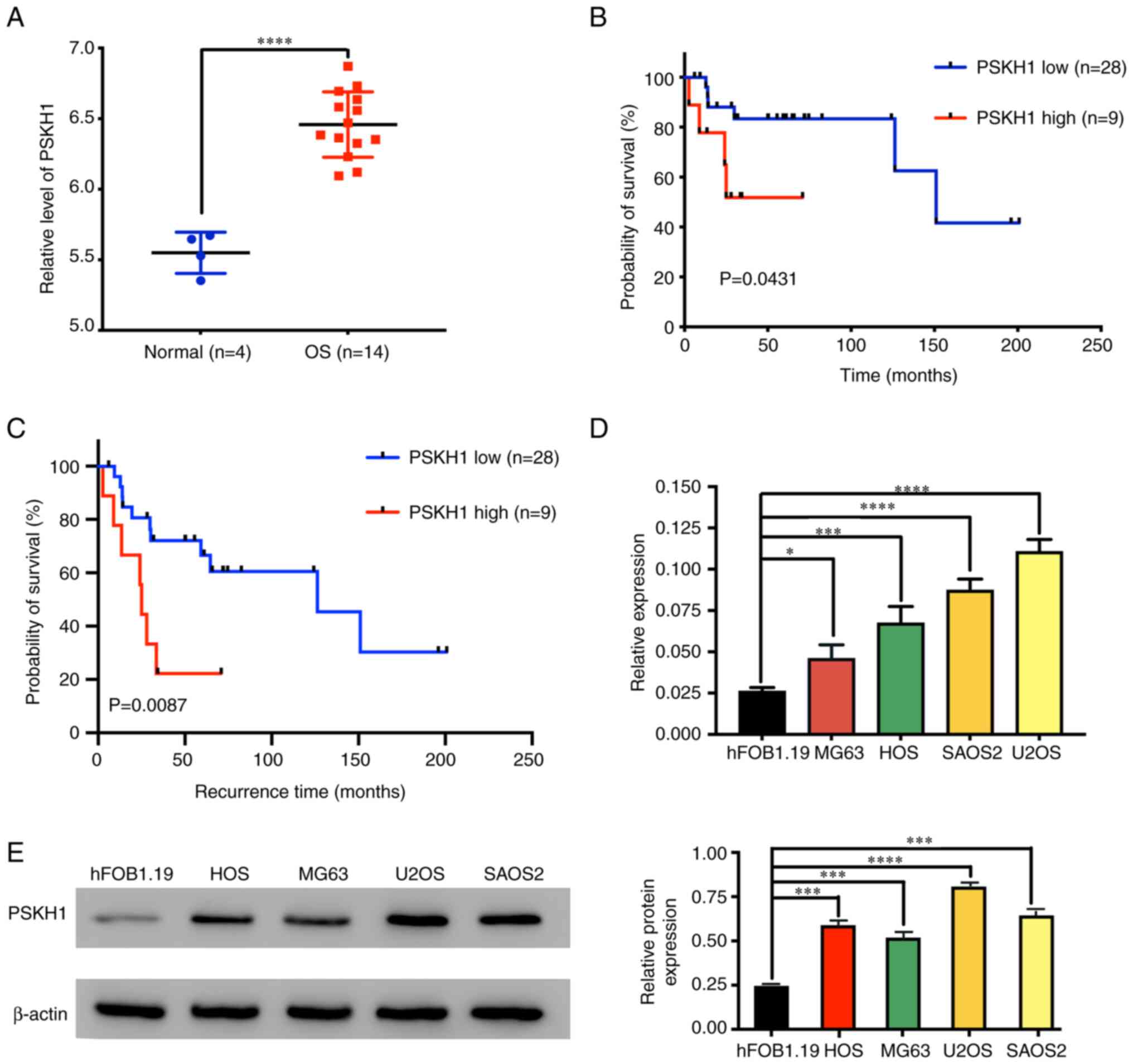
database. Expression levels of PSKH1 were significantly higher in
OS compared with normal bone tissue (Fig. 1A). PSKH1 expression was negatively
associated with overall and recurrence-free survival of patients
with OS (Fig. 1B and C).
Collectively, these data indicated that high PSKH1 expression
independently predicted poor survival for patients with OS.
Role of PSKH1 in OS cell lines
To understand how PSKH1 contributes to OS
development, PSKH1 expression levels were detected in OS cell lines
using RT-qPCR and western blotting with human osteoblast hFOB1.19
cells as control. The results showed upregulation of PSKH1 mRNA and
