Introduction
Melanoma is a highly malignant tumor that originates
from melanocytes. It occurs primarily in the skin, mucous
membranes, and viscera (1).
Malignant melanomas may arise from congenital or acquired benign
melanocytic nevi, malevolent dysplastic nevi, or may even develop
de novo (2). For the last 5
years, the incidence and mortality rates of melanoma have increased
on an annual basis, and the lethal age of melanoma is lower than
that of other solid tumors (3,4).
Melanoma can occur in all individuals of all ages, occurring more
often in men, and the mortality rate of male patients is higher
than that of females (5). Melanoma
is stratified as follows: Grade I, tumor cells are confined to the
epidermis above the basement membrane; Grade II, tumor cells have
broken through the basement membrane and have invaded the dermal
papillary layer; Grade III, the tumor cells fill the papillary
layer of the dermis and invade further downwards, but have not
reached the reticular layer of the dermis; Grade IV, the tumor
cells have invaded the dermal reticular layer; and Grade V, the
tumor cells have passed through the dermal reticular layer and
invaded the subcutaneous fat layer (6). There are generally no obvious symptoms
of occurrence during the earlier stages. During the later stages,
ulcerations, impaired healing, regional or distant lymph node
enlargement, and distant metastasis are observed (7). In addition to early surgical
resection, melanoma lacks specific treatment options, with a high
degree of malignancy, metastasis, and a poor prognosis (8,9).
However, the cause of melanoma is not fully understood. It is
generally hypothesized that several factors, such as race and
genetics, trauma and stimulation, sunlight, and immunity, amongst
others are all involved.
Bioinformatics can be used to study biological
problems using the methods of applied mathematics, informatics,
statistics, and computer science (10). The research materials and results of
bioinformatics analyses cover numerous types of biological data.
The methods typically involve sequence alignment, gene recognition,
gene recombination, protein structure prediction, gene expression,
protein response prediction, and evolutionary modeling (11).
Semaphorin 4D (SEMA4D) is a member of the Semaphorin
family of axon-directed molecules, also known as CD100,
hypothesized initially to be axon-directed factors affecting neural
development (12). In addition to
regulating axonal orientation, angiogenesis, and tumor metastasis,
SEMA4D also plays an essential role in the immune system. The Gene
Ontology (GO) annotations related to SEMA4D include signal receptor
binding and transmembrane signal receptor activity. An important
paralog of this gene is SEMA4B, which was discovered as a negative
regulator of the PI3K/AKT signaling pathway in breast cancer
(13). However, the relationship
between SEMA4D and melanoma is unclear.
In this study, bioinformatics analysis was used to
verify the potential role of SEMA4D in melanoma, and 272 melanoma
patients were recruited to study the impact of abnormal expression
of SEMA4D on the prognosis and survival time of melanoma patients,
with the aim of identifying the molecular mechanism involved.
Materials and methods
Expression of SEMA4D in a
database
Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/) was used
to analyze the expression of SEMA4D in melanoma tumors. GEPIA
generates box plots for comparing expression in several types of
cancer. The method used for differential analysis is
independent-samples t-test when two groups were compared, using the
disease state (Tumor or Normal) as the variable for calculating
differential expression.
Patients
A total of 272 patients diagnosed with melanoma and
liver cancer at the Fourth Hospital of Hebei Medical University
were selected for inclusion between March 2015 and June 2020.
The inclusion criteria were: Aged 18–80 years old,
diagnosed with melanoma, with normal cardiopulmonary function, and
normal coagulation.
The exclusion criteria were: Aged <18 years or
>80 years, required emergency surgery, or the patient and/or
their family did not agree to participate in the trial.
The Ethics Committee of the Fourth Hospital of Hebei
Medical University approved the present study (approval no.
FHBM2015014), and all patients signed informed consent.
Parameters assessed
Based on the clinical data, patients were classified
by sex (male/female), age (≤60/>60), tumor size (≤3cm/>3cm),
family history of cancer (no/yes), tumor grade (low/high), SEMA4D
expression (Low/High), tumor stage (low/high), and prognosis
(survival ≥30 months/survival <30 months). Patients' survival
times were recorded during follow-up. Patients were classified
according to Breslow depth (low/high), radial vs. vertical growth
phase (early/late), presence of ulceration (no/yes),
tumor-infiltrating lymphocyte numbers (low/high), lymphovascular
spread (no/yes), regression (no/yes), metastases plus (no/yes),
disease stage (mild/severe), discus melanin (low/high) and SEMA3B
expression (low/high).
RNA extraction
An RNA Extraction Kit (cat. no. G3013; Wuhan
Servicebio Technology Co., Ltd.) was used according to the
manufacturer's protocol. Pre-sample treatment: 1 ml whole blood was
taken, centrifuged at 80,000 × g for 5 min at 4°C, and the
supernatant was discarded. To the pellet, 3 ml erythrocyte lysate
reagent (cat. no. R1010; Beijing Solarbio Science & Technology
Co., Ltd.) was added, mixed, and placed at 4°C (or room
temperature) for 10 min, centrifuged again at 80,000 × g for 5 min
at 4°C, and the supernatant was discarded. Subsequently, red blood
cell lysate (1 ml; cat. no. R1010; Beijing Solarbio Science &
Technology Co., Ltd.) was added 1–2 times until the liquid was
clear, and then the precipitate was collected via centrifugation
(80,000 × g for 5 min at 4°C). Next, 1 ml RNA extract (cat. no.
G3013; Wuhan Servicebio Technology Co., Ltd.) was added and mixed
well by shaking. After pre-treatment, the supernatant was
centrifuged at 100,000 × g for 10 min at 4°C. Trichloromethane (250
µl) was added, after which the tube was turned upside down for 15
sec, mixed thoroughly, left to stand for 3 min, centrifuged at
100,000 × g at 4°C for 10 min, and 400 µl of the supernatant was
transferred into a new centrifuge tube. To this, isopropyl alcohol
(equivalent to 80% of the volume in the tube) was added and mixed
thoroughly., after which it was incubated at −20°C for 15 min.
After centrifugation at 100,000 × g at 4°C for 10 min, the white
precipitate at the bottom of the tube was the desired RNA. The
supernatant was removed and 1.5 ml 75% ethanol was added to wash
the precipitate. The mixture was again centrifuged at 100,000 × g
at 4°C for 5 min, after which the supernatant was obtained. The
centrifuge tube was placed on an ultra-clean platform for drying
for 3 min, after which 15 µl RNA solvent (cat. no. XY-TE-0129;
Shanghai Xuanya Biotechnology Co., Ltd.). This solution was
incubated at 55°C for 5 min. A Nanodrop 2000 was used to measure
RNA concentration and purity. The expression levels of the related
genes were detected by reverse transcription-quantitative
(RT-q)PCR.
RT-qPCR
Total RNA was extracted from the blood samples using
TRIzol® reagent (Beijing Biolab Technology Co., Ltd.)
and reverse transcribed into cDNA using a Servicebio® RT
First Strand cDNA synthesis kit (cat. no. G3330, Wuhan Servicebio
Biotechnology Co., Ltd.) for 60 min at 42°C, terminating the
reaction by heating at 70°C for 5 min. qPCR was performed in a
Light Cycler® 4800 System (Roche Diagnostics) with a
specific set of primers for the amplification of select hub genes.
