Introduction
Renal cell carcinoma (RCC) is one of the most common
malignancies among adults and is estimated to have caused 73,750
new cases and 14,830 deaths in the United States in 2020 (1). Clear cell (cc)RCC is the most common
histologic subtype of RCC, accounting for ~90% of RCC cases
(2). ccRCC is the leading cause of
death in the majority of patients with RCC and is responsible for
~3% of all adults diagnosed with cancer (3). The morbidity and mortality of ccRCC
are also increasing in the majority of countries (4). Despite remarkable advances in the
diagnosis and treatment of ccRCC in recent years, ccRCC remains one
of the deadliest cancers. In fact, metastasis at diagnosis occurs
in a large proportion of patients owing to the lack of
characteristic clinical symptoms (5). Approximately 30% of patients with
ccRCC developed recurrence and progression despite surgical
resection of the primary tumor (6,7). ccRCC
is a chemo- and radioresistant neoplasia, with limited alternative
treatment options available for therapeutic use (8). At present, novel therapeutic options
for ccRCC include anti-angiogenic agents (such as sorafenib,
sunitinib, pazopanib, axitinib and bevacizumab), mTOR inhibitors
(temsirolimus and everolimus) and immune checkpoint inhibitors
(nivolumab) (9,10). However, these drugs result in only
partial responses in few patients with ccRCC and a poor prognosis.
Therefore, there is an urgent need to identify new biomarkers for
accurate diagnosis of ccRCC and to explore its underlying
mechanisms.
The distal-less homeobox (DLX) gene family is a
cluster of homeobox genes comprising six different members
(DLX1-DLX6) (11). DLX is
postulated to serve a role in forebrain and craniofacial
development. In fact, according to previous studies, the DLX gene
family may be involved in early development and cell
differentiation, and is frequently dysregulated in neoplasms
(12,13). For instance, DLX1 may promote
progression and metastasis of prostate and ovarian cancer by
enhancing TGF-β/SMAD4 signaling (14,15).
DLX2 could counteract TGF-β-induced cell cycle arrest and apoptosis
to promote tumorigenesis (16), and
it is considered to be a marker of survival and disease progression
in prostate cancer (17); DLX3
regulates cell cycle progression and squamous tumor growth
(18). DLX5 is involved in the
growth and diffuse spreading of invasive glioma cells (19), and DLX4 serves a vital role in the
early development and differentiation of tumor cells (12). DLX4 may induce a megakaryocytic
transcriptional program by inducing IL-1β and NF-κB signaling
(20). DLX4 may induce expression
of the cell surface molecule CD44 in ovarian tumor cells by
stimulating IL-1β-mediated NF-κB activity, thereby promoting
tumor-mesothelial cell interactions and peritoneal metastasis of
ovarian cancer (21). Chen et
al (22) adapted the approach
of network-based molecular modeling of physiological behaviors to
specifically isolate physiological behaviors in alopecia areata
model that contribute to the recruitment of immune cells in
autoimmune disease. They found that DLX4 is expressed in the skin
and could induce recruitment of immune cells in alopecia areata
model. Therefore, they propose that DLX4 is a master regulator of
immune infiltration recruitment, and the loss of DLX4 expression
may contribute to immune evasion in cancer. These studies suggested
that aberrant expression of DLX gene family may be related to the
development and metastasis of tumors. However, the roles of DLX
gene family in ccRCC patients remain unclear.
In the present study, the expression levels of DLX
gene family in ccRCC were examined and its prognostic value and
correlations with pathological parameters were evaluated. Here,
differences in both overall survival and disease-free survival
(DFS) were selected to investigate the association with the
distributions of clinical phenotypes. DLX4 was selected for further
validation and its potential functional mechanisms were
explored.
Materials and methods
Data acquisition
mRNA expression data of six members of DLX gene
family in The Cancer Genome Atlas (TCGA) dataset (HTSeq Counts)
were acquired from University of California, Santa Cruz (UCSC) Xena
website (xenabrowser.net) (23);
the project number was TCGA-KIRC. The data were processed using
RSEM normalization methods and expressed as log2(x+1)
transformed. Clinical information of 533 patients, including age,
sex, pathological stage, pathological grade, TNM stage, overall
survival and survival status were obtained from UCSC Xena. DFS of
patients with ccRCC was obtained from the cBio Cancer Genomics
Portal (http://cbioportal.org).
Differential expression levels,
diagnostic efficiency and prognostic significances
The differential expression of six members of the
DLX gene family was compared between ccRCC and adjacent normal
renal tissues in TCGA dataset. The differential expression of DLX4
was also compared between the different clinical subgroups
(advanced vs. early stage, high vs. low grade, N0 vs. N1, M0 vs.
M1). Kaplan-Meier curves and log-rank tests were performed to
evaluate the prognostic significance, and receiver operating
characteristic (ROC) curves were generated to assess the diagnostic
efficiency (24). Univariate and
multivariate Cox hazard analyses were performed to assess the
overall survival and DFS of patients with ccRCC.
ccRCC tissue samples
A total of 40 pairs of ccRCC tumors and matched
normal renal tissues were retrieved from the Department of Urology
at the Second People's Hospital of Wuhu (Wuhu, China) and The First
Affiliated Hospital of Anhui Medical University (Hefei, China)
between May 2019 and May 2020. Adjacent normal renal tissues, ≥2 cm
away from the location of the tumor, were cut and collected.
Samples were placed in RNAlater stabilization solution (Invitrogen;
Thermo Fisher Scientific, Inc.) and frozen in liquid nitrogen.
These samples were evaluated by two pathologists to confirm the
histopathological results of ccRCC. Inclusion criteria were ccRCC,
and exclusion criteria were other types of RCC and benign renal
tumors. Written informed consent was provided by all patients. The
present study was approved by the Ethics Committee of Human
Research of The Second People's Hospital of Wuhu and The First
Affiliated Hospital of Anhui Medical University (approval no.
PJ2019-14-22). The study methodology conformed with the standards
of the Declaration of Helsinki.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from ccRCC tissue using the
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
method. The purity and concentration of total RNA solution were
detected using a NanoDrop spectrophotometer (NanoDrop Technologies;
Thermo Fisher Scientific, Inc.). Extracted RNA was reverse
transcribed into cDNA using a PrimeScript™ RT with gDNA Eraser kit
(Takara Bio, Inc.) according to the manufacturer's protocol. The
cDNA was subjected to qPCR by a SYBR Green Mix (Takara Bio, Inc.).
Thermocycling conditions were as follows: Initial denaturation at
95 for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for
34 sec, then final extension at 95°C for 15 sec, 60°C for 1 min and
95°C for 15 sec. A final reaction volume of 20 µl was used, and the
ABI 7500 Real-Time PCR System (Thermo Fisher Scientific, Inc.) was
used to measure the reactions. Relative gene expression of DLX4
were analyzed using the 2−ΔΔCq method (25). GAPDH was used as endogenous control
to normalize the relative expression of DLX4. The primers were
purchased from Sangon Biotech Co., Ltd., and the sequences were as
follows: DLX4 forward, 5′-CAGCACCTAAACCAGCGTTTC-3′ and reverse,
5′-GAGCTTCTTATACTTGGAGCGTT-3′; and GAPDH forward,
5′-GGGAGCCAAAAGGGTCAT-3′ and reverse,
5′-GAGTCCTTCCACGATACCAA-3′.
Immunohistochemistry
Paraffin-embedded tissue specimens were sliced into
4-µm-thick sections. The slides were placed in a 60°C thermostat
and baked for 20 min to dewax. Then, the slides were hydrated in
xylene I (30 min), xylene II (30 min), anhydrous ethanol (5 min)
twice and 95, 90, 80 and 70% ethanol (5 min each). Then, the slides
were heated at 100°C for 10 min in citric acid buffer (0.01 M, pH
6.0) for antigen retrieval. The slides were incubated in 3%
hydrogen peroxide solution (cat. no. SP 9000; Beijing Zhongshan
Jinqiao Biotechnology Co., Ltd.) for 15 min at room temperature and
washed three times in phosphate-buffered saline (PBS; pH 7.4). The
slides were blocked with 3% bovine serum albumin (cat. no. G5001;
Servicebio) for 1 h at room temperature, then incubated with
anti-DLX4 antibody (1:100; cat. no. DF3387; Affinity Biosciences,
Ltd.) overnight at 4°C. After three washes with PBS, the slides
were incubated with biotinylated goat anti-rabbit IgG (1:200; cat.
no. G23303; Servicebio) for 2 h at room temperature. Finally, DAB
(Beyotime Institute of Biotechnology) was used to detect the immune
complexes and the sections were counterstained with hematoxylin for
3 min at room temperature. The images were captured using a light
microscope (Olympus Corporation). DLX4 expression was measured
based on the intensity of immune staining (intensity score) with
ImageJ version 6.0 software (National Institutes of Health), as
previously described (26).
Gene set enrichment analysis
(GSEA)
Patients in TCGA data set were divided into high and
low expression groups according to the median values of DLX4 gene
expression. GSEA was conducted to explore the possible functional
mechanisms of DLX4 in ccRCC with GSEA 3.0 software (Broad
Institute; broad.mit.edu/gsea) (27). The reference gene set file was
h.all.v7.0.symbols.gmt; normalized P-value <0.05 and FDR
<0.25 were set as threshold values.
Functional enrichment analysis
Pearson correlation coefficients was calculated to
determine the association between DLX4 gene expression and genes in
TCGA dataset. Genes that strongly correlated with the expression
levels of DLX4 (correlation coefficient R>0.4) were obtained as
previously described (28). A total
of 559 positively correlated genes and 226 negatively correlated
genes were identified. Kyoto Encyclopedia of Genes and Genomes
(KEGG) and Gene Ontology (GO) enrichment analyses were performed
using The Database for Annotation, Visualization and Integrated
Discovery (https://david.ncifcrf.gov) to
investigate the possible molecular mechanisms of these selected
genes. The cutoff value was set at P<0.05.