protein in OS cells (Fig. 1D and
E). These results suggest that PSKH1 may be involved in the
development of OS.
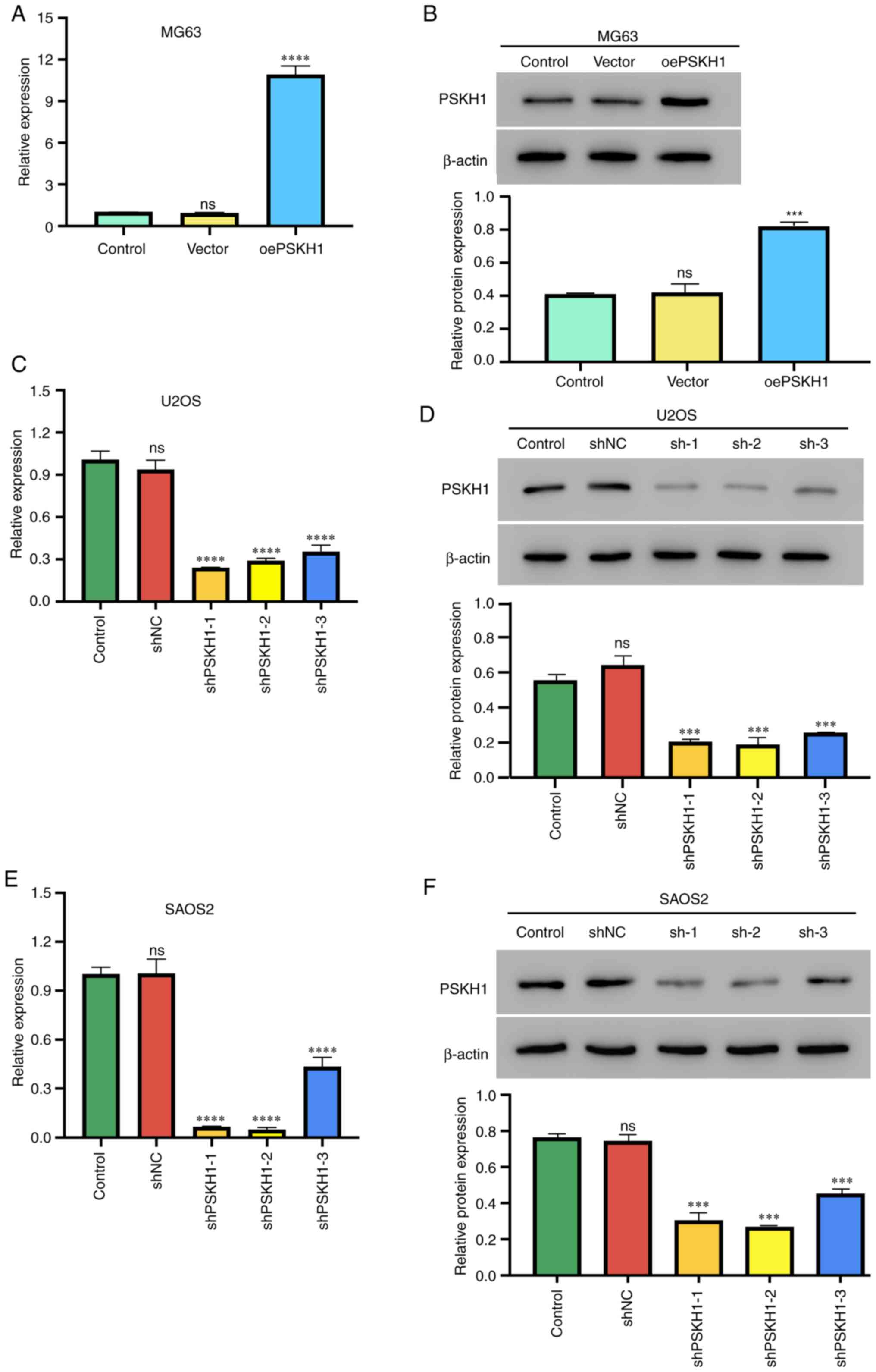
MG63 cells with relatively low expression of PSKH1
were infected with PSKH1 lentivirus or control virus and a stable
PSKH1 overexpressing cell line was successfully generated (Fig. 2A and B). PSKH1 was successfully
knocked down in U2OS and SAOS2 cell lines by infecting cells with
lenti-shPSKH1 and lenti-sh-control virus (Fig. 2C-F).
PSKH1 knockdown inhibits OS cell
migration and proliferation in vitro
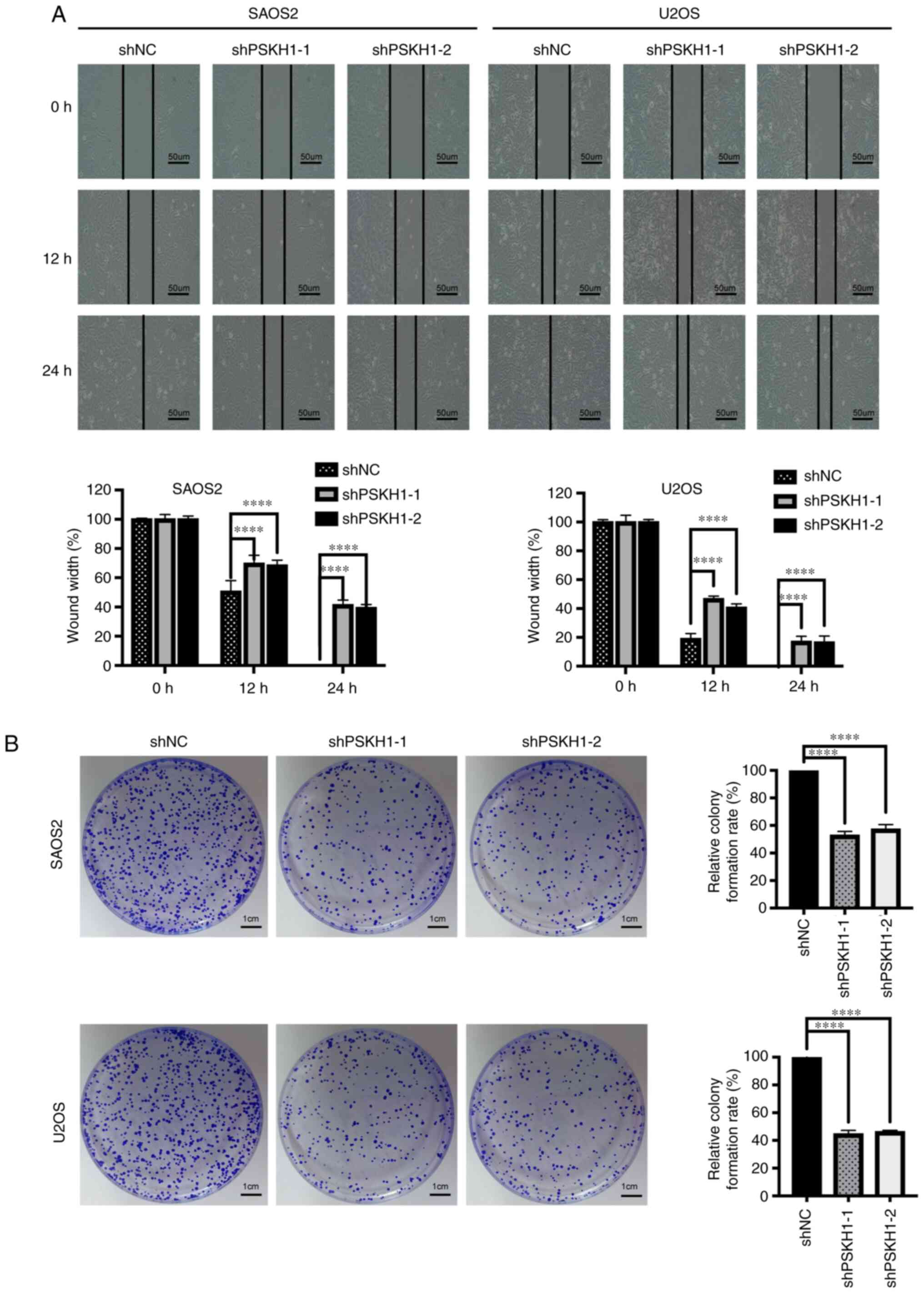
Knockdown of PSKH1 inhibited migration of U2OS and
SAOS2 cells (Fig. 3A). The role of