The thermocycling conditions used were: 95°C for 15 sec and 60°C
for 60 sec (a total of 30 cycles). The relative quantification
units (relative quantification=2−ΔΔCq, where Cq
represents quantification cycle values) of each sample were
calculated and presented as fold change of gene expression relative
to the control group. GAPDH was used as the endogenous control. The
sequences of the primers used were: SEMA4D forward,
TGAGCCAGACATCTACAACTACT and reverse, GAGTGCGTTCACAGCGAAGA; and
GAPDH forward, TGAAGGTCGGAGTGAACGGAT and reverse,
CGTTCTCAGCCTTGACCGTG.
Western blotting
Total protein from tissues was extracted and the
concentration was determined using the UV method (14). Next, one-quarter of the protein
sample (by volume) was added to 5× protein loading buffer
(reduced), and boiled at 100°C for 10 min. After cooling, the
samples were aliquoted and stored at −80°C until required. For
western blotting, protein (4 µg) was loaded on 12% SDS-gels,
resolved using SDS-PAGE, transferred to a PVDF membrane, blocked
using 5% skimmed milk at room temperature for 1 h, and incubated
with the primary antibody at 4°C overnight. The following day, the
membranes were washed with TBST three times (5 min/wash), incubated
with the HRP-conjugated rabbit secondary antibody (1:5,000; cat.
no. ab205718; Abcam) at room temperature for 1 h, and washed again
as above. Signals were visualized using chemiluminescence reagent.
The following antibodies were used: anti-Actin antibody (1:20,000;
cat. no. 66009-1-Ig; ProteinTech Group, Inc.), anti-SEMA4D antibody
(1:20,000; cat. no. 66582-1-Ig; ProteinTech Group, Inc.),
anti-SEMA3B antibody (1:5,000; cat. no. ab48197; Abcam). Actin was
used as the loading control.
Analysis of SEMA4D expression against
survival in patients with melanoma
Kaplan-Meier (K-M) survival analysis, also known as
Product-limit Estimate, is the most commonly used survival analysis
method, which is primarily used to estimate the survival rate of
patients and draw survival curves. A log-rank test was used to
compare survival.
Statistical analysis
Data are presented as percentages. Pearson
χ2 and Spearman's rank correlation coefficient analysis
were used to analyze clinical parameters and prognosis of melanoma
patients. Univariate and multivariate logistic regression analyses
were used to calculate odds ratios (ORs) of the prognostic
variables in melanoma patients. Univariate and multivariate Cox
proportional risk regression analyses were conducted to investigate
the correlation between the melanoma patients' survival time and
related factors. The receiver operating characteristic (ROC) curves
were obtained using MedCalc software (version 19.0.4; MedCalc
Software Ltd.). All other statistical analyses were performed in
SPSS version 21.0 (IBM Corp.). P<0.05 was considered to indicate
a statistically significant difference.
Results
Analysis of SEMA4D expression between
melanoma and normal tissues
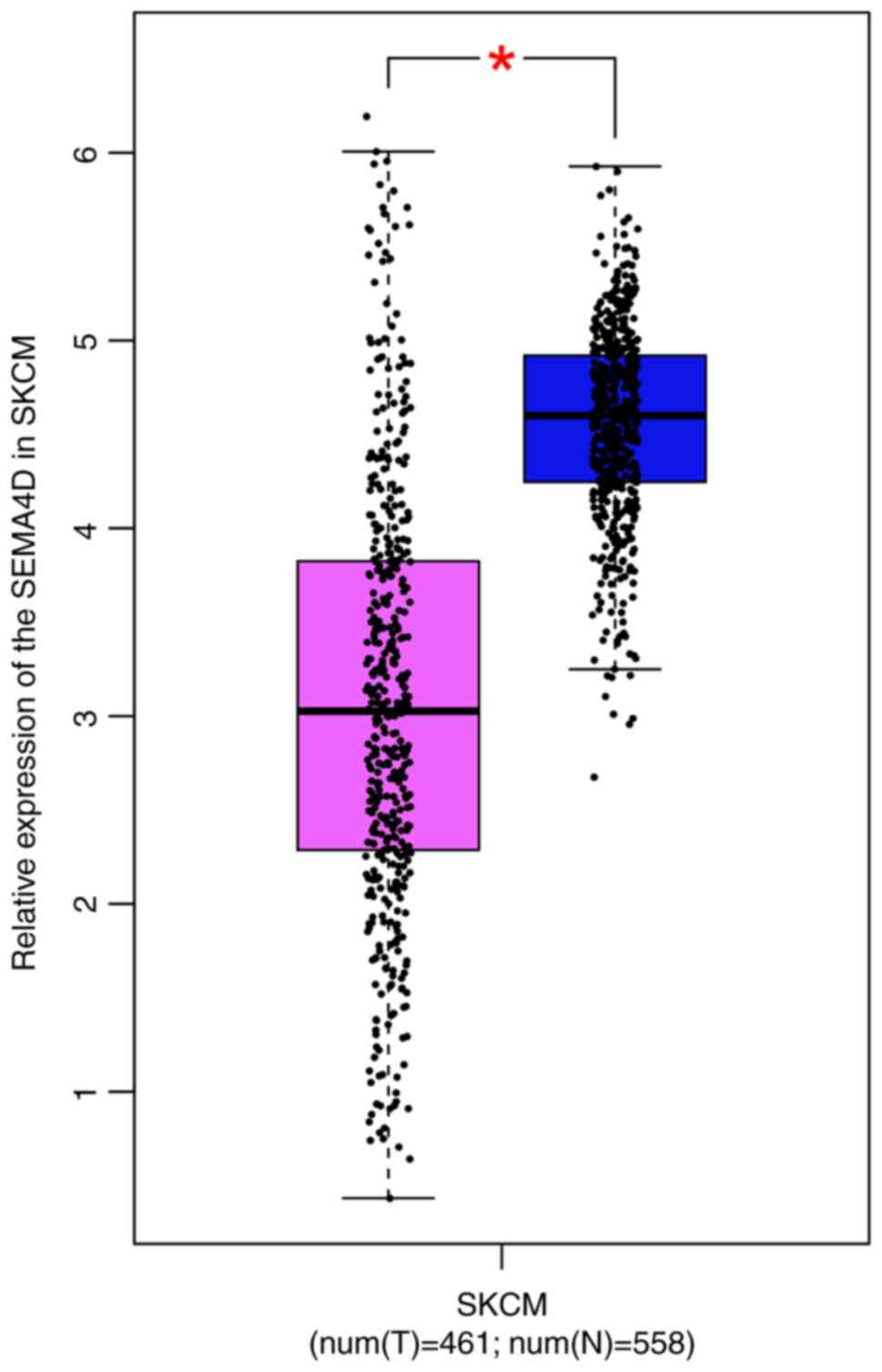
SEMA4D expression level in melanoma was
significantly lower than that in normal tissues in the GEPIA
database (Fig. 1).
Pearson χ2 analysis for
melanoma-related factors and patient prognosis
A Pearson χ2 test was used to summarize
the relationship between melanoma-related factors and patient
prognosis. Age (P=0.002), tumor grade (P=0.007), and SEMA4D
expression (P<0.001) were significantly associated with
prognosis. However, sex (P=0.800), tumor size (P=0.620), family
history (P=0.263), and tumor stage (P=0.592) were not significantly
associated with prognosis (Table
I).
 | Table I.Relevant characteristics of patients
with melanoma. |
Table I.
