Introduction
Liver cancer is one of the most common types of
malignant tumor and a leading cause of cancer-associated deaths
worldwide (1). Hepatitis B virus
(HBV) infection has been demonstrated to be an important risk
factor for high–risk liver cancer (2). Multiple studies have reported that HBV
contributes to the proliferation and metastasis of liver cells,
playing a crucial role in the high recurrence and mortality of
liver cancer (3–5). Despite improvements that have been made in
surgical resection and transplantation, the prognosis of patients
with liver cancer remains unsatisfactory with a 5–year survival
rate of <50% (6,7). Therefore, exploring the molecular
mechanism underlying the pathogenesis of HBV-associated liver
cancer has important clinical significance.
Non-coding RNAs (ncRNAs) are divided into small
ncRNAs with 18–200 nucleotides, known as microRNAs (miRNAs), and
long ncRNAs (lncRNAs) of between 200 nucleotides and 100 kb, which
represent the majority of human genome transcripts (8). Numerous studies have documented the
crucial roles of lncRNAs as regulators of viral replication or the
antiviral response in the pathogenesis and progression of
HBV-induced liver cancer (9,10). For
instance, HBV X protein-related long ncRNA MALAT1 has been shown to
promote cell metastasis via the upregulation of latent transforming
growth factor β binding protein 3 expression in liver cancer
(11). In another study, lncRNA
MAPKAPK5_AS1 was found to be upregulated in HBV-associated liver
cancer tissues, and facilitated the proliferation and G1/S
transition in HBV-positive liver cancer cells (12). Conversely, Deng et al
(13) found that another lncRNA,
F11-AS1, suppressed HBV-associated liver cancer progression via
interference with cellular physiology. Additionally, well-explored
lncRNAs, including DBH antisense RNA 1 (14), HOX transcript antisense intergenic
RNA (15) and X inactive specific
transcript (16) have been reported
to be potential prognostic biomarkers for patients with
HBV-associated liver cancer. Nevertheless, most of these studies
focused on a single lncRNA and comprehensive analysis of the lncRNA
signature of HBV-associated liver cancer is relatively limited
(17).
In the present study, the transcriptome expression
level data of samples from patients with liver cancer and HBV
infection were comprehensively analyzed on multiple platforms.
Based on the lncRNAs and genes identified to be associated with
HBV-related liver cancer, different optimization algorithms were
used to further screen the optimized lncRNA signatures. Following
the construction of a competing endogenous RNA (ceRNA) regulatory
network, functional experiments were performed to investigate the
biological role of characteristic lncRNAs. These experiments were
performed with the aim of identifying novel therapeutic targets for
the diagnosis and treatment of patients with HBV-associated liver
cancer.
Materials and methods
Data collection
Transcriptome expression profile data for
HBV-associated liver cancer were searched for in the Gene
Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) with the keywords
‘hepatitis’ and ‘liver cancer’. The final date covered by the
search was May 16, 2022, The following inclusion criteria were
used: i) Transcriptome expression profile data; ii) subjects were
skin solid tissue samples from patients with liver cancer; iii)
included hepatitis virus infection data; iv) contained normal
control specimens; v) sample size >100; vi) the detection
platform was capable of annotating a large number of lncRNAs. As a
result, two sets of transcriptome expression profile data obtained
using the GPL570 Affymetrix Human Genome U133 Plus 2.0 Array
platform were screened as follows: GSE121248, which contains 70
HBV-liver cancer samples and 37 control samples (18) as the training dataset, and GSE55092,
which contains 39 HBV-liver cancer samples and 81 control samples
(19) as the validation dataset. In
addition, the Ensembl genome browser 96 database (http://asia.ensembl.org/index.html) was used to
download detailed platform annotation information, including
probes, gene symbols and RNA types. The expression levels of
corresponding lncRNAs and mRNAs were obtained by reannotating the
detection probes downloaded from these two datasets.
In addition, transcriptome expression profile data
for patients with liver cancer were collected from The Cancer
Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) based on the Illumina
HiSeq 2000 RNA Sequencing platform as an independent validation
dataset. After the removal of samples without viral hepatitis
serology information, 60 HBV-liver cancer samples with survival
prognosis information and 46 control samples were obtained.
Screening of overlapped differentially
expressed RNAs (DERs)
The limma package of R3.6.1 version 3.34.7 (20) was utilized to screen the DERs,
including DElncRNAs and DEmRNAs, between the HBV-liver cancer and
normal control groups in GSE121248 and GSE55092. The cutoff value
for screening was |log2 fold change (FC)|>1.0 and FDR
(false discovery rate) <0.05. Then, the selected DERs were used
to draw a volcano plot and heatmap. A Venn diagram was created to
display the DERs that were present in both datasets. Using DAVID
version 6.8 (21,22) (https://david.ncifcrf.gov/), the DEmRNAs common to
both datasets were subjected to Gene Ontology (GO) function and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis with FDR <0.05 as the cutoff criterion.
Construction and validation of a
nomogram
The optimized lncRNA signatures were first screened
in the GSE121248 dataset using three types of optimization
algorithms, including recursive feature elimination (RFE) (23), least absolute shrinkage and
selection operator (LASSO) (24)
and random forest (RF) (25). By
comparing these results, the DElncRNAs common to all three analyses
were used as the final optimized lncRNA signatures. A concise
nomogram for predicting the survival of patients with HBV-liver
cancer was established in the training dataset using the rms
package version 5.1-2 in R3.6.1 language (26). Then, the R3.6.1 language rmda
package version 1.6 (https://cran.r-project.org/web/packages/rmda/index.html)
was used to perform decision curve analysis on individual optimized
lncRNA or combined optimized lncRNA signatures. Finally, the
nomogram model constructed based on the previously screened
optimized lncRNA signatures was verified as a diagnostic model in
the validation dataset GSE55092 and TCGA independent validation
dataset.
Prognostic performance of the
HBV-associated lncRNA signature in liver cancer
The obtained optimized lncRNA signatures were
subjected to univariate Cox regression analysis to screen the HBV
prognosis-associated lncRNA signatures from TCGA dataset. The
samples were divided into high and low expression groups according
to their expression median levels. The association between
different expression levels and survival prognosis was evaluated
using the Kaplan-Meier curve method with log-rank test.
Construction of a ceRNA network based
on the prognostic lncRNAs signature
According to the ceRNA hypothesis, a
lncRNA-miRNA-mRNA ceRNA interaction network was constructed as
follows: Liver cancer-associated miRNAs were mined from the
miR2Disease version 2022 database (27). By searching the DIANA-LncBase v2
database (28), the validated
relationships of the lncRNAs ST8SIA6-AS1 or LINC01093 with liver
cancer-related miRNAs were obtained with MitScore >0.6 as the
cutoff and the lncRNA-miRNA interactions were constructed. Then,
the corresponding mRNA targets of the liver cancer-associated
miRNAs were predicted by searching StarBase version 2.0 (29). Subsequently, the predicted mRNA
targets of the liver cancer-associated miRNAs were combined with
the consistently expressed overlapping DEmRNAs to obtain the final
miRNA-mRNA interactions. Finally, the lncRNA-miRNA-mRNA ceRNA
network was constructed based on lncRNA-miRNA and miRNA-mRNA
interactions and visualized using Cytoscape version 3.6.1 (30). To elucidate the functions of
important lncRNAs, KEGG pathway enrichment analysis was performed
on the DEmRNAs contained in the ceRNA network.
HBV-associated tissue samples
A total of 31 pairs of tumor tissues and adjacent
liver tissues were obtained from patients with HBV-associated liver
cancer (age range, 21–72 years; mean age, 42.5±7.8 years; 10
females and 21 males) at the Hospital for Infectious Diseases of
Pudong District (Shanghai, China) between August 2020 and September
2021. Prior to tissue collection, each patient provided written
informed consent for the use of their samples in scientific
research. The obtained tissues were immediately snap-frozen in
liquid nitrogen until further analysis. This study complied with
the guidelines of the Declaration of Helsinki and was approved by
the Ethics Committee of the Hospital for Infectious Diseases of
Pudong District (approval no. IDP-2020PD; 2020.8.16).
Cell transfection and culture
The HepG2 human hepatoblastoma cell line and the
HepG2.2.15 cell line, which was derived from HepG2 cells via the
integration of the full-length HBV genome into the cellular genome,
were purchased from American Type Culture Collection. The
identification of the two cell lines was confirmed using short
tandem repeat DNA profiling analysis. The cell lines were cultured
in DMEM (Thermo Fisher Scientific, Inc.) with 10% FBS, 100 U/ml
penicillin and 0.1% (w/v) streptomycin in a humidified incubator
with 5% CO2 at 37°C. The small interfering (si)RNAs
si-ST8SIA6-AS1 (5′-GCTCCTCCTTGCTCCAAAGAA-3′) and si-negative
control (si-NC; 5′-GCAGATCCTACCCGTCTCTAA-3′) and the plasmids
pcDNA3.1 and pcDNA3.1-LINC01093 were synthesized by Shanghai
GenePharma Co., Ltd. These oligonucleotides or plasmids (50 µg)
were transfected into HepG2.2.15 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 48 h, and then harvested immediately
for subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from tissues or cells using
TRIzol® reagent (cat. no. 10296010; Invitrogen; Themo
Fisher Scientific, Inc.). RT was performed using a PrimeScript RT
reagent kit with gDNA Eraser (Takara Bio, Inc.) following the
manufacturer's instructions. qPCR was carried out using
SYBR® Premix Ex Taq II (Takara Bio, Inc.) with the
following primer sequences: ST8SIA6-AS1 forward.