Estimation of the immune
microenvironment composition
To quantify the cellular composition of the immune
infiltrates in each patient in TCGA dataset, a set of metagenes,
including non-overlapping sets of genes that are representative of
28 specific immune cell subpopulations, was obtained from a
previous study (29). Subsequently,
single-sample GSEA was used to quantify the 28 types of immune
cells based on the metagenes. In the tumor microenvironment, immune
and stromal cells are the two main non-tumor components; these
components have been proposed to be valuable for tumor treatment
and prognosis (30). To assess the
tumor microenvironment associated with DLX4 expression levels, the
immune and the stromal scores (which reflect the infiltration
levels of non-tumor cells) for the TCGA dataset were calculated
using the ESTIMATE package (30).
The differences in immune cell composition and immune and stromal
scores were compared between the high and low DLX4 expression
groups, which were divided based on the median expression values.
In addition, the correlations between DLX4 expression levels and
immunosuppressive immune cells were evaluated.
Comparison of tumor mutation burden
and cytolytic activity
Tumor mutation burden (TMB) is defined as the total
amount of coding errors of somatic genes, base substitutions,
insertions or deletions detected per million bases (31). In the present study, 38 megabases
was used as the exon length. TMB was calculated as (the number of
variants)/(exon length) for each patient with ccRCC using Perl
scripts (32). The somatic mutation
status data of ccRCC samples (workflow type: VarScan2 Variant
Aggregation and Masking) were downloaded from the TCGA data portal
(https://portal.gdc.cancer.gov/repository) in May 2021.
The cytolytic activity scores were calculated as the geometric mean
of the granzyme A and perforin expression (13). The TMB scores and cytolytic activity
scores were also compared between high and low DLX4 expression
groups.
Statistical analysis
For equivalent variables with a normal distribution,
an unpaired Student's t-test was performed for comparisons between
the two groups, and one-way ANOVA followed by Tukey's post hoc test
was used for comparison between multiple groups. The paired
Student's t-test was used to compare paired samples. For
non-normally distributed variables, the Wilcoxon rank-sum test was
performed for comparisons between two groups. Pearson's
χ2 test was used to evaluate the distributions of
clinical factors between the high and low-DLX4 expression groups.
Log-rank tests were used for Kaplan-Meier curves to assess survival
differences. The correlations between DLX4 expression and tumor
immunosuppressive cells were evaluated using Pearson's correlation
analysis, and ‘general linear model’ method was used to fit curves.
P<0.05 was used to indicate a statistically significant
difference.
Results
Relative expression of DLX family
genes in ccRCC
To investigate the roles of DLX family genes in
ccRCC, the mRNA expression levels of six members of the DLX family
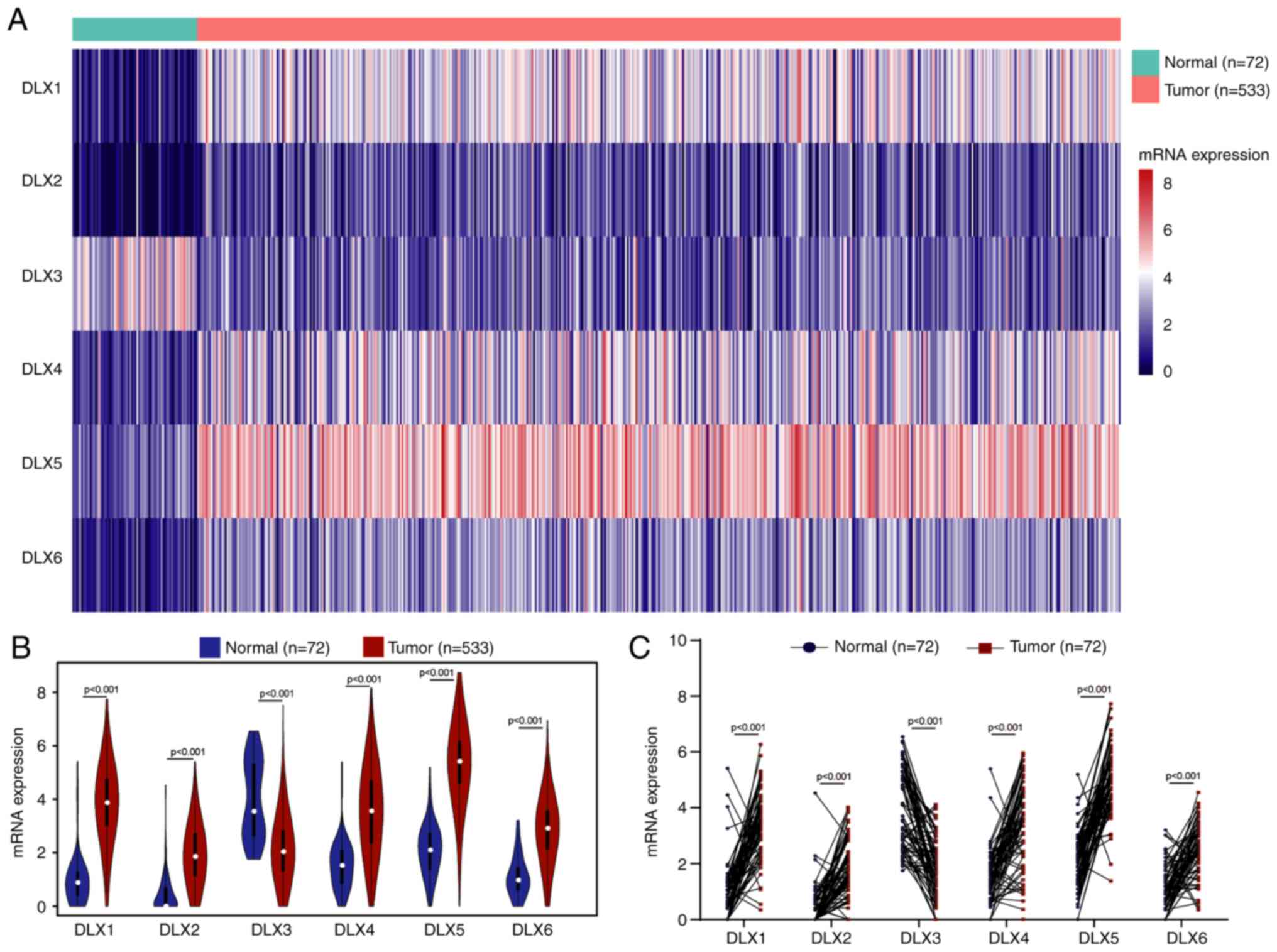
in the TCGA dataset were investigated. A heat map revealed the
overall expression levels of the six members of the DLX family
between normal and tumor samples in TCGA dataset (Fig. 1A). Furthermore, the expression
levels of six members were compared between normal and tumor
samples. Downregulated levels of DLX3 and upregulated levels of
DLX1, DLX2, DLX4, DLX5 and DLX6 were observed in tumor samples
compared with normal samples in TCGA dataset (Fig. 1B). The expression levels of paired
normal and tumor samples in TCGA dataset were also compared, which
revealed the same expression patterns (Fig. 1C).
Prognostic values of DLX family in
ccRCC
Patients in the TCGA data set were divided into high
and low expression group according to the median values of the mRNA
expression levels for each gene of the DLX family. A total of 511
patients with ccRCC with overall survival and 422 with DFS
information were included. The differences in overall and DFS
status for six members of DLX family were compared using the
χ2 test. The distribution for overall survival status
was found to be significantly different for DLX2 and DLX4, whereas
the distribution for DFS status was significantly different for
DLX2, DLX4 and DLX5 (Table I).