Relevant characteristics of patients
with melanoma.
|
|
| Prognosis, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Characteristic | n | Survival ≥30
months | Survival <30
months | P-value |
|---|
| Sex |
|
|
| 0.800 |
|
Male | 135 | 61 (22.4) | 74 (27.2) |
|
|
Female | 137 | 64 (23.5) | 73 (26.8) |
|
| Age, years |
|
|
| 0.002a |
|
≤60 | 99 | 58 (21.3) | 41 (15.1) |
|
|
>60 | 173 | 67 (24.6) | 106 (39.0) |
|
| Tumor size, cm |
|
|
| 0.620 |
| ≤3 | 137 | 65 (23.9) | 72 (26.5) |
|
|
>3 | 135 | 60 (22.1) | 75 (27.6) |
|
| Family history |
|
|
| 0.263 |
| No | 112 | 56 (20.6) | 56 (20.6) |
|
|
Yes | 160 | 69 (25.4) | 91 (33.5) |
|
| Tumor grade |
|
|
| 0.007a |
|
Low | 139 | 75 (27.6) | 64 (23.5) |
|
|
High | 133 | 50 (18.4) | 83 (30.5) |
|
| Semaphorin 4D
expression |
|
|
|
<0.001b |
|
Low | 138 | 29 (10.7) | 109 (40.1) |
|
|
High | 134 | 96 (35.3) | 38 (14.0) |
|
| Tumor stage |
|
|
| 0.592 |
|
Low | 141 | 67 (24.6) | 74 (27.2) |
|
|
High | 131 | 58 (21.3) | 73 (26.8) |
|
Spearman's rank correlation
coefficient analysis of melanoma-related factors and patient
prognosis
Spearman's rank correlation coefficient showed that
Age (ρ=0.192, P=0.001), tumor grade (ρ=0.164, P=0.007), SEMA4D
(ρ=−0.508, P<0.001) were significantly correlated with
prognosis. However, sex (ρ=−0.015, P=0.801), tumor size (ρ=0.030,
P=0.621), family history (ρ=0.068, P=0.264) and tumor stage
(ρ=0.033, P=0.593) had no significant correlation with prognosis
(Table II).
 | Table II.Relationship between the
characteristics of patients with melanoma and classification of the
patients' prognosis. |
Table II.
Relationship between the
characteristics of patients with melanoma and classification of the
patients' prognosis.
|
| Prognostic
value |
|---|
|
|
|
|---|
|
Characteristics | ρ | P-value |
|---|
| Sex | −0.015 | 0.801 |
| Age | 0.192 | 0.001b |
| Tumor size | 0.030 | 0.621 |
| Family history | 0.068 | 0.264 |
| Tumor grade | 0.164 | 0.007a |
| Semaphorin 4D | −0.508 |
<0.001b |
| Tumor stage | 0.033 | 0.593 |
Univariate logistic regression
analysis of prognosis and related factors in melanoma patients
Univariate logistic regression analysis was used to
determine the relationship between melanoma-related parameters and
prognosis, OR, and 95% confidence intervals (95% CI). Table III shows the OR and 95% CI values
of the subjects at the univariate logistic regression level; the
results show that age (OR=2.238, 95% CI: 1.353-3.703, P=0.003),
tumor grade (OR=1.945, 95% CI: 1.199-3.157, P=0.005) and SEMA4D
(OR=0.105, 95% CI: 0.060-0.184, P<0.001) were significantly
associated with prognosis. The prognosis of older patients was
worse than that of younger patients, the prognosis of patients with
a high tumor grade was worse than that of patients with a lower
tumor grade, and the prognosis of patients with low SEMA4D
expression levels was significantly worse than that of patients
with high SEMA4D levels. However, sex (OR=0.940, 95% CI:
0.584-1.515, P=0.0.791), tumor size (OR=1.128, 95% CI: 0.700-1.819,
P=0.609), family history (OR=1.319, 95% CI: 0.812-2.142, P=0.279)
and tumor stage (OR=1.140, 95% CI: 0.707-1.837, P=0.587) had no
significant association with prognosis (Table III).
 | Table III.Association between melanoma-related
parameters and prognosis based on univariate logistic regression
analysis. |
Table III.
Association between melanoma-related
parameters and prognosis based on univariate logistic regression
analysis.
|
|
| Prognosis |
|
|---|
|
|
|
|
|
|---|
| Characteristic | n | Odds ratio | 95% confidence
interval | P-value |
|---|
| Sex |
|
|
| 0.791 |
|
Male | 135 | 1 | 0.584-1.515 |
|
|
Female | 137 | 0.94 |
|
|
| Age, years |
|
|
| 0.003a |
|
≤60 | 99 | 1 | 1.353-3.703 |
|
|
>60 | 173 | 2.238 |
|
|
| Tumor size, cm |
|
|
| 0.609 |
| ≤3 | 137 | 1 | 0.700-1.819 |
|
|
>3 | 135 | 1.128 |
|
|
| Family history |
|
|
| 0.279 |
| No | 112 | 1 | 0.812-2.142 |
|
|
Yes | 160 | 1.319 |
|
|
| Tumor grade |
|
|
| 0.005a |
|
Low | 139 | 1 | 1.199-3.157 |
|
|
High | 133 | 1.945 |
|
|
| Semaphorin 4D
expression |
|
|
|
<0.001b |
|
Low | 138 | 1 | 0.060-0.184 |
|
|
High | 134 | 0.105 |
|
|
| Tumor stage |
|
|
| 0.587 |
|
Low | 141 | 1 | 0.707-1.837 |
|
|
High | 131 | 1.14 |
|
|
Multivariate logistic regression
analysis of the association between melanoma characteristics and
patient prognosis
Multivariate logistic regression was used to
describe the OR and 95% CI of the subjects at the multivariate
level. The results showed that the prognosis of older patients was
worse than that of younger patients (OR=2.254, 95% CI: 1.230-4.133,
P=0.009), and the prognosis of patients with low SEMA4D expression
was significantly worse than that of patients with high SEMA4D
expression levels (OR=0.106, 95% CI: 0.059-0.190, P<0.001).
While sex (OR=0.652, 95% CI: 0.361-1.177, P=0.156), tumor size
(OR=1.098, 95% CI: 0.622-1.939, P=0.746), family history (OR=1.200,
95% CI: 0.672-2.141, P=0.538), tumor grade (OR=1.677, 95% CI:
0.949-2.964, P=0.075) and tumor stage (OR=0.930, 95% CI:
0.524-1.650, P=0.805) had no significant association with prognosis
(Table IV).
 | Table IV.Relationship between melanoma-related
parameters and patient prognosis by multivariate logistic
regression analysis. |
Table IV.