5′-TCCTGATTCAGTGGCATGGT-3′ and reverse, 5′-AGGGTTTCTTCGGTCGTCAT-3′;
LINC01093 forward, 5′-CTCTTGCAAACCATGGGCAC-3′ and reverse,
5′-CATCTCCCAGTCGGGTTTCC-3′; GAPDH forward,
5′-GGTGAAGGTCGGAGTCAACG-3′ and reverse, 5′-GCATCGCCCCACTTGATTTT-3′.
The thermocycling conditions were 94°C for 30 sec, followed by 40
cycles of 94°C for 5 sec and 60°C for 30 sec. Relative expression
levels of target genes were calculated using the 2−ΔΔCq
method (31) with GAPDH as the
reference gene.
HBV replication analysis
The HBV DNA was extracted from cell supernatants
using a Column Viral DNAout kit (Tiandz, Inc.) according to the
manufacturer's instructions. HBV DNA levels were determined by qPCR
using QuantiFast SYBR® Green PCR Kit (Qiagen, Inc.) with
the following primer sequences: Forward, 5′-GCCTCATTTTGCGGATCACC-3′
and reverse, 5′-TGTCCCCATGCCTTTTCGAG-3′. Relative expression levels
of target genes were calculated using the 2−ΔΔCq method
(31) with GAPDH as the reference
gene.
Enzyme-linked immunosorbent assay
(ELISA)
In brief, the cell culture supernatants of
HepG2.2.15 cells were centrifuged at 250 × g for 5 min at 4°C and
kept −20°C until further analysis. Then, the viral protein
hepatitis B surface antigen (HBsAg) and hepatitis B e antigen
(HBeAg) in the culture supernatant were determined using
corresponding ELISA kits (cat. nos. CSB-E10089h and CSB-E13557h,
respectively; Cusabio Technology, LLC).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation capacity was assessed by
performing a CCK-8 assay. Briefly, HepG2.2.15 cells at a density of
5,000 cells per well were seeded into 96-well plates and cultured
overnight. At 0, 24, 48 and 72 h, 10 µl CCK-8 reagent (Dojindo
Laboratories, Inc.) was separately added into each well. After
incubation for another 2 h, the optical density (OD) of each well
was measured at 450 nm using a microplate reader (Bio-Rad
Laboratories, Inc.).
Migration and invasion assays
The in vitro migration assay was performed
using Transwell chambers (pore size, 8.0 µm; Corning Costar). For
invasion assays, the upper surface of the Transwell chamber was
precoated with Matrigel for 2 h at 37°C (Corning Costar) in
advance, while for migration assays no coating was applied.
Subsequently, ~5×104 HepG2.2.15 cells in serum-free
medium were placed in the upper Transwell chamber, while the lower
chamber was filled with complete medium. After 24 h incubation at
37°C, the transferred cells on the lower chamber were fixed with
70% ethanol for 30 min and stained with 0.2% crystal violet for 15
min at room temperature. The stained cells were photographed and
counted in five random fields under a light microscope (Olympus
Corporation).
Statistical analysis
GraphPad Prism software (version 8.0; GraphPad
Software, Inc.) was used to perform the statistical analysis of all
quantitative data. The data were expressed as the mean ± standard
deviation of three independent experiments. Differences between two
groups were assessed using paired Student's t-tests for comparing
tumor adjacent tissues and unpaired Student's t-tests for comparing
two cell groups. Comparisons among three groups were performed
using one-way ANOVA followed by Tukey post hoc tests. P<0.05 was
considered to indicate a statistically significant result.
Results
Identification of DERs and functional
enrichment analysis in patients with HBV-liver cancer
The differential analysis of HBV-liver cancer and
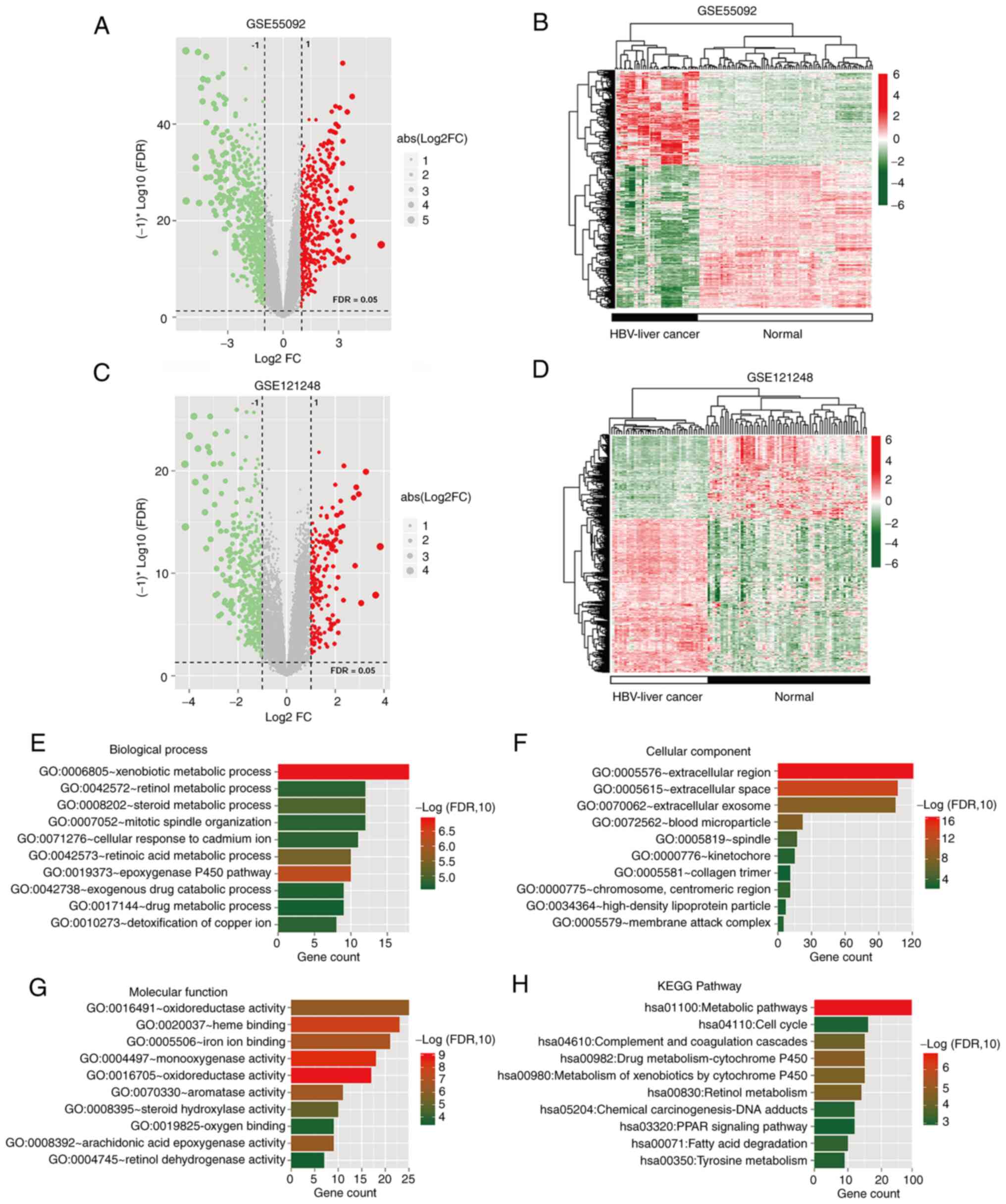
control groups with a cutoff value of |log2 FC|>1.0
and FDR <0.05 yielded 1,183 DERs (Table SI) in GSE55092 (Fig. 1A and B) and 666 DERs in GSE121248
(Fig. 1C and D). A total of 535
DERs were identified to be common to both datasets (Table SII); Venn diagrams showing the
overlap demonstrated that the DERs included 30 DElncRNAs, of which
14 were upregulated and 16 were downregulated, and 505 DEmRNAs of
which 164 were upregulated and 341 were downregulated (Fig. S1). Subsequently, functional
enrichment analysis was performed on the 505 overlapping DEmRNAs
(Table SIII). The 505 DEmRNAs were
significantly enriched in numerous GO terms, including 48
biological process (Fig. 1E), 18
cellular component (Fig. 1F) and 24
molecular function terms (Fig. 1G),
as well as 19 KEGG signaling pathways (Fig. 1H), of which the top 10 terms are
respectively displayed.