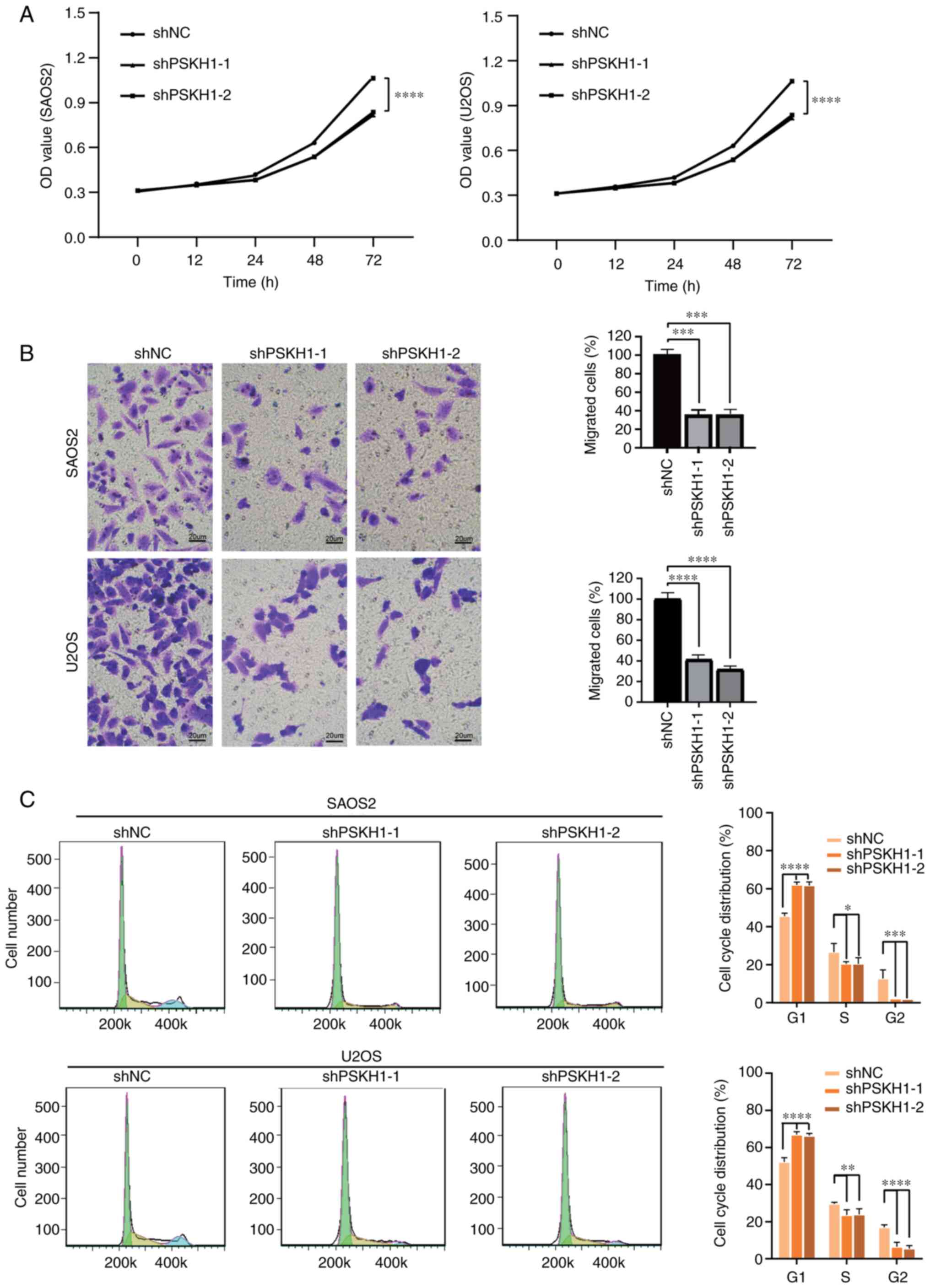
PSKH1 in tumorigenesis was investigated using CCK-8 and colony
formation assays. Silencing PSKH1 expression significantly
decreased proliferation of U2OS and SAOS2 cells (Fig. 4A), whereas cell migration and
proliferation were significantly increased in MG63 cells infected
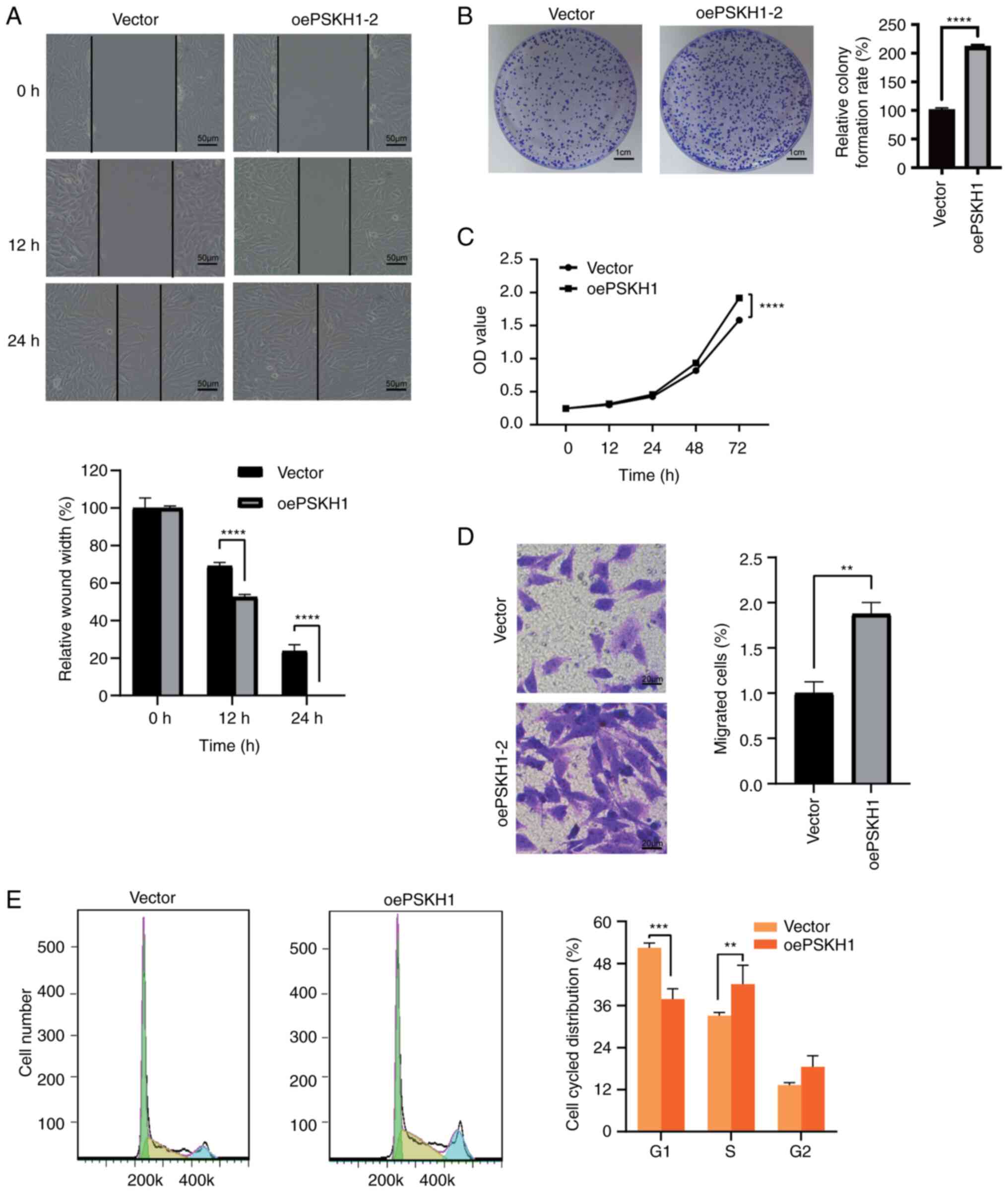
with PSKH1 lentivirus compared with NC (Fig. 5A and C). Silencing of PSKH1 by
shPSKH1 decreased the colony number of U2OS and SAOS2 cells
(Fig. 3B), whereas PSKH1
overexpression yielded the opposite result (Fig. 5B). Together, these results suggested
that PSKH1 knockdown inhibited OS cell migration and
proliferation.