Relationship between melanoma-related
parameters and patient prognosis by multivariate logistic
regression analysis.
|
| Prognostic
value |
|---|
|
|
|
|---|
|
Characteristics | Odds ratio | 95% confidence
interval | P-value |
|---|
| Sex | 0.652 | 0.361-1.177 | 0.156 |
| Age | 2.254 | 1.230-4.133 | 0.009a |
| Tumor size | 1.098 | 0.622-1.939 | 0.746 |
| Family history | 1.200 | 0.672-2.141 | 0.538 |
| Tumor grade | 1.677 | 0.949-2.964 | 0.075 |
| Semaphorin 4D | 0.106 | 0.059-0.190 |
<0.001b |
| Tumor stage | 0.930 | 0.524-1.650 | 0.805 |
Univariate Cox regression analysis of
the proportional risk of survival time in melanoma patients
Table V shows the
univariate Cox regression hazard ratios (HRs) and 95% CI values for
the melanoma patients. Older patients had lower survival times than
younger patients (HR=1.894, 95% CI: 1.412-2.541, P<0.001), the
survival time of melanoma patients with low SEMA4D expression
levels was significantly lower than that of patients with high
SEMA4D expression levels (HR=0.570, 95% CI: 0.431-0.755,
P<0.001). While sex (HR=1.203, 95% CI: 0.914-1.583, P=0.188),
tumor size (HR=1.033, 95% CI: 0.787-1.355, P=0.817), family history
(HR=0.931, 95% CI: 0.703-1.232, P=0.617), tumor grade (HR=1.291,
95% CI: 0.982-1.696, P=0.067) and tumor stage (HR=1.169, 95% CI:
0.889-1.538, P=0.264) were not significantly associated with
survival time (Table V).
 | Table V.Univariate Cox regression analysis of
melanoma-related characteristics on the survival time of
patients. |
Table V.
Univariate Cox regression analysis of
melanoma-related characteristics on the survival time of
patients.
|
|
| Survival time |
|---|
|
|
|
|
|---|
|
Characteristics | n | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Sex |
|
|
| 0.188 |
|
Male | 135 | 1 | 0.914-1.583 |
|
|
Female | 137 | 1.203 |
|
|
| Age, years |
|
|
|
<0.001a |
|
≤60 | 99 | 1 | 1.412-2.541 |
|
|
>60 | 173 | 1.894 |
|
|
| Tumor size, cm |
|
|
| 0.817 |
| ≤3 | 137 | 1 | 0.787-1.355 |
|
|
>3 | 135 | 1.033 |
|
|
| Family history |
|
|
| 0.617 |
| No | 112 | 1 | 0.703-1.232 |
|
|
Yes | 160 | 0.931 |
|
|
| Tumor grade |
|
|
| 0.067 |
|
Low | 139 | 1 | 0.982-1.696 |
|
|
High | 133 | 1.291 |
|
|
| Semaphorin 4D
expression |
|
|
|
<0.001a |
|
Low | 138 | 1 | 0.431-0.755 |
|
|
High | 134 | 0.57 |
|
|
| Tumor stage |