Construction and validation of the
nomogram
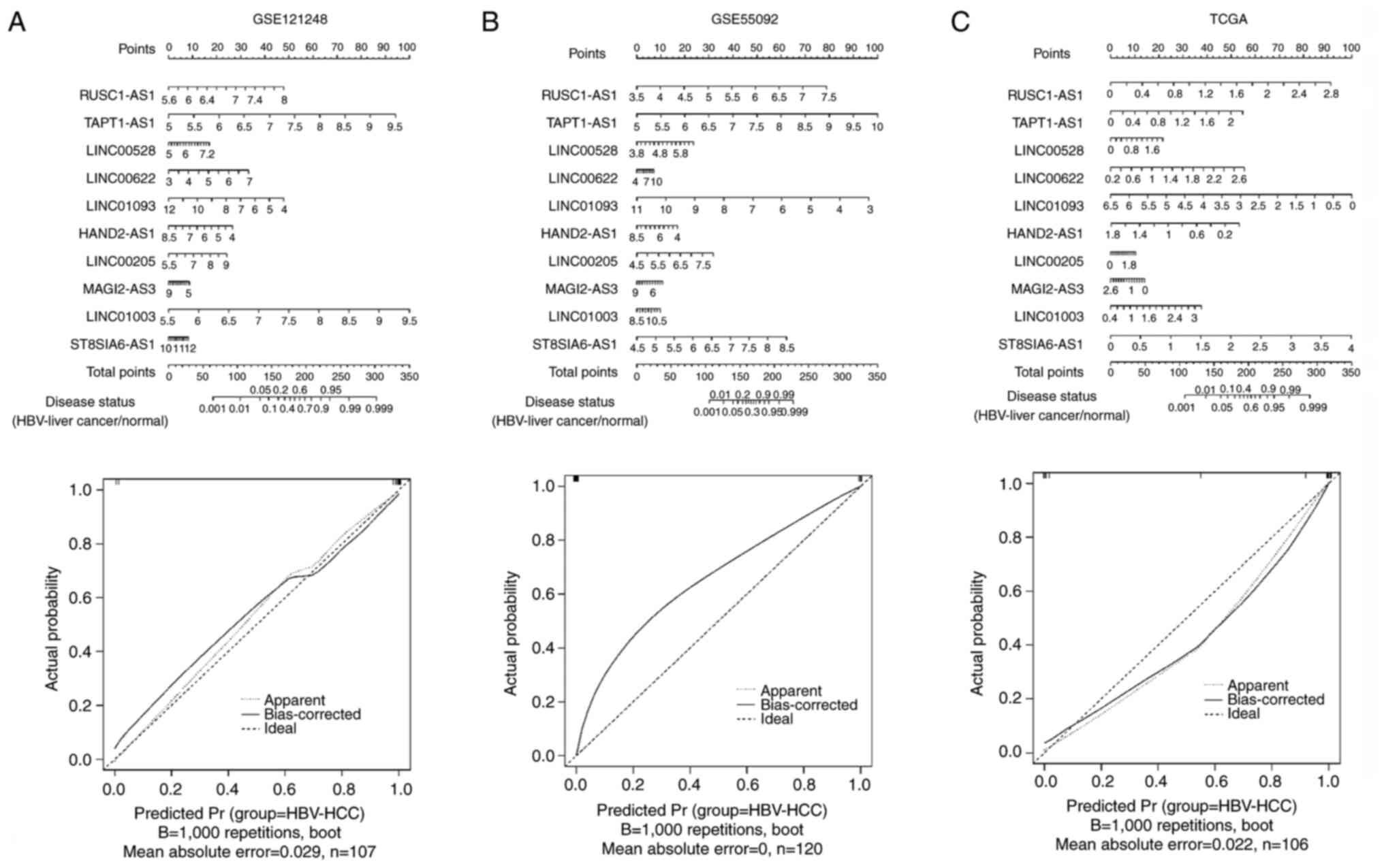
Using the 30 overlapping DElncRNAs, 13, 14 and 24
optimized lncRNA signatures were obtained using RFE, LASSO and RF
algorithms, respectively (Fig.
S2). After further screening, 10 optimized overlapping
DElncRNAs were obtained, namely heart and neural crest derivatives
expressed transcript 2 antisense RNA 1 (HAND2-AS1), LINC00622,
ST8SIA6-AS1, TAPT1-AS1, MAGI2-AS3, LINC00528, LINC00205, RUSC1-AS1,
LINC01093 and LINC01003. Next, a nomogram was constructed that
integrated the 10-DElncRNA-based signature to predict the disease
status of HBV-liver cancer in GSE121248 (Fig. 2A), GSE55092 (Fig. 2B) and TCGA (Fig. 2C) datasets. The corresponding
calibration plots indicated that the nomogram had good accuracy as
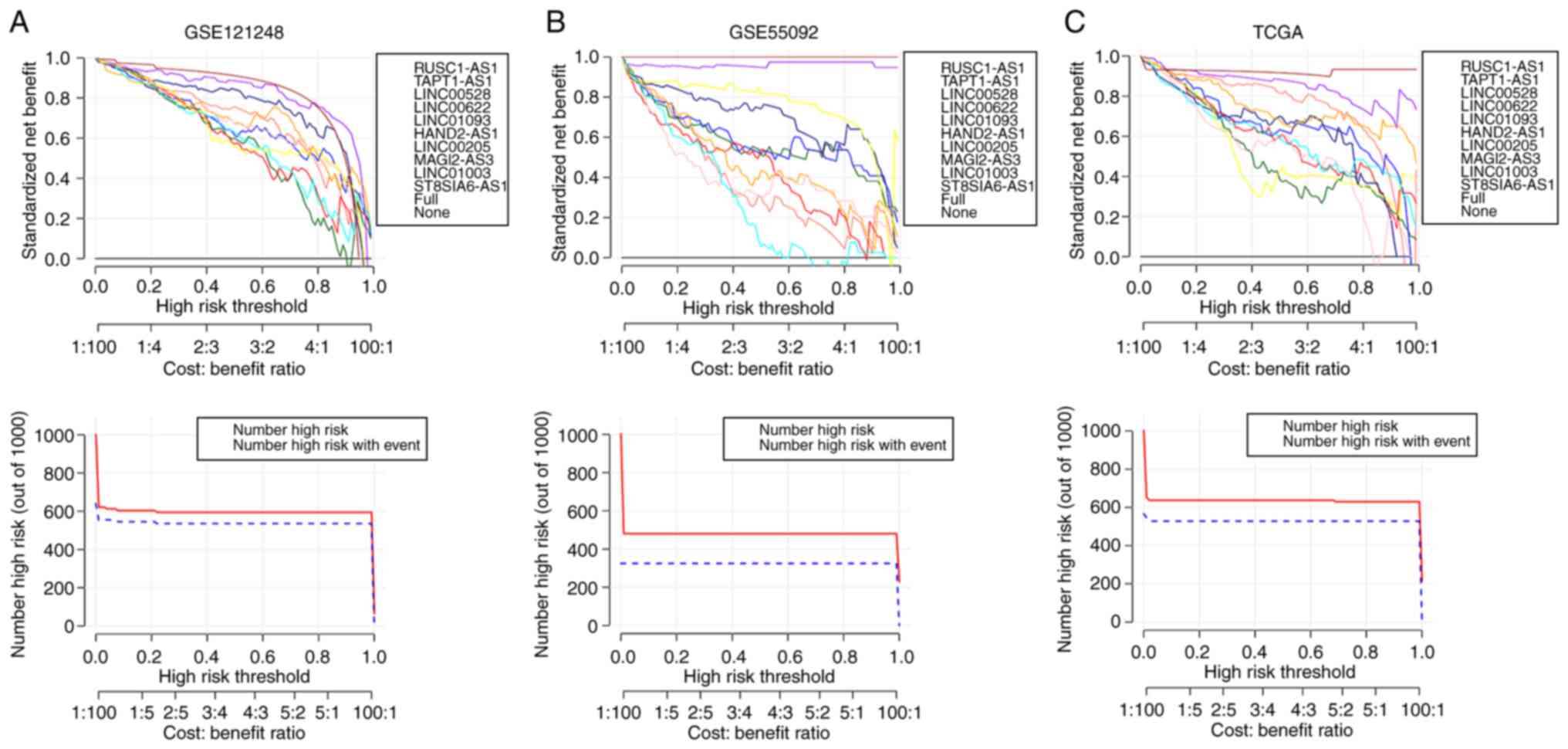
an ideal model for these three datasets. Moreover, the results from
decision curve analysis showed the data from training dataset
GSE121248 (Fig. 3A) were very
consistent with the results in validation dataset GSE55092
(Fig. 3B) and independent
validation dataset TCGA (Fig.
3C).