PSKH1 knockdown in OS cells inhibits
invasion and cell cycle transition in vitro
Transwell assay was performed to verify the effect
of PSKH1 on the invasion of OS cells. Silencing PSKH1 expression
decreased the number of invaded OS cells (Fig. 4B). However, PSKH1 overexpression
showed the opposite effect (Fig.
5D). The effect of PSKH1 on the cell cycle of OS cells was
investigated. Compared with the empty vector group, silencing of
PSKH1 led to a significant increase in the number of cells in the
G1 phase, accompanied by a decrease in the proportion of cells in
the S and G2 phase (Fig. 4C).
Overexpression of PSKH1 was found to decrease the number of cells
in G1 phase, with a corresponding increase in the proportion of
cells in the S phase (Fig. 5E).
PSKH1 expression levels were positively associated with OS cell
proliferation, invasion and cell cycle progression.
PSKH1 downregulation inhibits tumor
growth in vivo
The effect of PSKH1 on proliferation of OS cells was
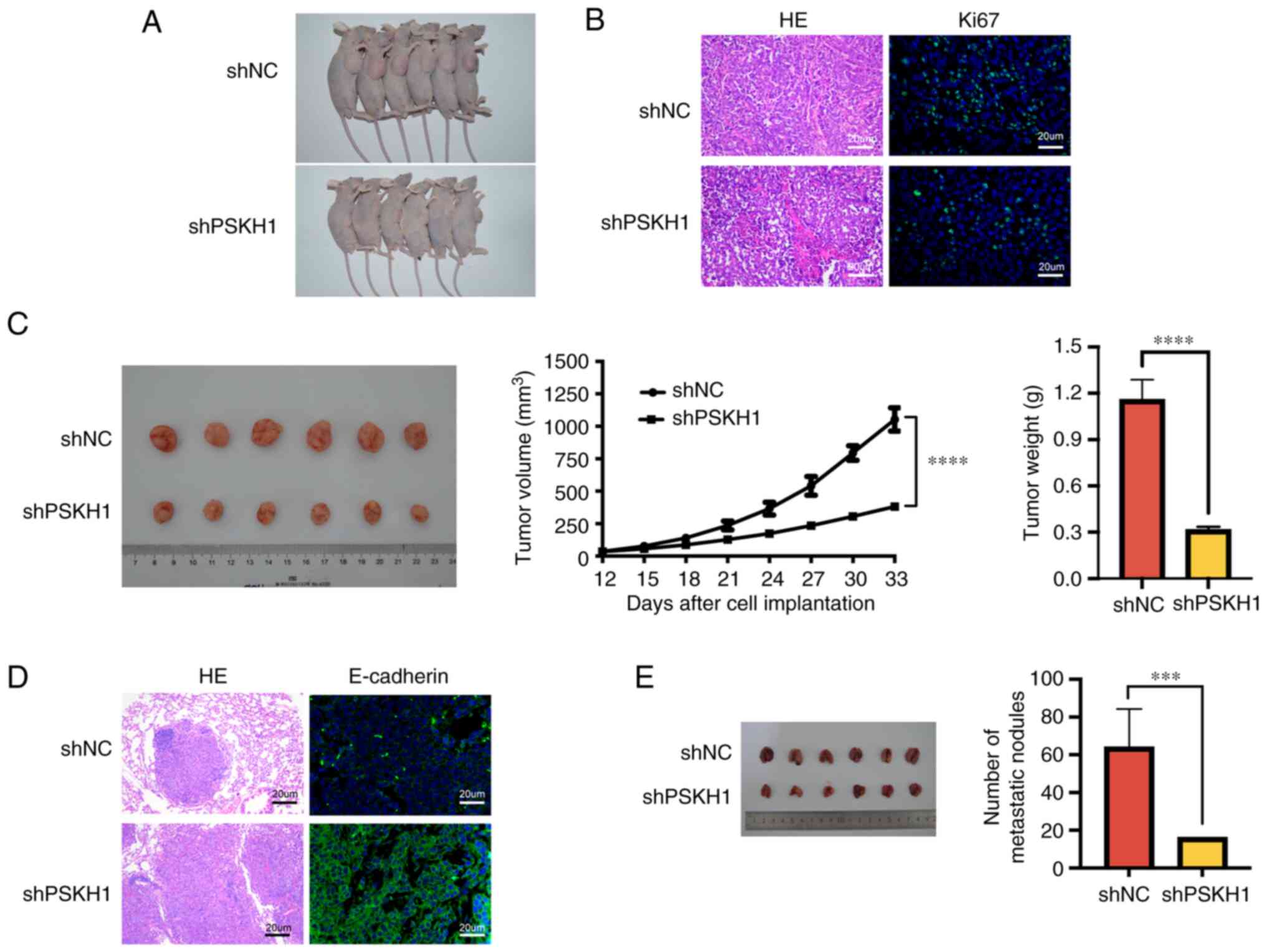
assessed using a mouse xenograft model. PSKH1 knockdown notably
inhibited tumor growth (Fig. 6A).
Immunofluorescence analysis revealed that Ki67 expression was
decreased in shPSKH1 group (Fig.