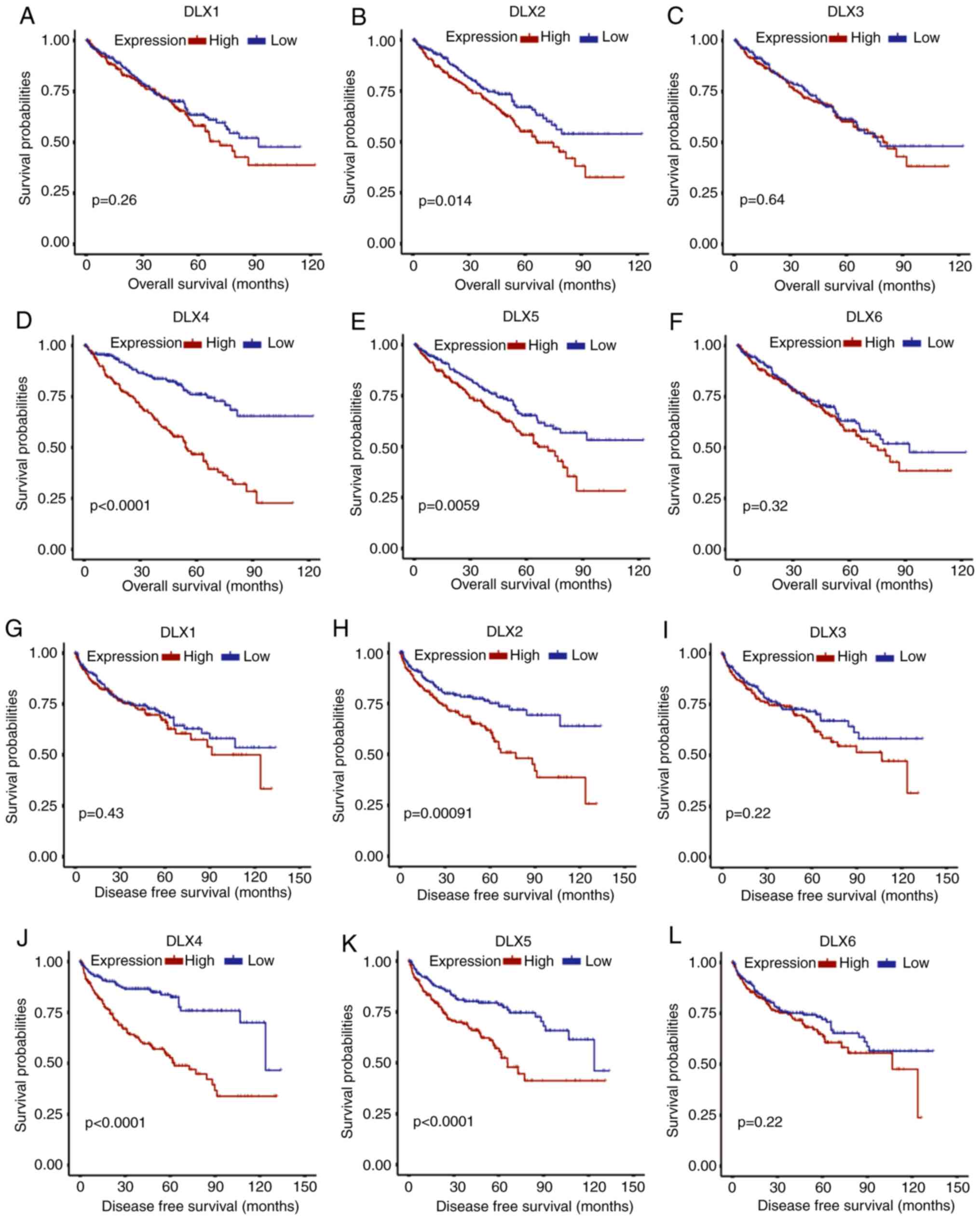
Kaplan-Meier curves were used to evaluate the prognostic
significances of the DLX family in ccRCC. Kaplan-Meier curves for
overall survival revealed poor prognosis in high DLX2 expression
group (Fig. 2B), DLX4 (Fig. 2D) and DLX5 (Fig. 2E) for patients with ccRCC. However,
the overall survival rates between the high and low expression
group of DLX1 (Fig. 2A), DLX3
(Fig. 2C) and DLX6 (Fig. 2F) were not significantly different.
DFS was also compared between the high and low expression groups. A
significantly lower DFS rate was identified in the high expression
group of DLX2 (Fig. 2H), DLX4
(Fig. 2J) and DLX5 (Fig. 2K) relative to the low expression
group. However, the DFS rate was not significantly different
between DLX1 (Fig. 2G), DLX3
(Fig. 2I) and DLX6 (Fig. 2L). The expression of DLX2 and DLX4
was revealed to be associated with overall and DFS in ccRCC
patients in both survival distribution difference assessment and
Kaplan-Meier curves. Difference in either overall survival or DFS
were not identified for several genes, including DLX3 and DLX5.
Therefore, DLX2 and DLX4 were selected for subsequent analyses.
 | Table I.Comparison of overall survival and
DFS between different expression levels of DLX1-6. |
Table I.
Comparison of overall survival and
DFS between different expression levels of DLX1-6.
|
| Overall
survival | DFS |
|---|
|
|
|
|
|---|
| Variable | χ2 | P-value | χ2 | P-value |
|---|
| DLX1 (high vs.
low) | 2.530 | 0.112 | 0.121 | 0.728 |
| DLX2 (high vs.
low) | 9.286 | 0.002 | 10.939 | <0.001 |
| DLX3 (high vs.
low) | 0.388 | 0.533 | 1.424 | 0.233 |
| DLX4 (high vs.
low) | 41.010 | <0.001 | 26.160 | <0.001 |
| DLX5 (high vs.
low) | 2.420 | 0.120 | 7.146 | 0.008 |
| DLX6 (high vs.
low) | 0.992 | 0.319 | 0.964 | 0.326 |
Association between
clinicopathological characteristics and DLX2 or DLX4
To investigate the association of DLX2 and DLX4 with
the distributions of clinical phenotypes, patients were divided
into two subgroups according to the median mRNA expression levels.
A high proportion of males, advanced pathological stage, higher
grade, higher T stage, N1 stage, and M1 stage were identified in
the high expression group of DLX4 (Table II); however, no significant
difference was identified for age distribution (Table II). In addition, the expression of
DLX2 was not shown to be associated with the clinicopathological
variables examined for patients with ccRCC (Table II). Therefore, DLX4 was selected
for further studies. Univariate Cox regression analysis showed that
the expression level of DLX4 was a risk factor for overall survival
(Table III) and DFS (Table IV) in ccRCC patients. Based on
multivariate Cox hazard analysis, the expression level of DLX4 was
an independent risk variable for the overall survival of ccRCC
patients (Table III) and DFS
(Table IV) after integration with
multiple clinicopathological characteristics. These findings
indicated that the expression level of DLX4 was as an independent
prognostic predictor for ccRCC patients, which may provide a
supplement for clinical factors.
 | Table II.Association between DLX2 or DLX4 mRNA
expression and clinicopathological characteristics of patients with
clear cell renal cell carcinoma. |
Table II.
Association between DLX2 or DLX4 mRNA
expression and clinicopathological characteristics of patients with
clear cell renal cell carcinoma.
|
| DLX2 mRNA
expression | DLX4 mRNA
expression |
|---|
|
|
|
|
|---|
| Clinicopathological
characteristic | High (n=266) | Low (n=267) | χ2 | P-value | High (n=266) | Low (n=267) | χ2 | P-value |
|---|
| Age, years (mean ±
SD) | 60.27±12.12 | 60.98±12.18 |
| 0.5 | 59.84±11.9 | 61.41±12.36 |
| 0.136 |
| Sex |
|
| 0 | 1 |
|
| 18.977 | <0.001 |
|
Male | 172 | 173 |
|
| 185 | 160 |
|
|
|
Female | 94 | 94 |
|
| 81 | 107 |
|
|
| Stage |
|
| 6.97 | 0.138 |
|
| 41.738 | <0.001 |
| I | 124 | 143 |
|
| 103 | 164 |
|
|
| II | 24 | 33 |
|
| 23 | 34 |
|
|
|
III | 69 | 54 |
|
| 78 | 45 |
|
|
| IV | 48 | 35 |
|
| 60 | 23 |
|
|
|
Unknown | 1 | 2 |
|
| 2 | 1 |
|
|
| Grade |
|
| 2.39 | 0.664 |
|
| 49.563 | <0.001 |
| G1 | 7 | 7 |
|
| 3 | 11 |
|
|
| G2 | 107 | 122 |
|
| 87 | 142 |
|
|
| G3 | 109 | 97 |
|
| 113 | 93 |
|
|
| G4 | 40 | 36 |
|
| 61 | 15 |
|
|
|
Unknown | 3 | 5 |
|
| 2 | 6 |
|
|
| T stage |
|
| 5.483 | 0.14 |
|
| 34.486 | <0.001 |
| T1 | 126 | 147 |
|
| 108 | 165 |
|
|
| T2 | 32 | 37 |
|
| 31 | 38 |
|
|
| T3 | 101 | 79 |
|
| 118 | 62 |
|
|
| T4 | 7 | 4 |
|
| 9 | 2 |
|
|
| N stage |
|
| 0.061 | 0.806 |
|
| 5.254 | 0.022 |
|
N0/Nx | 259 | 258 |
|
| 253 | 264 |
|
|
| N1 | 7 | 9 |
|
| 13 | 3 |
|
|
| M stage |
|
| 1.531 | 0.216 |
|
| 17.329 | <0.001 |
|
M0/Mx | 221 | 233 |
|
| 209 | 245 |
|
|
| M1 | 45 | 34 |
|
| 57 | 22 |
|
|
 | Table III.Univariate and multivariate Cox
hazard analyses of DLX4 mRNA expression levels for overall survival
of patients (n=511). |
Table III.
Univariate and multivariate Cox
hazard analyses of DLX4 mRNA expression levels for overall survival
of patients (n=511).
|
| Univariate
analysis | Multivariate
analysisa |
|---|
| Clinicopathological
characteristic |
|
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age, years | 1.702 | 1.243-2.332 | <0.001 | 1.54 | 1.122-2.112 | 0.007 |
| ≤60
(n=258) |
|
|
|
|
|
|
| >60
(n=253) |
|
|
|
|
|
|
| Sex | 1.024 | 0.743-1.409 | 0.886 |
|
|
|
| Female
(n=176) |
|
|
|
|
|
|
| Male
(n=335) |
|
|
|
|
|
|
| T stage | 1.839 | 1.571-2.152 | <0.001 | 1.306 | 1.083-1.576 | 0.005 |
| T1 or
T2 (n=326) |
|
|
|
|
|
|
| T3 or
T4 (n=185) |
|
|
|
|
|
|
| N stage | 3.579 | 1.885-6.798 | <0.001 | 1.835 | 0.952-3.537 | 0.069 |
| N0 or
NX (n=497) |
|
|
|
|
|
|
| N1
(n=14) |
|
|
|
|
|
|
| M stage | 4.488 | 3.262-6.175 | <0.001 | 2.527 | 1.749-3.654 | <0.001 |
| M0 or
MX (n=433) |
|
|
|
|
|
|
| M1
(n=78) |
|
|
|
|
|
|
| Grade | 1.618 | 1.357-1.929 | <0.001 | 1.241 | 1.028-1.5 | 0.025 |
| G1 or
G2 (n=235) |
|
|
|
|
|
|
| G3 or
G4 (n=276) |
|
|
|
|
|
|
| DLX4
expression | 2.841 | 2.027-3.982 | <0.001 | 1.885 | 1.320-2.691 | <0.001 |
| Low
(n=255) |
|
|
|
|
|
|
| High
(n=256) |
|
|
|
|
|
|
 | Table IV.Univariate and multivariate Cox
hazard analyses of DLX4 mRNA level for DFS of patients (n=422). |
Table IV.