Prognostic performance of the
HBV-related lncRNA signature in liver cancer
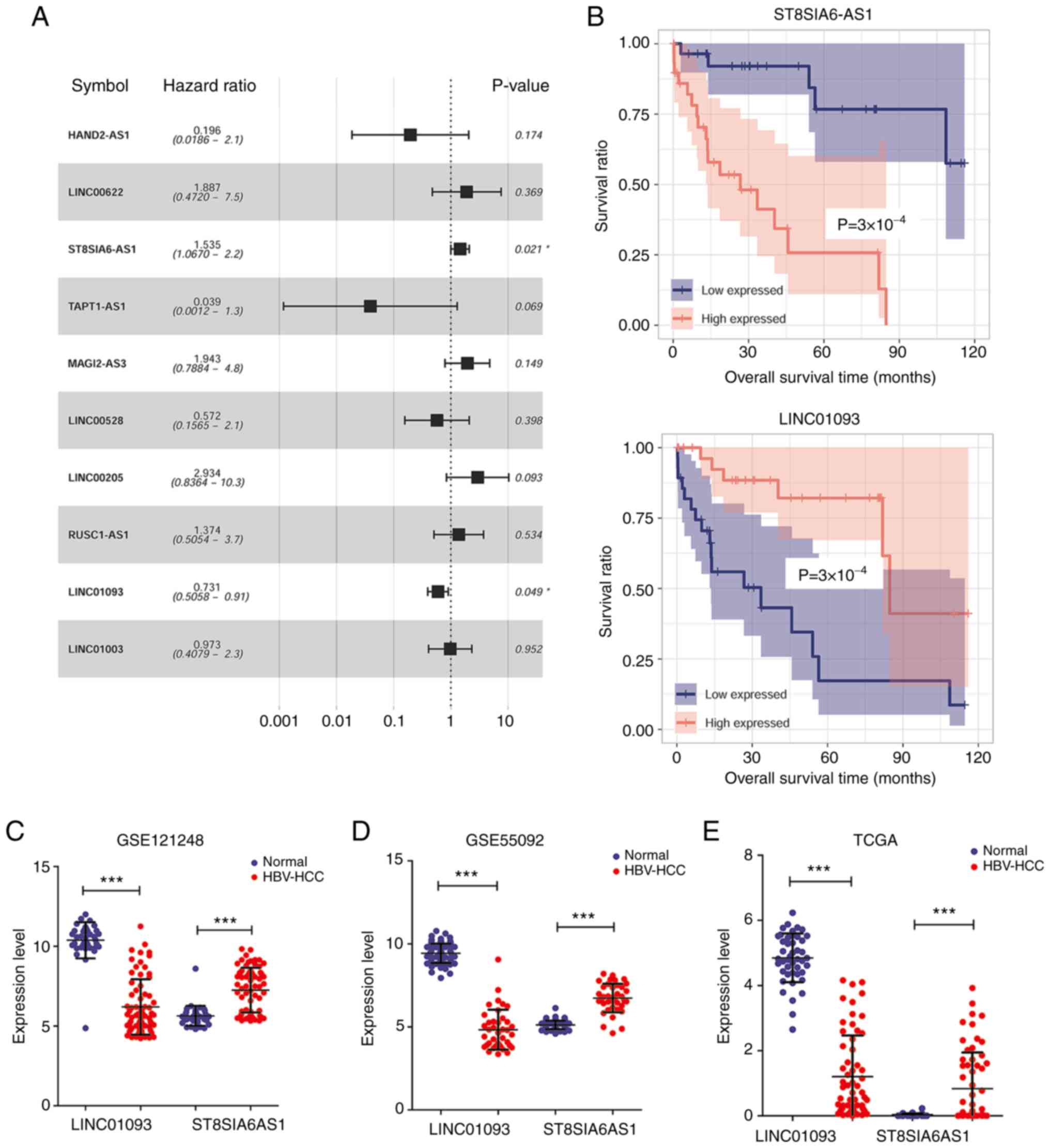
The survival prognosis information in TCGA dataset
with univariate Cox regression analysis identified two lncRNAs that
are associated with HBV-liver cancer prognosis, namely ST8SIA6-AS1
and LINC01093 with P<0.05, as displayed by forest plot (Fig. 4A). Kaplan-Meier survival analysis of
TCGA dataset was performed to further assess the capability of
these two lncRNAs to predict the prognosis of HBV-liver cancer. As
shown in Fig. 4B, patients with
HBV-liver cancer with downregulation of ST8SIA6-AS1 or upregulation
of LINC01093 presented higher overall survival rates. As shown in
Fig. 4C-E, ST8SIA6-AS1 was
upregulated while LINC01093 was downregulated in HBV-liver cancer
samples compared with normal controls in the GSE121248, GSE55092
and TCGA datasets.
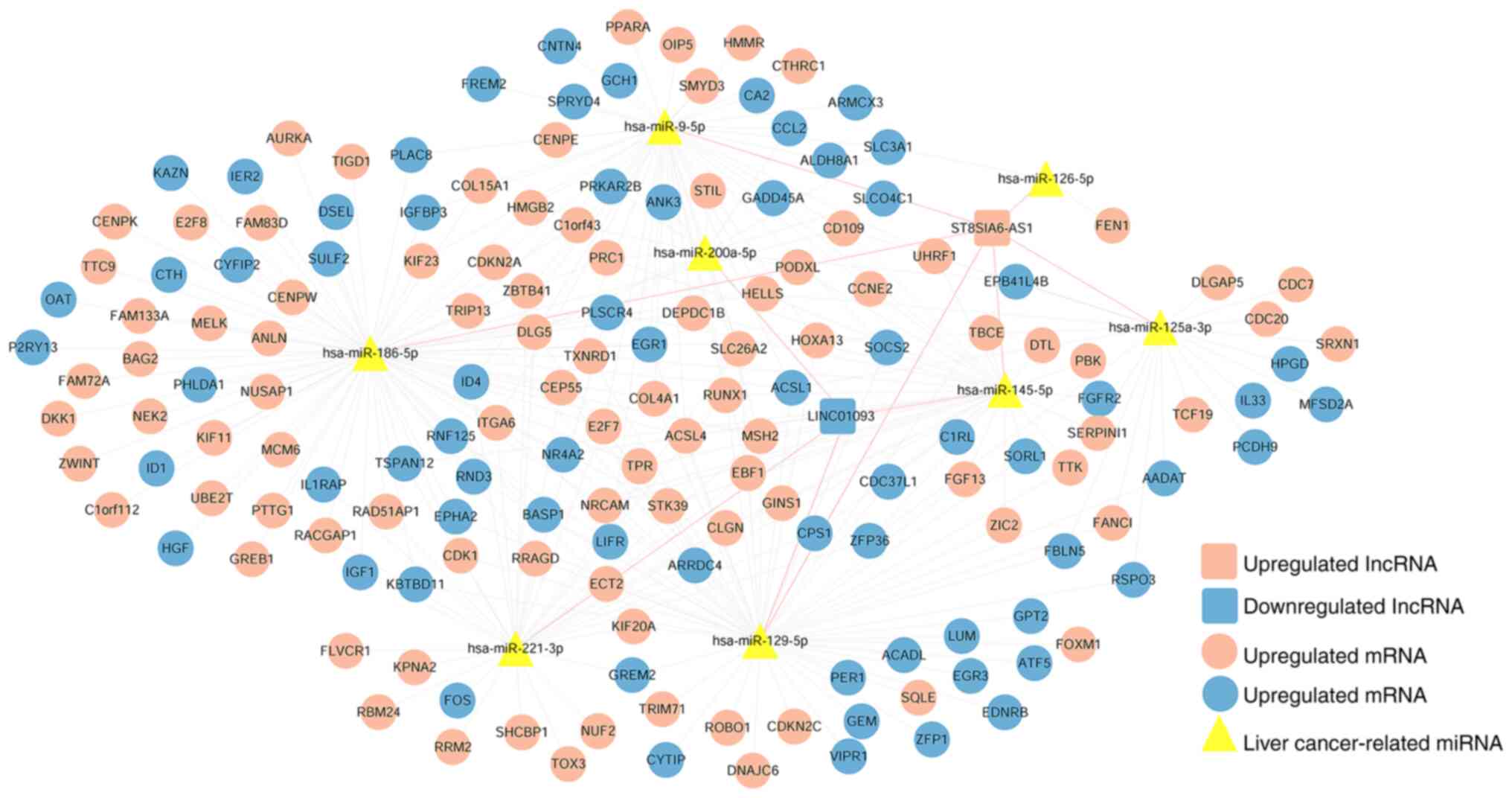
Construction of a ceRNA network based
on the prognostic lncRNAs signature
Guided by the ceRNA hypothesis, 106
disease-associated miRNAs were identified, of which 77 miRNAs were
associated with liver cancer (Table
SIV). Using the DIANA-LncBasev2 database, the interactions of
ST8SIA6-AS1 and LINC01093 with liver cancer-associated miRNAs were
searched, and 11 highly linked lncRNA-miRNA interactions involving
8 liver cancer-associated miRNAs were obtained, which are shown in
Table SV. The predicted mRNA
targets of 8 liver cancer-associated miRNAs and 505 DEmRNAs were
identified, and 285 miRNA-mRNA interactions were obtained (Table SVI). Finally, the ceRNA network was
constructed; it comprised 172 interactions, and included 2 lncRNAs,
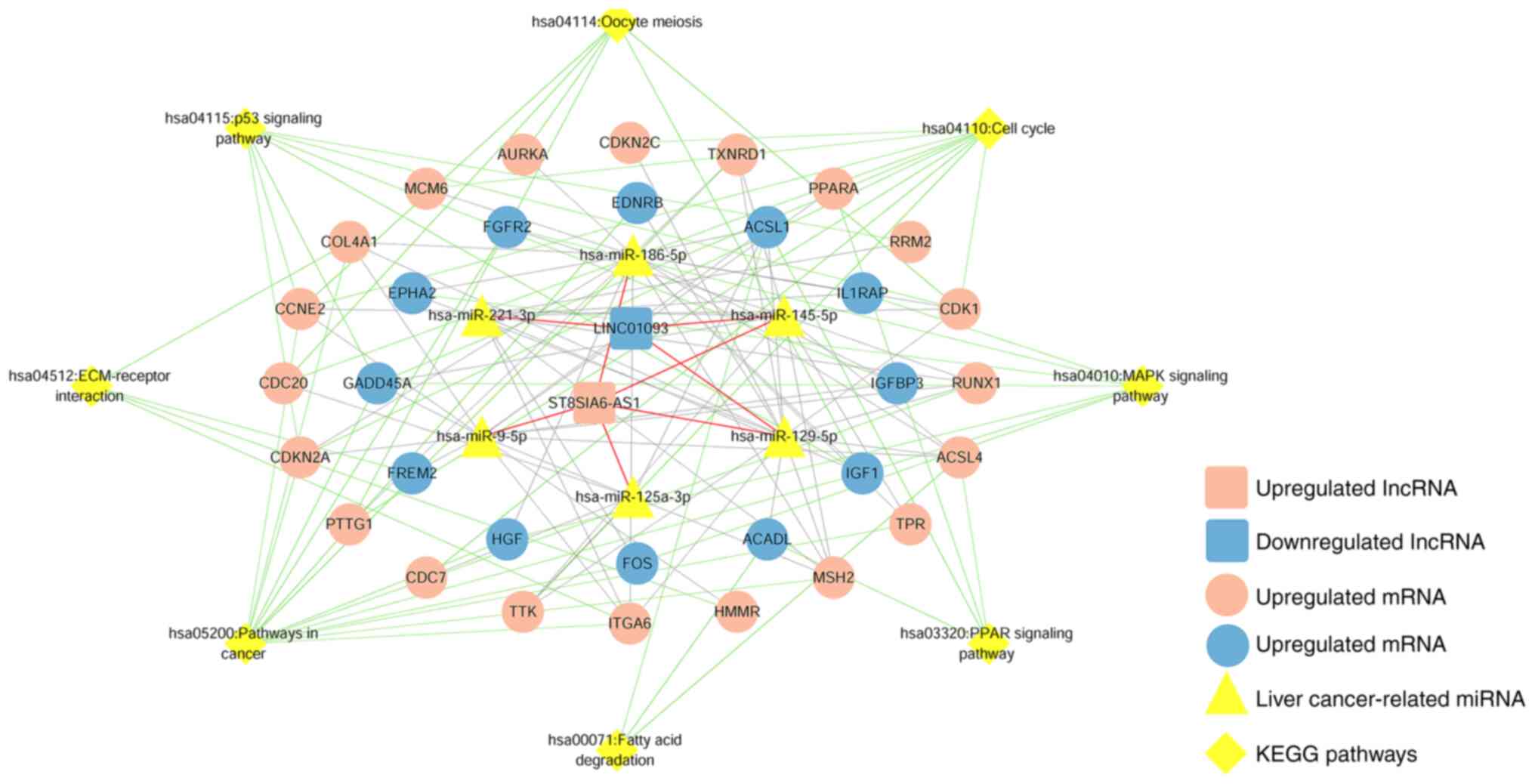
8 liver cancer-associated miRNAs and 162 DEmRNAs (Fig. 5). In addition, KEGG pathway