|
|
| 0.264 |
|
Low | 141 | 1 | 0.889-1.538 |
|
|
High | 131 | 1.169 |
|
|
Multivariate Cox regression analysis
of the proportional risk of survival time in melanoma patients
All factors were included in the Cox regression
model to control for the influence of confounding factors.
Multivariate Cox proportional regression analysis showed that the
survival time of older patients was lower than that of younger
patients (HR=1.778, 95% CI: 1.301-2.430, P<0.001), and the
survival time of melanoma patients with low SEMA4D expression
levels was significantly lower than that of patients with high
SEMA4D expression levels (HR=0.641, 95% CI: 0.473-0.867, P=0.004).
However, sex (HR=0.936, 95% CI: 0.693-1.265, P=0.669), tumor size
(HR=1.050, 95% CI: 0.797-1.384, P=0.727), family history (HR=0.947,
95% CI: 0.714-1.256, P=0.705), tumor grade (HR=1.317, 95% CI:
0.997-1.738, P=0.052) and tumor stage (HR=1.150, 95% CI:
0.870-1.519, P=0.327) were not significantly associated with
survival time (Table VI).
 | Table VI.Influence of melanoma-related
characteristics on patient survival time based on multivariate Cox
regression analysis. |
Table VI.
Influence of melanoma-related
characteristics on patient survival time based on multivariate Cox
regression analysis.
|
| Survival time |
|---|
|
|
|
|---|
|
Characteristics | Hazard ratio | 95% confidence
interval | P-value |
|---|
| Sex | 0.936 | 0.693-1.265 | 0.669 |
| Age | 1.778 | 1.301-2.430 |
<0.001b |
| Tumor size | 1.050 | 0.797-1.384 | 0.727 |
| Family history | 0.947 | 0.714-1.256 | 0.705 |
| Tumor grade | 1.317 | 0.997-1.738 | 0.052 |
| Semaphorin 4D | 0.641 | 0.473-0.867 | 0.004a |
| Tumor stage | 1.150 | 0.870-1.519 | 0.327 |
ROC curve
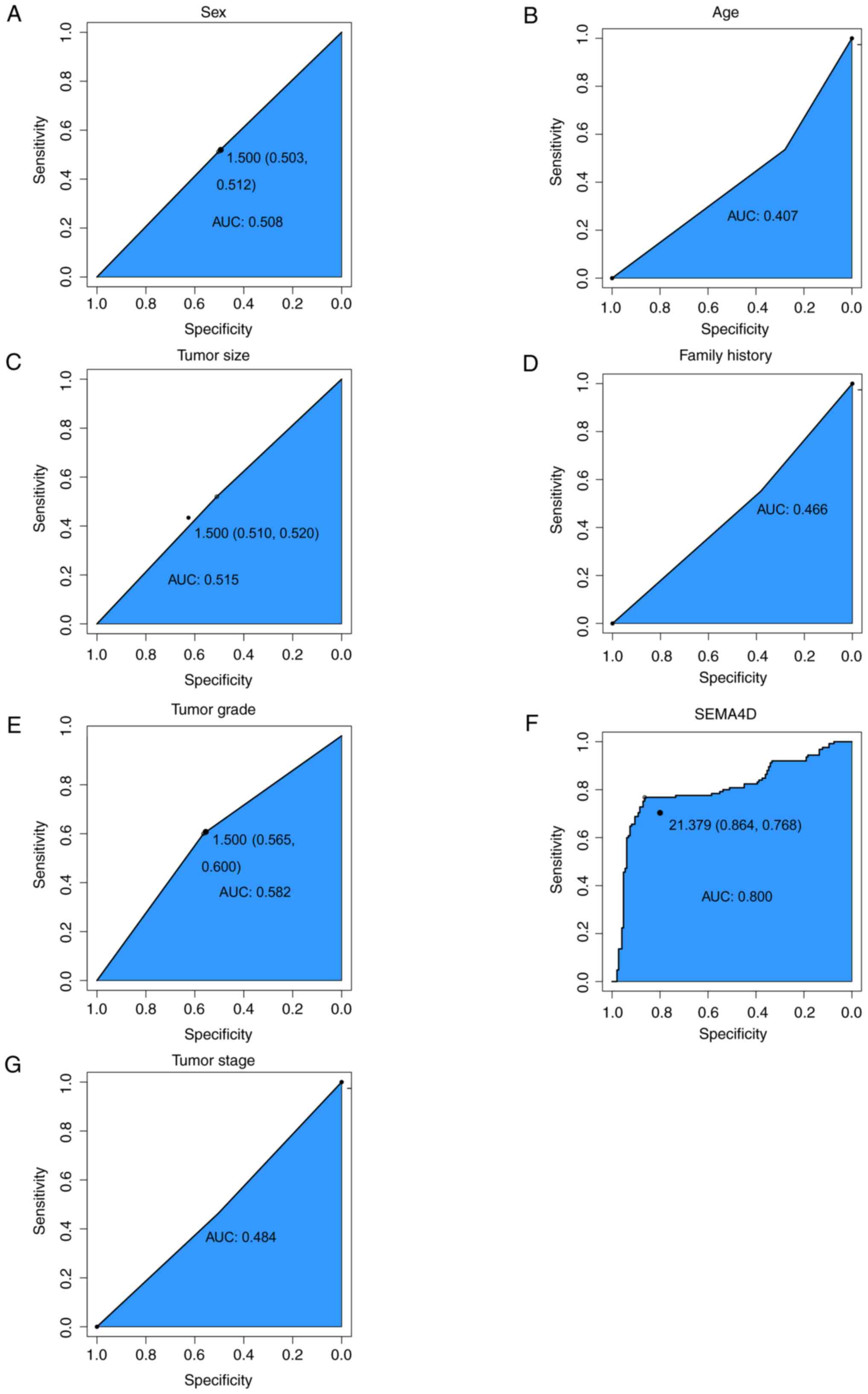
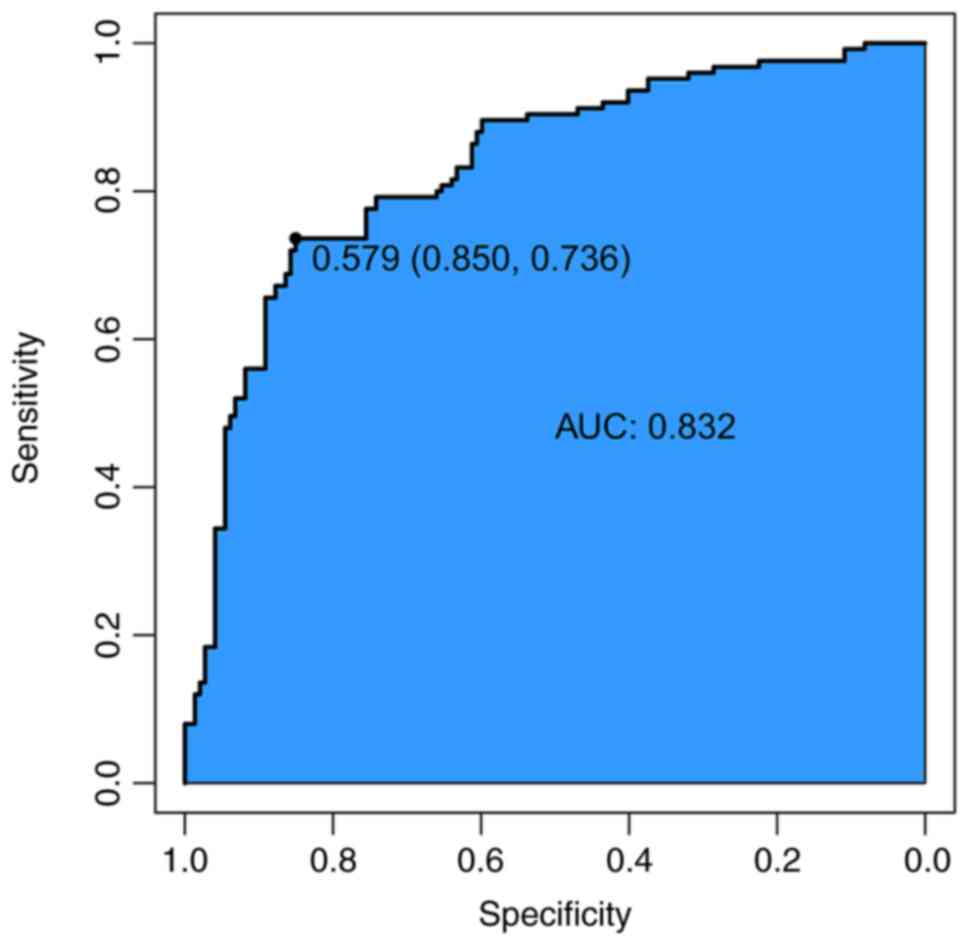
ROC curves indicated that SEMA4D was sensitive
(80.95%) and specific (85.36%) in predicting melanoma and was
associated with a higher risk of melanoma (area under the
curve=0.800, P<0.05) (Figs. 2
and 3).
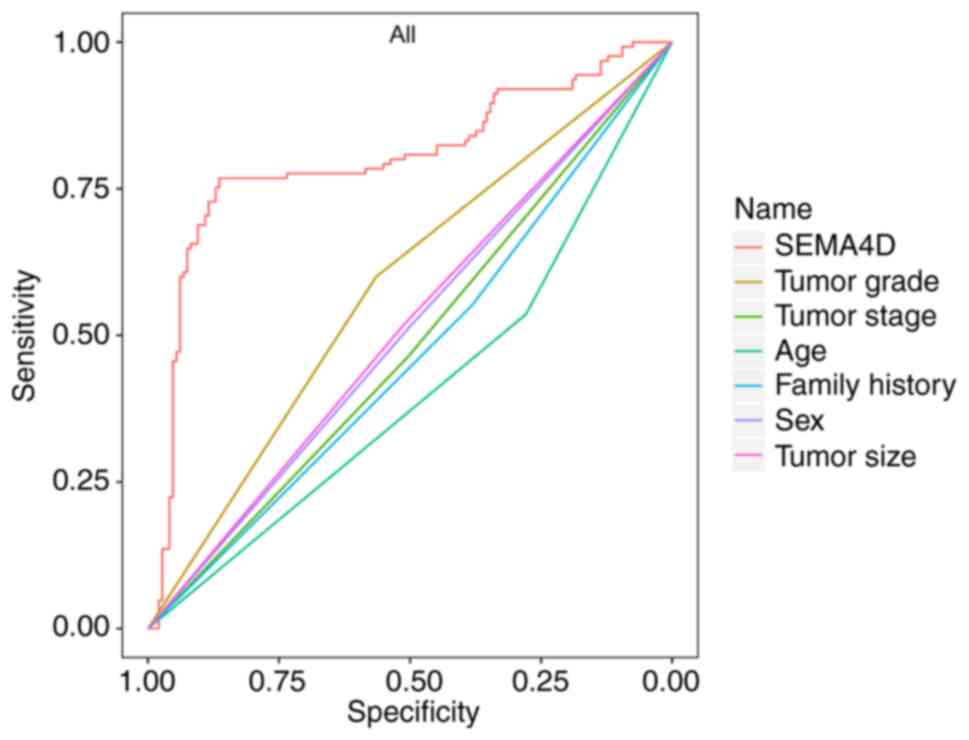
The ROC curve indicated that the combined influence
of all patient-related factors (SEMA4D, tumor grade, tumor stage,
age, family history, sex and tumor size) were sensitive (85.42%)
and specific (89.75%) for the prediction of melanoma (area under
the curve=0.832) (Fig. 4).