Univariate and multivariate Cox
hazard analyses of DLX4 mRNA level for DFS of patients (n=422).
|
| Univariate
analysis | Multivariate
analysisa |
|---|
| Clinicopathological
characteristic |
|
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) | 1.366 | 0.959-1.945 | 0.084 |
|
|
|
| ≤60
(n=230) |
|
|
|
|
|
|
| >60
(n=192) |
|
|
|
|
|
|
| Sex | 1.429 | 0.962-2.123 | 0.077 |
|
|
|
| Female
(n=142) |
|
|
|
|
|
|
| Male
(n=280) |
|
|
|
|
|
|
| T stage | 4.527 | 3.134-6.539 | <0.001 | 1.396 | 1.131-1.726 | 0.002 |
| T1 or
T2 (n=283) |
|
|
|
|
|
|
| T3 or
T4 (n=139) |
|
|
|
|
|
|
| N stage | 5.955 | 2.990-11.861 | <0.001 | 2.729 | 1.346-5.536 | 0.005 |
| N0 or
NX (n=410) |
|
|
|
|
|
|
| N1
(n=12) |
|
|
|
|
|
|
| M stage | 8.537 | 5.882-12.398 | <0.001 | 5.176 | 3.415-7.845 | <0.001 |
| M0 or
MX (n=371) |
|
|
|
|
|
|
| M1
(n=51) |
|
|
|
|
|
|
| Grade | 1.832 | 1.491-2.252 | <0.001 | 2.301 | 1.306-3.157 | 0.002 |
| G1 or
G2 (n=208) |
|
|
|
|
|
|
| G3 or
G4 (n=214) |
|
|
|
|
|
|
| DLX4
expression | 3.011 | 2.040-4.444 | <0.001 | 1.887 | 1.245-2.860 | 0.003 |
| Low
(n=211) |
|
|
|
|
|
|
| High
(n=211) |
|
|
|
|
|
|
The relationships between clinical variables and
DLX4 expression levels were investigated in the TCGA dataset. The
expression levels of DLX4 were significantly higher in stage
3&4 vs. 1&2 (Fig. 3A and
D), grade 3&4 vs. 1&2 (Fig.
3B and E) and T 3&4 vs. T 1&2 (Fig. 3C and F). In addition, high
expression levels of DLX4 were observed in N1 vs. N0 stage
(Fig. 3G), M1 vs. M0 stage
(Fig. 3H) and dead vs. alive
(Fig. 3I). These results suggested
that the expression levels of DLX4 were highly associated with
tumor malignancy.
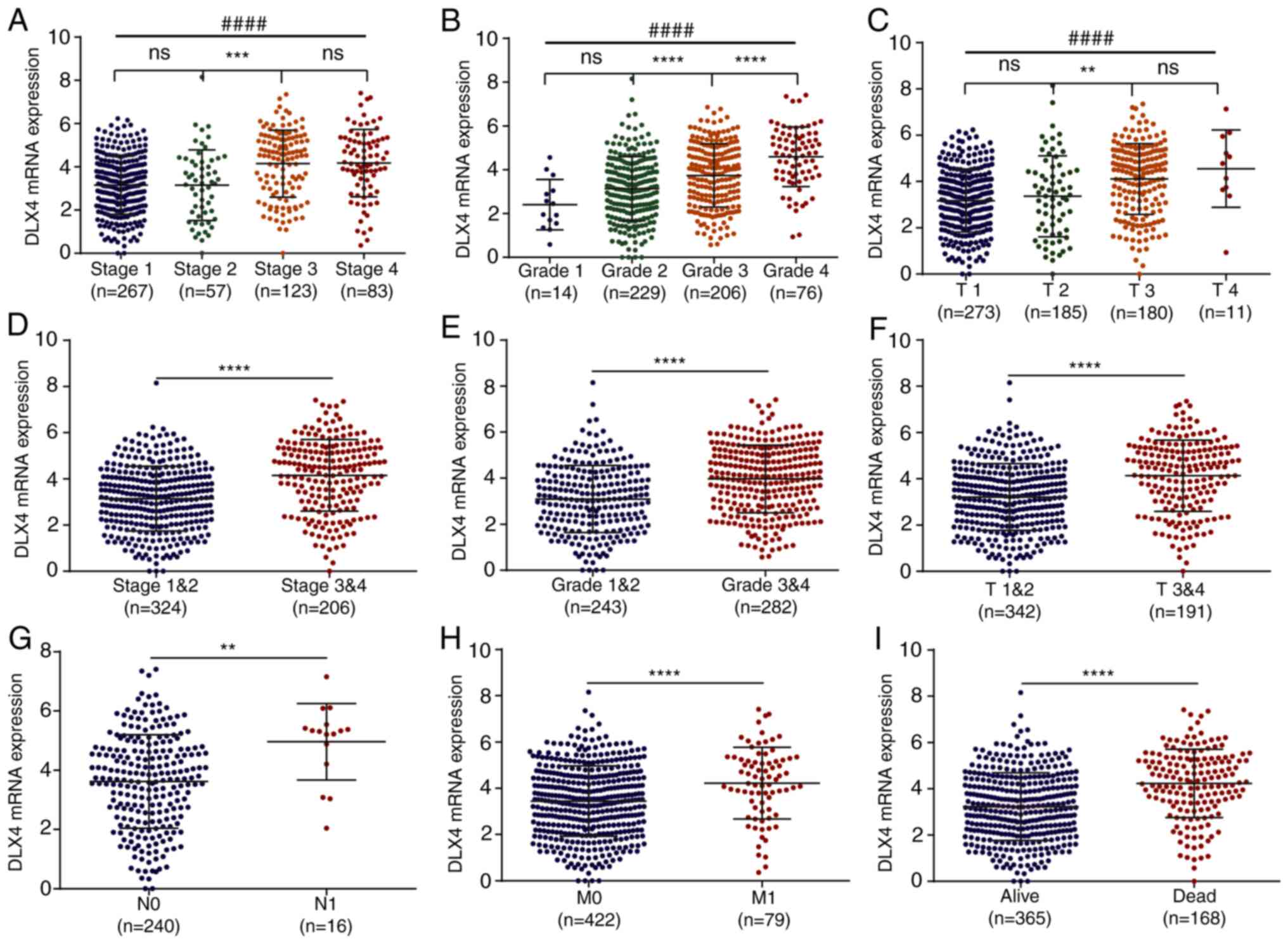 | Figure 3.Association with clinicopathological
variables for DLX2 and DLX4. Comparisons of the expression levels
of DLX4 for different subgroups of patients from (A) pathological
stage 1 to stage 4, (B) pathological grade 1 to grade 4; and (C)
tumor stage T1 to T4 (C) Comparisons of the expression levels of
DLX4 for patients (D) between pathological stages 1 and 2 and
stages 3 and 4, (E) between pathological grades 1 and 2 and grades
3 and 4, (F) between tumor stages T1 and T2 and stages T3 and T4,
(G) between lymphatic metastasis N0 and N1, (H) between distant
metastasis M0 and M1, and (I) between alive and dead. **P<0.01,
***P<0.001, ****P<0.0001 and ####P<0.0001. DLX,
distal-less homeobox; ns, not significant. |
Diagnostic and prognostic value of
DLX4 mRNA expression in ccRCC
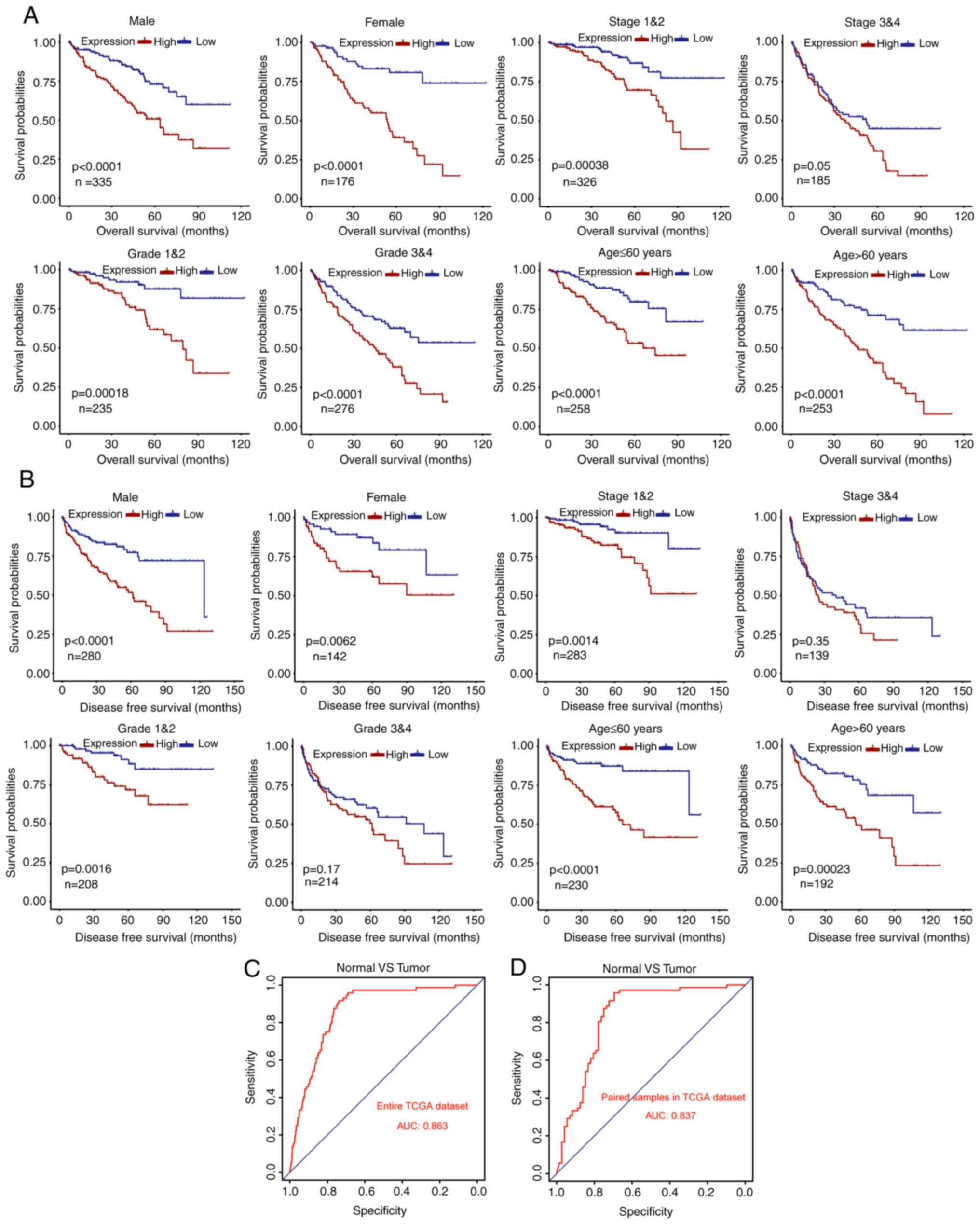
Based on the prognostic values of DLX4 for the
overall and DFS of patients with ccRCC in TCGA dataset, it was
sought to further confirm the prognostic significances of DLX4 in
different subgroups of patients. A worse overall survival rate was
revealed in the high DLX4 expression group compared with the low
expression group for male, female, stage I and II, stage III and
IV, grade 1 and grade 2, grade 3 and grade 4, low age (≤60 years)
and elderly (>60 years) subgroups (Fig. 4A). In addition, a worse DFS rate was
identified in high compared with low expression group for male,
female, stage I and II, grade 1 and grade 2, high age (>60
years) and low age (≤60 years) subgroups (Fig. 4B). However, DFS was not
significantly different between stage III and IV and grade 3 and
grade 4 subgroups. These results suggested that DLX4 may be a
potential prognostic biomarker for ccRCC. To determine whether the
expression of DLX4 has diagnostic values in ccRCC, ROC curves were
generated and AUCs were calculated to determine the diagnostic
efficiency. DLX4 could sufficiently differentiate ccRCC from normal
tissues with an AUC of 0.863 in the entire TCGA dataset (Fig. 4C). In addition, the diagnostic
values of DLX4 expression levels were evaluated for paired normal
and adjacent tumor samples in TCGA dataset, with an AUC of 0.837
(Fig. 4D). These results suggested
that DLX4 may be a potential biomarker for ccRCC patients with
favorable diagnostic performance.