enrichment analysis showed that the 162 DEmRNAs were significantly
enriched in 8 KEGG signaling pathways (Fig. S3) and details of the pathways are
displayed in Table I. By combining
the DEmRNAs with the associated KEGG signaling pathways, a ceRNA
regulatory network was constructed, as shown in Fig. 6.
 | Table I.Significant KEGG pathways associated
with differentially expressed mRNAs in the competitive endogenous
RNA network. |
Table I.
Significant KEGG pathways associated
with differentially expressed mRNAs in the competitive endogenous
RNA network.
| KEGG terms | Count | P-value | Genes |
|---|
| hsa04110: Cell
cycle | 10 |
4.32×10−6 | CDC20, CDKN2C,
PTTG1, CCNE2, CDKN2A, GADD45A, CDK1, TTK, CDC7, MCM6 |
| hsa04115: p53
signaling pathway | 7 |
8.62×10−5 | RRM2, CCNE2,
CDKN2A, GADD45A, IGFBP3, CDK1, IGF1 |
| hsa05200: Pathways
in cancer | 14 |
2.32×10−3 | CDKN2A, GADD45A,
TXNRD1, HGF, FOS, IGF1, RUNX1, EDNRB, CCNE2, MSH2, COL4A1, TPR,
ITGA6, FGFR2 |
| hsa04114: Oocyte
meiosis | 6 |
1.01×10−2 | CDC20, PTTG1,
CCNE2, CDK1, IGF1, AURKA |
| hsa03320: PPAR
signaling pathway | 4 |
3.96×10−2 | ACADL, ACSL1,
ACSL4, PPARA |
| hsa04512:
ECM-receptor interaction | 4 |
4.59×10−2 | COL4A1, ITGA6,
HMMR, FREM2 |
| hsa00071: Fatty
acid degradation | 3 |
4.69×10−2 | ACADL, ACSL1,
ACSL4 |
| hsa04010: MAPK
signaling pathway | 7 |
4.75×10−2 | GADD45A, HGF, FOS,
IL1RAP, IGF1, FGFR2, EPHA2 |
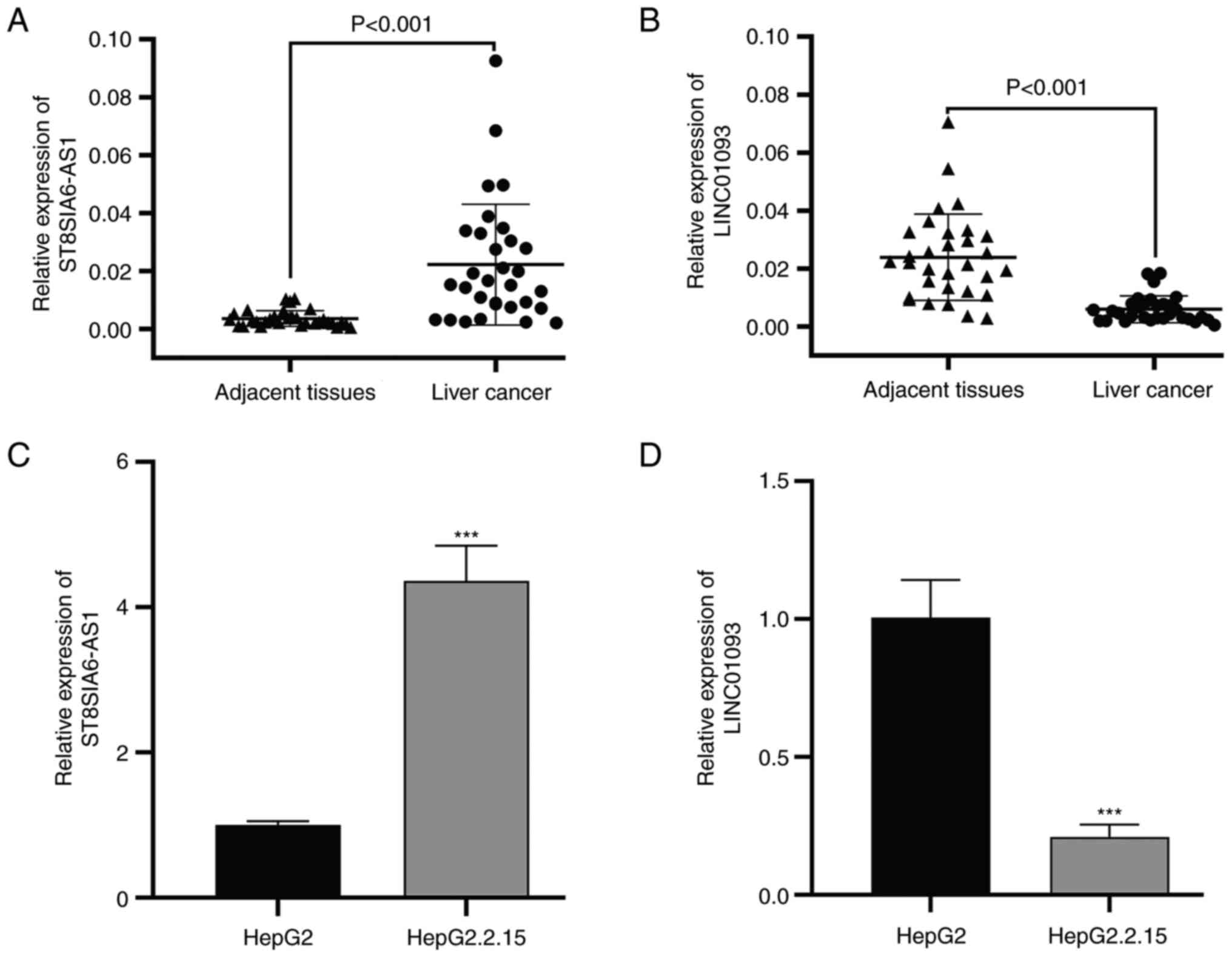
Validation of selected lncRNA
expression
To validate the accuracy of the data obtained by the
bioinformatics analysis, the expression levels of ST8SIA6-AS1 and
LINC01093 were measured using RT-qPCR in HBV-liver cancer samples
collected from patients. The results showed that ST8SIA6-AS1
(Fig. 7A) was significantly
upregulated, while LINC01093 (Fig.