6B). PSKH1 knockdown tumors had significantly smaller volume
and weight than control tumors (P<0.01; Fig. 6C), indicating that expression of
PSKH1 promoted tumor growth in solid tumors. Overall, these results
suggested that PSKH1 knockdown inhibited OS tumor growth in
vivo.
PSKH1 knockdown suppresses pulmonary
metastasis in vivo
A lung metastasis mouse model was also established
by injecting shNC- and shPSKH1-U2OS cells into the tail veins of
nude mice. Mice were then sacrificed 8 weeks after injection to
examine metastatic nodes in the lung. E-cadherin expression was
significantly decreased in the shPSKH1 group (Fig. 6D and E). PSKH1 decreased the number
of metastatic tumor nodes in mice. Collectively, these results
indicated that PSKH1 downregulation inhibited lung metastasis.
PSKH1 serves an oncogenic role in OS
cells via the p38 pathway
The potential mechanisms involved in OS pathogenesis
were investigated. GSEA was conducted on OS and normal samples in
the ArrayExpress database. PSKH1 expression was significantly
associated with the p38 pathway (Fig.
7A). The p38 pathway regulates cell invasion and migration
(20). To confirm GSEA results, the
association between PSKH1, p38 and p-p38 was investigated in
vitro. Overexpression of PSKH1 led to an elevation in the
expression of p-p38 and an increase in the p-p38/p38 ratio in MG63
cells (Fig. 7B). Conversely, the
silencing of PSKH1 through shPSKH1 resulted in a reduction of the
expression of p-p38, resulting in a decrease in the ratio of
p-p38/p38 in U2OS and SAOS2 cells (Fig.
7C). The effect of PSKH1 on proliferation, invasion and
migration of OS cells via the p38 pathway was investigated
in vitro. MG63 cells transfected with PSKH1 overexpression
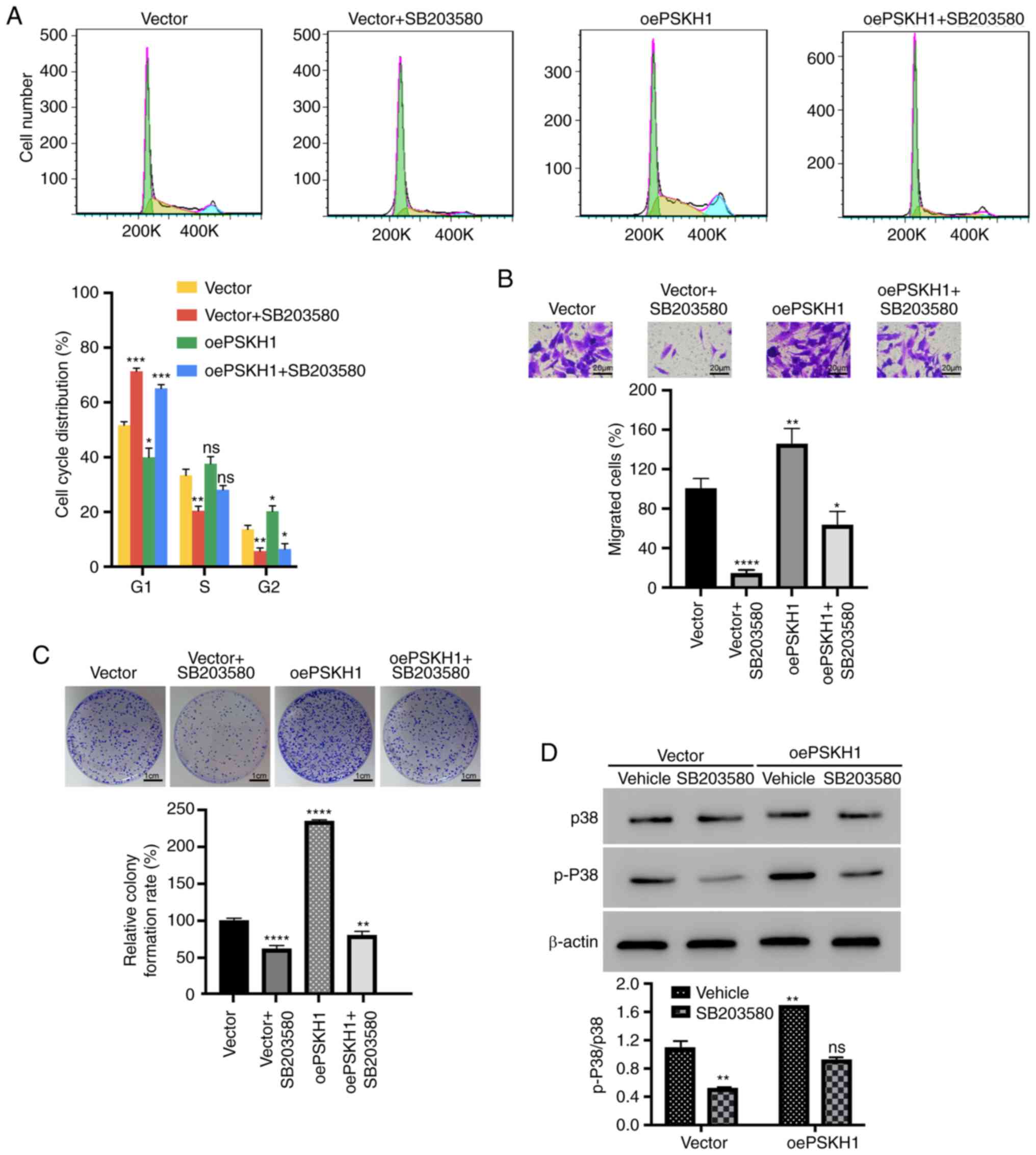
vector were treated with a p38 inhibitor, SB203580. The
overexpression of PSKH1 had a noticeable impact in promoting the
proliferation and migration of established osteosarcoma (OS) cells.
However, the treatment with SB203580 effectively attenuated the
migration and proliferation in comparison to the cells in the PSKH1
overexpression group (Fig. 7D and
E). Additionally, PSKH1 overexpression also significantly
increased the colony formation and invasion of OS cells, as well as
the proportion of cells in the S phase. However, the treatment with
SB203580 effectively reversed the cell cycle progression, invasion,
and proliferation in comparison to the cells in the PSKH1
overexpression group (Fig. 8A-C).
Furthermore, SB203580 also effectively restored the ratio of
p-p38/p38 in established OS cells, which had been induced by the
PSKH1 overexpression (Fig. 8D).
Together, these results suggested that PSKH1 served an oncogenic
role in the development of OS, potentially by activating the p38
pathway.