DLX4 is highly expressed in ccRCC
tissues
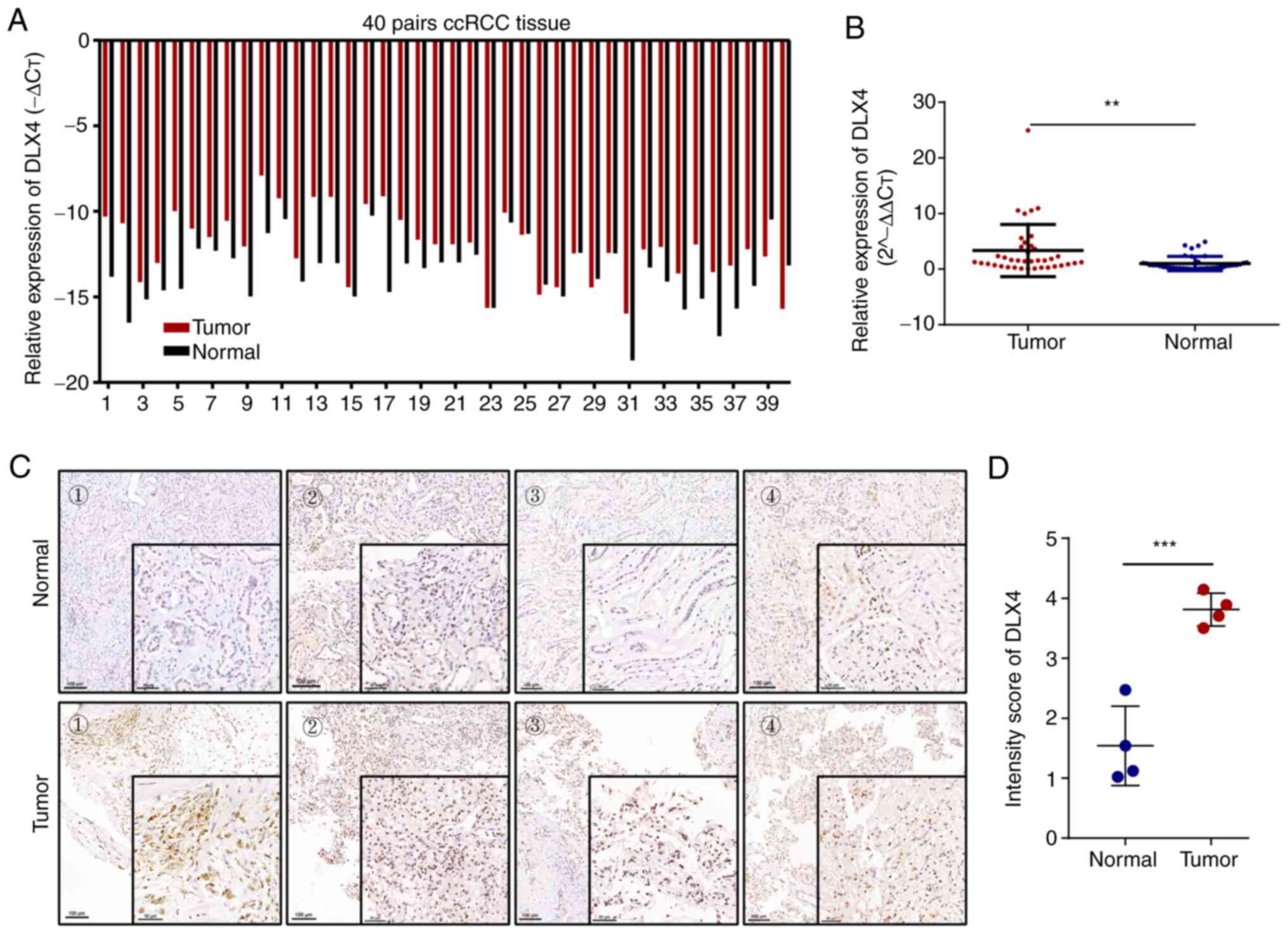
To further confirm the expression levels of DLX4 in
ccRCC vs. normal renal tissue, RT-qPCR and immunohistochemistry
were used to verify the results of the TCGA databases at the mRNA
and protein levels. The results revealed higher DLX4 mRNA
expression levels in ccRCC samples compared with matched normal
renal samples (Fig. 5A and B).
Furthermore, the expression levels of DLX4 in ccRCC samples and the
matched normal renal samples were detected using
immunohistochemistry, which revealed higher protein levels of DLX4
in ccRCC tissues (Fig. 5C and D).
These results indicated that DLX4 was highly expressed in ccRCC
tissues, which aligns with the TCGA database analysis results
aforementioned.
Biological pathogenesis of DLX4 in
ccRCC
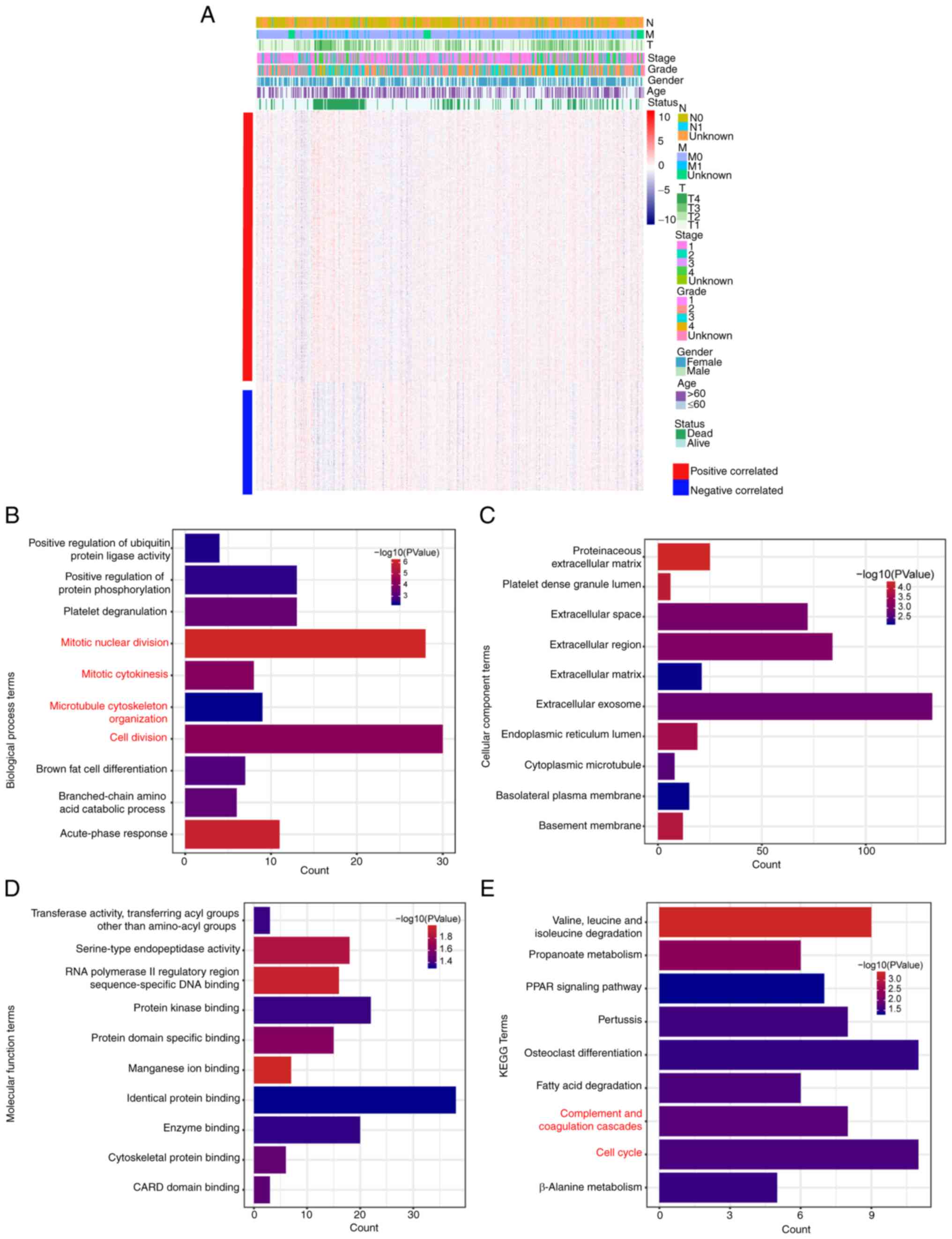
To investigate the functional mechanism of DLX4 in
ccRCC, genes that are highly associated with the expression levels
of DLX4 in ccRCC were first selected; of which 328 positively
related genes and 210 negatively associated genes were selected
(Fig. 6A). Functional enrichment
analysis of these genes was carried out to determine the potential
mechanisms. The GO terms, including ‘positive regulation of
ubiquitin protein ligase activity’, ‘positive regulation of protein
phosphorylation’, ‘mitotic nuclear division’, ‘microtubule
cytoskeleton organization’, and ‘cell division’ were enriched for
biological process (Fig. 6B);
‘extracellular region’, ‘endoplasmic reticulum lumen’,
‘extracellular exosome’, and ‘cytoplasmic microtubule’ were
enriched for cellular component (Fig.