7B) was significantly downregulated in the HBV-liver cancer
tissues compared with the adjacent liver tissues. Consistent with
this, the ST8SIA6-AS1 expression level was significantly
upregulated in HepG2.2.15 cells compared with HepG2 cells (Fig. 7C), and the LINC01093 expression
level was significantly downregulated in HepG2.2.15 cells when
compared with HepG2 cells (Fig.
7D).
Investigation on the functional role
of ST8SIA6-AS1 and LINC01093 in HBV-expressing liver cancer cells
in vitro
The functional roles of ST8SIA6-AS1 and LINC01093
were investigated by performing loss-of-function and
gain-of-function assays, respectively in HBV-expressing liver
cancer cells. First, RT-qPCR confirmed the transfection efficiency
of si-ST8SIA6-AS1 (Fig. 8A) and
pcDNA3.1-LINC01093 (Fig. 8B) in
HepG2.2.15 cells. In terms of HBV infection, the number of copies
of HBV DNA in the supernatant of HepG2.2.15 cells was diminished
after ST8SIA6-AS1 knockdown (Fig.
8C) or LINC01093 overexpression (Fig. 8D). ELISA results showed that the
levels of viral factors HBsAg (Fig. 8E
and F) and HBeAg (Fig. 8G and
H) were significantly decreased in HepG2.2.15 cells after
ST8SIA6-AS1 knockdown or LINC01093 overexpression. The ST8SIA6-AS1
downregulation and LINC01093 upregulation each lowered the
HepG2.2.15 cell proliferation rate, as indicated by lower OD values
after 72 h (Fig. 8I). In addition,
the results of Transwell assays indicated that numbers of migrated
and invaded cells were significantly reduced after ST8SIA6-AS1
knockdown (Fig. 8J) or LINC01093
overexpression (Fig. 8K) in
HepG2.2.15 cells.
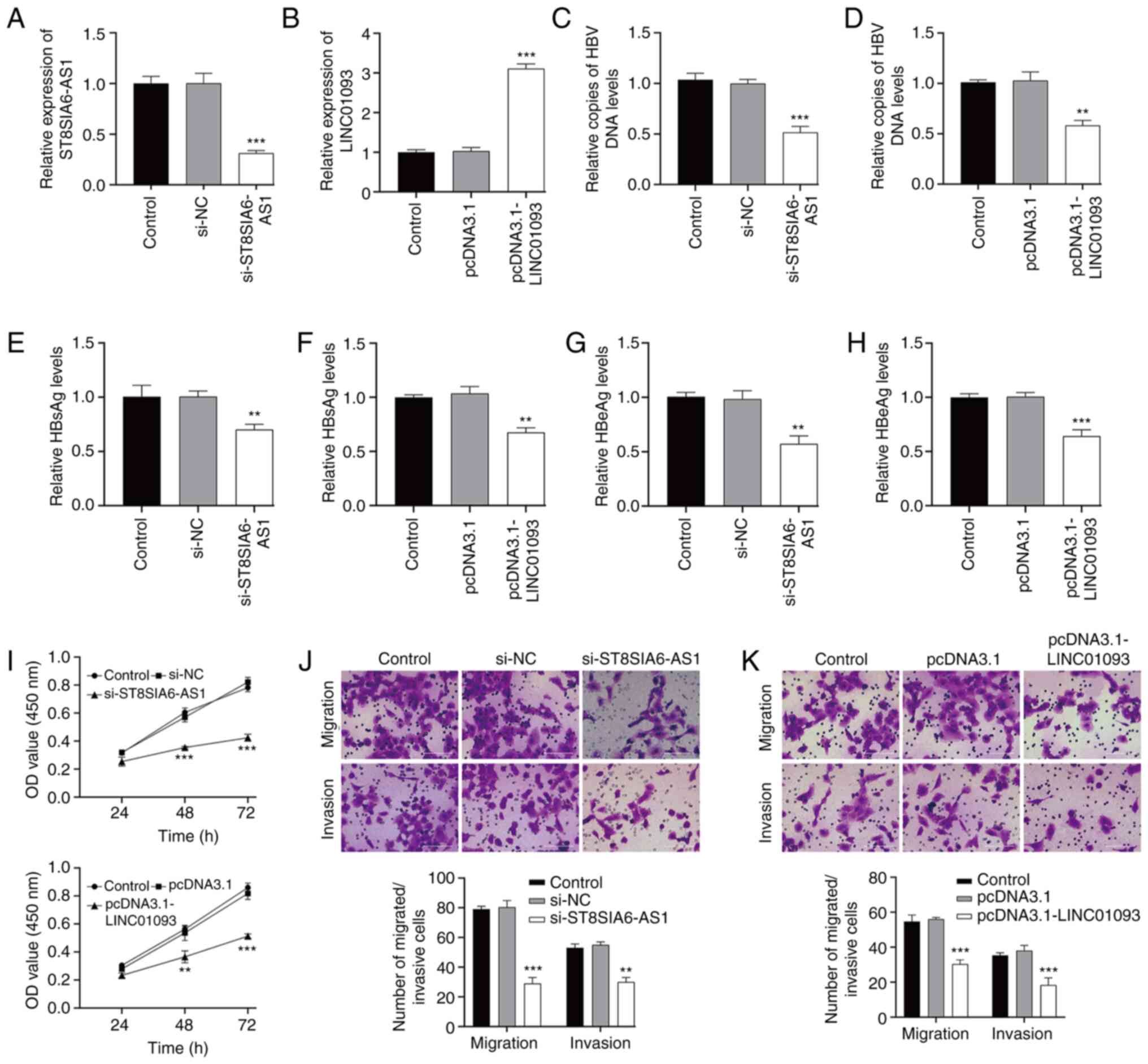 | Figure 8.Investigation of the functional role
of ST8SIA6-AS1 and LINC01093 in HBV-expressing liver cancer cells
in vitro. HepG2.2.15 cells were transfected with
si-ST8SIA6-AS1 and si-NC or pcDNA3.1-LINC01093 and pcDNA3.1,
respectively. RT-qPCR was used to determine the expression levels
of (A) ST8SIA6-AS1 and (B) LINC01093 in transfected HepG2.2.15
cells. (C and D) qPCR detected copies of HBV DNA in culture
supernatants of the transfected HepG2.2.15 cells. ELISAs were used
to measure levels of viral factors in the cell culture
supernatants; specifically, HBsAg in (E) ST8SIA6-AS1 knockdown and
(F) LINC01093 overexpressing cells, and HBeAg in (G) ST8SIA6-AS1
knockdown and (H) LINC01093 overexpressing cells. (I) Cell Counting
Kit-8 assay was used to evaluate OD value of the transfected
HepG2.2.15 cells at 450 nm. Transwell assays were used to determine
the numbers of migrated and invaded cells in the (J) ST8SIA6-AS1
knockdown and (K) LINC01093 overexpressing cells. Data are
presented as the mean ± SD. **P<0.01, ***P<0.001 vs. si-NC or
pcDNA3.1. HBV, hepatitis B virus; si, small interfering; NC,
negative control; RT-qPCR, reverse transcription-quantitative PCR;
HBsAg, hepatitis B surface antigen; HBeAg, hepatitis B e antigen;
OD, optical density. |
Discussion
To the best of current knowledge, persistent HBV
infection is the leading cause of liver cancer (32), which is considered to promote the
initiation and progression of liver cancer by integrating into the
host genome (33). Numerous studies
have elucidated the crucial role of lncRNAs in HBV-associated liver
cancer (34). In the present study,
three gene expression datasets were analyzed, in which 30 DElncRNAs
and 505 DEmRNAs were identified in HBV-associated liver cancer
samples compared with controls. GO and KEGG enrichment analyses
demonstrated these DEmRNAs were associated with various metabolic
processes, ‘extracellular region’, ‘oxidoreductase activity’, ‘cell
cycle’ and ‘PPAR signaling pathway’. Cancer is clearly associated
with abnormal metabolic processes, and enzymes that contribute to
altered retinol metabolism have been shown to participate in the
development of liver cancer (35).
Cell cycle deregulation is associated with the initiation of cancer
through the mutation of proteins with key roles at different stages
of the cell cycle (36). Previous
studies have shown that peroxisome proliferator-activated receptors
(PPARs) are nuclear receptors that function as transcription
factors and regulate physiological activities including invasion,
immune tolerance, metabolism and inflammation (37,38).