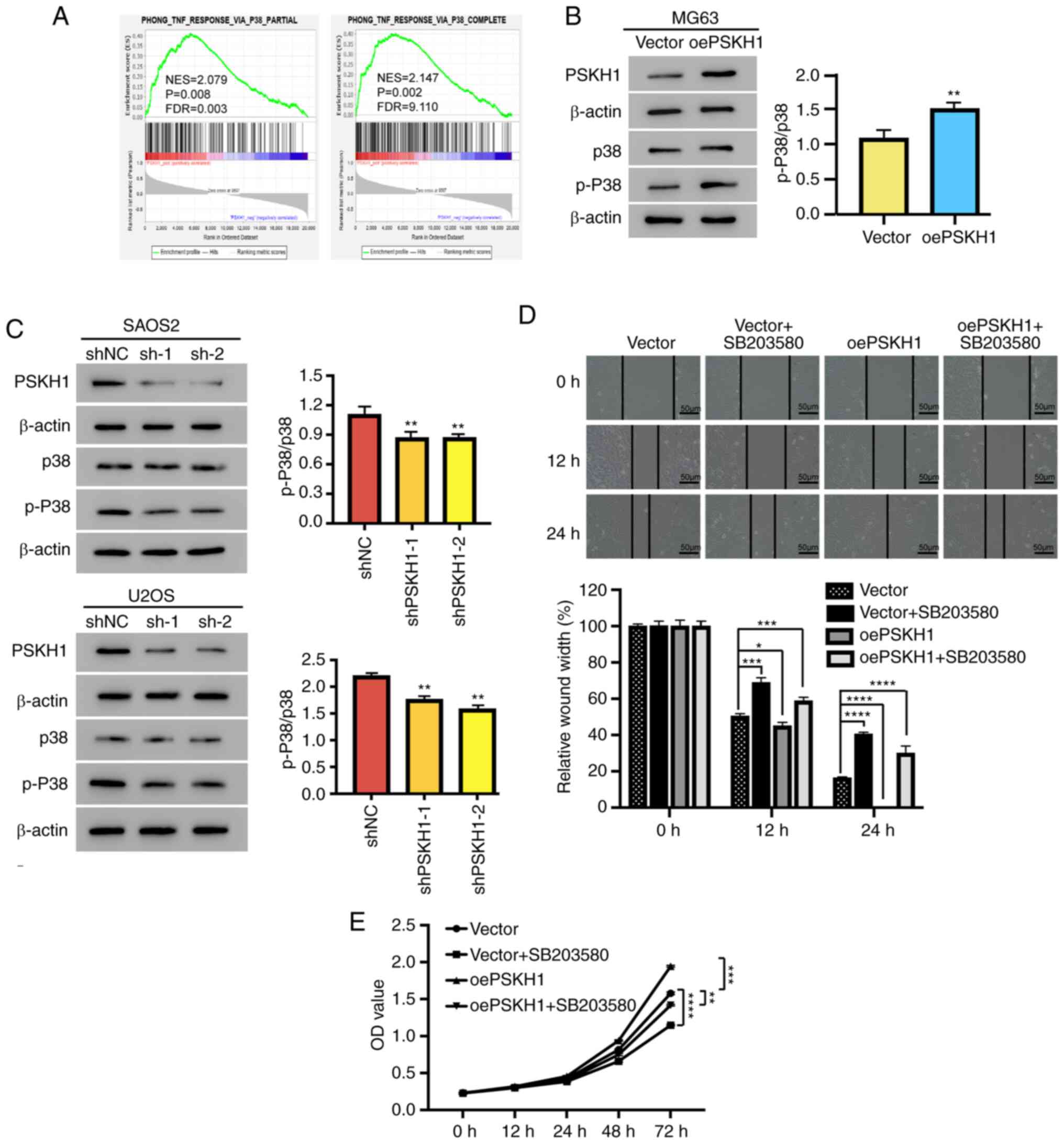 | Figure 7.PSKH1 promotes OS cell proliferation
by activating the p38 signaling pathway. The p38 pathway inhibitor
SB203580 (10 µmol/l) was added to PSKH1-overexpressing OS cells and
their controls for 24 h. (A) Association between PSKH1 expression
and the p38-independent signaling pathway. (B) Protein expression
of p-p38/p38 in MG63 cells with ectopic expression of PSKH1 and
vector control (**P<0.01 vs. vector). (C) Protein expression of
p-p38/p38 in U2OS and SAOS2 cells transfected with lenti-shPSKH1 or
lenti-shNC. β-actin served as an internal control
(**P<0.01 vs. shNC). (D) Wound healing in MG63 cells, with
ectopic expression of PSKH1 after treatment with SB203580. Scale
bar, 50 um (*P<0.05, ***P<0.001; ****P<0.0001 vs. vector).
(E) Growth curve of MG63 cells with ectopic expression of PSKH1 and
vector control after treatment with SB203580 (**P<0.01,
***P<0.001; ****P<0.0001 vs. vector). PSKH1, protein serine
kinase H1; sh, short hairpin; NC, negative control; OS,
osteosarcoma; p-, phosphorylated; OD, optical density; oe,
overexpression; FDR, false discovery rate; NES, normalized
enrichment score. |
Discussion
The p38/MAPK-mediated signaling pathway is activated
in many types of humoral tumor, including OS, and is implicated in
the progression of tumors (17,36,37).
MAPK is key for cell proliferation in gastric cancer (38,39).
Cancer-associated fibroblast produces interleukin-32, which
stimulates invasion and metastasis of breast cancer cells via
integrin β3/p38/MAPK signaling (40,41).
In the present study, overexpression of PSKH1
significantly increased p-p38 expression. The p38 inhibitor
SB203580 effectively rescued proliferation, migration, and invasion
in PSKH1 overexpression OS cells. The present results were
consistent with previous findings (42,43).
The present study investigated the effect of PSKH1 in OS cell lines
via phosphorylation of p38/MAPK. Further study is necessary to
determine the effect of p38/MAPK signaling pathway in vivo.
The present findings demonstrated that the p38/MAPK signaling
pathway may be responsible for PSKH1 function in OS.
OS is a prevalent form of malignant bone tumor that
primarily affects children and adolescents with a median age of 16
years (44). The tumor is commonly
located in the long bones of the extremities, including the tibia,
femur, and humerus. Despite the combination of traditional
treatments, including wide resection, radiotherapy, and
chemotherapy, providing a 5-year survival rate of 60 to 70% for
non-metastatic OS patients (45),
the prognosis for metastatic patients remains unfavorable with a
high rate of recurrence and low survival rate of nearly 20%
(46,47). Despite the efficacy of these
treatments, the challenges of metastasis and relapse persist
(48–50) and the underlying mechanisms of cell
proliferation, migration, and invasion in OS remain elusive. Thus,
identifying the molecular mechanisms underlying invasion and
metastasis of OS cells may provide novel therapeutic targets.