6C) and ‘protein domain specific binding’, ‘identical protein
binding’, ‘RNA polymerase II regulatory region sequence-specific
DNA binding’, ‘protein kinase binding’, and ‘enzyme binding’ were
enriched for molecular function (Fig.
6D). The terms ‘cell cycle’, ‘PPAR signaling pathway’ and
‘fatty acid degradation’ were enriched for KEGG pathways (Fig. 6E). In addition, the terms that are
more closely related to the cell cycle and complement and
coagulation cascades were selected and highlighted in red in
Fig. 6.
Patients with ccRCC in the TCGA dataset were divided
into high and low expression groups based on median expression
level of DLX4. GSEA was then performed to determine the statistical
significance of the biological pathways associated with the
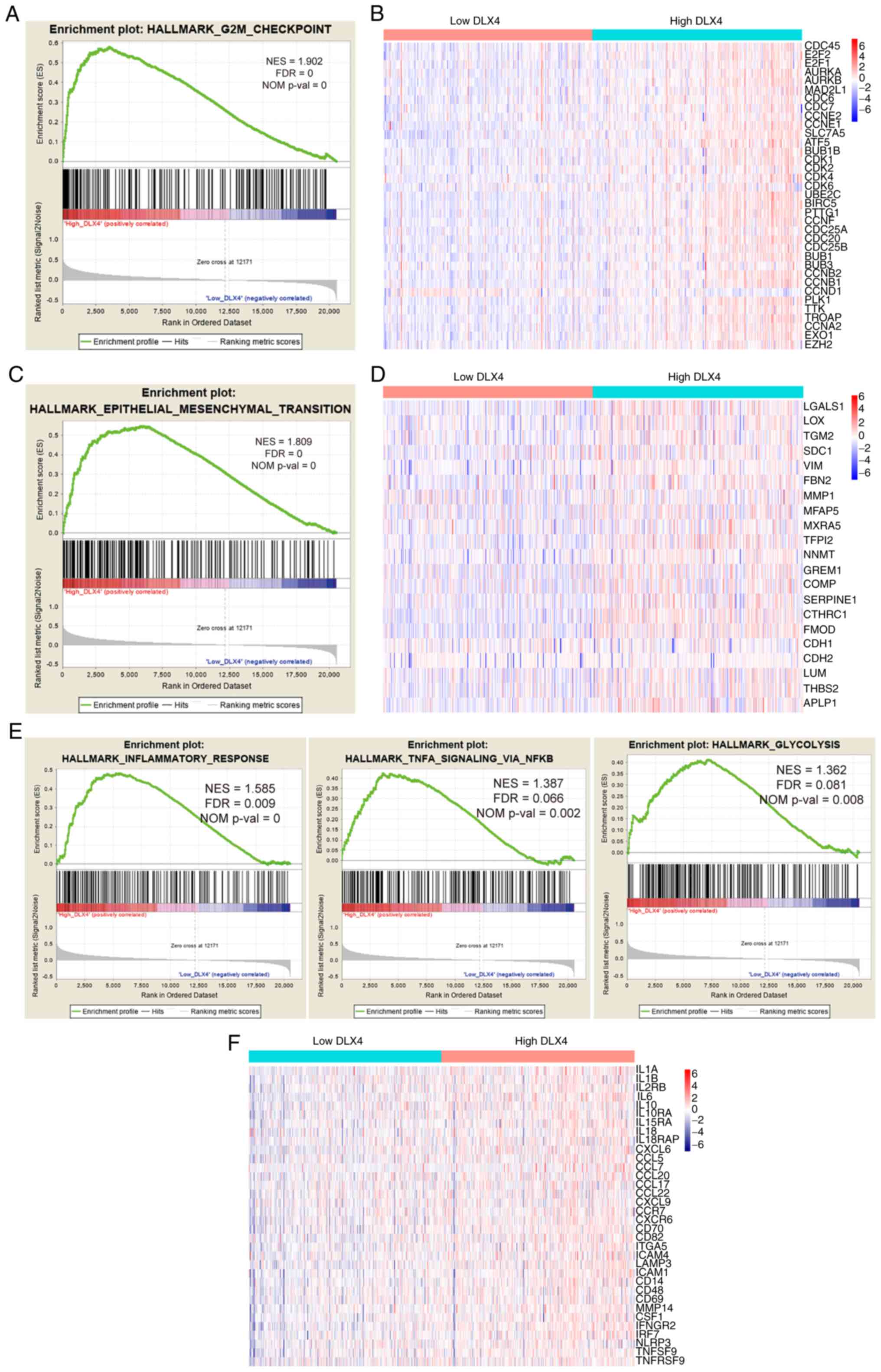
expression levels of DLX4 (Fig. 7A, C
and E). The results revealed that the expression of DLX4 was
associated with biological pathways related to G2M checkpoint,
epithelial-mesenchymal transition (EMT), glycolysis, inflammatory
response and TNFα signaling via NF-κB. The distributions of cell
cycle-related genes for high and low DLX4 expression groups are
presented in Fig. 7B. Based on the
heat map, patients in the high DLX4 expression group exhibited a
trend of higher expression levels of cell cycle-related genes
compared with the low DLX4 expression group. Genes involved in EMT
were expressed at notably higher expression levels in the high DLX4
expression group (Fig. 7D). The
distributions of the inflammatory molecules, including chemokines,
cytokines and their receptors, between the high and low expression
groups of DLX4 were also compared (Fig.
7F). A trend towards higher expression of inflammatory
molecules was identified in patients in the high DLX4expression
group. These findings suggested that cell cycle-related pathways,
EMT and inflammatory response may be a potential mechanism of DLX4
in ccRCC tumorigenesis.