For example, a study revealed that the activation of PPAR-α was
involved in the tumor.promoting effects of 4.phenylbutyric acid in
a liver cancer.inducing environment (39).
Feature selection along with three optimization
algorithms were employed to identify 10 optimal DElncRNA-based
signatures consisting of HAND2-AS1, LINC00622, ST8SIA6-AS1,
TAPT1-AS1, MAGI2-AS3, LINC00528, LINC00205, RUSC1-AS1, LINC01093
and LINC01003. HAND2-AS1 has been reported to be required for the
self-renewal maintenance of liver cancer stem cells to initiate
liver cancer development (40).
Furthermore, HAND2-AS1 overexpression has been shown to inhibit
cancer cell proliferation in liver cancer via the downregulation of
runt-related transcription factor 2 expression (41). ST8SIA6-AS1 has been reported to be a
cancer-associated lncRNA that facilitates cell proliferation and
resistance to apoptosis in liver cancer cells (42). Consistent with this, Zhang et
al (43) demonstrated that the
downregulation of ST8SIA6-AS1 suppressed liver cancer cell
proliferation, migration and invasion in vitro and
restrained liver cancer tumorigenesis in vivo. MAGI2-AS3 has
been found to be downregulated in the plasma of patients with
early-stage liver cancer, while the overexpression of MAGI2-AS3
exerts suppressive effects on the proliferation and migration of
liver cancer cells (44.46). LINC00205, a bidirectional lncRNA,
located at human chromosome 21q22.3, has been characterized as an
oncogenic molecule that contributes to cell proliferation in liver
cancer (47,48). Liu et al (49) recently reported that the knockdown
of RUSC1-AS1 inhibited liver cancer cell progression, including
proliferation, migration and invasion, and suppressed tumorigenesis
in vivo. LINC01093, as a recently identified liver-specific
lncRNA, has been shown to suppress liver cancer cell proliferation
and metastasis in vitro and in vivo (50). However, studies on the involvement
of HAND2-AS1, TAPT1-AS1, LINC00528 and LINC01003 in liver cancer
are lacking.
Based on the survival prognosis information in TCGA
dataset, ST8SIA6-AS1 and LINC01093 were further screened as
HBV-liver cancer prognosis-associated lncRNAs. Patients whose
HBV-liver cancer had low ST8SIA6-AS1 or high LINC01093 expression
levels had an improved overall survival rate. In vitro
experiments confirmed that ST8SIA6-AS1 was upregulated in
HBV-expressing liver cancer cells, and the knockdown of ST8SIA6-AS1
in these cells significantly suppressed cell proliferation,
migration and invasion, as well as HBV expression and replication.
Consistent with these data, Qin et al (51) revealed that the upregulation of
ST8SIA6-AS1 is associated with higher tumor stages and metastasis,
and thus has potential as a diagnostic biomarker for liver cancer
progression. Although the role of ST8SIA6-AS1 in HBV-liver cancer
cells has not been reported previously, to the best of our
knowledge, the promotive effects of ST8SIA6-AS1 on cell
proliferation, migration and invasion have already been elucidated
in liver cancer cells (52,53). Consistent with the present data
showing that LINC01093 was downregulated in HBV-expressing liver
cancer cells and suppressed proliferation and mobility, Mou et
al (54) identified LINC01093
as an optimal diagnostic and prognostic lncRNA biomarker for
HBV-associated liver cancer. The previously reported suppressive
effects of LINC01093 on liver cancer cell proliferation and
metastasis (50) support the
finding that LINC01093 acts a tumor suppressor in HBV-liver cancer
cells. Furthermore, the ceRNA network based on ST8SIA6-AS1 and
LINC01093 was constructed in the present study, which included 8
liver cancer-miRNAs and 162 DEmRNAs.
The present study has some limitations as follows:
i) Large HBV-liver cancer cohorts are required to confirm the
prognostic values of the lncRNA signature; ii) further wet
experiments are necessary to verify the mechanisms indicated by the
ceRNA network, for example luciferase assays with overexpression or
silencing, or co-expression (coimmunoprecipitation); iii) in
vivo experiments are essential to validate the functional role
of ST8SIA6-AS1 and LINC01093; iv) further mechanistic analysis is
required to confirm the oncogenic or suppressive roles of these two
lncRNAs.
In summary, 10 optimal DElncRNA-based signatures of
HBV-associated liver cancer were constructed based on microarray
datasets, which created a nomogram with good accuracy as an ideal
model. ST8SIA6-AS1 and LINC01093 were further identified as lncRNAs
that are associated with HBV-liver cancer prognosis and further
analyses demonstrated that they may play important roles in the
progression of HBV-liver cancer. Collectively, these data may
provide an understanding of the regulatory mechanisms of lncRNAs in
the development of HBV-associated liver cancer and help the
identification of potential targets for its treatment.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Cultivation of Academic Leaders
in Pudong New Area (grant no. PWRd2021-20) and Shanghai Traditional
Chinese Medicine Capacity Building Project for the Prevention and
Treatment of Infectious Diseases (grant no. ZYYB-NLPY-04).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XW conceived the design of the study. JX and HZ
acquired the data and conducted the statistical analysis. YF
collected tissue samples and participated in the interpretation of
the data. XL and XW analyzed and interpreted the data, and drafted
and revised the manuscript. All authors read and approved the final
manuscript. XW and JX confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
All human tissues were obtained in accordance with
the Declaration of Helsinki (1975) and approved by the Ethics
Committee of Hospital for Infectious Diseases of Pudong District
(Shanghai, China; approval no. IDP-2020PD; 2020.8.16). Each patient
provided written informed consent for the use of their samples in
scientific research.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Erratum. Global cancer statistics 2018:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 70:3132020. View Article : Google Scholar
|
|
2
|
Yang F, Ma L, Yang Y, Liu W, Zhao J, Chen
X, Wang M, Zhang H, Cheng S, Shen F, et al: Contribution of
hepatitis B virus infection to the aggressiveness of primary liver
cancer: A clinical epidemiological study in Eastern China. Front
Oncol. 9:3702019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu YC, Lu LF, Li CJ, Sun NK, Guo JY,
Huang YH, Yeh CT and Chao CCK: Hepatitis B virus X protein induces
RHAMM-dependent motility in hepatocellular carcinoma cells via
PI3K-Akt-Oct-1 signaling. Mol Cancer Res. 18:375–389. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kang I, Kim JA, Kim J, Lee JH, Kim MJ and
Ahn JK: Hepatitis B virus X protein promotes epithelial-mesenchymal
transition of hepatocellular carcinoma cells by regulating SOCS1.
BMB Rep. 55:220–225. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lei Y, Xu X, Liu H, Chen L, Zhou H, Jiang
J, Yang Y and Wu B: HBx induces hepatocellular carcinogenesis
through ARRB1-mediated autophagy to drive the G(1)/S cycle.
Autophagy. 17:4423–4441. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
European Association for the Study of the
Liver. Electronic address, . simpleeasloffice@easloffice.eu;
European Association for the Study of the Liver: EASL clinical
practice guidelines: Management of hepatocellular carcinoma. J
Hepatol. 69:182–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li LM, Chen C, Ran RX, Huang JT, Sun HL,
Zeng C, Zhang Z, Zhang W and Liu SM: Loss of TARBP2 drives the
progression of hepatocellular carcinoma via miR-145-SERPINE1 axis.
Front Oncol. 11:6209122021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiu L, Wang T, Xu X, Wu Y, Tang Q and Chen
K: Long non-coding RNAs in hepatitis B virus-related hepatocellular
carcinoma: Regulation, functions, and underlying mechanisms. Int J
Mol Sci. 18:25052017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fortes P and Morris KV: Long noncoding
RNAs in viral infections. Virus Res. 212:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stojic L, Lun ATL, Mascalchi P, Ernst C,
Redmond AM, Mangei J, Barr AR, Bousgouni V, Bakal C, Marioni JC, et
al: A high-content RNAi screen reveals multiple roles for long
noncoding RNAs in cell division. Nat Commun. 11:18512020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hou Z, Xu X, Fu X, Tao S, Zhou J, Liu S
and Tan D: HBx-related long non-coding RNA MALAT1 promotes cell
metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am
J Cancer Res. 7:845–856. 2017.PubMed/NCBI
|
|
12
|
Tao L, Li D, Mu S, Tian G and Yan G:
LncRNA MAPKAPK5_AS1 facilitates cell proliferation in hepatitis B
virus-related hepatocellular carcinoma. Lab Invest. 102:494–504.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deng Y, Wei Z, Huang M, Xu G, Wei W, Peng
B, Nong S and Qin H: Long non-coding RNA F11-AS1 inhibits
HBV-related hepatocellular carcinoma progression by regulating
NR1I3 via binding to microRNA-211-5p. J Cell Mol Med. 24:1848–1865.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM,
Hu YW, Lin L, Chen J, Zheng L and Wang Q: HBx-related long
non-coding RNA DBH-AS1 promotes cell proliferation and survival by
activating MAPK signaling in hepatocellular carcinoma. Oncotarget.