Previous studies have shown that PSKH1 is an autophosphorylating
human protein involved in colon cancer (10,12).
miR-566 directly targets PSKH1 and overexpression of PSKH1 reverses
the biological effects of miR-566 (12). However, the role of PSKH1 in the
proliferation, migration, and invasion of OS cells remains
unresolved. The present findings demonstrated that PSKH1 may serve
as a prognostic biomarker and a promising therapeutic target for
OS. Gene expression profiling showed that high expression of PSKH1
in OS tissue was associated with poor prognosis. Furthermore, high
levels of PSKH1 were detected in OS cell lines. The role of PSKH1
in OS cells was also investigated. Knockdown of PSKH1 suppressed OS
cell proliferation, migration and invasion in vitro. The
present in vivo data support the role of PSKH1 in the
development of OS. These findings are consistent with previous
reports on colorectal cancer (10,12).
The present findings indicated that PSKH1 may have an oncogenic
role in the development of OS. A limitation of the present study
was that the research was performed only in three OS cell lines.
Future studies should investigate the role of PSKH1 and p38 in more
OS cell lines, as well as other types of cancer.
In summary, the present data indicated that PSKH1
expression was associated with OS prognosis and PSKH1 may play an
oncogenic role in OS via the p38/MAPK pathway. Therefore, PSKH1 may
serve as a potential novel therapeutic target for OS.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81802721, 82001309) and Fundamental
Research Funds for the Central Universities of China (grant no.
22120210570).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XFZ, YY and XZZ conceived and designed the study.
XFZ, ZYW and YY performed the experiments; FY, ZYW and CJ analyzed
and interpreted data. XFZ drafted the manuscript. XFZ and YY
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved (approval no. 2020-DW-007) by
the Ethics Committee for Animal Experiments of Tongji Hospital,
Shanghai, China. Animals were handled according to The Guide for
the Care and Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
American Cancer Society, . Cancer Facts
& Figures 2020. Am Cancer Soc. 1–52. 2020.
|
|
2
|
Meltzer PS and Helman LJ: New horizons in
the treatment of osteosarcoma. N Engl J Med. 385:2066–2076. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Corre I, Verrecchia F, Crenn V, Redini F
and Trichet V: The osteosarcoma microenvironment: A complex but
targetable ecosystem. Cells. 9:9762020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gill J and Gorlick R: Advancing therapy
for osteosarcoma. Nat Rev Clin Oncol. 18:609–624. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Belayneh R, Fourman MS, Bhogal S and Weiss
KR: Update on osteosarcoma. Curr Oncol Rep. 23:712021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shoaib Z, Fan TM and Irudayaraj JMK:
Osteosarcoma mechanobiology and therapeutic targets. Br J
Pharmacol. 179:201–217. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sheng G, Gao Y, Yang Y and Wu H:
Osteosarcoma and metastasis. Front Oncol. 11:7802642021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brede G, Solheim J and Prydz H: PSKH1, a
novel splice factor compartment-associated serine kinase. Nucleic
Acids Res. 30:5301–5309. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brede G, Solheim J, Tröen G and Prydz H:
Characterization of PSKH1, a novel human protein serine kinase with
centrosomal, golgi, and nuclear localization. Genomics. 70:82–92.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim ST, Ahn TJ, Lee E, Do IG, Lee SJ, Park
SH, Park JO, Park YS, Lim HY, Kang WK, et al: Exploratory biomarker
analysis for treatment response in KRAS wild type metastatic
colorectal cancer patients who received cetuximab plus irinotecan.
BMC Cancer. 15:7472015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Whitworth H, Bhadel S, Ivey M, Conaway M,
Spencer A, Hernan R, Holemon H and Gioeli D: Identification of
kinases regulating prostate cancer cell growth using an RNAi
phenotypic screen. PLoS One. 7:e389502012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Zhang S, Yin J and Xu R: MiR-566
mediates cell migration and invasion in colon cancer cells by
direct targeting of PSKH1. Cancer Cell Int. 19:3332019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu H, Zou D, Ni N, Zhang S, Zhang Q and
Yang L: Overexpression of NCAPG in ovarian cancer is associated
with ovarian cancer proliferation and apoptosis via p38 MAPK
signaling pathway. J Ovarian Res. 15:982022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu M, Zhou J, Liu X, Feng Y, Yang W, Wu
F, Cheung OK, Sun H, Zeng X, Tang W, et al: Targeting
monocyte-intrinsic enhancer reprogramming improves immunotherapy
efficacy in hepatocellular carcinoma. Gut. 69:365–379. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Puri PL, Wu Z, Zhang P, Wood LD, Bhakta
KS, Han J, Feramisco JR, Karin M and Wang JY: Induction of terminal
differentiation by constitutive activation of p38 MAP kinase in
human rhabdomyosarcoma cells. Genes Dev. 14:574–584. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu-Lee LY, Yu G, Lee YC, Lin SC, Pan J,
Pan T, Yu KJ, Liu B, Creighton CJ, Rodriguez-Canales J, et al:
Osteoblast-secreted factors mediate dormancy of metastatic prostate
cancer in the bone via activation of the
TGFβRIII-p38MAPK-pS249/T252RB pathway. Cancer Res. 78:2911–2924.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Comes F, Matrone A, Lastella P, Nico B,
Susca FC, Bagnulo R, Ingravallo G, Modica S, Lo Sasso G, Moschetta
A, et al: A novel cell type-specific role of p38alpha in the
control of autophagy and cell death in colorectal cancer cells.
Cell Death Differ. 14:693–702. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Igea A and Nebreda AR: The stress kinase
p38α as a target for cancer therapy. Cancer Res. 75:3997–4002.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kumar VB, Lin SH, Mahalakshmi B, Lo YS,
Lin CC, Chuang YC, Hsieh MJ and Chen MK: Sodium danshensu inhibits
oral cancer cell migration and invasion by modulating p38 signaling
pathway. Front Endocrinol (Lausanne). 11:5684362020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gerdes J, Li L, Schlueter C, Duchrow M,
Wohlenberg C, Gerlach C, Stahmer I, Kloth S, Brandt E and Flad HD:
Immunobiochemical and molecular biologic characterization of the
cell proliferation-associated nuclear antigen that is defined by
monoclonal antibody Ki-67. Am J Pathol. 138:867–873.