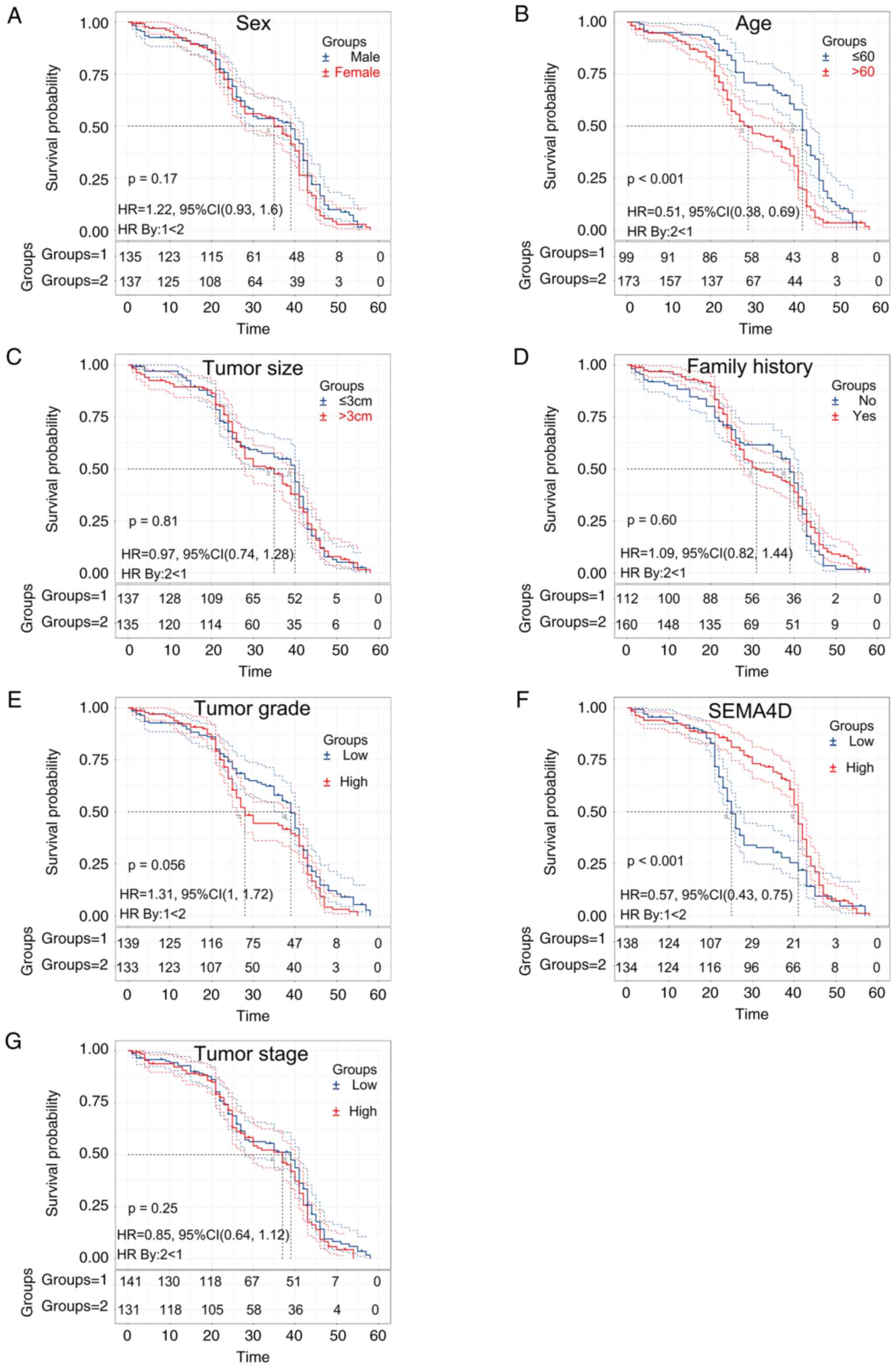
Survival analysis of factors
associated with melanoma
Analysis of the patient survival curves showed that
melanoma patients with lower SEMA4D expression had a shorter
survival time compared with those with above median levels of
SEMA4D expression levels (Fig.
5).
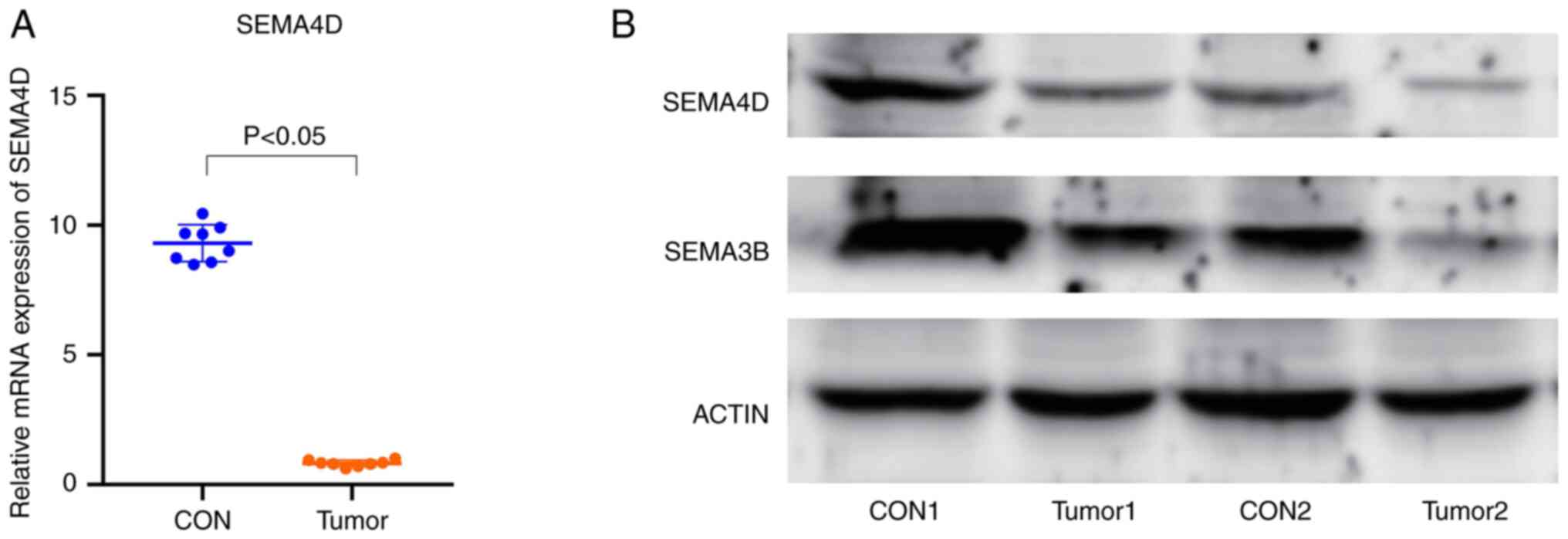
RT-qPCR and western blotting analysis
of SEMA4D expression
Using RT-qPCR to compare SEMA4D expression in normal
tissues and melanoma tissues, it was shown that the expression
levels of SEMA4D were significantly lower in melanoma tissues
(Fig. 6A). Furthermore, the results
of western blotting confirmed these results at the protein level.
That is, SEMA4D protein expression was downregulated in the
melanoma tissues compared with the normal tissues. There was no
significant difference in the expression of SEMA3B between the
melanoma tissues and normal tissues (Fig. 6B).