Immune cell infiltration associated
with DLX4 expression
Based on the associations of DLX4 expression with
inflammatory and immune responses, the associations between the
expression levels of DLX4 and immune cell infiltration were
investigated by examining the differences in the expression
profiles of 28 immune cell types separated into high and low DLX4
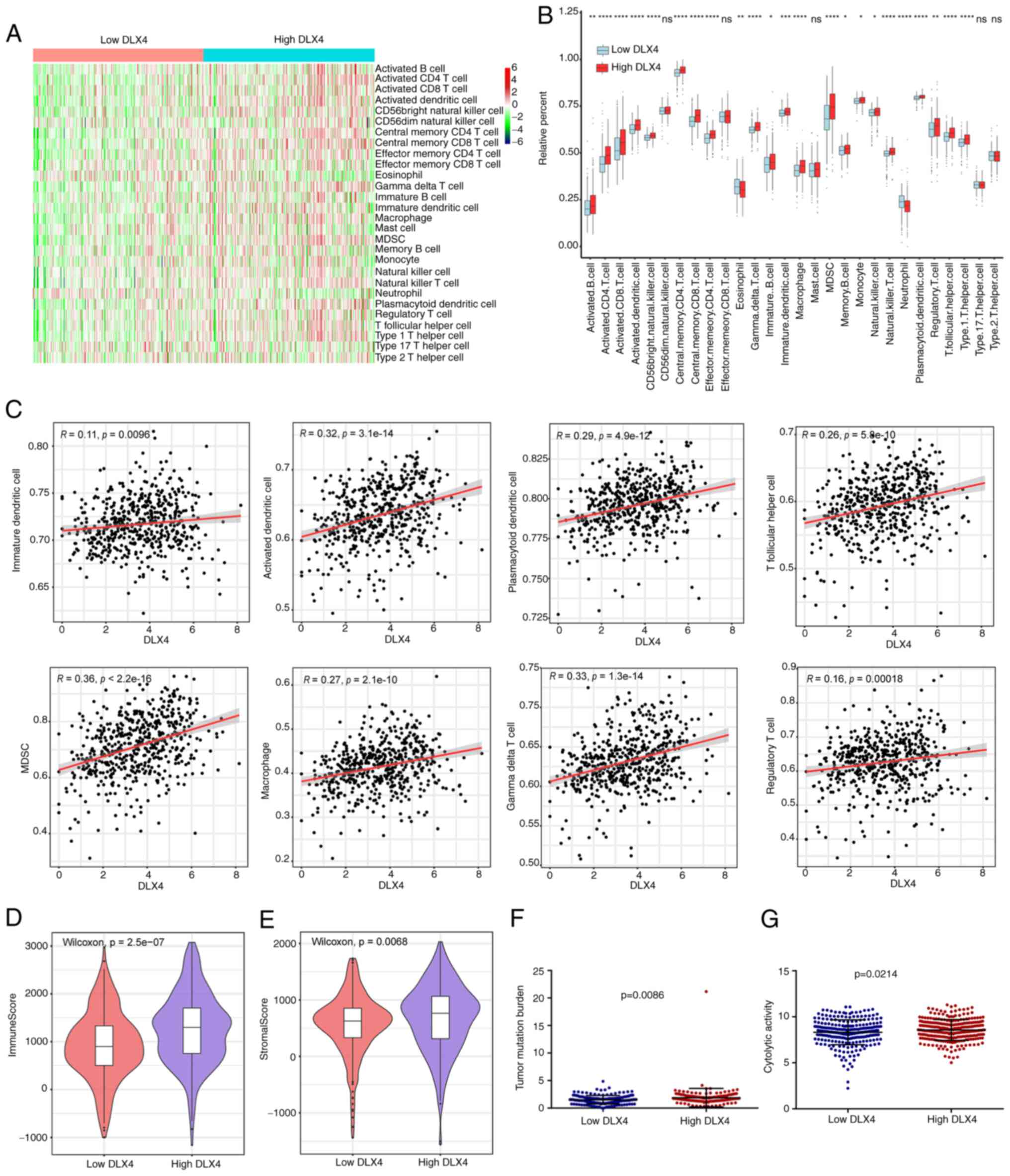
expression groups of DLX4 (Fig. 8A and
B). Patients with high DLX4 expression in TCGA dataset had a
high percentage of immune cells, including immature dendritic
cells, activated dendritic cells, gamma delta T cells, macrophages,
myeloid-derived suppressor cells (MDSCs), plasmacytoid dendritic
cells, regulatory T cells and T follicular helper cells. In
addition, a positive correlation was determined between the
expression levels of DLX4 and tumor immunosuppressive cells,
including immature dendritic cells, activated dendritic cells,
gamma delta T cells, macrophages, MDSCs, plasmacytoid dendritic
cells, regulatory T cells and T follicular helper cells (Fig. 8C). The immune and stromal scores for
TCGA cohort were calculated using the ESTIMATE algorithm. Patients
in the high DLX4 expression group had significantly higher immune
and stromal scores compared with the low DLX4 expression group
(Fig. 8D and E, respectively).
Furthermore, patients in the high DLX4 expression group had
significantly higher TMB and cytolytic activity scores compared
with those in the DLX4 low expression group (Fig. 8F and G, respectively).
Discussion
RCC is one of the most lethal urological
malignancies with high tumor heterogeneity (33); ccRCC is the most common histological
subtype. Prevalence of ccRCC has steadily increased (34), placing a heavy burden on health and
property of individuals. Biomarkers that could be used to
accurately diagnose or predict patient prognosis are thus urgently
required. In the present study, bioinformatics methods were used to
explore the functions of six members of the DLX gene family in
ccRCC in multiple public databases. DLX4, that may serve a more
important role in patients with ccRCC. Subsequently, the diagnostic
and prognostic values, the association with clinical factors and
the potential functional mechanisms of DLX4 were systematically
investigated. The associations between DLX4 expression and the
tumor immune microenvironment were systematically evaluated.
Members of the DLX gene family contain a homeobox
that could encode genes expressed in the head and limbs of the
developing fruit fly. The DLX gene family includes six different
members, DLX1-DLX6, and is hypothesized to serve vital roles in
forebrain and craniofacial development (35). Recently, multiple studies have
demonstrated different expression patterns of the DLX gene family
in malignant tissues compared with non-malignant tissues (12). Several members of the DLX gene
family are involved in early development and cell differentiation
and are frequently dysregulated in cancer (12,13,36).
At present, the roles of the DLX gene family in ccRCC remain
unclear. Therefore, the present study examined the expression
levels of the six members of the DLX gene family in ccRCC. Based on
the results, the expression levels of the six members of the DLX
gene family were different between tumor and normal tissues. These
findings suggested that the DLX gene family may play vital roles in
the tumorigenesis and development of ccRCC. A previous study
revealed that DLX2 could counteract TGF-β induced cell cycle arrest
and apoptosis to promote tumorigenesis (16). DLX2 has also been considered to be a
marker of survival and disease progression in prostate cancer
(17). DLX5 is involved in the
growth and metastasis of invasive glioma cells (19). Genes with prognostic values and
those associated with clinical factors have more clinical
significance. In the present study, it was demonstrated that the
DLX2 and DLX4 genes have prognostic value for overall survival and
DFS of patients with ccRCC, indicating their potential for use as
prognostic biomarkers for ccRCC. In addition, the expression of
DLX4 was revealed to be closely associated with clinical factors in
ccRCC patients. However, further studies may be necessary to
examine the differential expression and functional phenotype of all
these genes.
DLX4 proteins, first identified in human placental
tissue, is involved in the development and maturation of the
placenta. The human DLX4 gene encodes three functionally different
protein isoforms: β protein 1 (BP1), DLX7 and unidentified DLX4
(13). DLX7 has rarely been
studied, and recent research has mainly focused on BP1; thus, BP1
is also called DLX4 (13). The
expression of DLX4 in different types of cancer and the
characteristics of malignant behavior have been reported. For
example, high expression of DLX4 has been verified in various
cancers, including leukemia, breast, prostate, liver, endometrial
and ovarian, and this high expression levels may promote tumor
progression (37,38). In endometrial cancer, DLX4
overexpression leads to the upregulation of genes related to
proliferation, metastasis and cell cycle to promote cell
proliferation and migration, and is associated with poor prognosis
(37). In inflammatory breast
cancer, DLX4 promotes tumor progression, invasion and metastasis
(39). In breast cancer, DLX4
drives the expression of twist to promote epithelial to mesenchymal
transition, cancer migration, invasion and metastasis (40). In prostate cancer, DLX4 is an
important upstream factor in the carcinogenic pathway of prostate
cancer, which may contribute to tumor progression and invasion
(41). In ovarian cancer, DLX4
promotes peritoneal metastasis of tumor cells by stimulating
IL-1β-mediated NF-κB activity (21). In choriocarcinoma, DLX4 may be
involved in the survival of human choriocarcinoma cells by
inhibiting apoptosis (42). In the
present study, the roles and potential mechanisms of DLX4 in ccRCC
were investigated. A recent study showed that DLX4 contributed to
the proliferation and migration of ccRCC via EMT pathways (43). The present study further extends the
above research conclusions. Accordingly, DLX4 was revealed to be an
independent risk factor and a potential diagnostic and prognostic
biomarker for ccRCC. In the development of ccRCC, cancer-related
biological pathways, including the cell cycle, EMT and immune
response, were significantly altered. Cell cycle-related gene
expression signatures are a marker of highly proliferative cells
and, thus, widely regarded as a biomarkers of aggressive malignancy
and poor prognosis (44). In the
present study, it was revealed that cell cycle-related genes were
significantly higher in the high DLX4 expression group, suggesting
that DLX4 may promote tumor progression by changing the cell cycle.
EMT is characterized by the loss of epithelial characteristics
and/or gain of mesenchymal characteristics, which are key
components in metastasis formation (45). EMT is a highly complex biological
process, and numerous factors promote and inhibit EMT. In the
present study, the EMT pathway was enriched, and genes correlated
with EMT exhibited increased expression levels in the high
DLX4expression group, suggesting that DLX4 may promote tumor
progression through EMT. More detailed studies are required to
verify the potential functional mechanisms of DLX4 in ccRCC.
ccRCC is one of the most immune-infiltrated tumors
and rapidly responds to immunotherapy (46). The level of immune cell infiltration
is a key factor in determining the effect of immunotherapy and
prognosis in patients with ccRCC. Therefore, exploring the tumor
immune microenvironment and identifying biomarkers associated with
immunotherapeutic response have important clinical significance.
Previous studies identified DLX4 as a master regulator of immune
infiltration recruitment and have proposed the possibility that the
expression of DLX4 may affect immune evasion in cancer (21,22).
In the present study, it was revealed that immune and inflammatory
pathways were significantly enriched in the high DLX4 expression
group. DLX4 expression was positively correlated with the
infiltration ratio of tumor immunosuppressive cells, suggesting
that DLX4 may serve a role in tumor immunosuppression. These
results were consistent with those that revealed that the worse
prognosis of ccRCC patients was due to the tumor immune
microenvironment of ccRCC being dominated by suppressed and
dysfunctional cells (47). However,
more detailed studies were required to verify the roles of DLX4 in
the tumor immune microenvironment.
The present study found that DLX4 may be a potential
diagnostic and prognostic biomarker that promotes tumor progression
in ccRCC. High expression of DLX4 in ccRCC was also demonstrated
and was DLX4 was revealed to be associated with the malignant
characteristics of ccRCC. Furthermore, DLX4 expression was
associated with the tumor immunosuppressive microenvironment.
However, the present study has certain limitations. First, the
associations between DLX4 and ccRCC biological behaviors must be
confirmed in vivo and in vitro. Furthermore, the
underlying molecular mechanisms of DLX4 facilitating ccRCC must be
thoroughly investigated.
In conclusion, the present study demonstrated that
high expression levels of DLX4 in ccRCC were associated with poor
overall survival and DFS, as well as high tumor stage and grade.