1991.PubMed/NCBI
|
|
22
|
Gerdes J, Lemke H, Baisch H, Wacker HH,
Schwab U and Stein H: Cell cycle analysis of a cell
proliferation-associated human nuclear antigen defined by the
monoclonal antibody Ki-67. J Immunol. 133:1710–1715. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rioux-Leclercq N, Turlin B, Bansard J,
Patard J, Manunta A, Moulinoux JP, Guillé F, Ramée MP and Lobel B:
Value of immunohistochemical Ki-67 and p53 determinations as
predictive factors of outcome in renal cell carcinoma. Urology.
55:501–505. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krishna OH, Kayla G, Abdul Aleem M,
Malleboyina R and Reddy Kota R: Immunohistochemical expression of
Ki67 and p53 in wilms tumor and its relationship with tumor
histology and stage at presentation. Patholog Res Int.
2016:61239512016.PubMed/NCBI
|
|
25
|
Dudderidge TJ, Stoeber K, Loddo M,
Atkinson G, Fanshawe T, Griffiths DF and Williams GH: Mcm2,
Geminin, and KI67 define proliferative state and are prognostic
markers in renal cell carcinoma. Clin Cancer Res. 11:2510–2517.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi W, Hu J, Zhu S, Shen X, Zhang X, Yang
C, Gao H and Zhang H: Expression of MTA2 and Ki-67 in
hepatocellular carcinoma and their correlation with prognosis. Int
J Clin Exp Pathol. 8:13083–13089. 2015.PubMed/NCBI
|
|
27
|
Kankuri M, Söderström KO, Pelliniemi TT,
Vahlberg T, Pyrhönen S and Salminen E: The association of
immunoreactive p53 and Ki-67 with T-stage, grade, occurrence of
metastases and survival in renal cell carcinoma. Anticancer Res.
26:3825–3833. 2006.PubMed/NCBI
|
|
28
|
Zeng M, Zhou J, Wen L, Zhu Y, Luo Y and
Wang W: The relationship between the expression of Ki-67 and the
prognosis of osteosarcoma. BMC Cancer. 21:2102021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Evangelisti C, Paganelli F, Giuntini G,
Mattioli E, Cappellini A, Ramazzotti G, Faenza I, Maltarello MC,
Martelli AM, Scotlandi K, et al: Lamin A and Prelamin A counteract
migration of osteosarcoma cells. Cells. 9:7742020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Niu G, Ye T, Qin L, Bourbon PM, Chang C,
Zhao S, Li Y, Zhou L, Cui P, Rabinovitz I, et al: Orphan nuclear
receptor TR3/Nur77 improves wound healing by upregulating the
expression of integrin β4. FASEB J. 29:131–140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi B, Xue M, Wang Y, Wang Y, Li D, Zhao X
and Li X: An improved method for increasing the efficiency of gene
transfection and transduction. Int J Physiol Pathophysiol
Pharmacol. 10:95–104. 2018.PubMed/NCBI
|
|
35
|
Xiang B, Geng R, Zhang Z, Ji X, Zou J,
Chen L and Liu J: Identification of the effect and mechanism of
Yiyi Fuzi Baijiang powder against colorectal cancer using network
pharmacology and experimental validation. Front Pharmacol.
13:9298362022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi D, Wu F, Mu S, Hu B, Zhong B, Gao F,
Qing X, Liu J, Zhang Z and Shao Z: LncRNA AFAP1-AS1 promotes
tumorigenesis and epithelial-mesenchymal transition of osteosarcoma
through RhoC/ROCK1/p38MAPK/Twist1 signaling pathway. J Exp Clin
Cancer Res. 38:3752019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fan MK, Zhang GC, Chen W, Qi LL, Xie MF,
Zhang YY, Wang L and Zhang Q: Siglec-15 promotes tumor progression
in osteosarcoma via DUSP1/MAPK pathway. Front Oncol. 11:7106892021.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia Y, Khoi PN, Yoon HJ, Lian S, Joo YE,
Chay KO, Kim KK and Jung YD: Piperine inhibits IL-1β-induced IL-6
expression by suppressing p38 MAPK and STAT3 activation in gastric
cancer cells. Mol Cell Biochem. 398:147–156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Niu J, Yan T, Guo W, Wang W, Zhao Z, Ren
T, Huang Y, Zhang H, Yu Y and Liang X: Identification of potential
therapeutic targets and immune cell infiltration characteristics in
osteosarcoma using bioinformatics strategy. Front Oncol.
10:16282020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wen S, Hou Y, Fu L, Xi L, Yang D, Zhao M,
Qin Y, Sun K, Teng Y and Liu M: Cancer-associated fibroblast
(CAF)-derived IL32 promotes breast cancer cell invasion and
metastasis via integrin β3-p38 MAPK signalling. Cancer Lett.
442:320–332. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ni S, Li J, Qiu S, Xie Y, Gong K and Duan
Y: KIF21B expression in osteosarcoma and its regulatory effect on
osteosarcoma cell proliferation and apoptosis through the PI3K/AKT
pathway. Front Oncol. 10:6067652021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiang QG, Xiong CF and Lv YX: Kin17
facilitates thyroid cancer cell proliferation, migration, and
invasion by activating p38 MAPK signaling pathway. Mol Cell
Biochem. 476:727–739. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fang Y, Yang J, Zu G, Cong C, Liu S, Xue
F, Ma S, Liu J, Sun Y and Sun M: Junctional adhesion molecule-like
protein promotes tumor progression and metastasis via p38 signaling
pathway in gastric cancer. Front Oncol. 11:5656762021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Anderson ME: Update on survival in
osteosarcoma. Orthop Clin North Am. 47:283–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Saraf AJ, Fenger JM and Roberts RD:
Osteosarcoma: Accelerating progress makes for a hopeful future.
Front Oncol. 8:42018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Z, Wang Z, Li B, Wang S, Chen T and
Ye Z: Innate immune cells: A potential and promising cell
population for treating osteosarcoma. Front Immunol. 10:11142019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen C, Xie L, Ren T, Huang Y, Xu J and
Guo W: Immunotherapy for osteosarcoma: Fundamental mechanism,
rationale, and recent breakthroughs. Cancer Lett. 500:1–10. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Whelan JS and Davis LE: Osteosarcoma,
chondrosarcoma, and chordoma. J Clin Oncol. 36:188–193. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gianferante DM, Mirabello L and Savage SA:
Germline and somatic genetics of osteosarcoma-connecting aetiology,
biology and therapy. Nat Rev Endocrinol. 13:480–491. 2017.
View Article : Google Scholar : PubMed/NCBI
|