Discussion
In this study, it was shown that the prognosis of
melanoma patients was significantly correlated with age, tumor
grade, and SEMA4D expression. Spearman correlation coefficient
analysis showed that age, tumor grade, and SEMA4D expression were
significantly correlated with prognosis. Univariate logistic
regression analysis showed that age and tumor grade, and SEMA4D
expression were significantly correlated with prognosis. Older
patients, a higher tumor grade, and lower SEMA4D expression levels
were associated with a poor prognosis. Multivariate logistic
regression analysis showed that older patients had a poorer
prognosis, and patients with low SEMA4D expression had a
significantly worse prognosis than patients with high SEMA4D
expression. Univariate Cox regression analysis showed that the
survival time of older patients was lower than that of younger
patients, and the survival time of patients with low SEMA4D
expression was significantly lower than that of patients with high
SEMA4D expression. Multivariate Cox regression analysis showed that
the survival time of older patients was lower than that of younger
patients, and the survival time of melanoma patients with low
SEMA4D expression was significantly lower than that of patients
with high SEMA4D expression.
Melanoma is the most aggressive and deadly form of
skin cancer (15). The production
of a variety of pro-inflammatory cytokines secreted by macrophages,
T lymphocytes and B lymphocytes in the tumor microenvironment
encourages the survival and growth of tumor cells (16). Studies have shown that melanoma
often has noticeable inflammatory reactions in the
histopathological examination, and skin inflammation significantly
promotes the growth of melanoma (17–19).
Inflammation is a potential biomarker for stratified immunotherapy
and targeted therapy in patients with melanoma (20). In the inflammatory tumor
microenvironment of melanoma, immune cells, extracellular matrix
proteins, cytokines, and other factors affect the progression of
melanoma (21,22). Melanoma-related inflammation
involves multiple regulatory pathways (20), and inflammation and immune response
are critical to developing and treating melanoma.
SEMA4D is a protein-coding gene that is
physiologically expressed on the surfaces of T cells, activated B
cells, mature dendritic cells, macrophages, neutrophils, and
natural killer cells (23). SEMA4D
is involved in several processes, including positive regulation of
phosphatidylinositol three kinase signaling, neuronal projection
development, and regulation of phosphate metabolism. SEMA4D plays a
crucial role in axonal orientation in the nervous system,
activation of T and B cells in the immune system, and regulation
through various signal transduction pathways (24).
SEMA4D is the first signaling element to play a role
in the immune system (25). It
exists in a soluble functional form that can bind to multiple
receptors involved in immune regulation and inflammatory responses
(26). SEMA4D shows varying effects
on the inflammatory phenotype of different cell types (27,28).
The SEMA4D protein is a transmembrane protein expressed on T cells,
and platelets, amongst other cells. Activating T cells and
platelets results in the cleavage and release of active soluble
fragments of SEMA4D during the activation process, and it may also
be present in its soluble form following proteolytic cleavage
during cell activation (29).
SEMA4D promotes pro-inflammatory cytokine production in various
cells by binding to its plexin receptor (30). Cholangitis, primary sclerosis, and
autoimmune vasculitis are diseases connected to SEMA4D via pathways
including nervous system development and semaphore connections
(31). SEMA4D has also been
reported to induce proinflammatory cytokine production and is
involved in endothelial inflammation and vascular dysfunction
(32). SEMA4D is involved in
platelet and neutrophil activation, angiogenesis, and cancer
metastasis (33,34). Other studies have shown that SEMA4D
promotes bladder cancer proliferation and metastasis by activating
the PI3K/AKT pathway (35).
SEMA4D is inextricably linked to inflammation. Thus,
when SEMA4D is abnormally expressed, it induces an inflammatory
response, which is typically associated with the development and
progression of cancer and is one of the initiation processes by
which cells enter the tumor microenvironment through specific
cytokines called chemokines (36).
Inflammation also plays a decisive role in the initiation,
promotion, malignant transformation, invasion, and metastasis of
tumor development (37). Several
cancers form at a site of infection, chronic irritation, and
inflammation, and inflammatory cells have a powerful influence on
tumor development (38).
Inflammatory cells and the chemokines and cytokines they produce
affect the entire tumor organ and regulate growth, migration, and
differentiation of all cell types in the tumor microenvironment,
including tumor cells, fibroblasts, and endothelial cells (39,40).
Therefore, SEMA4D expression levels may play a role in the
occurrence and development of melanoma through an inflammatory
response.
The present study has some limitations. Although
clinical data have been examined and analyzed, the molecular
mechanisms by which SEMA4D expression levels affect melanoma
prognosis and survival have not been validated in animal models.
Therefore, future studies should focus on animal experiments to
explore the molecular pathway and mechanism of SEMA4D in
melanoma.
In conclusion, SEMA4D expression levels were shown
to be significantly correlated with the prognosis and survival time
of melanoma patients. Low SEMA4D expression is associated with a
poorer prognosis and reduced survival times in melanoma patients.
As a potential molecular marker of poor survival and prognosis of
melanoma, the low expression of SEMA4D provides a novel direction
for identifying the molecular mechanism underlying the development
and progression of melanoma.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author upon
reasonable request.
Authors' contributions
XL and ZZ conceived and designed the study. CZ, WY,
RW and YZ participated in data analysis and interpretation. WY and
SL completed the experiments. SL and RW drafted and revised key
theories in the paper. YZ and ZZ answered academic questions. ZZ
made substantial contributions to the conception and design of the
study. ZZ and WY confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The Ethics Committee of the Fourth Hospital of Hebei
Medical University approved the present study (approval no.
FHBM2015014), and all patients signed informed consent.
Patient consent for publication
All patients and their families were informed in
writing and consented to publication of this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dzwierzynski WW: Melanoma risk factors and
prevention. Clin Plast Surg. 48:543–550. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ahmed B, Qadir MI and Ghafoor S: Malignant
Melanoma: Skin cancer-diagnosis, prevention, and treatment. Crit
Rev Eukaryot Gene Expr. 30:291–297. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruno W, Dalmasso B, Barile M, Andreotti
V, Elefanti L, Colombino M, Vanni I, Allavena E, Barbero F, Passoni
E, et al: Predictors of germline status for hereditary melanoma: 5
years of multi-gene panel testing within the Italian Melanoma
Intergroup. ESMO Open. 7:1005252022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ismail H, Helby J, Hölmich LR, H Chakera
A, Bastholt L, Klyver H, Sjøgren P, Schmidt H, Schöllhammer L,
Nordestgaard BG and Bojesen SE: Genetic predisposition to long
telomeres is associated with increased mortality after melanoma: A
study of 2101 melanoma patients from hospital clinics and the
general population. Pigment Cell Melanoma Res. 34:946–954. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo W, Wang H and Li C: Signal pathways of
melanoma and targeted therapy. Signal Transduct Target Ther.
6:4242021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Craig S and Virós A: New biomarkers
improve stratification of patients with melanoma. Br J Dermatol.