6:33791–33804. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang H, Diab A, Fan H, Mani SKK,
Hullinger R, Merle P and Andrisani O: PLK1 and HOTAIR accelerate
proteasomal degradation of SUZ12 and ZNF198 during hepatitis B
virus-induced liver carcinogenesis. Cancer Res. 75:2363–2374. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Yin G, Bian H, Yang J, Zhou P, Yan
K, Liu C, Chen P, Zhu J, Li Z and Xue T: LncRNA XIST upregulates
TRIM25 via negatively regulating miR-192 in hepatitis B
virus-related hepatocellular carcinoma. Mol Med. 27:412021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu H, Zhao P, Jin X, Zhao Y, Chen Y, Yan
T, Wang J, Wu L and Sun Y: A 9lncRNA risk score system for
predicting the prognosis of patients with hepatitis B viruspositive
hepatocellular carcinoma. Mol Med Rep. 20:573–583. 2019.PubMed/NCBI
|
|
18
|
Wang SM, Ooi LL and Hui KM: Identification
and validation of a novel gene signature associated with the
recurrence of human hepatocellular carcinoma. Clin Cancer Res.
13:6275–6283. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Melis M, Diaz G, Kleiner DE, Zamboni F,
Kabat J, Lai J, Mogavero G, Tice A, Engle RE, Becker S, et al:
Viral expression and molecular profiling in liver tissue versus
microdissected hepatocytes in hepatitis B virus-associated
hepatocellular carcinoma. J Transl Med. 12:2302014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protocols. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deist TM, Dankers F, Valdes G, Wijsman R,
Hsu IC, Oberije C, Lustberg T, van Soest J, Hoebers F, Jochems A,
et al: Machine learning algorithms for outcome prediction in
(chemo) radiotherapy: An empirical comparison of classifiers. Med
Phys. 45:3449–3459. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goeman JJ: L1 penalized estimation in the
Cox proportional hazards model. Biom J. 52:70–84. 2010.PubMed/NCBI
|
|
25
|
Tolosi L and Lengauer T: Classification
with correlated features: Unreliability of feature ranking and
solutions. Bioinformatics. 27:1986–1994. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu J, Zhang H, Li L, Hu M, Chen L, Xu B
and Song Q: A nomogram for predicting overall survival in patients
with low-grade endometrial stromal sarcoma: A population-based
analysis. Cancer Commun (Lond). 40:301–312. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang Q, Wang Y, Hao Y, Juan L, Teng M,
Zhang X, Li M, Wang G and Liu Y: miR2Disease: A manually curated
database for microRNA deregulation in human disease. Nucleic Acids
Res. 37:D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ridruejo E: Does hepatitis B virus therapy
reduce the risk of hepatocellular carcinoma? Expert Opin Drug Saf.
14:439–451. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bisceglie AMD: Hepatitis B and
hepatocellular carcinoma. Hepatology. 49:S56–S60. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chauhan R and Lahiri N: Tissue- and
serum-associated biomarkers of hepatocellular carcinoma. Biomarkers
Cancer. 8:37–55. 2016.PubMed/NCBI
|
|
35
|
Pettinelli P, Arendt BM, Teterina A,
McGilvray I, Comelli EM, Fung SK, Fischer SE and Allard JP: Altered
hepatic genes related to retinol metabolism and plasma retinol in
patients with non-alcoholic fatty liver disease. PLoS One.
13:e02057472018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liping X, Jia L, Qi C, Liang Y, Dongen L
and Jianshuai J: Cell Cycle Genes are potential diagnostic and
prognostic biomarkers in hepatocellular carcinoma. Biomed Res Int.
2020:62061572020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee CH, Olson P and Evans RM: Minireview:
Lipid metabolism, metabolic diseases, and peroxisome
proliferator-activated receptors. Endocrinology. 144:2201–2207.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Oyefiade A, Erdman L, Goldenberg A, Malkin
D, Bouffet E, Taylor MD, Ramaswamy V, Scantlebury N and Law N: PPAR
and GST polymorphisms may predict changes in intellectual
functioning in medulloblastoma survivors. J Neurooncol. 142:39–48.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen SZ, Ling Y, Yu LX, Song YT, Chen XF,
Cao QQ, Yu H, Chen C, Tang JJ, Fan ZC, et al: 4-phenylbutyric acid
promotes hepatocellular carcinoma via initiating cancer stem cells
through activation of PPAR-α. Clin Transl Med. 11:e3792021.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Zhu P, Luo J, Wang J, Liu Z, Wu W,
Du Y, Ye B, Wang D, He L, et al: LncRNA HAND2-AS1 promotes liver
cancer stem cell self-renewal via BMP signaling. EMBO J.
38:e1011102019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jing GY, Zheng XZ and Ji XX: lncRNA
HAND2-AS1 overexpression inhibits cancer cell proliferation in
hepatocellular carcinoma by downregulating RUNX2 expression. J Clin
Lab Anal. 35:e237172021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fei Q, Song F, Jiang X, Hong H, Xu X, Jin
Z, Zhu X, Dai B, Yang J, Sui C and Xu M: LncRNA ST8SIA6-AS1
promotes hepatocellular carcinoma cell proliferation and resistance
to apoptosis by targeting miR-4656/HDAC11 axis. Cancer Cell Int.
20:2322020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang X, Xu S, Hu C, Fang K, Zhou J, Guo
Z, Zhu G and Li L: LncRNA ST8SIA6-AS1 promotes hepatocellular
carcinoma progression by regulating MAGEA3 and DCAF4L2 expression.
Biochem Biophys Res Commun. 533:1039–1047. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yin Z, Ma T, Yan J, Shi N, Zhang C, Lu X,
Hou B and Jian Z: LncRNA MAGI2-AS3 inhibits hepatocellular
carcinoma cell proliferation and migration by targeting the
miR-374b-5p/SMG1 signaling pathway. J Cell Physiol.
234:18825–18836. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fang G, Wang J, Sun X, Xu R, Zhao X, Shao
L, Sun C and Wang Y: LncRNA MAGI2-AS3 is downregulated in the
distant recurrence of hepatocellular carcinoma after surgical
resection and affects migration and invasion via ROCK2. Ann
Hepatol. 19:535–540. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wei H, Tang Q, Wang A, Zhang Y, Qin Z, Li
W, Xu Z, Wang J and Pu J: lncRNA MAGI2-AS3 exerts antioncogenic
roles in hepatocellular carcinoma via regulating the
miR-519c-3p/TXNIP Axis. J Oncol. 2021:55473452021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang L, Wang Y, Sun J, Ma H and Guo C:
LINC00205 promotes proliferation, migration and invasion of HCC
cells by targeting miR-122-5p. Pathol Res Pract. 215:1525152019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cheng T, Yao Y, Zhang S, Zhang XN, Zhang
AH, Yang W and Hou CZ: LINC00205, a YY1-modulated lncRNA, serves as
a sponge for miR-26a-5p facilitating the proliferation of
hepatocellular carcinoma cells by elevating CDK6. Eur Rev Med
Pharmacol Sci. 25:6208–6219. 2021.PubMed/NCBI
|
|
49
|
Liu C, Tang L, Xu M, Lin Y, Shen J, Zhou
L, Ho L, Lu J and Ai X: LncRNA RUSC1-AS1 contributes to the
progression of hepatocellular carcinoma cells by modulating
miR-340-5p/CREB1 axis. Am J Transl Res. 13:1022–1036.
2021.PubMed/NCBI
|
|
50
|
He J, Zuo Q, Hu B, Jin H, Wang C, Cheng Z,
Deng X, Yang C, Ruan H, Yu C, et al: A novel, liver-specific long
noncoding RNA LINC01093 suppresses HCC progression by interaction
with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett.
450:98–109. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Qin SJ, Zhou HZ, Xu NS, Yang HC and Chen
PX: The diagnostic value of serum ST8SIA6-AS1 as biomarker in
hepatocellular carcinoma. Clin Lab. 66:doi:
10.7754/Clin.Lab.2020.200231. 2020. View Article : Google Scholar
|
|
52
|
Kuai J, Zheng L, Yi X, Liu Z, Qiu B, Lu Z
and Jiang Y: ST8SIA6-AS1 promotes the development of hepatocellular
carcinoma cells through miR-338-3p/NONO axis. Digestive Liver Dis.
53:1192–1200. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang B, Liu Z, Liu J, Cao K, Shan W, Wen
Q and Wang R: Long non-coding RNA ST8SIA6-AS1 promotes the
migration and invasion of hypoxia-treated hepatocellular carcinoma
cells through the miR-338/MEPCE axis. Oncol Rep. 45:73–82. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mou Y, Wang D, Xing R, Nie H, Mou Y, Zhang
Y and Zhou X: Identification of long noncoding RNAs biomarkers in
patients with hepatitis B virus-associated hepatocellular
carcinoma. Cancer Biomark. 23:95–106. 2018. View Article : Google Scholar : PubMed/NCBI
|