DLX4 expression was also found to be associated with the tumor
immunosuppressive microenvironment. Collectively, the present data
suggested that DLX4 is a promising prognostic indicator and a
specific diagnostic biomarker for ccRCC. Accordingly, DLX4 may be
considered a potential therapeutic target for ccRCC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LST conceived and designed the experiments. JW and
LJT. acquired and analyzed the data and wrote the manuscript. YL,
HL and XS collected the tissue samples and performed genetic
studies. All authors read and approved the final manuscript. LJT
and JW confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Human Research of The Second People's Hospital of Wuhu
and The First Affiliated Hospital of Anhui Medical University
(approval no. PJ2019-14-22). Written informed consent was provided
by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu J, Latif S and Wei S: Metastatic renal
cell carcinoma presenting as gastric polyps: A case report and
review of the literature. Int J Surg Case Rep. 3:601–604. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Geissler K, Fornara P, Lautenschlager C,
Holzhausen HJ, Seliger B and Riemann D: Immune signature of tumor
infiltrating immune cells in renal cancer. Oncoimmunology.
4:e9850822015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu W, Atkins MB and McDermott DF:
Checkpoint inhibitor immunotherapy in kidney cancer. Nat Rev Urol.
17:137–150. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mantia CM and McDermott DF: Vascular
endothelial growth factor and programmed death-1 pathway inhibitors
in renal cell carcinoma. Cancer. 125:4148–4157. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Oostveen J, Bijl J, Raaphorst F,
Walboomers J and Meijer C: The role of homeobox genes in normal
hematopoiesis and hematological malignancies. Leukemia.
13:1675–1690. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abate-Shen C: Deregulated homeobox gene
expression in cancer: Cause or consequence? Nat Rev Cancer.
2:777–785. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lou Y, Fallah Y, Yamane K and Berg PE:
BP1, a potential biomarker for breast cancer prognosis. Biomark
Med. 12:535–545. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chan DW, Hui WW, Wang JJ, Yung MM, Hui LM,
Qin Y, Liang RR, Leung TH, Xu D, Chan KK, et al: DLX1 acts as a
crucial target of FOXM1 to promote ovarian cancer aggressiveness by
enhancing TGF-β/SMAD4 signaling. Oncogene. 36:1404–1416. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun B, Fan Y, Yang A, Liang L and Cao J:
MicroRNA-539 functions as a tumour suppressor in prostate cancer
via the TGF-β/Smad4 signalling pathway by down-regulating DLX1. J
Cell Mol Med. 23:5934–5948. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yilmaz M, Maass D, Tiwari N, Waldmeier L,
Schmidt P, Lehembre F and Christofori G: Transcription factor Dlx2
protects from TGFβ-induced cell-cycle arrest and apoptosis. EMBO J.
30:4489–4499. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Green WJ, Ball G, Hulman G, Johnson C, Van
Schalwyk G, Ratan HL, Soria D, Garibaldi JM, Parkinson R, Hulman J,
et al: KI67 and DLX2 predict increased risk of metastasis formation
in prostate cancer-a targeted molecular approach. Br J Cancer.
115:236–242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Palazzo E, Kellett M, Cataisson C, Gormley
A, Bible PW, Pietroni V, Radoja N, Hwang J, Blumenberg M, Yuspa SH
and Morasso MI: The homeoprotein DLX3 and tumor suppressor p53
co-regulate cell cycle progression and squamous tumor growth.
Oncogene. 35:3114–3124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu B, Wang Q, Wang YA, Hua S, Sauvé CG,
Ong D, Lan ZD, Chang Q, Ho YW, Monasterio MM, et al: Epigenetic
activation of WNT5A drives glioblastoma stem cell differentiation
and invasive growth. Cell. 167:1281–1295.e18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trinh BQ, Barengo N, Kim SB, Lee JS,
Zweidler-McKay PA and Naora H: The homeobox gene DLX4 regulates
erythro-megakaryocytic differentiation by stimulating IL-1β and
NF-κB signaling. J Cell Sci. 128:3055–3067. 2015.PubMed/NCBI
|
|
21
|
Haria D, Trinh BQ, Ko SY, Barengo N, Liu J
and Naora H: The homeoprotein DLX4 stimulates NF-κB activation and
CD44-mediated tumor-mesothelial cell interactions in ovarian
cancer. Am J Pathol. 185:2298–2308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen J, Cerise J, Jabbari A, Clynes R and
Christiano A: Master regulators of infiltrate recruitment in
autoimmune disease identified through network-based molecular
deconvolution. Cell Syst. 1:326–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hinrichs AS, Raney BJ, Speir ML, Rhead B,
Casper J, Karolchik D, Kuhn RM, Rosenbloom KR, Zweig AS, Haussler D
and Kent WJ: UCSC data integrator and variant annotation
integrator. Bioinformatics. 32:1430–1432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sing T, Sander O, Beerenwinkel N and
Lengauer T: ROCR: Visualizing classifier performance in R.
Bioinformatics. 21:3940–3941. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qu Y, Zhao R, Zhang H, Zhou Q, Xu F, Zhang
X, Xu W, Shao N, Zhou S, Dai B, et al: Inactivation of the
AMPK-GATA3-ECHS1 pathway induces fatty acid synthesis that promotes
clear cell renal cell carcinoma growth. Cancer Res. 80:319–333.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ge S, Hua X, Chen J, Xiao H, Zhang L, Zhou
J, Liang C and Tai S: Identification of a costimulatory
molecule-related signature for predicting prognostic risk in
prostate cancer. Front Genet. 12:6663002021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Charoentong P, Finotello F, Angelova M,
Mayer C, Efremova M, Rieder D, Hackl H and Trajanoski Z: Pan-cancer
immunogenomic analyses reveal genotype-immunophenotype
relationships and predictors of response to checkpoint blockade.
Cell Rep. 18:248–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshihara K, Shahmoradgoli M, Martinez E,
Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chalmers ZR, Connelly CF, Fabrizio D, Gay
L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J,
et al: Analysis of 100,000 human cancer genomes reveals the
landscape of tumor mutational burden. Genome Med. 9:342017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abida W, Cheng ML, Armenia J, Middha S,
Autio KA, Vargas HA, Rathkopf D, Morris MJ, Danila DC, Slovin SF,
et al: Analysis of the prevalence of microsatellite instability in
prostate cancer and response to immune checkpoint blockade. JAMA
Oncol. 5:471–478. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Znaor A, Lortet-Tieulent J, Laversanne M,
Jemal A and Bray F: International variations and trends in renal
cell carcinoma incidence and mortality. Eur Urol. 67:519–530. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Leibovich BC, Lohse CM, Crispen PL,
Boorjian SA, Thompson RH, Blute ML and Cheville JC: Histological
subtype is an independent predictor of outcome for patients with
renal cell carcinoma. J Urol. 183:1309–1315. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Panganiban G and Rubenstein JL:
Developmental functions of the Distal-less/Dlx homeobox genes.
Development. 129:4371–4386. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Popovici C, Leveugle M, Birnbaum D and
Coulier F: Homeobox gene clusters and the human paralogy map. FEBS
Lett. 491:237–242. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang L, Wan Y, Jiang Y, Zhang Z, Shu S,
Cheng W and Lang J: Overexpression of BP1, an isoform of Homeobox
Gene DLX4, promotes cell proliferation, migration and predicts poor
prognosis in endometrial cancer. Gene. 707:216–223. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu M, Yang Y, Shi Y, Wang D, Wei X, Zhang
N and Niu R: Expression level of beta protein 1 mRNA in Chinese
breast cancer patients: A potential molecular marker for poor
prognosis. Cancer Sci. 99:173–178. 2008.PubMed/NCBI
|
|
39
|
Man YG, Schwartz A, Levine PH, Teal C and
Berg PE: BP1, a putative signature marker for inflammatory breast
cancer and tumor aggressiveness. Cancer Biomark. 5:9–17. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang L, Yang M, Gan L, He T, Xiao X,
Stewart MD, Liu X, Yang L, Zhang T, Zhao Y and Fu J: DLX4
upregulates TWIST and enhances tumor migration, invasion and
metastasis. Int J Biol Sci. 8:1178–1187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schwartz AM, Man YG, Rezaei MK, Simmens SJ
and Berg PE: BP1, a homeoprotein, is significantly expressed in
prostate adenocarcinoma and is concordant with prostatic
intraepithelial neoplasia. Mod Pathol. 22:1–6. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun Y, Lu X, Yin L, Zhao F and Feng Y:
Inhibition of DLX4 promotes apoptosis in choriocarcinoma cell
lines. Placenta. 27:375–383. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun G, Ge Y, Zhang Y, Yan L, Wu X, Ouyang
W, Wang Z, Ding B, Zhang Y, Long G, et al: Transcription factors
BARX1 and DLX4 contribute to progression of clear cell renal cell
carcinoma via promoting proliferation and epithelial-mesenchymal
transition. Front Mol Biosci. 8:6263282021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Otto T and Sicinski P: Cell cycle proteins
as promising targets in cancer therapy. Nat Rev Cancer. 17:93–115.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pallasch F and Schumacher U: Angiotensin
Inhibition, TGF-β and EMT in cancer. Cancers. 12:27852020.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Şenbabaoğlu Y, Gejman RS, Winer AG, Liu M,
Van Allen EM, de Velasco G, Miao D, Ostrovnaya I, Drill E, Luna A,
et al: Tumor immune microenvironment characterization in clear cell
renal cell carcinoma identifies prognostic and
immunotherapeutically relevant messenger RNA signatures. Genome
Biol. 17:2312016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vuong L, Kotecha R, Voss M and Hakimi A:
Tumor Microenvironment dynamics in clear-cell renal cell carcinoma.
Cancer Discov. 9:1349–1357. 2019. View Article : Google Scholar : PubMed/NCBI
|