182:5–6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun L and Arbesman J: Canonical signaling
pathways in melanoma. Clin Plast Surg. 48:551–560. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Phoon YP, Tannenbaum C and Diaz-Montero
CM: Immunobiology of Melanoma. Clin Plast Surg. 48:561–576. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Skudalski L, Waldman R, Kerr PE and
Grant-Kels JM: Melanoma: An update on systemic therapies. J Am Acad
Dermatol. 86:515–524. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen C, Hou J, Tanner JJ and Cheng J:
Bioinformatics methods for mass spectrometry-based proteomics data
analysis. Int J Mol Sci. 21:28732020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fu Y, Ling Z, Arabnia H and Deng Y:
Current trend and development in bioinformatics research. BMC
Bioinformatics. 21 (Suppl 9):S5382020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu Q, Cai P, Yu Y, Liu Z, Chen G and Zeng
Z: Sema4D correlates with tumour immune infiltration and is a
prognostic biomarker in bladder cancer, renal clear cell carcinoma,
melanoma and thymoma. Autoimmunity. 54:294–302. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Jian W, Luo Q and Fang L:
CircSEMA4B inhibits the progression of breast cancer by encoding a
novel protein SEMA4B-211aa and regulating AKT phosphorylation. Cell
Death Dis. 13:7942022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hughes AJ and Herr AE: Microfluidic
Western blotting. Proc Natl Acad Sci USA. 109:21450–21455. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Castañeda-Reyes ED, Perea-Flores MJ,
Davila-Ortiz G, Lee Y and Gonzalez de Mejia E: Development,
characterization and use of liposomes as amphipathic transporters
of bioactive compounds for melanoma treatment and reduction of skin
inflammation: A review. Int J Nanomedicine. 15:7627–7650. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pereira J, Bessa C, Matos P and Jordan P:
Pro-Inflammatory cytokines trigger the overexpression of
tumour-related splice variant RAC1B in polarized colorectal cells.
Cancers (Basel). 14:13932022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ohno F, Nakahara T, Kido-Nakahara M, Ito
T, Nunomura S, Izuhara K and Furue M: Periostin links skin
inflammation to melanoma progression in humans and mice. Int J Mol
Sci. 20:1692019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rossi N, Lee KA, Bermudez MV, Visconti A,
Thomas AM, Bolte LA, Björk JR, de Ruijter LK, Newton-Bishop J,
Harland M, et al: Circulating inflammatory proteins associate with
response to immune checkpoint inhibition therapy in patients with
advanced melanoma. EBioMedicine. 83:1042352022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Zhang Y, Chen S, Liu W, Lin Y,
Zhang H and Yu F: Non-Steroidal Anti-Inflammatory Drugs (NSAIDs)
sensitize melanoma cells to MEK inhibition and inhibit metastasis
and relapse by inducing degradation of AXL. Pigment Cell Melanoma
Res. 35:238–251. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karlsson MJ, Costa Svedman F, Tebani A,
Kotol D, Höiom V, Fagerberg L, Edfors F, Uhlén M, Egyhazi Brage S
and Maddalo G: Inflammation and apolipoproteins are potential
biomarkers for stratification of cutaneous melanoma patients for
immunotherapy and targeted therapy. Cancer Res. 81:2545–2555. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalaora S, Nagler A, Wargo JA and Samuels
Y: Mechanisms of immune activation and regulation: Lessons from
melanoma. Nat Rev Cancer. 22:195–207. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Chen Z, Zhang A, Gupte AA and
Hamilton DJ: The role of calcium signaling in melanoma. Int J Mol
Sci. 23:10102022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rajabinejad M, Asadi G, Ranjbar S, Afshar
Hezarkhani L, Salari F, Gorgin Karaji A and Rezaiemanesh A:
Semaphorin 4A, 4C, and 4D: Function comparison in the autoimmunity,
allergy, and cancer. Gene. 746:1446372020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie J, Wang Z and Wang W: Semaphorin 4D
induces an imbalance of Th17/Treg cells by activating the Aryl
hydrocarbon receptor in ankylosing spondylitis. Front Immunol.
11:21512020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Zhang WS, Tang ZH, Ye DD, Su S,
Zhang SM and Qiu J: Anti-inflammatory effects of the immobilization
of SEMA4D on titanium surfaces in an endothelial cell/macrophage
indirect coculture model. Biomed Mater. 17:0150052021. View Article : Google Scholar
|
|
26
|
Younis RH, Ghita I, Elnaggar M,
Chaisuparat R, Theofilou VI, Dyalram D, Ord RA, Davila E, Tallon
LJ, Papadimitriou JC, et al: Soluble Sema4D in plasma of head and
neck squamous cell carcinoma patients is associated with underlying
non-inflamed tumor profile. Front Immunol. 12:5966462021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Maleki KT, Cornillet M and Björkström NK:
Soluble SEMA4D/CD100: A novel immunoregulator in infectious and
inflammatory diseases. Clin Immunol. 163:52–59. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chapoval SP, Vadasz Z, Chapoval AI and
Toubi E: Semaphorins 4A and 4D in chronic inflammatory diseases.
Inflamm Res. 66:111–117. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Willner N, Goldberg Y, Schiff E and Vadasz
Z: Semaphorin 4D levels in heart failure patients: A potential
novel biomarker of acute heart failure. ESC Heart Fail. 5:603–609.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Movila A, Mawardi H, Nishimura K, Kiyama
T, Egashira K, Kim JY, Villa A, Sasaki H, Woo SB and Kawai T:
Possible pathogenic engagement of soluble Semaphorin 4D produced by
γδT cells in medication-related osteonecrosis of the jaw (MRONJ).
Biochem Biophys Res Commun. 480:42–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu Y, Zhou Y, Di C, Zhao C, Chen J, Su W,
Wu Q, Wu M, Su X and Xia Z: Increased airway epithelial
cell-derived exosomes activate macrophage-mediated allergic
inflammation via CD100 shedding. J Cell Mol Med. 25:8850–8862.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu JH, Li YN, Chen AQ, Hong CD, Zhang CL,
Wang HL, Zhou YF, Li PC, Wang Y, Mao L, et al: Inhibition of
Sema4D/PlexinB1 signaling alleviates vascular dysfunction in
diabetic retinopathy. EMBO Mol Med. 12:e101542020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lontos K, Adamik J, Tsagianni A, Galson
DL, Chirgwin JM and Suvannasankha A: The role of semaphorin 4D in
bone remodeling and cancer metastasis. Front Endocrinol (Lausanne).
9:3222018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nishide M and Kumanogoh A: The role of
semaphorins in immune responses and autoimmune rheumatic diseases.
Nat Rev Rheumatol. 14:19–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu JJ, Su YW, Wang CJ, Li DF and Zhou L:
Semaphorin 4D promotes the proliferation and metastasis of bladder
cancer by activating the PI3K/AKT pathway. Tumori. 105:231–242.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Singh N, Baby D, Rajguru JP, Patil PB,
Thakkannavar SS and Pujari VB: Inflammation and cancer. Ann Afr
Med. 18:121–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schmitt M and Greten FR: The inflammatory
pathogenesis of colorectal cancer. Nat Rev Immunol. 21:653–667.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Iyengar NM, Gucalp A, Dannenberg AJ and
Hudis CA: Obesity and cancer mechanisms: Tumor microenvironment and
inflammation. J Clin Oncol. 34:4270–4276. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Landskron G, De la Fuente M, Thuwajit P,
Thuwajit C and Hermoso MA: Chronic inflammation and cytokines in
the tumor microenvironment. J Immunol Res. 2014:1491852014.
View Article : Google Scholar : PubMed/NCBI
|